Abstract
GW182 family proteins interact directly with Argonaute proteins and are required for miRNA-mediated gene silencing in animal cells. The domains of the GW182 proteins have recently been studied to determine their role in silencing. These studies revealed that the middle and C-terminal regions function as an autonomous domain with a repressive function that is independent of both the interaction with Argonaute proteins and of P-body localization. Such findings reinforce the idea that GW182 proteins are key components of miRNA repressor complexes in metazoa.
Keywords: Argonaute, GW182, miRNAs, mRNA decay, P-bodies, RBD, RRM, TNRC6A
INTRODUCTION
MicroRNAs are genome-encoded small RNAs that post-transcriptionally regulate gene expression and play critical roles in a wide range of important biological processes including cell growth, division and differentiation, and organism metabolism and development. About 500–1000 miRNA genes exist in vertebrates and plants, and 100 in invertebrates, and each miRNA is predicted to have target sites in hundreds of mRNAs, suggesting that miRNAs regulate a large proportion of the transcriptome (Bushati and Cohen 2007; Carthew and Sontheimer 2009).
To exert their regulatory function, miRNAs associate with Argonaute proteins (AGOs) to form RNA-induced silencing complexes (miRISCs), which repress translation and/or trigger degradation of target mRNAs (Eulalio et al. 2008a; Filipowicz et al. 2008; Carthew and Sontheimer 2009). The proteins required for miRNA biogenesis (i.e., Drosha and Dicer) are well characterized, as are the Argonaute proteins involved in the effector step of silencing. In addition, genetic screens and biochemical purifications in diverse organisms identified additional factors required for miRNA expression and function. Some of these factors are species, target, or miRNA specific; others play a general role in miRNA-mediated gene silencing and are conserved in diverse organisms (for review, see Carthew and Sontheimer 2009; Kim et al. 2009). Among proteins that play a general role, those in the GW182 family have emerged as key components of miRNA repressive complexes in animal cells (for review, see Ding and Han 2007; Eulalio et al. 2007a, 2008a).
The precise molecular function of GW182 proteins in the miRNA pathway is not fully understood; yet, recent studies have provided new insights into their role in silencing (Chekulaeva et al. 2009; Eulalio et al. 2009a,b; Lazzaretti et al. 2009; Miyoshi et al. 2009; Takimoto et al. 2009; Zipprich et al. 2009). This review summarizes recent findings and discusses the discrepancies and novel questions arising from these studies.
The GW182 protein family
GW182 was originally identified in human cells as the antigen recognized by serum from a patient suffering from motor and sensory neuropathy. This serum stained discrete cytoplasmic domains and allowed the identification and cloning of an uncharacterized human protein, termed GW182 because of its molecular weight and the presence of glycine and tryptophan repeats (either GW, WG, or GWG motifs, termed GW-repeats hereafter) (Eystathioy et al. 2002). GW182-containing foci became known as GW bodies (GWBs). Subsequent studies showed that GWBs coincide with mRNA-processing bodies or P-bodies (Eystathioy et al. 2002, 2003).
In independent studies, GW182 proteins were isolated as proteins that interact with Argonaute proteins and are required for miRNA-mediated gene silencing. These studies included genetic screens in C. elegans, RNAi screens in D. melanogaster, and biochemical purifications of Argonaute complexes in human cells (Ding et al. 2005; Jakymiw et al. 2005; Liu et al. 2005a; Meister et al. 2005; Rehwinkel et al. 2005; Behm-Ansmant et al. 2006a,b; Eulalio et al. 2008b).
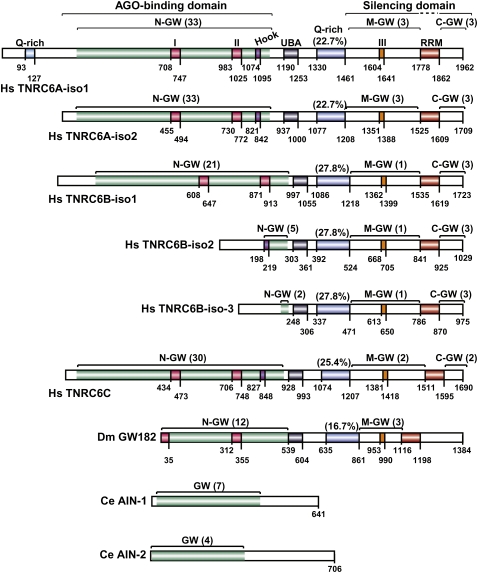
Three GW182 paralogs exist in vertebrates (TNRC6A/GW182, TNRC6B, and TNRC6C), a single ortholog in insects (GW182), and no orthologs in fungi (Behm-Ansmant et al. 2006a). The vertebrate and insect proteins share a common domain organization characterized by two annotated structural domains: a central ubiquitin associated (UBA)-like domain and a C-terminal RNA recognition motif (RRM). These domains are embedded in regions predicted to be unstructured (Fig. 1; Behm-Ansmant et al. 2006a; for review, see Ding and Han 2007; Eulalio et al. 2007a).
FIGURE 1.
Domain organization of GW182 proteins. (Hs) Homo sapiens; (Dm) Drosophila melanogaster. (N-GW, M-GW, C-GW) N-terminal, middle, and C-terminal GW-repeat-containing regions, respectively (the number of GW-repeats for each region is indicated in brackets). The white sectors within the N-terminal regions do not contain GW-repeats. UBA (gray box): ubiquitin associated-like domain; Q-rich (blue box): region rich in glutamine; RRM (red box): RNA recognition motif. Fuchsia boxes I and II: two conserved motifs within the N-terminal GW repeats. Violet box: Ago-hook motif. Orange box: conserved motif III (DUF). Numbers underneath the protein schematic represent amino acid positions at fragment boundaries for each protein. The AGO-binding and silencing domains are indicated. C. elegans (Ce) AIN-1 and AIN-2 proteins are included for comparison.
The unstructured regions include both two or three distinctive blocks of glycine-tryptophan repeats (termed: the N-terminal, middle-, and C-terminal GW-repeat regions), as well as a glutamine-rich (Q-rich) region located between the UBA-like domain and the RRM (Fig. 1; Eystathioy et al. 2002; Behm-Ansmant et al. 2006a; for review, see Ding and Han 2007; Eulalio et al. 2007a). For different GW182 family members, the number of GW-repeats in the N-, middle-, and C-terminal regions varies; nevertheless, for all proteins most GW-repeats lie within the N-terminal region, whereas the middle and C-terminal regions have fewer or no repeats (Fig. 1).
