Abstract
More than 130 mutations in human mitochondrial tRNA (mt-tRNA) genes have been correlated with a variety of neurodegenerative and neuromuscular disorders. Their molecular impacts are of mosaic type, affecting various stages of tRNA biogenesis, structure, and/or functions in mt-translation. Knowledge of mammalian mt-tRNA structures per se remains scarce however. Primary and secondary structures deviate from classical tRNAs, while rules for three-dimensional (3D) folding are almost unknown. Here, we take advantage of a myopathy-related mutation A7526G (A9G) in mt-tRNAAsp to investigate both the primary molecular impact underlying the pathology and the role of nucleotide 9 in the network of 3D tertiary interactions. Experimental evidence is presented for existence of a 9-12-23 triple in human mt-tRNAAsp with a strongly conserved interaction scheme in mammalian mt-tRNAs. Mutation A7526G disrupts the triple interaction and in turn reduces aspartylation efficiency.
Keywords: mammalian mitochondria, tRNA, structure probing in solution, pathology-related mutations, mitochondrial disorder
INTRODUCTION
The human mitochondrial (mt) genome codes for 13 proteins, 2 rRNAs, and 22 tRNAs. The 13 proteins are subunits of the respiratory chain complexes and contribute to ATP synthesis. The 22 tRNA genes are hotspots for mutations, which are either polymorphisms (neutral consequences) or related to diseases (see the MITOMAP database) (Ingman and Gyllensten 2006). Understanding the molecular impact of mutations on the multiple facets of tRNA biogenesis, structure, and function has been tackled by several complementary in vitro and in vivo studies. The mosaic of results collected so far favors diverse molecular impacts, with, in some cases, cumulative effects (Wallace et al. 1999; Florentz et al. 2003; Jacobs 2003; DiMauro 2006; Shapira 2006). Due to the lack of knowledge on structure/function relationships of mammalian mt-tRNAs, already recognized as “bizarre” at the primary and secondary structural levels (e.g., Helm et al. 2000), pathologic or polymorphic incidence of a mutation can still not be predicted (Florentz and Sissler 2001; Florentz et al. 2003; Levinger et al. 2004; McFarland et al. 2004). Knowledge on three-dimensional (3D) folding rules is scarce.
Human mt-tRNAAsp is a typical representative of the structural degeneration of mammalian mt-tRNAs. Like 14 out of the 22 mt-tRNAs, it has a biased A-, U-, and C-rich nucleotide content. It can fold into a theoretical secondary cloverleaf structure but with small D- and T-loops, and several conserved nucleotides that would be expected to induce and stabilize the 3D fold (Helm et al. 2000) characteristic of classical tRNAs (Juehling et al. 2009) appear to be absent. The gene for this tRNA is affected by mutation A7526G in a patient with a mitochondrial myopathy (Seneca et al. 2005). The molecular impact of this mutation is unknown but raises an interesting structural question. Indeed, mutation A7526G generates a nucleotide transition at a position of potential structural importance for the folding and the 3D structure of the RNA, namely position 9 (classical tRNA numbering, Juehling et al. 2009). Investigation of this mutation should assist not only in gaining a better understanding of the initial molecular impact responsible for pathology but also in gaining a better knowledge of rules governing human mt-tRNA 3D structures.
Here, we present a mutagenic analysis involving functional and structural studies in solution on human mt-tRNAAsp derivatives. Enzymatic structural probing on both wild-type and mutant in vitro transcripts reveals a primary impact of mutation A7526G on structure, which subsequently hampers aminoacylation properties of mt-tRNAAsp. Compensatory mutations restore both activity and structure and strongly support the existence of a tertiary interaction between nucleotide 9 and base-pair 12-23 within the D-stem. Compilation of sequences within the specifically dedicated database “Mamit-tRNA” (Pütz et al. 2007) extends the existence of this tertiary interaction to 97.8% of all known mammalian mt-tRNAAsp sequences and to a large group of additional mammalian mt-tRNAs.
