Abstract
Recent large-scale transcriptome analyses have revealed that large numbers of noncoding RNAs (ncRNAs) are transcribed from mammalian genomes. They include small nuclear RNAs (snRNAs), small nucleolar RNAs (snoRNAs), and longer ncRNAs, many of which are localized to the nucleus, but which have remained functionally elusive. Since ncRNAs are only known to exist in mammalian species, established experimental systems, including the Xenopus oocyte system and yeast genetics, are not available for functional analysis. RNA interference (RNAi), commonly used for analysis of protein-coding genes, is effective in eliminating cytoplasmic mRNAs, but not nuclear RNAs. To circumvent this problem, we have refined the system for knockdown of nuclear ncRNAs with chemically modified chimeric antisense oligonucleotides (ASO) that were efficiently introduced into the nucleus by nucleofection. Under optimized conditions, our system appeared to degrade at least 20 different nuclear ncRNA species in multiple mammalian cell lines with high efficiency and specificity. We also confirmed that our method had greatly improved knockdown efficiency compared with that of the previously reported method in which ASOs are introduced with transfection reagents. Furthermore, we have confirmed the expected phenotypic alterations following knockdown of HBII295 snoRNA and U7 snRNA, which resulted in a loss of site-specific methylation of the artificial RNA and the appearance of abnormal polyadenylated histone mRNA species with a concomitant delay of the cell cycle S phase, respectively. In summary, we believe that our system is a powerful tool to explore the biological functions of the large number of nuclear ncRNAs with unknown function.
Keywords: noncoding RNA, snoRNA, U7 snRNA, knockdown, nucleus, antisense oligonucleotide
INTRODUCTION
In mammalian cells, many small noncoding RNAs are localized to the cell nucleus, where they play important roles in various steps of gene expression. Subsets of small nuclear RNAs (snRNAs) constitute a large ribonucleoprotein complex called the spliceosome that excises introns from pre-mRNA (Tycowski et al. 2006). U7 snRNA participates in the creation of an unusual 3′ end formation in nonpolyadenylated histone mRNAs. Metazoan replication-dependent histone mRNA is the only eukaryotic mRNA that lacks a poly(A) tail; instead, processed histone mRNA remains stable with a stem–loop structure at its 3′ end. U7 snRNA interacts with a purine-rich element downstream from the stem–loop structure, where a 5′ terminal portion of U7 snRNA forms a base-pairing interaction (Dominski and Marzluff 2007). Histone gene transcription and subsequent RNA processing events occur at the Cajal body, where some histone gene loci frequently attach, and where U7 snRNA is localized (Frey and Matera 1995).
The second class of nuclear-localized small ncRNAs, small nucleolar RNAs (snoRNAs), are well characterized; snoRNAs are predominantly localized to the nucleolus, where the majority are mainly involved in ribosome biogenesis. U3 is the most abundant snoRNA (1 × 105/cell) possessing a trimethyl guanosine (TMG) cap at its 5′ terminus, and it is involved in 5′ETS processing of pre-rRNA. U8 and U13 are similar and less abundant snoRNAs with TMG caps that are believed to participate in pre-rRNA processing (Tycowski et al. 2006). The remaining hundreds of snoRNAs, with a few exceptions, lack cap structures at their 5′ termini and are encoded in the introns of various host genes. snoRNA is processed from the excised intron as a result of splicing (Filipowicz and Pogacic 2002; Tycowski et al. 2006). Most intronic snoRNAs are classified into two groups, box C/D snoRNA and box H/ACA snoRNA, which are involved in site-specific ribose methylation and pseudouridylation of pre-rRNA, respectively (Kiss et al. 2006; Tycowski et al. 2006). Both classes of snoRNA form base pairings with substrate RNA via their short single-stranded guide sequences, which specify the sites to be modified. Subsets of snoRNA-like RNAs are predominantly localized to the Cajal body, and are thereby called scaRNA (Cajal body-specific small RNA), where they are involved in the modification of U-snRNA transcribed by RNA polymerase II (Kiss et al. 2006; Tycowski et al. 2006). Large-scale analysis of small RNAs, so-called RNomic analysis, has revealed that some snoRNAs lack any sequence complementarity to known stable RNAs, including rRNAs and snRNAs, suggesting the existence of novel snoRNA targets, and, therefore, this class of snoRNAs is termed “orphan snoRNAs” (Cavaillé et al. 2000; Jády and Kiss 2000; Bachellerie et al. 2002; Hüttenhofer et al. 2002).
The function of snRNA in vertebrates has been mainly studied by recapitulation in in vitro systems (e.g., in vitro splicing systems and in vitro histone mRNA processing systems). Complementary research has been performed in a Xenopus oocyte system, where antisense oligonucleotides were microinjected into the nucleus to degrade specific snRNAs, or to block their action (Pan and Prives 1988; Peculis and Steitz 1993; Tycowski et al. 1994). Since no one has successfully recapitulated snoRNA action in vitro, the Xenopus system is currently the sole system used to explore snoRNA function in vertebrates. This is one reason why the function of orphan snoRNAs is still largely unknown, since most mammalian orphan snoRNAs are not conserved in amphibians.
Many researchers have used RNA interference (RNAi) to explore gene function. However, RNAi targets have been restricted to cytoplasmic mRNAs, since the RNAi machinery is thought to be exclusively present in the cytoplasm of mammalian cells. Recently, it was reported that an abundant nuclear 7SK RNA was successfully knocked down by introduced siRNA, suggesting that RNAi can be a potent tool for analysis of nuclear RNAs (Robb et al. 2005). The mechanism by which cytoplasmic RISC targets and exclusively degrades nuclear 7SK RNA is still largely unknown. Except for a few reports, the majority of nuclear ncRNAs are not susceptible to siRNA, and, therefore, a more efficient knockdown method is required to explore their functions.
