Abstract
MicroRNAs (miRNAs) are a recently discovered class of small noncoding RNAs and are implicated in an increasing number of biological processes. To examine whether miRNAs might contribute to sexual differentiation, we performed expression profiling of miRNAs in mouse embryonic gonads with the use of a highly sensitive cloning method, mRAP. Our profiling data revealed substantial differences in the miRNA repertoire between male and female gonads at embryonic (E) day 13.5 (E13.5), suggesting that such differentially expressed miRNAs may function in sexual differentiation. Female-specific miRNAs included miR-29b, which also has been known to be expressed in a similar sex-dependent manner in the gonads of chicken embryos, suggestive of a conserved role in gonadogenesis. Transcripts of the human genes for the de novo methyltransferases DNMT3A and DNMT3B have been identified as targets of miR-29b, and we found that mouse miR-29b also negatively regulates Dnmt3a and Dnmt3b expression in luciferase reporter assays. We also found that miR-29b is expressed in mouse primordial germ cells (PGCs) at E13.5 and that its expression is up-regulated in a female-specific manner between E13.5 and E17.5, when male-specific de novo methylation of the PGC genome is known to occur. Our data thus suggest that miR-29b may play an important role in female gonadal development by targeting Dnmt3a and Dnmt3b and thereby modulating methylation of genomic DNA in PGCs.
Keywords: mRAP, microRNA, mouse, chicken, gonad, primordial germ cell
INTRODUCTION
MicroRNAs (miRNAs) are short noncoding RNAs of 20–24 nucleotides (nt) that negatively regulate protein production from target mRNAs. They function by interacting with their target mRNAs through incomplete base-pairing to the 3′ untranslated region (3′UTR) (Filipowicz 2005; Hammond 2005; Hannon 2002; Mattick and Makunin 2005) and thereby either trigger degradation of the target mRNAs or suppress their translation. MicroRNAs have been identified in a wide range of organisms, including plants and animals (Carrington and Ambros 2003; Bartel 2004). Many miRNAs are conserved throughout evolution, but substantial diversity is also apparent for some miRNAs even between closely related species (Berezikov et al. 2006a).
Expression of miRNAs has been shown to be tightly regulated in a developmental stage-dependent, as well as in an organ-dependent, manner (Aboobaker et al. 2005; Wienholds et al. 2005; Ason et al. 2006; Kloosterman et al. 2006; Takada et al. 2006a), suggesting that they may play important roles in embryonic development and tissue organization. The miRNAs miR-1 and miR-124, for example, are specifically expressed in muscle and the central nervous system, respectively, in zebrafish, medaka, mouse, and fly, suggesting that the function of these miRNAs is conserved across animal phyla (Kloosterman and Plasterk 2006).
Although the precise function of most miRNAs remains unclear, some have been shown to contribute to a variety of biological phenomena, including intracellular signaling, apoptosis, metabolism, cardiogenesis, myogenesis, and brain development (Kloosterman and Plasterk 2006). Essential roles for miRNAs in animal development have been revealed by analysis of cells deficient in Dicer, an enzyme required for the production of miRNAs from their precursors. Dicer null mutant mice thus die in utero at embryonic (E) day 7.5 (E7.5) and lack stem cell compartments (Bernstein et al. 2003). Mice with conditional ablation of this gene have also revealed that Dicer (and, therefore, probably also miRNAs) is required for morphogenesis of the limb (Harfe et al. 2005), skin (Andl et al. 2006; Yi et al. 2006), and lung epithelium (Harris et al. 2006). However, the genes targeted by miRNAs to achieve their effects remain largely unknown.
The heterogametic pairing of the sex chromosomes, X and Y, results in male development in mammals, whereas females are the heterogametic sex (ZW) in birds. Despite this difference in sex determination, several important genes for sexual differentiation are expressed in a similar manner in the gonads of both mammals and birds, suggesting the existence of a shared mechanism for this process. In mammals, the gonads first emerge as bipotential organs that subsequently develop into testes or ovaries depending on whether Sry, a sex-determining gene on the Y chromosome, is expressed or not (Gubbay et al. 1990; Koopman et al. 1991).
Identification of a function for miRNAs in mammalian gonadal development would be facilitated by characterization of miRNA expression profiles in developing gonads. However, most current technologies for measurement of miRNA expression either require substantial amounts of RNA for analysis (in the case of conventional cloning methods) or are unable to examine unknown miRNA species (in the case of microarray- or stem–loop-based detection methods). Neither of these types of approach is, therefore, suitable for extensive miRNA profiling of the gonads, for which only small quantities of tissue are usually available and many unknown miRNAs may be present.
We have recently developed a highly sensitive cloning procedure for miRNAs (Mano and Takada 2007; Takada et al. 2006a; Takada and Mano 2007). This procedure, designated miRNA amplification profiling (mRAP), allows the isolation of tens of thousands of miRNA clones from small quantities of starting material (even from as few as 10,000 cells). Coupling of mRAP to a computational pipeline in order to detect or predict miRNAs thus represents an optimal means for quantitative measurement of miRNA expression profiles in small amounts of tissue or clinical specimens. We have now applied such technology to mouse gonads in order to identify sex-dependent expression of miRNAs.
RESULTS
miRNA expression profiling of mouse embryonic gonads
To identify miRNAs expressed differentially between male and female mouse gonads at E13.5, we constructed small RNA-derived cDNA libraries by the mRAP protocol from these tissues. Totals of 672 and 1440 cDNA concatamers were sequenced, resulting in the identification of 1153 and 1480 small RNA sequences, for male and female embryonic gonads, respectively. Totals of 180 and 184 of these sequences from male and female gonad libraries, respectively, matched known miRNAs (Table 1). In addition, three candidates for novel miRNAs (miR-143*, miR-715*, miR-689*) were identified, each of which is present in the hairpin structure of known miRNAs. The remaining small RNA sequences likely represent RNA degradation products.