RESULTS AND DISCUSSION
Predicted impact of mutation A7526G (A9G) on mt-tRNAAsp structure
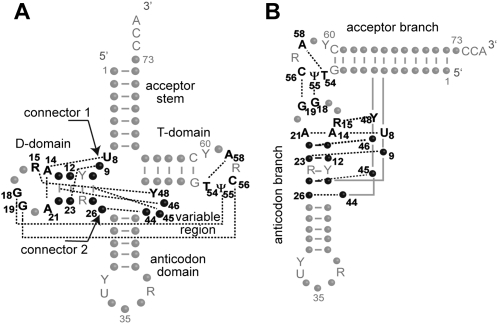
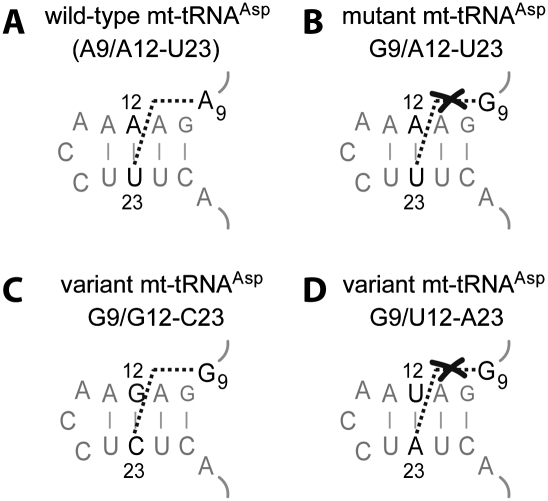
In classical tRNAs (i.e., from bacteria, archae, and cytosol of eukaryotes), nine tertiary interactions sustain the 3D fold (Fig. 1), three in and between D- and T-loops, and six forming the core of the structure (Kim et al. 1974; Westhof et al. 1985; for review, see Giegé et al. 1993; Dirheimer et al. 1995). Myopathy-related mutation A7526G (Seneca et al. 2005) affects nucleotide at position 9 in tRNAAsp, potentially part of the 3D network. Mutation A9G (mutant G9/A12-U23) would break the link between residues 9 and 23 present in the wild-type triple A9/A12-U23. To investigate this hypothesis, the structure as well as the function of wild-type and mutant G9/A12-U23 mt-tRNAAsp were compared. Solution structures were investigated by enzymatic probing while the aminoacylation properties were established in the presence of recombinant human mt aspartyl-tRNA synthetase (mt-AspRS). Two additional variants, tRNAAspG9/G12-C23 (with a double transition in the D-stem) and variant tRNAAspG9/U12-A23 (with a double transversion) were also studied.
FIGURE 1.
Tertiary interaction network in canonical tRNAs. (A) Secondary cloverleaf structure. (B) Tertiary “L-shaped” structure. Structural domains are indicated, and the tertiary interaction network is emphasized by dotted lines and boldface characters (for review, see Giegé et al. 1993; Dirheimer et al. 1995). Nucleotide positions involved in the nine tertiary interactions are either strictly conserved, semiconserved (Y for pyrimidine, R for purine), or not conserved (black dots). Other strictly conserved or semiconserved nucleotides are indicated by gray letters. Numbering is according to Juehling et al. (2009). Residues 8, 9, and 26 form connectors between domains. Residue 9 forms a triple by interaction with residue 23 of base-pair 12-23.
Comparative structural investigation
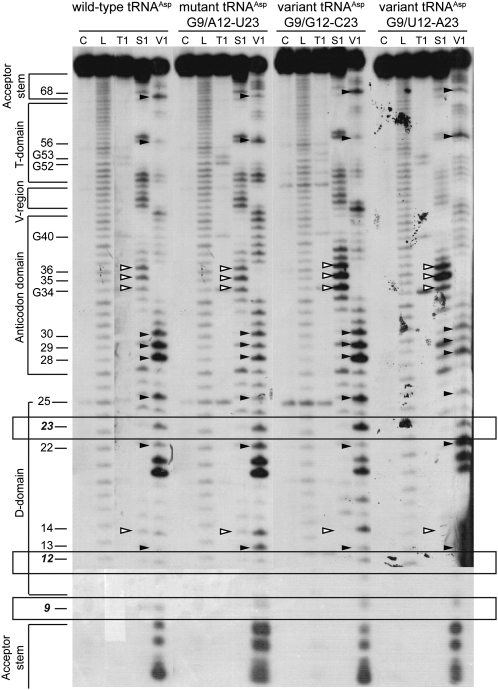
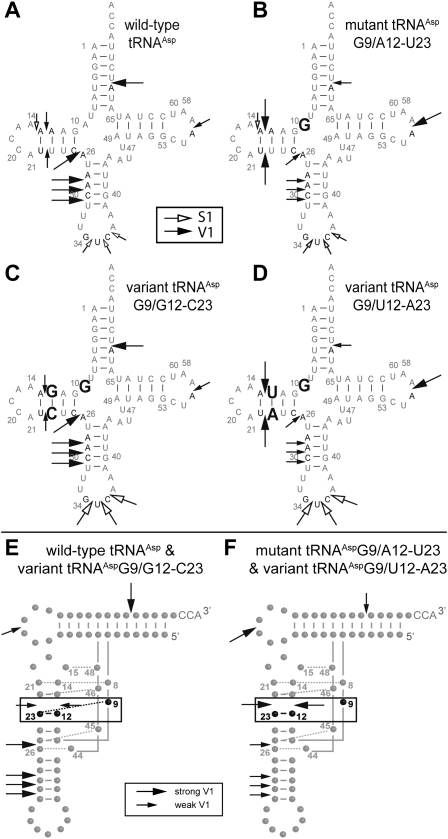
Structural probing of the transcripts was performed with nuclease S1 (cleaves single-stranded regions) and ribonuclease V1 (cleaves double-stranded or highly structured regions) (Giegé et al. 1999) on 5′-end labeled tRNAs. Typical cleavage profiles are shown in Figure 2. They fit with cloverleaf structures of the RNAs, stems being cleaved by ribonuclease V1 and loops by nuclease S1. Interestingly, only subtle but significant differences in cleavage intensities appear. There is no general structural breakdown for the pathology-related mutant. Differences in profiles are highlighted in Figure 3, A–D. None of the phosphodiester bonds located 3′ of residues 9, 12, and 23, are cleaved by nuclease S1, but all are cleaved by ribonuclease V1. This is in agreement with involvement of the three nucleotides in a higher-order structure, in wild type as well as in each of the three other molecules. Focus on base-pair 12-23 in the D-stem reveals stronger sensitivity to V1 ribonuclease in mutant mt-tRNAAspG9/A12-U23 and in variant mt-tRNAAspG9/G12-C23, as compared with the cleavage pattern in wild-type tRNAAsp and variant G9/U12-A23. This is in favor of a local structural effect of mutation A9G