Here, we describe an improved method for antisense oligonucleotide-mediated degradation of specific small nuclear RNAs in mammalian cultured cells. Since this system is able to knock down multiple classes of nuclear-localized ncRNAs, including snRNA, telomerase RNA, box C/D-, and box H/ACA snoRNAs, as well as mRNA-like long ncRNAs in various mammalian cultured cells, it is a valuable tool to accelerate the functional analysis of nuclear localized ncRNA in mammalian cultured cells.
RESULTS
Optimization of conditions for antisense oligonucleotide-mediated snoRNA knockdown
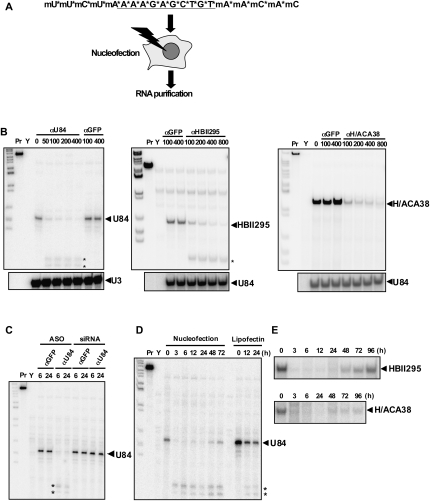
To determine the optimal conditions for nuclear ncRNA knockdown, we utilized U84 snoRNA, an orphan box C/D snoRNA with unknown targets that is localized to the nucleolus. Counterparts of this orphan snoRNA are found only in mammalian species, and not in amphibians or yeast (Jády and Kiss 2000); therefore, its function could not be analyzed utilizing previously available methods. We used phosphorothioate-modified chimeric antisense oligonucleotides (ASOs) to knock down nuclear U84 snoRNA. We designed ASOs that were 20-nucleotides (nt) long and comprised of 2′-O-methoxyribonucleotide segments of 5 nt at both termini and a deoxynucleotide segment containing 10 central nucleotides. All of the phosphate backbones were converted by phosphorothioate (Fig. 1A). The ASOs were directly introduced into HeLa cell nuclei by nucleofection (Fig. 1A). An ASO with sequence complementarity to the putative guide sequence upstream U84 snoRNA boxD (Fig. 1A, αU84) efficiently degraded U84 snoRNA (Fig. 1B, left). Another box C/D snoRNA, HBII295, and a box H/ACA snoRNA, H/ACA38, were successfully knocked down with ASOs designed to have homology with the putative guide sequences in the respective snoRNAs (Fig. 1B, middle and right). Titration of ASO concentrations for nucleofection revealed that the introduction of 100 pmol of oligonucleotide into 1 × 106 cells consistently decreased snoRNA levels to <20% of basal levels, and raising the oligonucleotide concentration to 400 pmol further decreased levels (to <10% of basal) of three different snoRNAs (Fig. 1B). Importantly, the control oligonucleotide containing the unrelated sequence (αGFP) did not affect snoRNA levels (see Fig. 1B, lanes αGFP), and each of the ASOs specifically affected the target snoRNAs, with no effects on the levels of untargeted snoRNA (Fig. 1B, lower panels). Our time course experiments revealed that almost complete elimination of target snoRNA had occurred within 3 h and was maintained for at least 48 h (Fig. 1D,E). Two putative degradation products were detected by an RNase protection assay (RPA) after knockdown of U84 snoRNA (Fig. 1B,D, asterisks), suggesting that an endogenous endonuclease, likely RNase H, recognized and cleaved the snoRNA-ASO hybrid. Meanwhile, the synthesized siRNA with the corresponding sequence did not affect U84 snoRNA levels, despite the fact that it was administered to cells under nucleofection conditions that had been optimized in canonical RNAi experiments. The ineffectiveness of siRNA was confirmed in another snoRNA, HBII295 (Fig. 2A). Lipofectin reagent, which was previously reported to be superior for the administration of ASOs into cultured cells (Vickers et al. 2003) and other transfection reagents (lipofectamine 2000 and oligofectamine) have been used for the knockdown of many long nuclear ncRNAs with ASOs (e.g., Prasanth et al. 2005; Shamovsky et al. 2006; Mariner et al. 2008). However, transfected ASOs have apparently been less effective in knocking down U84 snoRNA and other small nuclear RNAs (Fig. 1D; data not shown), indicating that our method has greatly improved efficiency compared with these previously characterized methods.
FIGURE 1.
(A) Schematic of snoRNA knockdown in human cultured cells. The chimeric ASO (αU84) targeting U84 snoRNA is shown. The five terminal nucleotides on each end are 2′-O-methoxyethyl nucleotides indicated as “mN,” where N is the nucleotide. The phosphothioate backbones are indicated with asterisks. Oligonucleotides were introduced into human cultured cells using a nucleofector device. Total RNA was prepared and used for RPA. (B) Oligonucleotide dose-dependency in the knockdown of three snoRNAs (U84, HBII295, and H/ACA38) as measured by RPA. Numbers shown above the blot represent oligonucleotide amounts (pmol) used for nucleofection. Pr and Y represent the RNA probe and control yeast RNA used for RPA, respectively. (*) Putative degradation products. (C) Effects of ASOs and siRNAs. ASOs and siRNAs with complementarity to GFP (αGFP) or U84 (αU84) were administered by nucleofection. Total RNA was prepared at 6 or 24 h after nucleofection (as shown above the panel). (D,E) Time course of knockdown efficacy of αU84 (D) and αHBII295 and αH/ACA38 (E). Numbers above the blots represent incubation time (hours) after nucleofection. U84 levels are shown after oligonucleotide administration with the lipofectin reagent (three right lanes in D).
FIGURE 2.