TABLE 1.
Profiling of miRNAs by mRAP in mouse embryonic gonads at E13.5
Identification of miRNAs that are expressed in a sex-dependent manner at E13.5
We have previously shown that mRAP cloning frequency reflects relative miRNA abundance in cells, provided that sufficient numbers of small RNA-derived cDNAs are sequenced (Takada et al. 2006a; S Takada and H Mano, unpubl.). In the current data set, many miRNAs were found to be expressed in a sex-dependent manner. For instance, the most abundant miRNA in both sexes, miR-29b, is preferentially expressed (by a factor of ∼2) in the female gonad (Table 1).
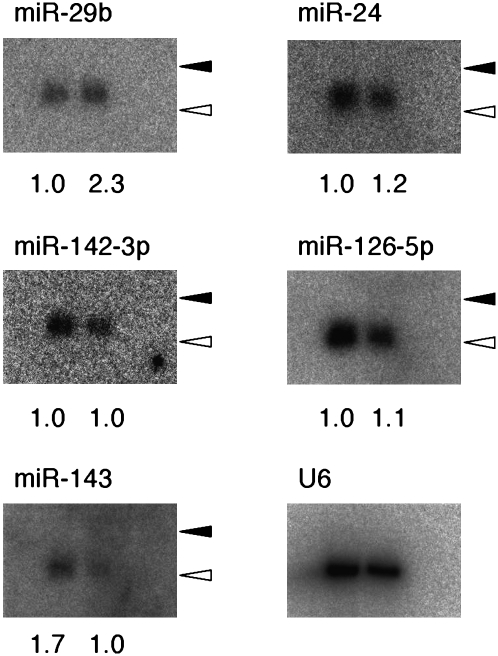
To confirm such sexually differential expression of miRNAs, we performed Northern blot analysis. The abundance of miR-29b normalized by that of U6 RNA was 2.3 arbitrary units (U) in the female gonad and 1.0 U in the male gonad at E13.5 (Fig. 1), consistent with the mRAP data (read counts of 73.0% and 30.4% for female and male, respectively) (Table 1). In contrast, the fourth most abundant miRNA, miR-143, was found to be preferentially expressed in male gonads by both Northern blot analysis (1.7 versus 1.0 U in male and female, respectively) (Fig. 1) and mRAP analysis (8.70% versus 0.51%) (Table 1), although the male-to-female ratio differed between the two approaches. The sex-related expression of other miRNAs (miR-24, miR-142-3p, miR-126-5p) revealed by mRAP was not detected by Northern analysis (Fig. 1). Together, these data thus showed that certain miRNAs are expressed in the gonads in a sex-dependent manner.
FIGURE 1.
Validation of mRAP data by Northern blot analysis. Fractions containing small RNAs (0.5–0.8 μg per lane) isolated from (left lanes) male or (right lanes) female mouse gonads at E13.5 were subjected to Northern analysis with LNA probes specific for the indicated miRNAs or U6 RNA. The signal intensity for each miRNA normalized by that of U6 RNA is shown at the bottom of each lane. (Closed and open arrowheads) The positions corresponding to 24 and 19 nt, respectively.
Expression patterns of miR-29b and miR-143 during sexual differentiation
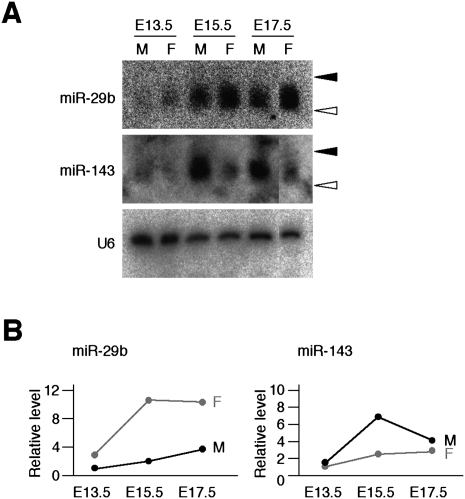
With the use of Northern blot analysis, we next examined the expression profiles of the sex-specific miRNAs miR-29b and miR-143 during mouse gonadogenesis. In the female gonad, expression of miR-29b was increased at E15.5 compared with that at E13.5, and the increased level of expression was maintained through E17.5 (Fig. 2). In the male gonad, the relative abundance of miR-29b increased gradually from E13.5 to E17.5 but was consistently lower than that in stage-matched female gonads, suggesting that miR-29b may play different roles in male and female gonads.
FIGURE 2.
Sex-dependent expression profiles of miR-29b and miR-143 in mouse embryos. (A) Fractions containing small RNAs isolated from male (M) or female (F) mouse gonads at E13.5, E15.5, or E17.5 were subjected to Northern blot analysis with probes specific for miR-29b, miR-143, or U6 RNA. (Closed and open arrowheads) The positions corresponding to 24 and 19 nt, respectively. (B) The hybridization signal intensity for each miRNA in A was normalized by that of U6 RNA and plotted against embryonic stage.
The expression of miR-143 in the male gonad increased from E13.5 to E15.5 but decreased to an intermediate level at E17.5 (Fig. 2). In contrast, the relative amount of miR-143 increased gradually between E13.5 and E17.5 in the female gonad, suggesting that this miRNA might have a specific function in male gonadogenesis at E15.5.