on base-pair 12-23. Furthermore, intensity of cleavages is affected at a distance from the mutation as well in the acceptor stem, the T-domain, and the anticodon arm as within the D-domain. The four tRNAs investigated sort into two sets based on their nuclease susceptibilities, wild-type and variant mt-tRNAAspG9/G12-C23 on one hand, and mutant mt-tRNAAspG9/A12-U23 and variant mt-tRNAAspG9/U12-A23 on the other. The first tRNAs (Fig. 3A,C) show strong V1-cuts within the acceptor stem (residue 68), anticodon arm (residues 28, 29, and 30), and residue 25 (next to connector 2), as well as weak V1-cuts within the T-loop (residue 56). The second tRNAs (Fig. 3B,D) exhibit the opposite strength in cleavage patterns, namely, weak V1-cuts within the acceptor stem (residue 68), anticodon arm (residues 28, 29, and 30), and residue 25 and a strong V1-cut within the T-loop (residue 56). The opposite ribonuclease V1 reactivity within the two sets is emphasized on 3D folds (Fig. 3E,F) and suggests differential overall flexibilities. Perturbations within the 9/12-23 interaction influence stacking interactions downstream of the anticodon arm, upstream of base pair 12-23, and within the T-loop and acceptor stem. In conclusion, 1) pathology-related mutation A9G in connector 1 influences the overall plasticity of the tRNA without breakdown of the secondary or tertiary structure, but with subtle local and long-range effects, and 2) effects of mutation A9G can be compensated by a double transition at base-pair 12-23 in the D-stem.
FIGURE 2.
Enzymatic structure probing of different human mt-tRNAAsp transcripts. tRNAs were 5′-labeled and treated with different probes as indicated, cleavage products were separated on 12% polyacrylamide/8M urea gels, and tRNAs were revealed by autoradiography. Probing was conducted at 25°C using nuclease S1 and ribonuclease V1. (C) Control; (L) alkaline ladder; (T1) denaturing ribonuclease T1. Secondary structural domains are indicated. Nucleotides that display different reactivities within the four transcripts are highlighted by white (nuclease S1) and black (ribonuclease V1) arrowheads. Bands corresponding to cleavages at residues 9, 12, and 23 are enclosed in rectangles.
FIGURE 3.
Differential sensitivity of mt-tRNAAsp derivatives to enzymatic probes. (A–D) Cloverleaf structures. Mutated residues are shown in boldface type for each variant. Only differential cleavages between the four molecules are indicated. Black and white arrows stand for cleavages by ribonuclease V1 and nuclease S1, respectively. Large and small arrows correspond to strong and weak cleavages, respectively. (E,F) L-shaped folds. Mt-tRNAAsp transcripts fall into two structural sets according to their reactivity to ribonuclease V1. (E) tRNAs active in aminoacylation and with unchanged reactivity to enzymatic probes as compared with wild type. (F) tRNAs with altered structures and impaired aminoacylation properties in comparison with wild-type tRNA. The differential reactivities toward ribonuclease V1 between the two sets are highlighted. Residue 9 and base-pair 12-23 are boxed and indicated in boldface type. Other predicted tertiary interactions in the core of the 3D structure are indicated by dotted gray lines. Numbering is according to Pütz et al. (2007).