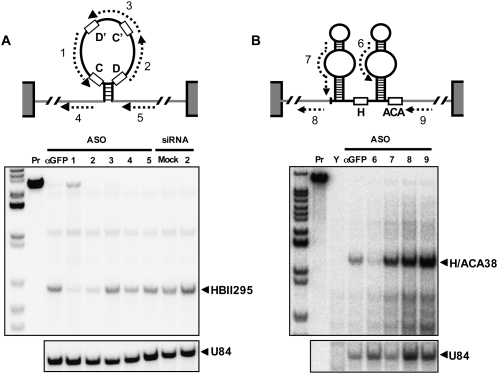
Screening of oligonucleotide positions for efficient knockdown of HBII295 (A) and H/ACA38 (B). The positions of the designed ASOs are indicated by dashed arrows. The canonical snoRNA box sequences are shown by open boxes. The gray lines and gray boxes represent host introns and host exons, respectively.
Optimization of antisense oligonucleotide design
We designed our ASOs using the putative guide sequence upstream of box D in U84 snoRNA, since guide sequences are typically exposed in their single-stranded form. We then assessed the effectiveness of oligonucleotides complementary to other regions within snoRNA and the flanking host introns. HBII295 box C/D snoRNA was successfully knocked down with oligos 1 and 2, both of which were targeted to the putative guide sequences upstream of box D′ and box D, respectively. However, oligo 3, complementary to the sequence spanning the box C′/D′ internal core, and oligos 4 and 5, both of which are complementary to the flanking host intron, were ineffective. Only oligo 6, which targets the bulged loop corresponding to the guide sequence, was able to knock down H/ACA38 snoRNA. These results indicate that the putative guide sequences maintain a single-stranded conformation and, therefore, that ASOs were able to access them.
Knockdown specificity
We next assessed the knockdown specificity of ASOs. Mut1–Mut3 ASOs, which respectively possess one to three mismatched nucleotides to the U84 guide sequence (Fig. 3A), were tested for their knockdown capabilities. As shown in Figure 3C, Mut1 with one nucleotide mismatch decreased the U84 snoRNA level to 35% of basal (cf. αU84: 11%), however, Mut2 and Mut3 were much less effective, indicating that the presence of more than one mismatched nucleotide significantly reduces knockdown ability. To confirm these findings, two box C/D snoRNAs (U83 and U84) with closely related guide sequences were employed as natural knockdown targets. U83 and U84 share a common host gene (BAT1), and are highly related (Jády and Kiss 2000). The putative guide sequences of the two snoRNAs, which contain three nucleotide mismatches to one another, were targeted by ASOs (Fig. 3B). Introduction of the αU84 oligonucleotide led to only a slight decrease in U83 snoRNA (Fig. 3C, lower panel), but it almost completely abolished U84 snoRNA (Fig. 3C, upper panel), supporting our observation that the knockdown was highly specific.
FIGURE 3.
(A) Knockdown specificity was examined using three αU84 derivative oligonucleotides. Mismatched nucleotides are underlined. (B) The putative guide sequences of U83 and U84 snoRNAs are shown. Mismatched nucleotides are underlined. αU84 sequence underscored by an arrow is complementary to the U84 guide sequence. (C) RPA for detection of U84 and U83 snoRNA.
Application to other nuclear ncRNAs
The refined method with optimized conditions was applied to attempt to knock down several snoRNAs. We selected 16 intronic orphan snoRNAs, 13 of which were box C/D-type (Fig. 4A) and three of which were box H/ACA-type (Fig. 4C), from the snoRNABase (http://www-snorna.biotoul.fr/index.php) and three abundant capped snoRNAs (Fig. 4B, U3, U8, U13) as knockdown targets. The ASOs were designed to be complementary to the respective putative guide sequences for the intronic snoRNAs. For the capped snoRNAs, the ASOs were designed based on the sequences of the unstructured 5′ extensions that directly interact with pre-rRNAs. The knockdown conditions were then carried out as follows: 400 pmol of oligonucleotides were nucleofected into 1 × 106 HeLa cells, followed by preparation of total RNAs after 24 h. Each ASO efficiently degraded both box C/D and box H/ACA intronic orphan snoRNAs to <20% of basal (Fig. 4A,C). Capped U3, U8, and U13 were also efficiently degraded (Fig. 4B), and resulted in the production of shorter fragments that appear to correspond to the 3′ terminal box C/D core structures (asterisks in Fig. 4B).
FIGURE 4.
Knockdown of multiple nuclear ncRNAs. Intronic box C/D snoRNA (A), trimethyl capped boxC/D snoRNA (B), intronic box H/ACA snoRNA (C), nucleoplasmic snRNA, U7 snRNA and telomerase RNA (hTR) (D), and an mRNA-like ncRNA, HoxA11 antisense RNA (E), were subjected to knockdown with ASOs (#1 and #2). RPA was employed to detect ncRNA in A, C, and D. Trimethylated boxC/D snoRNA was detected by Northern blot hybridization (B), and HoxA11 antisense RNA was quantified by qRT–PCR (D). RNA prepared from HeLa cells treated with either GFP control (−) or knockdown oligonucleotide (+) was used for RPA and Northern blotting.
Next, the same method was applied to knock down other small nuclear RNAs. U7 snRNA and telomerase RNA (hTR), which are mainly localized to the Cajal body, were utilized as knockdown targets. Both U7 snRNA and hTR possess putative single-stranded regions that form base pairings with histone pre-mRNA and telomere DNA, respectively, and therefore, the single-stranded regions can be targets for ASOs. Both U7 and hTR were efficiently degraded by the corresponding ASOs (Fig. 4D).
We next attempted to knock down a longer mRNA-like ncRNA. We found that antisense noncoding transcript from the human HoxA11 locus (HoxA11 antisense) (Hsieh-Li et al. 1995) is mainly localized to the nucleoplasmic fraction of HeLa cells (data not shown). We designed ASOs for this locus, and nucleofection experiments revealed that one oligonucleotide (oligo #2) reduced the level of HoxA11 antisense RNA to 20% of basal (Fig. 4D). We also found that nine nuclear polyadenylated ncRNAs could be successfully knocked down using this method (Sasaki et al. 2009). Furthermore, we confirmed that this knockdown method was workable in nine human cancer cell lines (A549, HEK293, HeLa, HPB-ALL, Jurkat, LNCap, MCF7, SK-N-SH, and T24), one human fibroblast cell line (MRC5), and one mouse cell line (NIH3T3) (data not shown). Taken together, these results show that our refined method of nuclear RNA knockdown is convincingly versatile for functional analyses of multiple classes of nuclear ncRNAs in mammalian cultured cells.