Evolutionary conservation of miR-29b and miR-143 expression
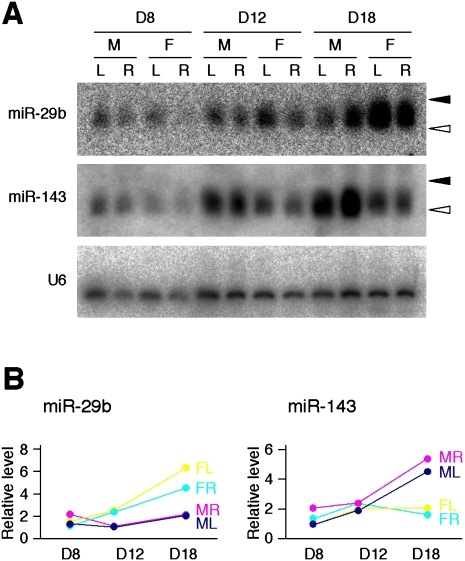
If miR-29b or miR-143 contributes directly to sex-dependent differentiation of the gonads in mice, their expression profiles might be expected to be evolutionarily conserved. This has been, indeed, shown in the cases for Sox9, Amh, and FoxL2, all of which are essential for sexual differentiation of the gonads (Carre-Eusebe et al. 1996; Kent et al. 1996; Morais da Silva et al. 1996; Loffler et al. 2003). We therefore examined the expression of miR-29b and miR-143 in chicken embryonic gonads during sexual differentiation with Northern blot analysis using male and female chicken embryonic gonads. The right and left gonads were treated separately, because there is a left–right asymmetry associated with the chicken gonadal differentiation. The relative abundance of miR-29b was greater in female gonads than in male gonads of chicken embryos at days 12 and 18 (Fig. 3), an expression profile similar to that apparent for mouse gonads (Fig. 2). In contrast, the expression of miR-143 was greater in male than in female chicken gonads at day 18 (Fig. 3). The preferential expression of miR-143 in male gonads was thus confirmed in chicken in a similar pattern to that in mouse (Fig. 2).
FIGURE 3.
Sex-specific up-regulation of miR-29b and miR-143 expression in chicken embryos. (A) Fractions containing small RNAs isolated from male (M) or female (F) chicken gonads at day (D) 8, 12, or 18 were subjected to Northern blot analysis with probes specific for miR-29b, miR-143, or U6 RNA. (L) Left gonad; (R) right gonad. (Closed and open arrowheads) The positions corresponding to 24 and 19 nt, respectively. (B) The hybridization signal intensity for each miRNA in A was normalized by that of U6 RNA and plotted against embryonic stage. (ML) Male left gonad; (MR) male right gonad; (FL) female left gonad; (FR) female right gonads.
Identification of cell types in which miR-29b and miR-143 are expressed
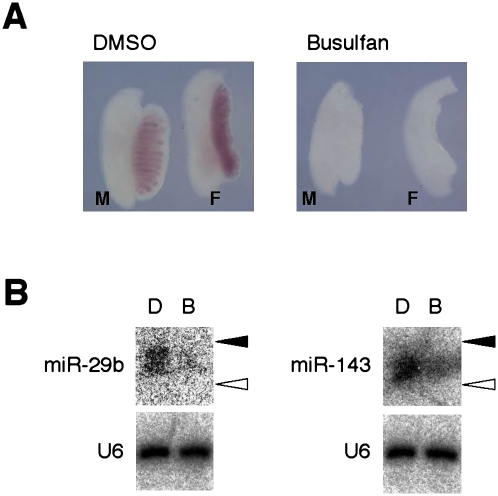
We next investigated which cells express miR-29b or miR-143 in the gonads of mouse embryos. To determine whether such expression is restricted to somatic or germ cells, we performed Northern blot analysis with RNA isolated from embryos exposed in utero to busulfan, which is known to eliminate germ cells (Forsberg and Olivecrona 1966). Depletion of germ cells in busulfan-treated mouse gonads was confirmed by measurement of the expression of Oct4 that is restricted to primordial germ cells (PGCs) in both testis and ovary at E13.5 (Pesce et al. 1998). Whole-mount in situ hybridization thus revealed the presence of Oct4 mRNA in E13.5 mouse gonads exposed to DMSO vehicle but not in those exposed to busulfan (Fig. 4A).
FIGURE 4.
Expression of miR-29b and miR-143 in PGCs of mouse embryos at E13.5. (A) Male (M) and female (F) embryonic gonads were recovered at E13.5 from dams injected with DMSO or busulfan and were subjected to whole-mount in situ hybridization with a probe specific for Oct4 mRNA. (B) Fractions containing small RNAs isolated from female or male gonads treated as in A were subjected to Northern blot analysis with probes specific for miR-29b or miR-143, respectively. The same blots were also subjected to hybridization with a probe specific for U6 RNA. (Closed and open arrowheads) The positions corresponding to 24 and 19 nt, respectively. Embryos exposed to (D) DMSO or (B) busulfan.
Northern blot analysis with an LNA probe for miR-29b detected a discrete signal with RNA recovered from DMSO-treated female gonads, whereas the signal was mostly lost with RNA from those treated with busulfan (Fig. 4B), indicating that miR-29b is expressed exclusively in PGCs in female embryonic gonads. Although miR-143 was also found to be preferentially expressed in PGCs, a substantial amount of this miRNA remained in busulfan-treated male gonads, suggesting that miR-143 may be expressed in both somatic cells and PGCs of the testis.