Ricochet impact on functional properties
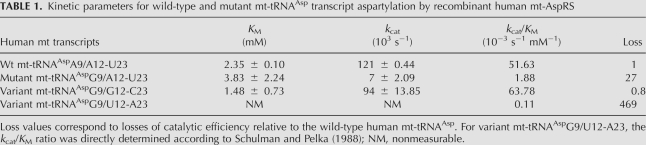
Aminoacylation properties (kcat and KM parameters) of the four transcripts were determined by aspartylation assays in the presence of human mt-AspRS (Table 1). As anticipated, A9G mutation leads to a significant 27-fold loss of catalytic efficiency (kcat/KM) with kcat value decreased 17-fold and KM value increased 1.6-fold. Variant tRNAAspG9/G12-C23 behaves similar to wild-type mt-tRNAAsp (with kcat and KM values in the same range, i.e., KM of 1–2 μM and kcat of 100 × 10−3 s-1). Variant tRNAAspG9/U12-A23 is a poor substrate for mt-AspRS, with an estimated 470-fold loss in aminoacylation efficiency. Interestingly, the kinetic properties for aspartylation overlap with the structural features, with efficient aminoacylation of wild-type tRNAAsp and variant tRNAAspG9/G12-C23 but not mutant tRNAAspG9/A12-U23 and variant tRNAAspG9/U12-A23. Conserved KM values are consistent with retention of a global 3D structure for all these molecules and their ability to be recognized by mt-AspRS. Differences between kcat values reflect an effect of the mutation on catalysis. This is linked to changes in the overall plasticity of the molecules (as a long-range consequence of the perturbation within the core of the tRNA rather than to perturbed binding [Giegé 2008]).
TABLE 1.
Kinetic parameters for wild-type and mutant mt-tRNAAsp transcript aspartylation by recombinant human mt-AspRS
Residues 9-12-23 are partners in a tertiary interaction in mammalian mt-tRNAs
The present data show that introduction of mutation A9G into tRNAAsp leads to both structural perturbation and decrease in aminoacylation efficiency. It is possible to recover both structure and function with a second site mutation in the D-stem by replacing base-pair A12-U23 by G12-C23. This reveals a link between the three nucleotides (Fig. 4A,C). This link is however broken within pathology-related mutant and within variant G9/U12-A23 for which no compensatory effects where found (Fig. 4B,D). In classical tRNAs, nucleotide 9 interacts with nucleotide 23 through their Hoogsteen faces (i.e., N7-face of purines, C6-face of pyrimidines) (Kim et al. 1974; Westhof et al. 1985). A trans-Hoogsteen interaction is possible between A9 and A23, as well as between G9 and C23, but not between G9 and U23 (Leontis et al. 2002).
FIGURE 4.
Tolerated nucleotide combinations for positions 9-12-23 in human mt-tRNAAsp variants. The subdomain shown includes connectors 1 and 2 and the D-stem. (A) Wild-type sequence. (B) Pathology-related mutant A9G. (C) Sequence of variant G9/G12-C23 with a double transition in the D-stem. (D) Sequence of variant G9/U12-A23 with a double transversion in the D-stem.
Analysis of the 136 available mammalian mt-tRNAAsp sequences within the “Mamit-tRNA” database (Pütz et al. 2007) allows a deeper insight into the conservation of residues 9, 12, and 23. Nucleotide 9 is strictly conserved (100%) as an A residue. The combination of nucleotides A9/A12-U23 (as in wild-type human mt-tRNAAsp) is present in 97.8% of sequences, while in the 2.2% of remaining cases, the combination A9/U12-A23 is found. Further compilation of residues 9, 12, and 23 in the other tRNA families within the database (except tRNASer(AGY) and tRNASer(UCN), both having important deviating secondary structure features) (Watanabe et al. 1994; Hayashi et al. 1997) reveals that 12 tRNA families share local sequence features with mt-tRNAAsp. All possess A9 and either A12-U23 or U12-A23 (results not shown). Interestingly, three families [Ile, Tyr, Leu(UUR)] possess a strongly conserved G residue at position 9 and either A12-U23 or U12-A23 pairs, the combinations present in the pathology-related tRNAAsp mutant and in the noncompensating variant studied herein. The natural existence of associations of nucleotides at positions 9, 12, and 23, experimentally found to be nonoptimal in the frame of tRNAAsp, supports the importance of the structural neighborhood of this triple. As seen in Figure 1B, residues 9/12-23 are stacked in the middle of the core domain in classical tRNAs. The existence of these neighboring tertiary interactions in mammalian mt-tRNAs needs to be investigated in a systematic manner.