Validation of functional snoRNA knockdown by detecting the loss of ribose 2′-O-methylation activity
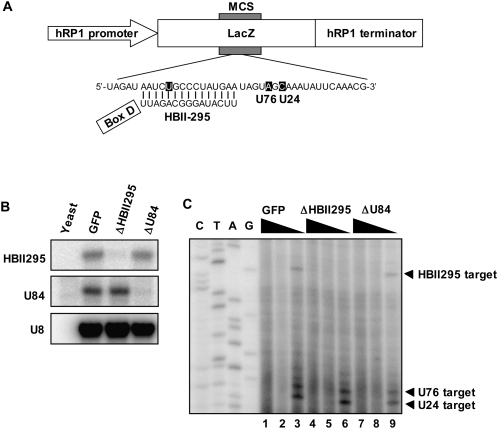
Canonical box C/D snoRNAs direct 2′-O-methylation of pre-rRNAs. The potential role of at least three orphan snoRNAs in site-specific 2′-O-methylation of specific RNAs had been previously examined using artificial RNA polymerase I transcripts. The transcripts were derived from a transfected plasmid construct in which the snoRNA target sequences were inserted between the mouse RNA polymerase I promoter and the terminator. Site-specific methylation was detected at the respective designated sites for cognate orphan snoRNAs in the artificial RNA polymerase I transcripts (Jády and Kiss 2000; Vitali et al. 2005); however, it was unclear whether the methylation was directed by endogenous orphan snoRNAs. Here, we carried out similar experiments using a newly constructed expression plasmid driven by human RNA polymerase I (h-pol I) (Fig. 5A). We attempted to show that endogenous orphan snoRNAs direct site-specific ribose methylation, which can be confirmed by verifying the elimination of a specific orphan snoRNA with ASOs and the loss of 2′-O-methylation of the target site. The transfected plasmid construct possessed a human RNA polymerase I expression cassette in which a 15-nt segment complementary to the putative guide sequence of HBII295 and two target sites of canonical U76 and U24 boxC/D snoRNAs (as controls) were inserted into the multicloning site (MCS) in the LacZ gene sequence (phRP1-HBII295) (Fig. 5A). phRP1-HBII295 was transfected into HeLa cells, and the methylation state of the expressed pol I transcripts was monitored by primer extension. We also confirmed that the artificial pol I transcripts were predominantly localized to the nucleolus by RNA-FISH, indicating that it is a methylation substrate guided by box C/D snoRNAs (data not shown). In the presence of a low concentration of dNTPs, ribose-methylated nucleotides interfere with the passage of reverse transcriptase, which results in the stop of transcription 1 nt before or at the modified nucleotide (Maden et al. 1995). As shown in Figure 5C (lane 3), a stop signal was detected 1 nt upstream of the U residue that is predicted to be 2′-O-methylated by endogenous HBII295 snoRNPs. The stop signal disappeared when HBII295 snoRNA was eliminated by the introduced ASO (Fig. 5C, lane 6), but it was not affected by the U84 snoRNA ASO (Fig. 5C, lane 9). The other sites designed for methylation by the U76 and U24 canonical snoRNAs were also determined to have been methylated (Fig. 5C); however, the stop signals were unchanged when either the HBII295 or U84-targeted ASO was introduced. Taken together, our data demonstrate that HBII295 orphan snoRNA has the ability to direct site-specific ribose methylation. At the same time, we have proven that our ASO-mediated snoRNA knockdown specifically eliminates the in vivo function of the target nuclear ncRNA molecule.
FIGURE 5.
U84 and HBII295 box C/D orphan snoRNA guide site-specific ribose methylation of artificial RNA in human cells. (A) Schematic structure of the ph-pol1 expression construct used for transfection of human cells. The RNA polymerase I promoter and terminator, and a lacZ gene portion with a multiple cloning site (MCS) are indicated. The synthetic DNA fragment was inserted into the MCS. The artificial transcript includes three potential methylation sites (shown by white letters) targeted by HBII295, U76, and U24. The potential base pairing between the guide sequence of HBII295 and the artificial transcript is shown. The HBII295 boxD sequence is underlined. (B) An RPA confirmed the specific degradation of snoRNAs with ASOs for GFP control, HBII295, and U84 (GFP, ΔHBII295, and ΔU84, respectively, shown above the lanes). (C) Primer extension mapping of 2′-O-methylated nucleotides. A 5′-end-labeled oligonucleotide primer was annealed with RNA extracted from GFP, ΔHBII295, and ΔU84 cells (shown above the lanes), and extended with AMV reverse transcriptase in the presence of 1, 0.04, or 0.004 mM dNTPs (indicated as black triangles above the lanes). Lanes C, T, A, and G are dideoxy sequencing reactions performed on the ph-pol1 expression construct.
Cellular and molecular analyses of phenotypic alterations caused by nuclear ncRNA knockdown
We next used U7snRNA to investigate the phenotypic alterations caused by nuclear ncRNA knockdown. U7 snRNA is known to be involved in the 3′ end formation of histone mRNA; therefore, we expected that efficient knockdown of U7 snRNA (>95% knockdown, see Fig. 4D) would lead to defects in histone mRNA maturation. In these experiments, we monitored phenotypic alterations at both the cellular and molecular levels. First, the alteration of cellular phenotypes was monitored by flow cytometry (FACS). HeLa cells were synchronized with a standard double thymidine block. The ASO against either U7 snRNA or control (GFP) was administered to cell cycle arrested cells at G1/S for 12 h, and then the cell cycle was restarted by the removal of thymidine from the medium, followed by monitoring of cell cycle progression by FACS. As shown in Figure 6A, U7 knockdown led to a marked alteration in cell cycle progression: a majority of control cells proceeded to the G2 phase at 6 h after the removal of thymidine (Fig. 6A, GFP), but 100% of ΔU7 cells remained in the S phase (Fig. 6A, ΔU7). Long-range monitoring revealed that the cells did eventually proceed to G2 after 10 h (Fig. 6A, ΔU7), indicating that U7 depletion resulted in a marked delay of S-phase progression.