The C. elegans genome encodes two proteins, AIN-1 and AIN-2, which interact with Argonaute proteins and are required for miRNA function (Ding et al. 2005; Zhang et al. 2007; Ding and Großhans 2009). AIN-1 and AIN-2 contain a small number of GW-repeats (seven and four, respectively), and although they are relatively glutamine rich, these proteins lack a defined Q-rich region (observed in the insect and vertebrate proteins) (Fig. 1). Furthermore, AIN-1 and AIN-2 lack the UBA-like and the RRM domains (Fig. 1; Ding et al. 2005; Behm-Ansmant et al. 2006a; Zhang et al. 2007). This lack of common domain architecture suggests AIN-1 and AIN-2 are not members of the GW182 protein family, but rather represent functional analogs.
Similarly, GW-repeats are found in divergent proteins from diverse organisms. For example, the A. thaliana proteins NRPD1b, a subunit of Polymerase IV and the SPT5-like transcription elongation factor, both contain C-terminal domains with multiple GW-repeats that mediate the interaction with AGO4 (El-Shami et al. 2007; Bies-Etheve et al. 2009). In S. pombe, a GW-repeat-containing motif in the Tas3 protein interacts with AGO1, thus forming the RITS complex (RNA-induced transcriptional silencing complex) (Partridge et al. 2007; Till et al. 2007). Although these proteins may employ a mode of interaction with AGOs similar to GW182 proteins, they are not members of the GW182-protein family.
GW182 proteins are required for miRNA repressive functions
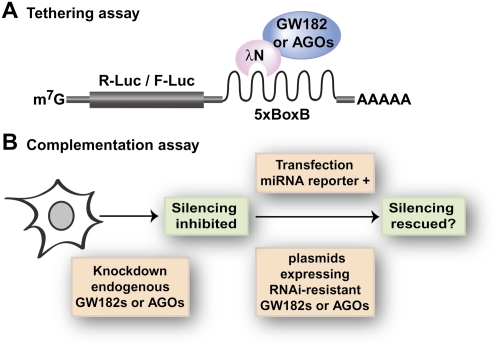
D. melanogaster contains a single GW182 family member (DmGW182), which simplifies the study of GW182 function (DmGW182 is also know as Gawky) (Schneider et al. 2006). We found that depleting GW182 suppresses silencing of miRNA targets, irrespective of whether they are translationally repressed or directed to degradation (Rehwinkel et al. 2005; Behm-Ansmant et al. 2006a,b; Eulalio et al. 2008b, 2009a,b,c). However, depleting DmGW182 does not affect expression levels of miRNAs or DmAGO1 (the Argonaute protein dedicated to the miRNA pathway in D. melanogaster) (Eulalio et al. 2008b, 2009a). Furthermore, when GW182 is directly tethered to an mRNA reporter (see Fig. 2 for an explanation of the tethering assay), reporter expression is silenced independently of Argonaute proteins, suggesting that GW182 plays a role in silencing at the effector step (Behm-Ansmant et al. 2006a,b). These and additional observations lead us to propose that target silencing by miRNAs is effected by a protein complex consisting minimally of an Argonaute and a GW182 protein (Eulalio et al. 2008b).
FIGURE 2.
Functional assays. (A) Tethering assay. This assay involves expressing λN-fusions of wild-type or mutant Argonaute or GW182 proteins, or protein fragments, together with a firefly or renilla luciferase reporter mRNA containing five BoxB hairpins (5xBoxB) in its 3′ UTR. The λN-peptide binds with high affinity to the BoxB hairpins. If the fused protein exhibits silencing activity, then luciferase levels are reduced relative to the λN-peptide alone. Firefly or renilla luciferase expression levels are normalized to a transfection control (renilla or firefly luciferase, respectively) without the BoxB elements. (B) Complementation assay. Endogenous Argonaute or GW182 proteins are depleted using specific siRNAs (human or D. melanogaster cells) or long dsRNA targeting 5′ or 3′ UTRs of the corresponding mRNAs (D. melanogaster cells). Control or depleted cells are transfected with miRNA reporters and plasmids allowing the expression of wild-type or mutant Argonaute or GW182 proteins. These plasmids express the proteins from cDNAs made resistant to the siRNAs. In cells depleted of Argonaute or GW182 proteins, silencing is suppressed. The mutant proteins or fragments are tested for their ability to rescue silencing in these cells.
In other organisms, demonstrating that GW182 proteins play a role in silencing is hampered by the existence of multiple paralogs with partially redundant functions. The idea that human TNRC6A/GW182, TNRC6B, and TNRC6C are redundant is supported by the observation that these proteins associate with all four Argonaute proteins (AGO1–AGO4) and with a common set of miRNA targets (Jakymiw et al. 2005; Liu et al. 2005a,b; Meister et al. 2005; Landthaler et al. 2008; Lazzaretti et al. 2009; Lian et al. 2009; Takimoto et al. 2009; Zipprich et al. 2009). Moreover, in human cells, depleting TNRC6A, TNRC6B, or TNRC6C partially relieves silencing of siRNA and miRNA targets (Jakymiw et al. 2005; Liu et al. 2005a,b; Meister et al. 2005; Chu and Rana 2006; Landthaler et al. 2008; Zipprich et al. 2009). Functional redundancy was also demonstrated for the C. elegans proteins AIN-1 and AIN-2, in that both proteins interact with C. elegans Argonaute proteins 1 and 2 (ALG-1, ALG-2), and their codepletion suppresses silencing more efficiently than depleting each protein individually (Zhang et al. 2007; Ding and Großhans 2009). The conclusion of these collective results is that, in animal cells, GW182 proteins are essential components of the miRNA pathway.
The N-terminal GW-repeat region of GW182 proteins interacts with Argonaute proteins
That the GW182 N-terminal GW-repeat region plays a role in mediating the interaction with Argonautes was first demonstrated in D. melanogaster cells. Here, coimmunoprecipitation assays demonstrated that the N-terminal region of DmGW182 (residues 1–539) was necessary and sufficient for the protein to interact with AGO1 (Behm-Ansmant et al. 2006a). As mentioned above, a striking feature of this region is the presence of multiple GW-repeats. A subsequent study provided evidence that GW-repeats are critical for the interaction between human TNRC6B and AGO2 (Till et al. 2007). Furthermore, El-Shami et al. (2007) showed that the C-terminal domain of NRPD1b interacts with A. thaliana AGO4 via a region containing GW-repeats that are reminiscent of those found in GW182 proteins. A TNRC6A protein fragment containing GW-repeats also interacted with AGO4 (El-Shami et al. 2007), suggesting that for a variety of proteins, GW-repeats act as AGO-binding determinants (Behm-Ansmant et al. 2006a; El-Shami et al. 2007; Till et al. 2007).