Altered tRNA flexibility as the primary molecular effect of myopathy-related mutation A7526G in human mt-tRNAAsp
The present data on aminoacylation and global structure of the wild-type and mutant human mt-tRNAAsp reveal a primary mechanism of impact of the pathology-related mutation A7526G in vitro. The mutation alters the tRNA structural plasticity, which in turn hampers aminoacylation. Several cases have been reported in which structural impairments subsequently interfere with steps of the tRNA life-cycle (for review, see Florentz et al. 2003; Wittenhagen and Kelley 2003; Levinger et al. 2004). For example, mutation A4317G (A58G) in tRNAIle leads to an alternative T-stem structure and structural fragility of the tRNA incompatible with aminoacylation (Kelley et al. 2001). Mutation A3243G (A14G) in tRNALeu(UUR) leads to a two- to 10-fold loss in aminoacylation efficiency and either to homodimerization of the tRNA (Wittenhagen and Kelley 2002) or to a rearrangement within the D-stem and loop (Sohm et al. 2003). Mutation T3271C (T40C) in tRNALeu(UUR) destabilizes the anticodon domain, which interferes with binding of LeuRS (Wittenhagen et al. 2003). Pathological mutations C5877T (C15A) and T5874C (A22G) in human mt-tRNATyr trigger a structural destabilization in the D-loop that propagates toward the anticodon arm and leads to a loss of aminoacylation efficiency of 40- and 600-fold, respectively (Bonnefond et al. 2008). Mutation A10044G (A59G) induces a weaker interaction between the D- and the T-loops, which perturbs interaction of mt-tRNAGly with the CCA adding enzyme (Tomari et al. 2003).
The possibility that the structural perturbation introduced by mutation A7526G (A9G) in tRNAAsp has functional consequences outside the frame of aminoacylation remains to be tested. Pathology-related mutation could, for example, interfere with the local and/or global modification pattern of the affected tRNA, which in turn could affect its functional properties. Such situations were described for mutations A3243G (A14G) in the D-loop of tRNALeu(UUR) and A8344G (A55G) in the T-loop of tRNALys, which affect the modification pattern in the D-domain (Helm et al. 1999a) and dominantly in the anticodon loops (Yasukawa et al. 2001; Kirino et al. 2004). This hinders codon reading and inhibits translation. Alteration of the post-transcriptional modification patterns can also lead to accelerated degradation of tRNAs (Kadaba et al. 2004). Pathology-related mutation A7526G in mt-tRNAAsp affects a position often methylated in mt-tRNAs (Juehling et al. 2009). In human mt-tRNALys, m1A9 is a key modification for optimal folding into a functional structure. This modification hinders base-pairing of residue 9 with nucleotide 64 in the T-stem and shifts the dynamic equilibrium of a pre-tRNA toward the functional structure (Helm et al. 1998, 1999b; Voigts-Hoffmann et al. 2007; Kobitski et al. 2008). Methylation of residue 9 at atomic position 1 (a Watson–Crick position) in mt-tRNAAsp would not interfere with formation of the triple interaction (with residue 23 via a Hoogsteen interaction) and might not be hindered by the pathology-related mutation A9G. Indeed, the recent characterization of the human mt-RNaseP revealed its association with a G9 methyl-transferase activity, which might act conjointly to 5′-processing of the pre-tRNA (Holzmann et al. 2008).
Outlook
More and more examples of human mt-tRNA mutations with primary effects on tRNA structure are being reported, including mutation A7526G (A9G) in mt-tRNAAsp, which has further consequences for aminoacylation properties. Dissecting the molecular incidence of the pathology-related mutation in turn sheds light on structural features, that is, the existence of a triple interaction involving residues 9, 12, and 23 in human and mammalian mt-tRNAAsp. Fine-tuned structural knowledge on “bizarre” mt-tRNAs remains of fundamental necessity to allow understanding the precise molecular mechanisms of many pathology-related mt-tRNA mutations and to distinguish them from polymorphisms. Exploration of the molecular impact of pathology-related mutations will further help to decipher structural properties of mt-tRNAs, and vice versa, structural knowledge on mt-tRNAs will further help to unravel the impact of mutations.
MATERIALS AND METHODS
Human mitochondrial tRNA transcripts and aspartyl-tRNA synthetase
Human mt-tRNAAsp has been cloned previously (Bonnefond et al. 2005). Derivative plasmids expressing mutant mt-tRNAAspG9/A12-U23 and variants mt-tRNAAspG9/G12-C23 and mt-tRNAAspG9/U12-A23 were obtained with the QuikChange Site-Directed Mutagenesis Kit (Stratagene). In vitro transcriptions were performed as described (Bonnefond et al. 2005). Human mt-AspRS was previously cloned into pQE70 vector that introduces a (His)6-tag to the C-term of the expressed protein. Overproduction and purification steps were conducted as described (Bonnefond et al. 2005).