Target genes of miR-29b in mouse PGCs
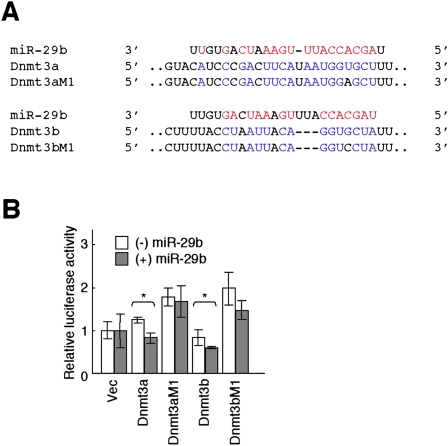
Our findings that miR-29b is expressed almost exclusively in PGCs of female gonads and that the expression profile of this miRNA is conserved between mouse and chicken suggests that miR-29b function is likely important for PGCs in female gonads. The genes for DNA methyltransferases 3A (DNMT3A) and 3B (DNMT3B) were recently shown to be direct targets of miR-29b, and overexpression of this miRNA results in a reduction in the global level of DNA methylation in the human genome (Fabbri et al. 2007). Given that mouse Dnmt3a and Dnmt3b are expressed in PGCs at E12.5–E17.5 (Lees-Murdock et al. 2005), these genes are candidates for regulation by miR-29b in mouse PGCs. Indeed, the TargetScan program (Lewis et al. 2003; Grimson et al. 2007), an in silico approach to the prediction of miRNA targets, indicated that both Dnmt3a and Dnmt3b are potential targets of this mouse miRNA (Fig. 5A). We, therefore, examined whether Dnmt3a or Dnmt3b is regulated by miR-29b in mouse.
FIGURE 5.
Identification of Dnmt3a and Dnmt3b as potential targets of miR-29b in mouse. (A) Potential target sites for miR-29b in the 3′UTRs of Dnmt3a and Dnmt3b mRNAs are shown aligned with the miR-29b sequence. The same 3′UTR sequences with a 1-bp mismatch (M1) were used as negative controls in luciferase reporter assays. Complementary bases between miR-29b (red) and the 3′UTRs (blue) are shown, respectively. (B) Luciferase reporter assays were performed with vectors containing DNA fragments corresponding to the putative wild-type or mutant target sites for miR-29b in the 3′UTRs of Dnmt3a or Dnmt3b mRNAs. The assays were performed in NIH3T3 cells cotransfected (or not) with the miR-29b precursor. Vec indicates cells transfected with the luciferase vector without Dnmt3 sequences. Firefly luciferase activity was normalized by that of Renilla luciferase, and the data are means ± SD from three independent experiments. (*) P < 0.05 for the indicated comparisons (Student's t-test).
To this end, we utilized a luciferase reporter assay. As a host cell line, using Northern blot analysis we searched for a cell line in which miR-29b does not exist; however, such a cell line was not found (data not shown). Since transfection efficiency of NIH3T3 is high, we used this cell line as a host cell. DNA fragments corresponding to portions of the 3′UTRs of Dnmt3a or Dnmt3b mRNAs containing the predicted miR-29b target sites were inserted into the 3′UTR of firefly luciferase cDNA, and the resulting reporter plasmids were introduced with or without miR-29b into NIH3T3 fibroblasts. The same target sequences but with a 1-base-pair (bp) mismatch were similarly inserted into the luciferase cDNA to yield negative control constructs (Fig. 5A). The luciferase activities of the constructs with the wild-type target regions of Dnmt3a or Dnmt3b were reduced compared with those of the corresponding mutant constructs (Fig. 5B), likely because miR-29b is expressed in NIH3T3 cells (data not shown). Furthermore, the luciferase activities of the wild-type constructs for Dnmt3a or Dnmt3b were reduced by cotransfection with miR-29b, whereas those of the mutant constructs were not (Fig. 5B). These results thus suggested that both genes may be targets of miR-29b in mouse.
DISCUSSION
We have determined the miRNA expression profiles of male and female embryonic mouse gonads at E13.5. Given that most conventional methods for cloning of miRNAs require >100 μg of total RNA (Lagos-Quintana et al. 2002), it would have been difficult to profile miRNA expression in embryonic gonads by such approaches with the small amounts of tissue available. We therefore adopted mRAP, a cloning- and sequencing-based method with a high sensitivity, for miRNA profiling in embryonic gonads.
We obtained a total of 380 miRNA-derived sequences, consisting of 374 reads for known miRNAs and six reads for previously unreported ones (four corresponding to miR-143*, one to miR-715*, and one to miR-689*). Although 2000 mRAP clones were sequenced in the present study, most of the miRNAs (both known and novel) in our data set were detected five or fewer times (Table 1), indicating that all miRNAs in the gonads may not yet have been identified. Extensive sequencing of mRAP clones may therefore result in the identification of additional novel hairpin structures containing unreported miRNA candidates.
We found that miR-29b is the most abundant miRNA in mouse gonads at E13.5. Northern blot analysis of PGC-depleted gonadal tissue further indicated that miR-29b is expressed almost exclusively in PGCs of female gonads (at least at E13.5). This finding contrasts with results of Hayashi et al. (2008) showing that miR-29b was virtually undetectable in mouse PGCs at this stage by RT-PCR analysis. This discrepancy may be attributable to the differences in methodology or in genetic background of the mice between the two studies.
Although the differences in mRAP cloning frequency between male and female gonads for some miRNAs were reflected in differences in expression as determined by Northern blot analysis, those for others were not. We have recently performed deep sequencing of mRAP clones from various mouse organs and found that the correlation between cloning frequency and Northern blot data was dependent on miRNA sequence (S Takada and H Mano, unpubl.). Even some miRNAs with a read number of >100,000 per organ were not detected by Northern blot analysis. The nucleotide sequence of miRNAs may thus greatly affect their sensitivity to detection by Northern blot analysis or by RT-PCR (a hybridization-based detection system). However, it remains possible that mRAP may have a cloning bias for some miRNAs.