More recent studies confirmed and extended these previous observations and showed that the N-terminal regions of human TNRC6A, TNRC6B, and TNRC6C interact with all four human Argonaute proteins (Lazzaretti et al. 2009; Lian et al. 2009; Takimoto et al. 2009; Zipprich et al. 2009). Intriguingly, Lian et al. (2009) showed that, in addition to the N-terminal region, the middle and C-terminal repeats of human TNRC6A interact with AGO2. For reasons that remain unclear, these results are inconsistent with those reported by us and others showing that for the D. melanogaster and human GW182s, GW-repeats outside of the N-terminal region do not contribute to Argonaute binding (Eulalio et al. 2009a; Lazzaretti et al. 2009; Takimoto et al. 2009; Zipprich et al. 2009). The simplest explanation for this discrepancy is that the middle and C-terminal regions exhibit low affinity for Argonaute proteins, owing to the small number of GW-repeats in these regions; therefore, detection of this interaction may be subject to experimental variations.
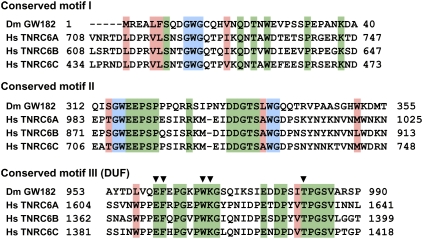
Early studies identified two highly conserved motifs (motifs I and II) within the N-terminal region of GW182 proteins (Fig. 3; Behm-Ansmant et al. 2006a,b). In addition, a GW-repeat-containing sequence in the N-terminal region of human TNRC6B (isoform 2) was identified by Till et al. (2007). This sequence lies downstream from motif II and is termed the AGO-hook. As its name implies, the AGO-hook is sufficient to mediate an interaction with human AGO2 in vitro; however, the AGO-hook is not conserved in all members of the GW182 family; it is missing from isoform 1 of TNRC6B (TNRC6B-iso1) and is not conserved in D. melanogaster GW182 (Fig. 1; Eulalio et al. 2009a).
FIGURE 3.
Sequence alignment of conserved motifs I, II, and III (DUF) from H. sapiens (Hs) TNRC6A–TNRC6C, and D. melanogaster (Dm) GW182. GW-repeats are shadowed in blue, invariant residues in green, and conserved residues are shadowed in salmon. Residues mutated in motif III (DUF) by Zipprich et al. (2009) are indicated by black inverted triangles above the sequence.
A priori, one would expect that the conserved interaction between GW182 and Argonaute proteins is mediated by the most conserved motifs. However, when this was analyzed in detail, different results were obtained for the different proteins. For DmGW182, motif I alone provides the major binding site for the interaction with DmAGO1, although the additional repeats strengthen the interaction (Eulalio et al. 2009a). Indeed, GW182 binding to AGO1 was abolished only when 10 out of 12 repeats in the N-terminal region were substituted with alanines (Eulalio et al. 2009a).
In contrast, in the context of full-length human TNRC6A, TNRC6B, and TNRC6C, deleting motifs I and II individually did not prevent them from interacting with AGO2 (Lazzaretti et al. 2009). The interaction was impaired when both motifs I and II were deleted from TNRC6C and further inhibited when the AGO-hook was also deleted (Lazzaretti et al. 2009). These results suggest that the conserved motifs I and II in TNRC6C provide major binding sites for the interaction with Argonaute proteins and that the additional repeats provide low affinity binding sites.
Deleting motifs I and II, plus the AGO-hook, in the context of full-length TNRC6A, only slightly reduced AGO2-binding. Similarly, only a slight reduction in AGO binding was observed by deleting motifs I and II in TNRC6B-iso1, which lacks the AGO-hook. The conclusion is that most of the N-terminal GW-repeats in TNRC6A and TNRC6B appear to contribute to the interaction with Argonaute proteins (Lazzaretti et al. 2009). Accordingly, Takimoto et al. (2009) found that three fragments of TNRC6A (overlapping with motif I, motif II, and the AGO-hook) were sufficient to interact with Argonaute proteins. The investigators also identified specific GW-repeats that, in the context of the isolated protein fragments, were required for this interaction; the relevance of these repeats was not tested in the context of full-length TNRC6A.
Previous work by El-Shami et al. (2007) and Till et al. (2007) showed the tryptophan residues in the GW-repeats are critical for AGO binding; indeed, binding was abolished when these residues were substituted with alananines or phenylalanines. Takimoto et al. (2009) has now confirmed the importance of tryptophan residues for the GW182–AGO interaction. Nevertheless, the context of the repeats also contributes to the strength of the interaction. Two lines of evidence support this conclusion. First, not all repeats contribute equally to the interaction (Eulalio et al. 2009a; Lazzaretti et al. 2009); second, mutation analysis showed substituting residues adjacent to the repeats can also affect the interaction (Till et al. 2007). In this context, the study by Lian et al. (2009) is again at odds with available data, as these investigators concluded that tryptophan residues in the repeats do not contribute to AGO binding. Certainly, GW-repeats are critical specificity determinants for GW182–AGO interaction, because when both tryptophan and glycine residues are substituted with alanines the interaction is lost (Eulalio et al. 2009a). A better understanding of how GW-repeats interact with Argonaute proteins awaits the structural analysis of GW182–AGO protein complexes.
Regardless of the discrepancies mentioned above, two major conclusions can be drawn from the present studies. One is that the N-terminal GW-repeat-containing region of GW182 proteins provides multiple binding sites for AGOs. We have termed this region the AGO-binding domain (Fig. 1). Second, several GW-repeats contribute to the interaction, and therefore the contribution of each repeat to the binding affinity can only be analyzed in the context of full-length proteins.
Takimoto et al. (2009) demonstrated that Argonaute proteins have a single binding site for GW182 proteins; in contrast, GW182 proteins can interact with multiple AGOs simultaneously. Why might this be? One possibility is that GW182 proteins bridge the interaction between multiple silenced mRNPs, promoting them to aggregate in RNP granules. Alternatively, the presence of multiple Argonaute-binding sites in GW182 proteins may explain the synergistic effects of miRNA-binding sites in close proximity to each other (between 8 and 40 nucleotides) (Grimson et al. 2007): One GW182 protein may stabilize and bind cooperatively to multiple Argonaute proteins on adjacent miRNA target sites.