Aminoacylation assays
Assays were performed as described (Bonnefond et al. 2005) in 50 mM HEPES-KOH (pH 7.5), 25 mM KCl, 12 mM MgCl2, 2.5 mM ATP, 0.2 mg/mL BSA, 1 mM spermine, 32 μM [3H]-aspartic acid (208 Gbq/mmol), transcript, and enzyme. Transcripts were renaturated by heating at 60°C for 60 sec in H2O and slow cooling to room temperature before aminoacylation performed at 25°C. Kinetic parameters kcat and KM were derived from Lineweaver-Burk plots obtained under adequate ranges of tRNA (100 nM to 4 μM) and mt-AspRS (12 nM to 500 nM). For mt-tRNAAspG9/U12-A23 variant, direct determination of the ratio kcat/KM was established as described in Schulman and Pelka (1988). Experimental errors in kcat and KM varied at most by 20%. Numerical values are averages of at least four independent experiments.
Structural mapping
Enzymatic probes specific for single-stranded (nuclease S1) and double-stranded or highly structured domains (RNase V1) were used. Labeling of transcript 5′-end and subsequent purification of tRNA molecules from polyacrylamide gels were carried out as described earlier (Helm et al. 1998). Before probing, transcripts were denaturated by heating at 60°C in H2O and slowly cooled down to room temperature. Enzymatic digestions were performed as described (Helm et al. 1998). Briefly, transcripts were incubated in 50 mM HEPES-KOH (pH 7.5), 10 mM MgCl2, and 25 mM KCl for 5 min at 25°C. For digestion with S1 nuclease, 1 mM ZnCl2 was added. Reaction mixtures (10 μL) contained 30,000 Cerenkov cpm of transcript supplemented with 2 μg of unlabeled Escherichia coli total tRNA and either 12.8 U of nuclease S1 or 3.6 × 10−2 U of RNase V1. Cleavage reactions were performed during 5 min at 25°C and were stopped by adding one volume of 0.6 M Na-acetate (pH 5.0), 3 mM EDTA, and 0.1 mg/mL E. coli total tRNA followed by ethanol precipitation. Alkaline ladders were performed by incubation of labeled transcripts (30,000 Cerenkov cpm) and 2 μg of total tRNA from E. coli in 50 mM NaHCO3 (pH 9.0), at 80°C for 8 min. Guanine ladders were obtained by incubation in 10 mM NaH2[C3H5O(COO)3] at pH 4.5, 3.2 M urea, 0.4 mM EDTA for 4 min at 60°C followed by addition of 2 U of T1 and a second incubation at 60°C for 4 min. Products of the probing reactions were analyzed on 12% polyacrylamide/8M urea denaturing gels. Controls without probes but supplemented with ZnCl2 were run in parallel.
ACKNOWLEDGMENTS
We thank R. Giegé and L. Levinger for critical reading of the manuscript. This work was supported by Centre National de la Recherche Scientifique (CNRS), Université Louis Pasteur, and Association Française pour les Myopathies (AFM). M.M. was supported by a fellowship from the Ministère de l'Enseignement Supérieur et de la Recherche.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1626109.
REFERENCES
- Bonnefond L, Fender A, Rudinger-Thirion J, Giegé R, Florentz C, Sissler M. Towards the full set of human mitochondrial aminoacyl-tRNA synthetases: Characterization of AspRS and TyrRS. Biochemistry. 2005;44:4805–4816. doi: 10.1021/bi047527z. [DOI] [PubMed] [Google Scholar]
- Bonnefond L, Florentz C, Giegé R, Rudinger-Thirion J. Decreased aminoacylation in pathology-related mutants of mitochondrial tRNATyr is associated with structural perturbations in tRNA architecture. RNA. 2008;14:641–648. doi: 10.1261/rna.938108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMauro S. Mitochondrial myopathies. Curr Opin Rheumatol. 2006;18:636–641. doi: 10.1097/01.bor.0000245729.17759.f2. [DOI] [PubMed] [Google Scholar]
- Dirheimer G, Keith G, Dumas P, Westhof E. Primary, secondary, and tertiary structures of tRNAs. In: Söll D, RajBhandary UL, editors. tRNA: Structure, Biosynthesis, and Function. American Society of Microbiology Press; Washington, DC: 1995. pp. 93–126. [Google Scholar]