FIGURE 6.
(A) Phenotype monitoring by detection of cell cycle progression after U7 snRNA knockdown. Synchronized HeLa cells were treated with either αGFP or αU7 oligonucleotides. The cell population in each cell cycle stage (G1: red, S: white, and G2/M: blue) was visualized at time points after retarting the cell cycle by counting cells stained with DAPI. (B) Schematic of the histone mRNA species detected by RPA. The dashed arrow represents the antisense RNA probe. The small box and large arrowhead represent the U7 binding site and processing site, respectively. The extended form appeared following knockdown of U7 snRNA. (C) RPA was used to detect histone H1C, H2A, H3B, H4B, and H4I mRNA in control (GFP) and U7 knockdown cells (ΔU7). The protected RNA species were visualized on an 8% polyacrylamide gel containing 7 M urea. The open and filled arrowheads correspond to the processed and extended mRNA species in B, respectively. (D) Characterization of extended histone mRNA species. An RPA to detect H2A mRNA was carried out with total RNA (lanes 1,2) and poly(A)+ RNA selected with oligo-dT beads (lanes 3,4). The extended (closed arrowhead) and processed (open arrowhead) mRNA was characterized as poly(A)+ and poly(A)− RNA, respectively. (E) Microarray analysis to detect genes regulated by U7 snRNA. Total RNA prepared from HeLa cells transfected with GFP control ASO, αU7 ASO, αU84 ASO, or water (as a control) was hybridized with a human oligo microarray (Agilent). A heat map of 96 transcripts whose signal intensities were more than twofold over- or underrepresented in GFP, ΔU7, or U84 relative to water control is shown. A total of 70 transcripts specifically altered in ΔU7 (shown by arrow) are listed as the candidate targets for control by U7 snRNA. The gene list is shown in Supplemental Table S1.
We next investigated the alteration of gene expression resulting from U7 snRNA knockdown. First, the 3′ terminal structures of five histone mRNAs (H1C, H2A, H3B, H4B, and H4I) were determined using RPAs (Fig. 6C, schematized in B). All five histone mRNAs were almost exclusively expressed as normally processed mRNA forms in control cells (Fig. 6C, open arrowheads, lanes GFP). The additional bands that represent extension downstream from the normal processing sites (see Fig. 6B) appeared specifically in ΔU7 cells (Fig. 6C, filled arrowheads). The appearance of extended mRNA forms was notable in H1C and H2A. Oligo dT-bead selection from total RNA revealed that the extended H2A mRNA species are polyadenylated (Fig. 6D). In addition, real time PCR analysis with oligo dT-bead selected RNA showed that polyadenylated mRNA levels are markedly elevated in H1, H2A, H2B, H3, and H4 histone mRNAs (data not shown). Thus, abnormal production of polyadenylated mRNA species in the absence of U7 snRNA is common in replication-dependent histone gene families. It is likely that the knockdown slowed normal 3′ end processing in ΔU7 cells, resulting in the production of aberrant 3′ extended and polyadenylated transcripts. To further investigate the global effects of U7 knockdown on the HeLa cell transcriptome, we performed a DNA microarray analysis to globally detect changes in whole human polyadenylated mRNA using RNAs prepared from control and ΔU7 HeLa cells. We identified 52 replication-dependent histone genes out of the 70 genes that were specifically over-represented at least twofold in U7 knockdown cells compared with control cells (Fig. 6E; Supplemental Table 1). This finding indicates that elimination of U7 snRNA results in global polyadenylation of replication-dependent histone mRNAs.
In summary, we found that U7 snRNA knockdown led to an alteration of cell cycle progression, a cellular phenotype, as well as alterations in histone mRNA processing, a molecular phenotype. These data showing phenotypic alterations as a result of U7 snRNA knockdown provide further evidence that our refined method is a useful tool for investigation of nuclear ncRNAs that may have multiple effects on cellular phenotypes.
DISCUSSION
We have refined the method for knockdown of nuclear-localized RNAs in cultured mammalian cells. By employing nucleofection, a technique that has been utilized for efficient administration of plasmids into the nucleus, the knockdown efficiency of ASO was greatly improved compared with standard transfection. We observed that fluorescently labeled ASOs introduced by nucleofection were indeed efficiently and rapidly transfected into HeLa cell nuclei (Supplemental Fig. 1). As a result, the introduced ASO is likely to hybridize with target RNAs that are normally cleaved with endogenous RNase H. We also confirmed that siRNA was not able to degrade snoRNAs, even using the same nucleotide sequences as potent ASOs (Figs. 1D, 2A). This result is consistent with the hypothesis that the RNAi machinery in mammalian cells is solely present in the cytoplasm, although several reports have argued that siRNA is capable of degrading specific nuclear RNAs and inducing transcriptional silencing (Morris et al. 2004; Robb et al. 2005; Kim et al. 2006). Indeed, the mechanism of siRNA action on nuclear RNAs has remained elusive. In our procedure, ASOs were able to knock down multiple nuclear RNAs, including nucleoplasmic ncRNA, nucleolar RNAs, and nuclear body-associated RNAs (this study; Sasaki et al. 2009), indicating that ASOs are delivered throughout the nuclear areas where RNaseH activity may be present.