The AGO-binding domain of GW182 proteins is also required for them to accumulate in P-bodies (see below), but whether this domain has additional functions in silencing remains controversial. Studies with human TNRC6A–TNRC6C showed that the AGO-binding domain has no silencing activity in tethering assays (Lazzaretti et al. 2009; Zipprich et al. 2009). In contrast, the AGO-binding domain of DmGW182 represses expression of bound transcripts in tethering assays (Chekulaeva et al. 2009). The repressive activity of this domain is observed even in cells depleted of AGO1 or GW182, showing that this domain has additional functionalities (Chekulaeva et al. 2009; see below).
The AGO-binding domain and the Q-rich region contribute to the accumulation of GW182 proteins in P-bodies
The Q-rich region is a common feature of GW182 family proteins; however, in addition to the glutamine residues, the composition of this region varies. In D. melanogaster, the Q-rich region is also enriched in asparagine residues (14.5%), but in the human proteins this is not the case. Notably, isoform-1 of TNRC6A exhibits an additional Q-rich region upstream of the AGO-binding domain (Fig. 1). As mentioned above, AIN-1 and AIN-2 do not contain a defined Q-rich region, but are overall Q-rich.
Evidence that the Q-rich region of DmGW182 plays a role in P-body localization stems from the observation that an N-terminal fragment of DmGW182 (containing the AGO-binding, the UBA-like domains, and the Q-rich region) was necessary and sufficient for localization to P-bodies, although none of these regions was sufficient on their own (Behm-Ansmant et al. 2006a). This protein fragment also promoted the accumulation of DmAGO1 in P-bodies, a process that depends on the interaction with GW182 in D. melanogaster cells (Behm-Ansmant et al. 2006a).
Current studies delineated more precisely the motifs required for GW182 to localize to P-bodies. In particular, the UBA-like domain of DmGW182 does not contribute to P-body localization (Eulalio et al. 2009a). In contrast, we showed that for DmGW182 to localize to P-bodies, it requires both the AGO-binding domain and the Q-rich region. In agreement with this, GW182 mutants that cannot interact with DmAGO1 also do not accumulate in P-bodies, despite the fact that these mutants have a Q-rich region. Conversely, a protein lacking the Q-rich region disperses throughout the cytoplasm, although this protein interacts with DmAGO1 (Eulalio et al. 2009a). For the human proteins, the requirements for P-body localization differ slightly from those observed for DmGW182. Nevertheless, in all cases, the interaction with the Argonaute proteins is required for P-body localization, whereas the Q-rich regions either contribute (human TNRC6A and TNRC6C) or play an essential role (human TNRC6B-iso1) (Lazzaretti et al. 2009).
GW182 accumulation in P-bodies is not required for silencing
Despite the role of the DmGW182 Q-rich region in P-body localization, this region is dispensable for silencing. Indeed, a DmGW182 protein lacking the Q-rich region can complement silencing, even if it does not localize to P-bodies (see Fig. 2 for an explanation of the complementation assay; Eulalio et al. 2009a). These findings are particularly relevant because they show that the silencing activity of DmGW182 does not correlate with its ability to localize to P-bodies.
Similarly (as discussed below), C-terminal fragments of human TNRC6A–TNRC6C that encompass protein sequences downstream from the Q-rich regions retain full silencing activity in tethering assays, although these fragments do not localize to P-bodies (Lazzaretti et al. 2009; Zipprich et al. 2009). Thus, together with previous studies (Chu and Rana 2006; Eulalio et al. 2007b), these observations add to the growing evidence that detectable P-bodies are not required for silencing, but rather represent a consequence of silencing (Eulalio et al. 2007a,b). In support of the idea that GW182 and Argonaute proteins detected in P-bodies may not represent the pool actively engaged in silencing is the observation that DmAGO1 mutants carrying four or six substitutions in the PAZ domain (which disrupt miRNA binding, and thus the interaction with the mRNA target) accumulate in P-bodies (Eulalio et al. 2009a; Miyoshi et al. 2009). Intriguingly, mutations in the PAZ domain of human AGO2 abolished P-body localization (PAZ9 and PAZ10 mutants) (Liu et al. 2005a); it is likely that this reflects species-specific differences, because the corresponding mutations in DmAGO1 did not prevent P-body localization (Eulalio et al. 2009a).
The UBA-like domain
A UBA-like domain was originally predicted only to be present in human TNRC6C (Behm-Ansmant et al. 2006a; for review, see Ding and Han 2007); however, using iterative PSI-BLAST searches, we predicted that a UBA homology domain exists in all members of the GW182 protein family (Behm-Ansmant et al. 2006a). UBA domains are small domains of about 40 residues, originally identified in proteins involved in ubiquitination (for review, see Buchberger 2002). These domains bind mono- or polyubiquitin chains on ubiquitinated partners. However, distantly related UBA domains (UBA-like domains) that might have evolved different functions have been described in proteins with no role in the ubiquitin–proteasome pathway (for review, see Buchberger 2002). It remains to be shown whether the UBA-like domains in GW182 proteins bind ubiquitin or have different functionalities. Furthermore, the role of this domain is not understood, and the present studies failed to provide evidence for a contribution of this domain to silencing (Chekulaeva et al. 2009; Eulalio et al. 2009a, Lazzareti et al. 2009; Zipprich et al. 2009). It is still possible, though, that this domain plays a role in silencing specific targets, or during specific stress conditions, or during a specific phase of the cell cycle.
The RRM domain
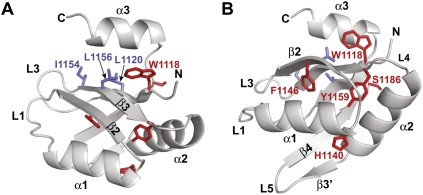
The RRM is highly conserved among GW182 proteins, and its presence was interpreted as indicative of RNA-binding activity (Eystathioy et al. 2002). It is unknown whether GW182 proteins bind RNA directly, but if they do, it is probably not through the RRM. Indeed, the structure of the RRM domain of DmGW182 revealed that it lacks RNA-binding features (Eulalio et al. 2009b). Although the domain adopts a canonical RRM fold, consisting of a four-stranded anti-parallel β-sheet packed against two α-helices (Fig. 4A,B; Eulalio et al. 2009b), an additional C-terminal α-helix (α3) shields the β-sheet surface, which in canonical RRMs is involved in RNA binding.