Both Dnmt3a and Dnmt3b are expressed in mouse PGCs of both sexes at E13.5 (Lees-Murdock et al. 2005), suggesting a possibility that DNA methylation of the genome is regulated by miR-29b. Methylation of cytosine residues is the only known direct epigenetic modification of mammalian genomic DNA and contributes to various biological phenomena including transposon silencing and genomic imprinting (Reik et al. 2001; Bird 2002). There are two main types of DNA methyltransferase: maintenance methyltransferases that methylate hemimethylated CpG sequences after DNA replication, and de novo methyltransferases that methylate cytosine residues of unmethylated CpG sequences, and which include DNMT3A and DNMT3B (Okano et al. 1998). Genome-wide demethylation occurs early during development and is complete around E13 to E14 in PGCs of both male and female mouse embryos (Monk et al. 1987; Kafri et al. 1992; Brandeis et al. 1993; Surani 1998; Tada et al. 1998). Re-methylation then takes place earlier in the male germ cells (from E15 to E16) (Kafri et al. 1992; Brandeis et al. 1993; Coffigny et al. 1999) than in the female cells (after birth, during the growth of oocytes) (Lucifero et al. 2002, 2004; Hiura et al. 2006). DNMT3A and DNMT3B were recently shown to mediate de novo methylation of differentially methylated genomic regions corresponding to imprinted genes and some repetitive elements in male germ cells (Kato et al. 2007). It is thus possible that the expression of these two enzymes is repressed by miR-29b in female germ cells, allowing an escape from male-type methylation of the genome and underlying female-type methylation.
MicroRNAs have been shown to contribute to the fine-tuning of physiological events or to function as molecular switches in cellular signaling (Lee et al. 1993; Wightman et al. 1993; Moss et al. 1997; Reinhart et al. 2000; Brennecke et al. 2003; Johnston and Hobert 2003; Sokol and Ambros 2005). In addition, some miRNAs function in a fail-safe mechanism to silence mRNAs that are unwanted in specific cell types (Hornstein et al. 2005; Cohen et al. 2006). It seems likely that miR-29b may function in such a mechanism to regulate methylation of the genome, given that the amount of Dnmt3a mRNA in PGCs is similar in male and female mouse embryos at E13.5 but is greater in male than in female PGCs at E15.5 and E17.5 (Iwahashi et al. 2007).
Further extensive sequencing of mRAP clones from the gonads will provide additional insight into the cell type-dependent and developmental stage-dependent expression profiles of miRNAs, and such information will likely contribute to understanding of the function of miRNAs in sex determination and differentiation.
MATERIALS AND METHODS
Mice and tissues
C57BL/6J mice (Mus musculus domesticus) were obtained from a local supplier (Japan SLC). The mice were allowed to mate naturally, and at noon of the day in which a vaginal plug was observed were considered to be E0.5. Gonads depleted of germ cells were prepared by intraperitoneal injection of pregnant females at E9.5 with 100 μL of a warmed solution (16 mg/mL) of busulfan (Sigma-Aldrich) in 50% dimethly sulfoxide (DMSO), and harvesting of the embryos occurred on E13.5 (Forsberg and Olivecrona 1966); as a control, dams were injected with 50% DMSO alone. The sex of each embryo was determined on the basis of the presence (male) or absence (female) of a testis cord.
Chickens and tissues
Fertilized chicken (Gallus gallus domestica) eggs were obtained from a local supplier (Saitama Experimental Animal Supply) and were maintained at 18°C until their transfer to an incubator at 37.8°C. Staging of chicken embryos was confirmed at dissection as described by Hamburger and Hamilton (1951). The gonads of each embryo were snap frozen, and the sex of the embryos was determined as described previously (Clinton et al. 2001; Takada et al. 2006b,c) with the use of a polymerase chain reaction (PCR)-based method performed with genomic DNA extracted from the hind limbs.
mRAP
The mRAP procedure was performed as described previously (Takada et al. 2006a; Mano and Takada 2007; Takada and Mano 2007), and miRNAs were identified from mRAP clones with the use of the computational pipeline developed by Berezikov et al. (2006b).
Northern blot analysis
A fraction of small RNA molecules (<200 nt) was prepared with the use of a mirVana miRNA Isolation Kit (Applied Biosystems), and portions of the fraction (0.5–0.8 μg per lane) were subjected to electrophoresis on a 15% polyacrylamide gel under denaturing conditions. The separated RNA molecules were transferred electrophoretically to a Hybond-N membrane (GE Healthcare UK) and were subjected to hybridization with the use of the ULTRAhyb-Oligo reagent (Applied Biosystems) and with 32P-labeled locked nucleic acid (LNA) probes corresponding to reverse complementary sequences of mature miRNAs. Signals were detected with a BAS-1500 image analyzer (Fuji Photo Film), and signal intensities were measured with the use of Image Gauge version 4.1 software (Fuji Photo Film). The signal intensities of the miRNAs were normalized to that of U6 RNA. Probes included:
mmu-miR-29bRCLNA (5′-AAcACtGAtTTcAAaTGgTGcTA-3′) for miR-29b;
mmu-miR-142-3pRCLNA (5′-CCaTAaAGtAGgAAaCAcTAcA-3′) for miR-142-3p;
mmu-miR-143RCLNA (5′-TGaGCtACaGTgCTtCAtCTcA-3′) for miR-143;
mmu-miR-24RCLNA (5′-CTgTTcCTgCTgAAcTGaGCcA-3′) for miR-24;
mmu-miR-126-5pRCLNA (5′-CgCGtACcAAAAGtAAtAAtG-3′) for miR-126-5p; and
U6AS (5′-AACGCTTCACGAATTTGCGT-3′) for U6 RNA.