- Florentz C, Sissler M. Disease-related versus polymorphic mutations in human mitochondrial tRNAs: Where is the difference? EMBO Rep. 2001;2:481–486. doi: 10.1093/embo-reports/kve111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florentz C, Sohm B, Tryoen-Tóth P, Pütz J, Sissler M. Human mitochondrial tRNAs in health and disease. Cell Mol Life Sci. 2003;60:1356–1375. doi: 10.1007/s00018-003-2343-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giegé R. Toward a more complete view of tRNA biology. Nat Struct Mol Biol. 2008;15:1007–1014. doi: 10.1038/nsmb.1498. [DOI] [PubMed] [Google Scholar]
- Giegé R, Puglisi J, Florentz C. tRNA structure and aminoacylation efficiency. Prog Nucleic Acid Res Mol Biol. 1993;45:129–206. doi: 10.1016/s0079-6603(08)60869-7. [DOI] [PubMed] [Google Scholar]
- Giegé R, Helm M, Florentz C. Chemical and enzymatic probing of RNA structure. In: Söll D, et al., editors. Prebiotic Chemistry, Molecular Fossils, Nucleosides, and RNA. Pergamon; Oxford, UK: 1999. pp. 63–80. [Google Scholar]
- Hayashi I, Yokogawa T, Kawai G, Ueda T, Nishikawa K, Watanabe K. Assignment of imino proton signals of G-C base pairs and magnesium ion binding: An NMR study of bovine mitochondrial tRNASer(GCU) lacking the entire D arm. J Biochem. 1997;121:1115–1122. doi: 10.1093/oxfordjournals.jbchem.a021703. [DOI] [PubMed] [Google Scholar]
- Helm M, Brulé H, Degoul F, Cepanec C, Leroux J-P, Giegé R, Florentz C. The presence of modified nucleotides is required for cloverleaf folding of a human mitochondrial tRNA. Nucleic Acids Res. 1998;26:1636–1643. doi: 10.1093/nar/26.7.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm M, Florentz C, Chomyn A, Attardi G. Search for differences in post-transcriptional modification patterns of mitochondrial DNA-encoded wild-type and mutant human tRNALys and tRNALeu(UUR) Nucleic Acids Res. 1999a;27:756–763. doi: 10.1093/nar/27.3.756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm M, Giegé R, Florentz C. A Watson–Crick base-pair disrupting methyl group (m1A9) is sufficient for cloverleaf folding of human mitochondrial tRNALys. Biochemistry. 1999b;38:13338–13346. doi: 10.1021/bi991061g. [DOI] [PubMed] [Google Scholar]
- Helm M, Brulé H, Friede D, Giegé R, Pütz J, Florentz C. Search for characteristic structural features of mammalian mitochondrial tRNAs. RNA. 2000;6:1356–1379. doi: 10.1017/s1355838200001047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzmann J, Frank P, Löffler E, Bennett KL, Gerner C, Rossmanith W. RNase P without RNA: Identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell. 2008;135:462–474. doi: 10.1016/j.cell.2008.09.013. [DOI] [PubMed] [Google Scholar]
- Ingman M, Gyllensten U. mtDB: Human Mitochondrial Genome Database, a resource for population genetics and medical sciences. Nucleic Acids Res. 2006;34:D749–D751. doi: 10.1093/nar/gkj010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs HT. Disorders of mitochondrial protein synthesis. Hum Mol Genet. 2003;12:R293–R301. doi: 10.1093/hmg/ddg285. [DOI] [PubMed] [Google Scholar]
- Juehling F, Moerl M, Hartmann RK, Sprinzl M, Stadler P, Puetz J. tRNAdb 2009: Compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009;37:D159–D162. doi: 10.1093/nar/gkn772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes & Dev. 2004;18:1227–1240. doi: 10.1101/gad.1183804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley SO, Steinberg SV, Schimmel P. Fragile T-stem in disease-associated human mitochondrial tRNA sensitizes structure to local and distant mutations. J Biol Chem. 2001;276:10607–10611. doi: 10.1074/jbc.M008320200. [DOI] [PubMed] [Google Scholar]
- Kim SH, Suddath FL, Quigley GJ, McPherson A, Sussman JL, Wang AHJ, Seeman NC, Rich A. Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. Science. 1974;185:435–440. doi: 10.1126/science.185.4149.435. [DOI] [PubMed] [Google Scholar]
- Kirino Y, Yasukawa T, Ohta S, Akira S, Ishihara K, Watanabe K, Suzuki T. Codon-specific translational defect caused by a wobble modification deficiency in mutant tRNA from a human mitochondrial disease. Proc Natl Acad Sci. 2004;101:15070–15075. doi: 10.1073/pnas.0405173101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobitski AY, Hengesbach M, Helm M, Nienhaus GU. Sculpting an RNA conformational energy landscape by a methyl group modification—a single-molecule FRET study. Angew Chem Int Ed Engl. 2008;47:4326–4330. doi: 10.1002/anie.200705675. [DOI] [PubMed] [Google Scholar]