FIGURE 4.
Ribbon representation of the D. melanogaster GW182 RRM showing the β-sheet surface normally involved in RNA binding (A) or the opposite helical surface (helix α1 and helix α2) (B). The C-terminal α-helix α3 covers the β-sheet surface, which in canonical RRMs is involved in RNA binding (Eulalio et al. 2009b). Side chains of aliphatic residues, which are normally aromatic in RNP1 and RNP2 motifs of canonical RRMs are shown as blue sticks. Residues corresponding to those mutated by Zipprich et al. (2009) in TNRC6C are shown as red sticks. Residues F1146 (F1543 in TNRC6C) and Y1159 (Y1556 in TNRC6C) are part of the hydrophobic core of the domain, whereas W1118 (W1515 in TNRC6C) contributes to hydrophobic interactions with α-helix α3.
Canonical RRM domains interact with RNA through stacking interactions between two RNA bases and the ring of conserved aromatic residues in the RNP motifs 1 and 2 (for review, see Maris et al. 2005). In GW182 RRM, the conserved aromatic residues in the RNP1 and RNP2 motifs are substituted with aliphatic residues (Fig. 4A, blue residues), indicating that even if the C-terminal α-helix could move away from the β-sheet, the exposed surface still lacks RNA-binding features. Moreover, the RRM lacks any positively charged surface patch that could mediate RNA binding. Finally, DmGW182 RRM lacks a detectable general RNA-binding affinity in vitro, suggesting that this domain does not bind RNA. Thus, the RRM may instead engage in protein–protein interactions through an unusual hydrophobic cleft exposed on the opposite face of the β-sheet (Eulalio et al. 2009b).
The conservation of critical structural residues indicates that the occlusion of the putative RNA-binding surface by the C-terminal α-helix and the presence of a hydrophobic cleft on the opposite face of the β-sheet are structural features conserved in RRM domains of vertebrate GW182 proteins (Eulalio et al. 2009b). Despite conservation, the RRM domain is not required for GW182 proteins to localize to P-bodies or to interact with either Argonaute proteins or with miRNAs (Chekulaeva et al. 2009; Eulalio et al. 2009a,b; Lazzaretti et al. 2009; Zipprich et al. 2009). Furthermore, in complementation assays, a DmGW182 protein lacking the RRM was impaired in silencing, but only for a subset of miRNA-target pairs (Eulalio et al. 2009b).
Zipprich et al. (2009) tested how mutations in the RRM affect TNRC6C in tethering assays (Fig. 4A,B, red residues). The mutations were tested in the context of a C-terminal fragment of TNRC6C, which is fully active in this assay. The authors showed that these mutations affect the activity of the protein fragment. The human and D. melanogaster RRM are highly conserved (59% sequence identity); thus, mutations on the human TNRC6C RRM can be easily mapped onto the structure of the D. melanogaster RRM, showing that some of the residues mutated by Zipprich et al. (2009) are part of the hydrophobic core of the protein (Fig. 4A,B red residues). In summary, the RRM is not strictly required for the silencing activity of GW182 proteins, although it is possible that this domain contributes to silencing of a subset of miRNA targets.
The middle and C-terminal regions of GW182 define a silencing domain
As mentioned above, our studies suggest that the UBA-like domain, the Q-rich region, and the RRM domain are not essential for the silencing activity of GW182 proteins. Some discrepancies arose, however, regarding the role of the middle and C-terminal regions.
Chekulaeva et al. (2009) observed that N-terminal fragments of DmGW182 (1–605 and 1–830; see Fig. 1) are sufficient to rescue silencing in cells depleted of endogenous GW182, albeit less efficiently than the full-length protein. These results suggest that the corresponding C-terminal fragments of the protein (i.e., 605–1384 and 830–1384) contribute to, but are not absolutely required for, silencing. In agreement with this, in tethering assays the C-terminal fragments of DmGW182 silence the expression of bound transcripts, indicating that they exhibit silencing activity (Chekulaeva et al. 2009).
Notably, the same N-terminal fragments of DmGW182 that restore silencing in complementation assays inhibit silencing in a dominant negative manner in control cells (Chekulaeva et al. 2009). Indeed, it was shown both in Drosophila cells and in extracts that recapitulate silencing in vitro, that protein fragments encompassing the GW182 AGO-binding domain inhibit silencing in a dominant negative manner (Till et al. 2007; Eulalio et al. 2008b; Iwasaki et al. 2009; Takimoto et al. 2009). The dominant negative effect is most likely explained by sequestration of Argonaute proteins into inactive complexes. Thus, how might DmGW182 N-terminal fragments rescue silencing when endogenous GW182 is depleted and also inhibit silencing when endogenous GW182 is present? Chekulaeva et al. (2009) suggested that DmGW182 N-terminal fragments contribute to the assembly of repressive complexes, but are less efficient than the full-length protein, and thereby their activity can only be detected in cells lacking endogenous GW182. In wild-type cells, in contrast, these fragments would interfere with the activity of endogenous GW182 by competing for binding to AGO1.
In contrast to the results reported by Chekulaeva et al. (2009), we observed that deleting either the middle or the C-terminal GW-repeat regions of DmGW182 impaired silencing in complementation assays (Eulalio et al. 2009a). Furthermore, if both of these regions were deleted, then the silencing activity of DmGW182 was completely abolished. This suggests that these sequences define a bipartite silencing domain separated by the RRM. The DmGW182 mutant that lacks both the middle and C-terminal regions interacts with AGO1 and miRNAs, and localizes to P-bodies; however, this DmGW182 mutant cannot rescue silencing, even though it contains the N-terminal residues identified by Chekulaeva et al. (2009) as being sufficient for silencing in both tethering and complementation assays. Clearly, additional analysis is required to resolve this discrepancy.