Orphan snoRNA was used to optimize the conditions for ASO design and introduction. So far, more than 100 snoRNAs with unknown targets have been identified. Although the functions of orphan snoRNAs are still unclear, they are believed to be involved in events occurring in the nucleolus, where they are exclusively localized. Our method provides a valuable tool to explore the biological function of orphan snoRNAs by knocking them down. The ASOs for snoRNAs were effective when they were designed using putative guide sequences, both for boxC/D and boxH/ACA snoRNAs (Fig. 2). As expected, the guide sequence remained single stranded, making it accessible to the ASOs. One of the ASOs designed based on the guide sequences of box H/ACA38 (oligo #2) was unable to degrade its target RNA, suggesting that that targeted bulged loop sequence might be structured or bound in some way that indicates that it is not likely to be a bona fide guide sequence. The ASOs that were targeted to the intronic sequence were not able to degrade snoRNAs. This may be due to the fact that endonucleolytic cleavage within introns might not affect the splicing efficiency of pre-mRNA, or because it partially bypasses the splicing-mediated snoRNA maturation pathway though exonucleolytic trimming. The former possibility is supported by evidence that intronic microRNA processing by Drosha does not significantly affect host gene expression (Kim and Kim 2007).
Knockdown of HBII295 boxC/D snoRNA and U7 snRNA led to the expected alterations at the molecular and cellular levels. Our knockdown experiment demonstrated that HBII295 was capable of guiding site-specific 2′-O-methylation as the canonical boxC/D snoRNAs do (Fig. 5). U83, U84, and MBII52 box C/D snoRNAs are believed to have the ability to methylate artificial RNA polymerase I transcripts (Jády and Kiss 2000; Vitali et al. 2005), and, therefore, orphan snoRNPs commonly exhibit methylation activity. Identification of the endogenous methylation targets of these orphan snoRNAs in the nucleolus is an obvious next step. We also found that U7 snRNAs were diminished to <10% of basal after knockdown with ASOs (Fig. 4D). As expected, the mRNA level of normally processed histone H1C was decreased, and an aberrant polyadenylated form was seen (Fig. 6C, H1C). This is consistent with our knowledge that U7 snRNA is essential for 3′ end processing of nonpolyadenylated histone mRNAs. It is also consistent with our hypothesis that defective 3′ end processing in ΔU7 cells leads to the extension of histone pre-mRNA downstream from the processing site, where the existing polyadenylation signal-like sequences may produce these polyadenylated histone mRNAs. In fact, a polyadenylation signal-like sequence (AAUAAA) was found upstream of the mapped polyadenylation site for the H2A gene (data not shown). The processed histone H2A, H3B, H4B, and H4I mRNAs were detected in ΔU7 cells at the same levels as in control cells (Fig. 6C). It is unlikely that the remaining U7 snRNA (<10% of basal) is fully capable of processing histone pre-mRNA, since we estimate that only 40 U7 snRNA molecules remained in each ΔU7 cell. This raises the possibility of the existence of a U7 snRNA independent pathway for the processing of histone mRNA. For example, a 3′-to-5′ exoribonuclease may process histone mRNA to form polyadenylated histone mRNAs, in which the stable stem–loop structure can interfere with exoribonucleolytic processing. Further analysis is needed to clarify the mechanism by which histone mRNAs are processed in ΔU7 cells. Our FACS analysis of synchronized HeLa cells revealed that U7 snRNA is required for normal progression through the cell cycle S phase. This cell cycle phase defect is likely caused by a partial defect in histone mRNA processing. Alternatively, it is an intriguing possibility that the appearance of aberrant polyadenylated histone mRNA somehow affects cell cycle progression.
In summary, our refined system for knockdown of nuclear ncRNAs is able to specifically target and degrade multiple nuclear ncRNAs, whose function has been difficult to analyze by other methods. Upon knockdown, microarray analysis can be used to detect alterations caused by the elimination of specific ncRNAs, and in some cases, the resulting phenotypic changes can be detected at the cellular level. There are large numbers of recently identified ncRNAs of unknown function. Since many of them are conserved only in mammalian species, our system represents a powerful tool to explore the biological function of these nuclear ncRNAs.
MATERIALS AND METHODS
RNA analyses
Total RNA was prepared using Sepasol (Nacalai). RPAs were performed as described (Hirose and Steitz 2001). A PCR fragment that covers each ncRNA region was cloned into pGEM-T Easy vector (Promega), followed by digestion with an appropriate restriction enzyme for in vitro transcription. Then, 3–10 μg of total RNA was hybridized with a 32P-labeled antisense RNA probe that was synthesized using T7 or SP6 RNA polymerase (TaKaRa). RNase A/T1 digestion excluded unhybridized single-stranded RNA probes. The protected RNA fragments were separated by polyacrylamide gel electrophoresis (PAGE) on an 8% gel with 7 M urea. Radioactive RNA bands were visualized using a Bioimaging analyzer BAS3000. For Northern blot hybridization, total RNA was run on a 8% PAGE gel with 7 M urea followed by electroblotting to a Hybond N+ nitrocellulose membrane (GE Healthcare Life Sciences). The blotted RNAs were covalently fixed to the membrane by UV cross-linking. Antisense RNA probes were synthesized using a DIG RNA Labeling kit (Roche), and hybridized in DIG Easy Hyb reagent at 60°C overnight. The membrane was washed, and bands were detected with the DIG Wash and Block Buffer Set (Roche). Reverse transcription–quantitative polymerase chain reaction (qRT-PCR) was performed as described (Ideue et al. 2007). Briefly, total RNA (1 μg) was reverse transcribed using the QuantiTect reverse transcription kit (Qiagen). Primers were designed using Primer3 software (http://www-genome.wi.mit.edu/ftp/distribution/software/) and purchased from Invitrogen. Aliquots of cDNA were subjected to real time PCR, performed using a LightCycler 480 SYBR Green I Master (Roche Diagnostics) according to the manufacturer's protocol. For each reaction, no amplification was observed when reverse transcriptase was omitted.