Here, upper- and lowercase letters designate DNA and LNA, respectively.
Whole-mount in situ hybridization
Whole-mount in situ hybridization was performed by the maleic acid buffer (MABT) method as previously described (Xu and Wilkinson 1998). A probe for Oct4 mRNA was prepared from total RNA isolated from E12.5 mouse embryos. The RNA was subjected to reverse transcription (RT), and the resulting cDNA was subjected to PCR with the primers mOct4probe1F (5′-GCCTTGCAGCTCAGCCTTAAGA-3′) and mOct4probe1R (5′-CCTCGCCCTCAGGAAAAGGGAC-3′). The amplification product, which corresponds to the probe described by Thomas et al. (1998), was cloned into the pGEM-T Easy vector (Promega) for production of the probe by in vitro transcription.
Luciferase assay
DNA fragments corresponding to a 981-bp portion of the 3′UTR of Dnmt3a or a 637-bp portion of the 3′UTR of Dnmt3b were amplified by PCR from C57BL/6J mouse genomic DNA. The primers wereDnmt3aAmpF (5′-ACTAGTGACTGAAACAAGAGAGTTA-3′) and Dnmt3aAmpR (5′-ACGCGTGGACCGGAGCTGCCATGTGC-3′) for Dnmt3a and Dnmt3bAmpF (5′-ACTAGTGGTACAAGGGCTGAAGTCC-3′) and Dnmt3bAmpR (5′-ACGCGTAAGGCAGTCTCTCCCCACAC-3′) for Dnmt3b, with the underlined sequences corresponding to recognition sites for restriction endonucleases. The PCR products were cloned into pGEM-T Easy and verified by nucleotide sequencing. Single nucleotide substitutions were introduced into the DNA sequences with the use of a QuikChange Multi Site-Directed Mutagenesis Kit (Stratagene) and the primers Dnmt3aM1 (5′-GACTTCATAATGGAGCTTTCAAAACAG-3′) for Dnmt3a and Dnmt3bM1 (5′-ACCTAATTACAGGTCCTATTTTATAG-3′) for Dnmt3b, with the underlined residues corresponding to the substituted bases. The insert of each clone was subcloned into the SpeI and MluI sites of the multiple cloning region of the pMIR-Report-luciferase vector (Applied Biosystems). The coding sequence and 3′UTR of the firefly luciferase cDNA as well as the insert of each of the resulting vectors were then subcloned into the BamHI and NotI sites of the pMXS vector (kindly provided by T. Kitamura, Institute of Medical Science, University of Tokyo).
NIH3T3 cells grown in 24-well plates were transfected with 50 μM miR-29b precursor (Pre-miR miRNA Precursor Molecule; Applied Biosystems), 0.6 μg of pMXS-based luciferase reporter vector, and 0.2 μg of a control Renilla luciferase vector (pRL-TK; Promega) in the presence of the Lipofectamine 2000 reagent (Invitrogen). Firefly and Renilla luciferase activities in cell lysates were assayed with the use of a Dual-Luciferase Reporter Assay System (Promega) 48 h after cell transfection, and the former activity was normalized by the latter.
ACKNOWLEDGMENTS
We thank the laboratory members for their discussions, as well as M. Otani, K. Nakamura, and S. Aoyagi for technical assistance. This work was supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to S.T.); a grant for Third-Term Comprehensive Control Research for Cancer from the Ministry of Health, Labor, and Welfare of Japan (to H.M.); and a grant for Scientific Research on Priority Areas “Applied Genomics” from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to H.M.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1418309.
REFERENCES
- Aboobaker AA, Tomancak P, Patel N, Rubin GM, Lai EC. Drosophila microRNAs exhibit diverse spatial expression patterns during embryonic development. Proc Natl Acad Sci. 2005;102:18017–18022. doi: 10.1073/pnas.0508823102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andl T, Murchison EP, Liu F, Zhang Y, Yunta-Gonzalez M, Tobias JW, Andl CD, Seykora JT, Hannon GJ, Millar SE. The miRNA-processing enzyme dicer is essential for the morphogenesis and maintenance of hair follicles. Curr Biol. 2006;16:1041–1049. doi: 10.1016/j.cub.2006.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ason B, Darnell DK, Wittbrodt B, Berezikov E, Kloosterman WP, Wittbrodt J, Antin PB, Plasterk RH. Differences in vertebrate microRNA expression. Proc Natl Acad Sci. 2006;103:14385–14389. doi: 10.1073/pnas.0603529103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Berezikov E, Thuemmler F, van Laake LW, Kondova I, Bontrop R, Cuppen E, Plasterk RH. Diversity of microRNAs in human and chimpanzee brain. Nat Genet. 2006a;38:1375–1377. doi: 10.1038/ng1914. [DOI] [PubMed] [Google Scholar]
- Berezikov E, van Tetering G, Verheul M, van de Belt J, van Laake L, Vos J, Verloop R, van de Wetering M, Guryev V, Takada S, et al. Many novel mammalian microRNA candidates identified by extensive cloning and RAKE analysis. Genome Res. 2006b;16:1289–1298. doi: 10.1101/gr.5159906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes & Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Brandeis M, Kafri T, Ariel M, Chaillet JR, McCarrey J, Razin A, Cedar H. The ontogeny of allele-specific methylation associated with imprinted genes in the mouse. EMBO J. 1993;12:3669–3677. doi: 10.1002/j.1460-2075.1993.tb06041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- Carre-Eusebe D, di Clemente N, Rey R, Pieau C, Vigier B, Josso N, Picard JY. Cloning and expression of the chick anti-Müllerian hormone gene. J Biol Chem. 1996;271:4798–4804. doi: 10.1074/jbc.271.9.4798. [DOI] [PubMed] [Google Scholar]