In contrast, analysis of human proteins gave clear-cut results. As mentioned above, the AGO-binding domain of human TNRC6A–TNRC6C has no or only a modest silencing activity in tethering assays, whereas the C-terminal fragments (starting at the UBA) are even more active than the full-length proteins in tethering assays (Lazzaretti et al. 2009; Zipprich et al. 2009). Furthermore, extended N-terminal fragments containing, in addition to the AGO-binding domain, the UBA-like domain, and the Q-rich region silence reporter expression, only 1.6- to 2.5-fold, whereas the corresponding C-terminal fragments retain full activity. In fact, fragments encompassing the middle and C-terminal GW-repeat regions and the RRM are sufficient for silencing in tethering assays (Lazzaretti et al. 2009; Zipprich et al. 2009). As mentioned above, Zipprich et al. (2009) showed that within this fragment mutations in the RRM have a modest effect. These results are consistent with those obtained with DmGW182, and suggest that the RRM contributes to, but it is not absolutely required for the silencing activity of this fragment. The conclusion of these studies is that the middle and C-terminal regions of GW182 proteins define a bipartite domain with autonomous silencing activity (Eulalio et al. 2009a; Zipprich et al. 2009). In this context, it will be interesting in the future to determine whether the C. elegans AIN-1 and AIN-2 proteins, which lack the middle and C-terminal regions, exhibit silencing activity independent of the interaction with Argonaute proteins (e.g., in tethering assays).
What are the features of this silencing domain? The middle and C-terminal GW-repeat-containing regions of GW182 proteins are not highly conserved, with the exception of a motif of about 40 residues (Figs. 1, 3, conserved motif III) within the middle region. This motif was termed DUF (domain of unknown function) (Zipprich et al. 2009), although secondary structure predictions suggest that this sequence is unstructured, and as such, does not represent an independent folding unit. Regardless, mutating the most conserved residues in this motif (Fig. 3, black inverted triangles) or deleting the entire motif did not affect the silencing activity of human TNRC6C or DmGW182 (Eulalio et al. 2009a; Zipprich et al. 2009). These observations are very intriguing and unexpected because they show that the sequences that contribute to silencing are the least conserved among GW182 proteins. The only feature these sequences have in common is the high content of serine residues, suggesting that the activity of GW182 proteins may be regulated by phosphorylation. Human TNRC6A is indeed phosphorylated (Eystathioy et al. 2002); however, it remains to be shown that phosphorylation plays a role in silencing.
GW182 silencing domain represses translation and promotes mRNA decay
Another controversial issue concerns the mechanism by which the silencing domains of GW182 proteins silence the expression of reporter mRNAs. As observed in tethering assays for the full-length proteins, these domains trigger both translational repression and degradation of bound mRNAs (Behm-Ansmant et al. 2006a,b; Li et al. 2008; Chekulaeva et al. 2009; Lazzareti et al. 2009; Zipprich et al. 2009). The extent of degradation caused by the silencing domains, or for that matter, by the full-length proteins, differs among the studies and may be explained by differences in cell lines. As shown before for DmGW182, we observed significant mRNA degradation that was at least in part attributed to deadenylation (Behm-Ansmant et al. 2006a,b; Lazzaretti et al. 2009). Zipprich et al. (2009) observed a much stronger effect on protein expression (10- to 20-fold) than on mRNA levels (threefold).
To what extent translational repression and mRNA decay contribute to silencing is a fundamental, unanswered question in the miRNA field (for review, see Eulalio et al. 2008a; Filipowicz et al. 2008; Carthew and Sontheimer 2009). The current view is that miRNAs trigger both translational repression and mRNA decay via deadenylation and decapping, and that these two effects contribute to silencing in a target-miRNA pair-specific manner (for review, see Eulalio et al. 2008a, Carthew and Sontheimer 2009). This dichotomy is also observed for the GW182 proteins (Behm-Ansmant et al. 2006a,b; Eulalio et al. 2008b, 2009c; Li et al. 2008; Chekulaeva et al. 2009; Lazzareti et al. 2009; Zipprich et al. 2009). In fact, the similarities between miRNA- and GW182-mediated silencing are striking: both can repress the expression of polyadenylated as well as nonadenylated mRNAs (Wu et al. 2006; Eulalio et al. 2008b, 2009c; Chekulaeva et al. 2009), indicating that in both cases deadenylation is not absolutely required for silencing, although it contributes to the extent to which a polyadenylated transcript is repressed.
GW182- and miRNA-binding sites in Argonaute proteins partially overlap
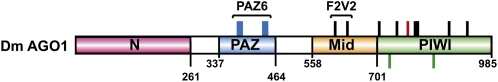
Recent studies have also aimed to establish which domains of Argonaute proteins are required for the interaction with GW182 proteins. Argonaute proteins are characterized by four domains: the N-terminal domain; the PAZ domain, which binds the 3′ end of miRNAs/siRNAs; the mid-domain, which provides a binding pocket for the 5′-phosphate of miRNAs/siRNAs; and the PIWI domain, which adopts an RNase H-like fold and has slicing activity in some, but not all AGOs (Fig. 5; for review, see Jinek and Doudna 2009).
FIGURE 5.
Domain organization of Drosophila melanogaster AGO1. N-terminal domain (fuchsia box); PAZ domain (blue box); Mid-domain (orange box) and PIWI domain (green box). Numbers underneath the protein schematic represent amino acid positions at fragment boundaries. Vertical lines above the protein schematic indicate the position of residues that when mutated affect miRNA-binding (blue, R386, K387, Y388, T446, Y447, L448), GW182-binding (red, F777), or both miRNA and GW182 binding (black) (see Eulalio et al. 2009a for additional information). Green lines below the protein schematic indicate the position of catalytic residues.
Earlier studies in D. melanogaster showed that the C-terminal PIWI domain of DmAGO1 was sufficient for the interaction with GW182 (Behm-Ansmant et al. 2006a); a study by Till et al. (2007) provided a detailed analysis of this interaction. Based on the structure of the archeal Archaeoglobus fulgidus PIWI protein (Ma et al. 2005), the investigators designed a series of 58 mutations across the middle and PIWI domains of human AGO2. Surprisingly, they found that AGO2 residues that were predicted to form the pocket that binds the 5′-end of miRNAs are also required for TNRC6B binding, suggesting that the AGO2 sites that bind the 5′-end of miRNAs and TNRC6B partially overlap. Importantly, Till et al. (2007) showed that miRNA and TNRC6B binding can occur simultaneously, and TNRC6B neither interferes nor enhances the AGO2–miRNA interaction. In agreement with this, the interaction between DmAGO1 and miRNAs remains unaffected in cells depleted of GW182 (Eulalio et al. 2009a).