Transfection of antisense oligonucleotides
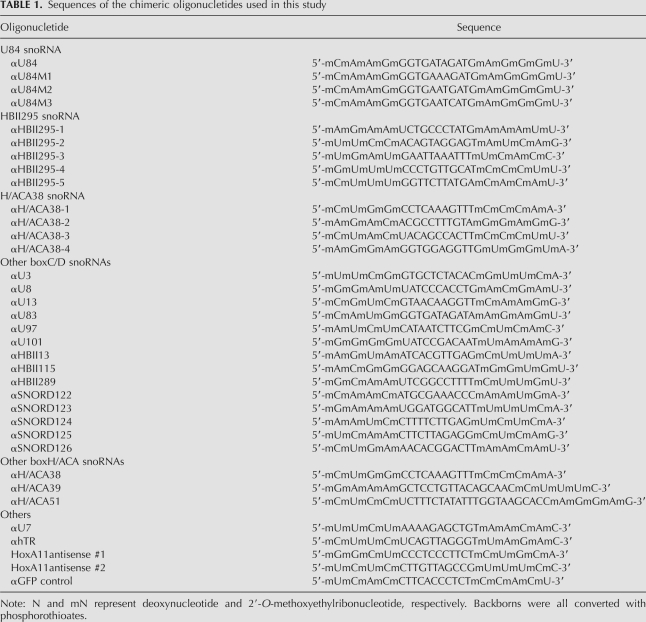
The ASOs (IDT) used for knockdown experiments were phosphorothioate converted at their backbones to increase stability. Five terminal nucleotides from the 5′ and 3′ ends were substituted with 2′-O-methoxyethyl ribonucleotides. Trypsinized HeLa cells (1 × 106 cells) were suspended in 100 μL of Solution R from the Cell Line Nucleofector Kit R (Amaxa Biosystems) and mixed with the ASOs (4 μM final concentration). Transfection was carried out in an electroporation cuvette using a Nucleofector instrument (Amaxa Biosystems). The transfected cells were transferred to fresh DMEM plus 10% FBS, incubated at 37°C and 5% CO2 for 24 h, and then the cells were harvested for RNA preparation. The ASOs were designed as complementary sequences to snRNA single-stranded guide sequences. The sequences of the ASOs used are provided in Table 1. For transfection of fluorescently labeled ASOs, the Cy3-labeled ASO (Cy3- αU84) was synthesized (IDT), and transfected into HeLa cells either by nucleofection or lipofection with Lipofectamine 2000 reagent (Invitrogen).
TABLE 1.
Sequences of the chimeric oligonucletides used in this study
Methylation site mapping by primer extension
Methylation site mapping was carried out according to the methods of Kiss and Jády (2004). A deoxyoligonucelotide primer (5′-GCGAATTCGAGCTCGGTAC-3′) was annealed with 10 μg of total RNA prepared from HeLa cells treated with multiple ASOs; 1 pmol of 5′ end-labeled primer was denatured at 95°C for 1 min and annealed at 43°C for 10 min in 50 mM Tris-HCl (pH 8.0), 50 mM KCl, 5 mM MgCl2, 5 mM DTT, and 50 ng/mL gelatin. The primer extention reaction was carried out in 50 mM Tris-HCl (pH 8.0), 50 mM KCl, 5 mM MgCl2, 5 mM DTT, and 50 ng/mL gelatin in the presence of 10 U of AMV reverse transcriptase, 40 U of RNasin (Promega), and 1, 0.04, or 0.004 mM dNTP at 43°C for 30 min. Template RNA was digested in 250 ng/mL RNase A at 43°C for 10 min. The extension products were resolved by electrophoresis on a 12.5% gel.
Synchronization of cell cycle
The cell cycle of HeLa cells was synchronized by release from either G1/S arrest or mitotic arrest. For G1/S arrest, the cells were treated with 2.5 mM thymidine for 14–16 h twice with a 9-h interval of growth in-between without the drug. The cells were then incubated in the absence of the drug for the indicated times and then harvested. Half of the cells were processed for FACS analysis of cell cycle distribution by DAPI staining, and the remaining half were used for RNA preparation. To construct a DNA content histogram, FACS data were analyzed by Cell Lab Quanta Sc software (Beckman Coulter).
DNA Microarray
HeLa cells were nucleofected with water, αGFP, αU7, or αU84, and incubated for 24 h, and then total RNA was prepared and labeled with Cy3. Samples were hybridized to a Human Oligo Microarray (G4112F, Agilent) according to the manufacturer's protocol. Arrays were scanned with a G2565BA Microarray Scanner System (Agilent), and the resulting data were analyzed using GeneSpring GX software (Agilent).
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Toutai Mituyama and the members of the Hirose laboratory for valuable discussions and help. This research was supported by a grant from the New Energy and Industrial Technology Development Organization (NEDO) (to T.H.), a grant from the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT) (to T.H.), and the Astellas Foundation for Research on Metabolic Disorders (to T.H.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1657609.