- Carrington JC, Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301:336–338. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- Clinton M, Haines L, Belloir B, McBride D. Sexing chick embryos: A rapid and simple protocol. Br Poult Sci. 2001;42:134–138. doi: 10.1080/713655025. [DOI] [PubMed] [Google Scholar]
- Coffigny H, Bourgeois C, Ricoul M, Bernardino J, Vilain A, Niveleau A, Malfoy B, Dutrillaux B. Alterations of DNA methylation patterns in germ cells and Sertoli cells from developing mouse testis. Cytogenet Cell Genet. 1999;87:175–181. doi: 10.1159/000015460. [DOI] [PubMed] [Google Scholar]
- Cohen SM, Brennecke J, Stark A. Denoising feedback loops by thresholding—A new role for microRNAs. Genes & Dev. 2006;20:2769–2772. doi: 10.1101/gad.1484606. [DOI] [PubMed] [Google Scholar]
- Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering C, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W. RNAi: The nuts and bolts of the RISC machine. Cell. 2005;122:17–20. doi: 10.1016/j.cell.2005.06.023. [DOI] [PubMed] [Google Scholar]
- Forsberg JG, Olivecrona H. The effect of prenatally administered Busulphan on rat gonads. Biol Neonat. 1966;10:180–192. doi: 10.1159/000240027. [DOI] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubbay J, Collignon J, Koopman P, Capel B, Economou A, Münsterberg A, Vivian N, Goodfellow P, Lovell-Badge R. A gene mapping to the sex-determining region of the mouse Y chromosome is a member of a novel family of embryonically expressed genes. Nature. 1990;346:245–250. doi: 10.1038/346245a0. [DOI] [PubMed] [Google Scholar]
- Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryo. J Morphol. 1951;88:49–92. [PubMed] [Google Scholar]
- Hammond SM. Dicing and slicing: The core machinery of the RNA interference pathway. FEBS Lett. 2005;579:5822–5829. doi: 10.1016/j.febslet.2005.08.079. [DOI] [PubMed] [Google Scholar]
- Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci. 2005;102:10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KS, Zhang Z, McManus MT, Harfe BD, Sun X. Dicer function is essential for lung epithelium morphogenesis. Proc Natl Acad Sci. 2006;103:2208–2213. doi: 10.1073/pnas.0510839103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Chuva de Sousa Lopes SM, Kaneda M, Tang F, Hajkova P, Lao K, O'Carroll D, Das PP, Tarakhovsky A, Miska EA, et al. MicroRNA biogenesis is required for mouse primordial germ cell development and spermatogenesis. PLoS One. 2008;3:e1738. doi: 10.1371/journal.pone.0001738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiura H, Obata Y, Komiyama J, Shirai M, Kono T. Oocyte growth-dependent progression of maternal imprinting in mice. Genes Cells. 2006;11:353–361. doi: 10.1111/j.1365-2443.2006.00943.x. [DOI] [PubMed] [Google Scholar]
- Hornstein E, Mansfield JH, Yekta S, Hu JK, Harfe BD, McManus MT, Baskerville S, Bartel DP, Tabin CJ. The microRNA miR-196 acts upstream of Hoxb8 and Shh in limb development. Nature. 2005;438:671–674. doi: 10.1038/nature04138. [DOI] [PubMed] [Google Scholar]
- Iwahashi K, Yoshioka H, Low EW, McCarrey JR, Yanagimachi R, Yamazaki Y. Autonomous regulation of sex-specific developmental programming in mouse fetal germ cells. Biol Reprod. 2007;77:697–706. doi: 10.1095/biolreprod.107.062851. [DOI] [PubMed] [Google Scholar]
- Johnston RJ, Hobert O. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature. 2003;426:845–849. doi: 10.1038/nature02255. [DOI] [PubMed] [Google Scholar]
- Kafri T, Ariel M, Brandeis M, Shemer R, Urven L, McCarrey J, Cedar H, Razin A. Developmental pattern of gene-specific DNA methylation in the mouse embryo and germ line. Genes & Dev. 1992;6:705–714. doi: 10.1101/gad.6.5.705. [DOI] [PubMed] [Google Scholar]
- Kato Y, Kaneda M, Hata K, Kumaki K, Hisano M, Kohara Y, Okano M, Li E, Nozaki M, Sasaki H. Role of the Dnmt3 family in de novo methylation of imprinted and repetitive sequences during male germ cell development in the mouse. Hum Mol Genet. 2007;16:2272–2280. doi: 10.1093/hmg/ddm179. [DOI] [PubMed] [Google Scholar]
- Kent J, Wheatley SC, Andrews JE, Sinclair AH, Koopman P. A male-specific role for SOX9 in vertebrate sex determination. Development. 1996;122:2813–2822. doi: 10.1242/dev.122.9.2813. [DOI] [PubMed] [Google Scholar]
- Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Kloosterman WP, Wienholds E, de Bruijn E, Kauppinen S, Plasterk RH. In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nat Methods. 2006;3:27–29. doi: 10.1038/nmeth843. [DOI] [PubMed] [Google Scholar]