Based on these mutations described by Till et al. (2007), we generated the corresponding mutations in DmAGO1 and tested their affinity for DmGW182, miRNAs, as well as the effect on P-body localization and the ability to complement silencing in cells depleted of endogenous DmAGO1 (Eulalio et al. 2009a). As expected, most mutations in the mid and PIWI domains that affect DmGW182 binding also prevent miRNA-binding (Fig. 5, black bars). In fact, the only mutations that disrupt binding to miRNAs, but not to DmGW182, are those located in the PAZ domain (Eulalio et al. 2009a; Miyoshi et al. 2009). Nevertheless, we could identify a single residue in DmAGO1 required for the interaction with DmGW182, but not with miRNAs (Fig. 5, red vertical line). However, a protein carrying a substitution of this residue retains a residual binding affinity for DmGW182, and attempts to completely abolish this interaction always resulted in loss of miRNA binding. The conclusion is that the binding sites for miRNA 5′-ends and GW182 proteins partially overlap, as reported by Till et al. (2007). However, these two binding activities are not interdependent and can be uncoupled from one another by specific mutations (Eulalio et al. 2009a).
In complementation assays, DmAGO1 mutants that lose the ability to interact either with both miRNAs and DmGW182, or only with miRNAs, do not rescue silencing as expected. DmAGO1 mutants that interact with miRNAs but are impaired in DmGW182 binding are also impaired in the complementation assay, indicating that DmAGO1 does indeed require the interaction with DmGW182 to mediate miRNA silencing. In support of this model, Lian et al. (2009) showed that a C-terminal fragment of human AGO2 (containing part of the mid domain and the PIWI domain) silences an mRNA reporter in tethering assays, and this activity requires the interaction with GW182.
Importantly, not all Argonaute proteins interact with GW182 proteins. For example, DmGW182 interacts with DmAGO1, but not DmAGO2 (Behm-Ansmant et al. 2006a; Iwasaki et al. 2009; Miyoshi et al. 2009). Also, the PIWI-like proteins do not interact with GW182 (Miyoshi et al. 2009). Thus, it appears that the interaction with GW182 proteins is restricted to the Argonaute proteins involved in the miRNA pathway, although, as mentioned above, other Argonaute proteins may interact with GW-repeats present in unrelated proteins, as is the case of A. thaliana AGO4 and S. pombe AGO1.
GW182 proteins act downstream from miRNA processing and loading
The present studies provide further evidence that GW182 proteins play an essential role in silencing, and although they do not provide an answer for what this role might be, they help to exclude some obvious possibilities. First, GW182 proteins are not required for miRNA biogenesis, because miRNA levels are not affected in cells depleted of GW182 (Eulalio et al. 2009a; Miyoshi et al. 2009). Second, GW182 proteins are not required to maintain Argonaute protein expression levels (Eulalio et al. 2008b; 2009a). Third, GW182 proteins are not required for miRNA to be loaded onto Argonaute proteins (Eulalio et al. 2009a; Miyoshi et al. 2009). Finally, the role of GW182 proteins in silencing is unrelated to its ability to localize to P-bodies or to direct Argonaute proteins to P-bodies, because GW182 mutants that do not localize to P-bodies can rescue silencing in cells lacking the endogenous protein (Eulalio et al. 2009a). Thus, GW182 proteins act downstream from miRNA processing and loading.
These findings raise an important question: which step of silencing is effected by GW182 proteins? One possibility is that GW182 proteins contribute to target recognition by AGOs, either by interacting with proteins bound to the mRNA target or by interfering with their binding, thereby increasing target accessibility. However, this cannot be the only function, because GW182 proteins possess silencing activity when they are artificially tethered to an mRNA reporter (Behm-Ansmant et al. 2006a,b; Eulalio et al. 2008b; Li et al. 2008; Chekulaeva et al. 2009; Lazzaretti et al. 2009; Zipprich et al. 2009). Therefore, it is likely that these proteins act downstream from target binding by interfering with the function of components of the translation machinery and facilitating the recruitment of general mRNA decay factors.
SUMMARY
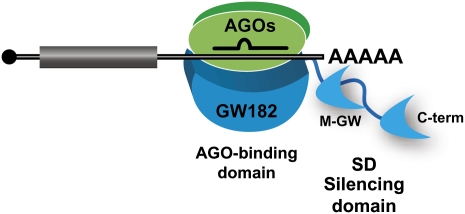
The present studies add to the growing support for the idea that miRNA silencing in animal cells is mediated by Argonaute proteins in complex with GW182 proteins. They further show that two domains in GW182 proteins play an essential role in silencing (Fig. 6). One is the N-terminal domain, which provides multiple binding sites for Argonaute proteins and also contributes to the assembly of silencing complexes in the D. melanogaster protein. The other is a bipartite C-terminal silencing domain, which promotes translational repression and decay of mRNA targets. Mechanistically, however, it remains unclear how GW182 proteins contribute to silencing. By identifying which GW182 domains have autonomous silencing activity we have a starting point from which to dissect their role in silencing, a question that will no doubt be resolved in the near future.
FIGURE 6.
Model for miRNA-mediated gene silencing. RISC is represented as a complex minimally including an Argonaute protein (green) and GW182 (cyan). GW182 proteins contain an N-terminal AGO-binding domain, which provides multiple binding sites for Argonaute proteins and a bipartite C-terminal silencing domain (SD), which promotes translational repression and decay of mRNA targets. Note that at least in D. melanogaster GW182, the AGO-binding domain exhibits strong silencing activity in tethering assays and complements silencing in cells lacking endogenous GW182, indicating that in addition to interacting with AGO1, this domain contributes to the assembly of repressive complexes (Chekulaeva et al. 2009). Overexpressing this domain alone in wild-type cells inhibits silencing in a dominant negative manner (Till et al. 2007; Eulalio et al. 2008b; Chekulaeva et al. 2009; Iwasaki et al. 2009; Takimoto et al. 2009).
ACKNOWLEDGMENTS
We are grateful to Witold Filipowicz for comments on the manuscript. This research is supported by the Max Planck Society, by grants from the Deutsche Forschungsgemeinschaft (DFG, FOR855, and the Gottfried Wilhelm Leibniz Program awarded to E.I.), and by the Sixth Framework Programme of the European Commission through the SIROCCO Integrated Project LSHG-CT-2006-037900.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1703809.