- Leontis NB, Stombaugh J, Westhof E. The non-Watson–Crick base pairs and their associated isostericity matrices. Nucleic Acids Res. 2002;30:3497–3531. doi: 10.1093/nar/gkf481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinger L, Mörl M, Florentz C. Mitochondrial tRNA 3′ end metabolism and human disease. Nucleic Acids Res. 2004;32:5430–5441. doi: 10.1093/nar/gkh884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarland R, Elson JL, Taylor RW, Howell N, Turnbull DM. Assigning pathogenicity to mitochondrial tRNA mutations: When ‘definitely maybe’ is not good enough. Trends Genet. 2004;20:591–596. doi: 10.1016/j.tig.2004.09.014. [DOI] [PubMed] [Google Scholar]
- Pütz J, Dupuis B, Sissler M, Florentz C. Mamit-tRNA, a database of mammalian mitochondrial tRNA primary and secondary structures. RNA. 2007;13:1184–1190. doi: 10.1261/rna.588407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman LH, Pelka H. Anticodon switching changes the identity of methionine and valine transfer RNAs. Science. 1988;242:765–768. doi: 10.1126/science.3055296. [DOI] [PubMed] [Google Scholar]
- Seneca S, Goemans N, Van Coster R, Givron P, Reybrouck T, Sciot R, Meulemans A, Smet J, Van Hove JL. A mitochondrial tRNA aspartate mutation causing isolated mitochondrial myopathy. Am J Med Genet A. 2005;137:170–175. doi: 10.1002/ajmg.a.30854. [DOI] [PubMed] [Google Scholar]
- Shapira AHV. Mitochondrial disease. Lancet. 2006;368:70–82. doi: 10.1016/S0140-6736(06)68970-8. [DOI] [PubMed] [Google Scholar]
- Sohm B, Frugier M, Brulé H, Olszak K, Przykorska A, Florentz C. Towards understanding human mitochondrial leucine aminoacylation identity. J Mol Biol. 2003;328:995–1010. doi: 10.1016/s0022-2836(03)00373-5. [DOI] [PubMed] [Google Scholar]
- Tomari Y, Hino N, Nagaike T, Suzuki T, Ueda T. Decreased CCA-addition in human mitochondrial tRNAs bearing a pathogenic A4317G or A10044G mutation. J Biol Chem. 2003;278:16828–16833. doi: 10.1074/jbc.M213216200. [DOI] [PubMed] [Google Scholar]
- Voigts-Hoffmann F, Hengesbach M, Kobitski AY, van Aerschot A, Herdewijn P, Nienhaus GU, Helm M. A methyl group controls conformational equilibrium in human mitochondrial tRNALys. J Am Chem Soc. 2007;129:13382–13383. doi: 10.1021/ja075520+. [DOI] [PubMed] [Google Scholar]
- Wallace D, Brown M, Lott M. Mitochondrial DNA variation in human evolution and disease. Gene. 1999;238:211–230. doi: 10.1016/s0378-1119(99)00295-4. [DOI] [PubMed] [Google Scholar]
- Watanabe Y-I, Kawai G, Yokogawa T, Hayashi N, Kumazawa Y, Ueda T, Nishikawa K, Hirao I, Miura K-I, Watanabe K. Higher-order structure of bovine mitochondrial tRNASer(UGA): Chemical modification and computer modeling. Nucleic Acids Res. 1994;22:5378–5384. doi: 10.1093/nar/22.24.5378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhof E, Dumas P, Moras D. Crystallographic refinement of yeast aspartic acid transfer RNA. J Mol Biol. 1985;184:119–145. doi: 10.1016/0022-2836(85)90048-8. [DOI] [PubMed] [Google Scholar]
- Wittenhagen LM, Kelley SO. Dimerization of a pathogenic human mitochondrial tRNA. Nat Struct Biol. 2002;9:586–590. doi: 10.1038/nsb820. [DOI] [PubMed] [Google Scholar]
- Wittenhagen LM, Kelley SO. Impact of disease-related mitochondrial mutations on tRNA structure and function. Trends Biochem Sci. 2003;28:605–611. doi: 10.1016/j.tibs.2003.09.006. [DOI] [PubMed] [Google Scholar]
- Wittenhagen LM, Roy MD, Kelley SO. The pathogenic U3271C human mitochondrial tRNALeu(UUR) mutation disrupts a fragile anticodon stem. Nucleic Acids Res. 2003;31:596–601. doi: 10.1093/nar/gkg131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasukawa T, Suzuki T, Ishii N, Ohta S, Watanabe K. Wobble modification defect in tRNA disturbs codon–anticodon interaction in a mitochondrial disease. EMBO J. 2001;20:4794–4802. doi: 10.1093/emboj/20.17.4794. [DOI] [PMC free article] [PubMed] [Google Scholar]