- Koopman P, Gubbay J, Vivian N, Goodfellow P, Lovell-Badge R. Male development of chromosomally female mice transgenic for Sry. Nature. 1991;351:117–121. doi: 10.1038/351117a0. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–737. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lees-Murdock DJ, Shovlin TC, Gardiner T, De Felici M, Walsh CP. DNA methyltransferase expression in the mouse germ line during periods of de novo methylation. Dev Dyn. 2005;232:992–1002. doi: 10.1002/dvdy.20288. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Loffler KA, Zarkower D, Koopman P. Etiology of ovarian failure in blepharophimosis ptosis epicanthus inversus syndrome: FOXL2 is a conserved, early-acting gene in vertebrate ovarian development. Endocrinology. 2003;144:3237–3243. doi: 10.1210/en.2002-0095. [DOI] [PubMed] [Google Scholar]
- Lucifero D, Mertineit C, Clarke HJ, Bestor TH, Trasler JM. Methylation dynamics of imprinted genes in mouse germ cells. Genomics. 2002;79:530–538. doi: 10.1006/geno.2002.6732. [DOI] [PubMed] [Google Scholar]
- Lucifero D, Mann MR, Bartolomei MS, Trasler JM. Gene-specific timing and epigenetic memory in oocyte imprinting. Hum Mol Genet. 2004;13:839–849. doi: 10.1093/hmg/ddh104. [DOI] [PubMed] [Google Scholar]
- Mano H, Takada S. mRAP, a sensitive method for determination of microRNA expression profiles. Methods. 2007;43:118–122. doi: 10.1016/j.ymeth.2007.04.006. [DOI] [PubMed] [Google Scholar]
- Mattick JS, Makunin IV. Small regulatory RNAs in mammals. Hum Mol Genet. 2005;14:R121–R132. doi: 10.1093/hmg/ddi101. [DOI] [PubMed] [Google Scholar]
- Monk M, Boubelik M, Lehnert S. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development. 1987;99:371–382. doi: 10.1242/dev.99.3.371. [DOI] [PubMed] [Google Scholar]
- Morais da Silva S, Hacker A, Harley V, Goodfellow P, Swain A, Lovell-Badge R. Sox9 expression during gonadal development implies a conserved role for the gene in testis differentiation in mammals and birds. Nat Genet. 1996;14:62–68. doi: 10.1038/ng0996-62. [DOI] [PubMed] [Google Scholar]
- Moss EG, Lee RC, Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997;88:637–646. doi: 10.1016/s0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet. 1998;19:219–220. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- Pesce M, Wang X, Wolgemuth DJ, Schöler H. Differential expression of the Oct-4 transcription factor during mouse germ cell differentiation. Mech Dev. 1998;71:89–98. doi: 10.1016/s0925-4773(98)00002-1. [DOI] [PubMed] [Google Scholar]
- Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Sokol NS, Ambros V. Mesodermally expressed Drosophila microRNA-1 is regulated by Twist and is required in muscles during larval growth. Genes & Dev. 2005;19:2343–2354. doi: 10.1101/gad.1356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surani MA. Imprinting and the initiation of gene silencing in the germ line. Cell. 1998;93:309–312. doi: 10.1016/s0092-8674(00)81156-3. [DOI] [PubMed] [Google Scholar]
- Tada T, Tada M, Hilton K, Barton SC, Sado T, Takagi N, Surani MA. Epigenotype switching of imprintable loci in embryonic germ cells. Dev Genes Evol. 1998;207:551–561. doi: 10.1007/s004270050146. [DOI] [PubMed] [Google Scholar]
- Takada S, Mano H. Profiling of microRNA expression by mRAP. Nat Protocols. 2007;2:3136–3145. doi: 10.1038/nprot.2007.457. [DOI] [PubMed] [Google Scholar]
- Takada S, Berezikov E, Yamashita Y, Lagos-Quintana M, Kloosterman WP, Enomoto M, Hatanaka H, Fujiwara S, Watanabe H, Soda M, et al. Mouse microRNA profiles determined with a new and sensitive cloning method. Nucleic Acids Res. 2006a;34:e115. doi: 10.1093/nar/gkl653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada S, Ota J, Kansaku N, Yamashita H, Izumi T, Ishikawa M, Wada T, Kaneda R, Choi YL, Koinuma K, et al. Nucleotide sequence and embryonic expression of quail and duck Sox9 genes. Gen Comp Endocrinol. 2006b;145:208–213. doi: 10.1016/j.ygcen.2005.08.009. [DOI] [PubMed] [Google Scholar]
- Takada S, Wada T, Kaneda R, Choi YL, Yamashita Y, Mano H. Evidence for activation of Amh gene expression by steroidogenic factor 1. Mech Dev. 2006c;123:472–480. doi: 10.1016/j.mod.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Thomas PQ, Brown A, Beddington RS. Hex: A homeobox gene revealing peri-implantation asymmetry in the mouse embryo and an early transient marker of endothelial cell precursors. Development. 1998;125:85–94. doi: 10.1242/dev.125.1.85. [DOI] [PubMed] [Google Scholar]
- Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Xu Q, Wilkinson D. In situ hybridization of mRNA with hapten labelled probes. In: Wilkinson D, editor. In situ hybridization: A practical approach. 2nd ed. Oxford University Press; Oxford, UK: 1998. pp. 87–106. [Google Scholar]
- Yi R, O'Carroll D, Pasolli HA, Zhang Z, Dietrich FS, Tarakhovsky A, Fuchs E. Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat Genet. 2006;38:356–362. doi: 10.1038/ng1744. [DOI] [PubMed] [Google Scholar]