REFERENCES
- Bachellerie JP, Cavaillé J, Hüttenhofer A. The expanding snoRNA world. Biochimie. 2002;84:775–790. doi: 10.1016/s0300-9084(02)01402-5. [DOI] [PubMed] [Google Scholar]
- Cavaillé J, Buiting K, Kiefmann M, Lalande M, Brannan CI, Horsthemke B, Bachellerie JP, Brosius J, Huttenhofer A. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc Natl Acad Sci. 2000;97:14311–14316. doi: 10.1073/pnas.250426397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z, Marzluff WF. Formation of the 3′ end of histone mRNA: Getting closer to the end. Gene. 2007;396:373–390. doi: 10.1016/j.gene.2007.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Pogacic V. Biogenesis of small nucleolar ribonucleoproteins. Curr Opin Cell Biol. 2002;14:319–327. doi: 10.1016/s0955-0674(02)00334-4. [DOI] [PubMed] [Google Scholar]
- Frey MR, Matera AG. Coiled bodies contain U7 small nuclear RNA and associate with specific DNA sequences in interphase human cells. Proc Natl Acad Sci. 1995;92:5915–5919. doi: 10.1073/pnas.92.13.5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose T, Steitz JA. Position within the host intron is critical for efficient processing of box C/D snoRNAs in mammalian cells. Proc Natl Acad Sci. 2001;98:12914–12919. doi: 10.1073/pnas.231490998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh-Li HM, Witte DP, Weinstein M, Branford W, Li H, Small K, Potter SS. Hoxa 11 structure, extensive antisense transcription, and function in male and female fertility. Development. 1995;121:1373–1385. doi: 10.1242/dev.121.5.1373. [DOI] [PubMed] [Google Scholar]
- Hüttenhofer A, Brosius J, Bachellerie JP. RNomics: Identification and function of small, non-messenger RNAs. Curr Opin Chem Biol. 2002;6:835–843. doi: 10.1016/s1367-5931(02)00397-6. [DOI] [PubMed] [Google Scholar]
- Ideue T, Sasaki YT, Hagiwara M, Hirose T. Introns play an essential role in splicing-dependent formation of the exon junction complex. Genes & Dev. 2007;21:1993–1998. doi: 10.1101/gad.1557907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jády BE, Kiss T. Characterization of the U83 and U84 small nucleolar RNAs: Two novel 2′-O-ribose methylation guide RNAs that lack complementarities to ribosomal RNAs. Nucleic Acids Res. 2000;28:1348–1354. doi: 10.1093/nar/28.6.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YK, Kim VN. Processing of intronic microRNAs. EMBO J. 2007;26:775–783. doi: 10.1038/sj.emboj.7601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Villeneuve LM, Morris KV, Rossi JJ. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat Struct Mol Biol. 2006;13:793–797. doi: 10.1038/nsmb1142. [DOI] [PubMed] [Google Scholar]
- Kiss T, Jády BE. Functional characterization of 2′-O-methylation and pseudouridylation guide RNAs. Methods Mol Biol. 2004;265:393–408. doi: 10.1385/1-59259-775-0:393. [DOI] [PubMed] [Google Scholar]
- Kiss T, Fayet E, Jády BE, Richard P, Weber M. Biogenesis and intranuclear trafficking of human box C/D and H/ACA RNPs. Cold Spring Harb Symp Quant Biol. 2006;71:407–417. doi: 10.1101/sqb.2006.71.025. [DOI] [PubMed] [Google Scholar]
- Maden BE, Corbett ME, Heeney PA, Pugh K, Ajuh PM. Classical and novel approaches to the detection and localization of the numerous modified nucleotides in eukaryotic ribosomal RNA. Biochimie. 1995;77:22–29. doi: 10.1016/0300-9084(96)88100-4. [DOI] [PubMed] [Google Scholar]
- Mariner PD, Walters RD, Espinoza CA, Drullinger LF, Wagner SD, Kugel JF, Goodrich JA. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol Cell. 2008;29:499–509. doi: 10.1016/j.molcel.2007.12.013. [DOI] [PubMed] [Google Scholar]
- Morris KV, Chan SW, Jacobsen SE, Looney DJ. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004;305:1289–1292. doi: 10.1126/science.1101372. [DOI] [PubMed] [Google Scholar]
- Pan ZQ, Prives C. Assembly of functional U1 and U2 human-amphibian hybrid snRNPs in Xenopus laevis oocytes. Science. 1988;241:1328–1331. doi: 10.1126/science.2970672. [DOI] [PubMed] [Google Scholar]
- Peculis BA, Steitz JA. Disruption of U8 nucleolar snRNA inhibits 5.8S and 28S rRNA processing in the Xenopus oocyte. Cell. 1993;73:1233–1245. doi: 10.1016/0092-8674(93)90651-6. [DOI] [PubMed] [Google Scholar]
- Prasanth KV, Prasanth SG, Xuan Z, Hearn S, Freier SM, Bennett CF, Zhang MQ, Spector DL. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249–263. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- Robb GB, Brown KM, Khurana J, Rana TM. Specific and potent RNAi in the nucleus of human cells. Nat Struct Mol Biol. 2005;12:133–137. doi: 10.1038/nsmb886. [DOI] [PubMed] [Google Scholar]
- Sasaki YTF, Ideue T, Sano M, Mituyama T, Hirose T. MENɛ/β noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci. 2009;106:2525–2530. doi: 10.1073/pnas.0807899106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamovsky I, Ivannikov M, Kandel ES, Gershon D, Nudler E. RNA-mediated response to heat shock in mammalian cells. Nature. 2006;440:556–560. doi: 10.1038/nature04518. [DOI] [PubMed] [Google Scholar]
- Tycowski KT, Shu M-D, Steitz JA. Requirement for intron-encoded U22 small nucleolar RNA in 18S ribosomal RNA maturation. Science. 1994;266:1558–1561. doi: 10.1126/science.7985025. [DOI] [PubMed] [Google Scholar]
- Tycowski KT, Kolev NG, Conrad NK, Fok V, Steitz JA. The ever-growing world of small nuclear ribonucleoproteins. In: Gesteland RF, et al., editors. The RNA World. 3rd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2006. pp. 327–368. [Google Scholar]
- Vickers TA, Koo S, Bennett CF, Crooke ST, Dean NM, Baker BF. Efficient reduction of target RNAs by small interfering RNA and RNase H-dependent antisense agents. A comparative analysis. J Biol Chem. 2003;278:7108–7118. doi: 10.1074/jbc.M210326200. [DOI] [PubMed] [Google Scholar]
- Vitali P, Basyuk E, Le Meur E, Bertrand E, Muscatelli F, Cavaillé J, Huttenhofer A. ADAR2-mediated editing of RNA substrates in the nucleolus is inhibited by C/D small nucleolar RNAs. J Cell Biol. 2005;169:745–753. doi: 10.1083/jcb.200411129. [DOI] [PMC free article] [PubMed] [Google Scholar]