REFERENCES
- Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes & Dev. 2006a;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm-Ansmant I, Rehwinkel J, Izaurralde E. MicroRNAs silence gene expression by repressing protein expression and/or by promoting mRNA decay. Cold Spring Harb Symp Quant Biol. 2006b;71:523–530. doi: 10.1101/sqb.2006.71.013. [DOI] [PubMed] [Google Scholar]
- Bies-Etheve N, Pontier D, Lahmy S, Picart C, Vega D, Cooke R, Lagrange T. RNA-directed DNA methylation requires an AGO4-interacting member of the SPT5 elongation factor family. EMBO Rep. 2009;10:649–654. doi: 10.1038/embor.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchberger A. From UBA to UBX: New words in the ubiquitin vocabulary. Trends Cell Biol. 2002;12:216–221. doi: 10.1016/s0962-8924(02)02269-9. [DOI] [PubMed] [Google Scholar]
- Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekulaeva M, Filipowicz W, Parker R. Multiple independent domains of dGW182 function in miRNA-mediated repression in Drosophila. RNA. 2009;15:794–803. doi: 10.1261/rna.1364909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CY, Rana TM. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol. 2006;4:e210. doi: 10.1371/journal.pbio.0040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Han M. GW182 family proteins are crucial for microRNA-mediated gene silencing. Trends Cell Biol. 2007;17:411–416. doi: 10.1016/j.tcb.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Ding XC, Großhans H. Repression of C. elegans microRNA targets at the initiation level of translation requires GW182 proteins. EMBO J. 2009;28:213–222. doi: 10.1038/emboj.2008.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Spencer A, Morita K, Han M. The developmental timing regulator AIN-1 interacts with miRISCs and may target the argonaute protein ALG-1 to cytoplasmic P bodies in C. elegans. Mol Cell. 2005;19:437–447. doi: 10.1016/j.molcel.2005.07.013. [DOI] [PubMed] [Google Scholar]
- El-Shami M, Pontier D, Lahmy S, Braun L, Picart C, Vega D, Hakimi MA, Jacobsen SE, Cooke R, Lagrange T. Reiterated WG/GW motifs form functionally and evolutionarily conserved ARGONAUTE-binding platforms in RNAi-related components. Genes & Dev. 2007;21:2539–2544. doi: 10.1101/gad.451207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Izaurralde E. P bodies: At the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol. 2007a;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E. P-body formation is a consequence, not the cause of RNA-mediated gene silencing. Mol Cell Biol. 2007b;27:3970–3981. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008a;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Izaurralde E. GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat Struct Mol Biol. 2008b;15:346–353. doi: 10.1038/nsmb.1405. [DOI] [PubMed] [Google Scholar]
- Eulalio A, Helms S, Fritzsch C, Fauser M, Izaurralde E. A C-terminal silencing domain in GW182 is essential for miRNA function. RNA. 2009a;15:1067–1077. doi: 10.1261/rna.1605509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Tritschler F, Buettner R, Weichenrieder O, Izaurralde E, Truffault V. The RRM domain in GW182 proteins contributes to miRNA-mediated gene silencing. Nucleic Acids Res. 2009b;37:2974–2983. doi: 10.1093/nar/gkp173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Nishihara T, Rehwinkel J, Fauser M, Izaurralde E. Deadenylation is a widespread effect of miRNA regulation. RNA. 2009c;15:21–32. doi: 10.1261/rna.1399509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eystathioy T, Chan EK, Tenenbaum SA, Keene JD, Griffith K, Fritzler MJ. A phosphorylated cytoplasmic autoantigen, GW182, associates with a unique population of human mRNAs within novel cytoplasmic speckles. Mol Biol Cell. 2002;13:1338–1351. doi: 10.1091/mbc.01-11-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eystathioy T, Jakymiw A, Chan EK, Seraphin B, Cougot N, Fritzler MJ. The GW182 protein colocalizes with mRNA degradation associated proteins hDcp1 and hLSm4 in cytoplasmic GW-bodies. RNA. 2003;9:1171–1173. doi: 10.1261/rna.5810203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki S, Kawamata T, Tomari Y. Drosophila Argonaute1 and Argonaute2 employ distinct mechanisms for translational repression. Mol Cell. 2009;34:58–67. doi: 10.1016/j.molcel.2009.02.010. [DOI] [PubMed] [Google Scholar]
- Jakymiw A, Lian S, Eystathioy T, Li S, Satoh M, Hamel JC, Fritzler MJ, Chan EK. Disruption of GW bodies impairs mammalian RNA interference. Nat Cell Biol. 2005;7:1267–1274. doi: 10.1038/ncb1334. [DOI] [PubMed] [Google Scholar]
- Jinek M, Doudna JA. A three-dimensional view of the molecular machinery of RNA interference. Nature. 2009;457:405–412. doi: 10.1038/nature07755. [DOI] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- Landthaler M, Gaidatzis D, Rothballer A, Chen PY, Soll SJ, Dinic L, Ojo T, Hafner M, Zavolan M, Tuschl T. Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. RNA. 2008;14:2580–2596. doi: 10.1261/rna.1351608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzaretti D, Tournier I, Izaurralde E. The C-terminal domains of human TNRC6A, TNRC6B, and TNRC6C silence bound transcripts independently of the Argonaute proteins. RNA. 2009;15:1059–1066. doi: 10.1261/rna.1606309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Lian SL, Moser JJ, Fritzler ML, Fritzler MJ, Satoh M, Chan EK. Identification of GW182 and its novel isoform TNGW1 as translational repressors in Ago2-mediated silencing. J Cell Sci. 2008;121:4134–4144. doi: 10.1242/jcs.036905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian SL, Songqing Li S, Abadal GX, Pauley BA, Fritzler MJ, Chan EKL. The C-terminal half of human Ago2 binds to multiple GW-rich regions of GW182 and requires GW182 to mediate silencing. RNA. 2009;15:804–813. doi: 10.1261/rna.1229409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Rivas FV, Wohlschlegel J, Yates JR, III, Parker R, Hannon GJ. A role for the P-body component GW182 in microRNA function. Nat Cell Biol. 2005a;7:1261–1266. doi: 10.1038/ncb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol. 2005b;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JB, Yuan YR, Meister G, Pei Y, Tuschl T, Patel DJ. Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature. 2005;434:666–670. doi: 10.1038/nature03514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maris C, Dominguez C, Allain FH-T. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;272:2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Peters L, Chen PY, Urlaub H, Lührmann R, Tuschl T. Identification of novel Argonaute-associated proteins. Curr Biol. 2005;15:2149–2155. doi: 10.1016/j.cub.2005.10.048. [DOI] [PubMed] [Google Scholar]
- Miyoshi K, Okada TN, Siomi H, Siomi MC. Characterization of miRNA-RISC loading complex and miRNA-RISC formed in the Drosophila miRNA pathway. RNA. 2009;15:1282–1291. doi: 10.1261/rna.1541209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge JF, DeBeauchamp JL, Kosinski AM, Ulrich DL, Hadler MJ, Noffsinger VJ. Functional separation of the requirements for establishment and maintenance of centromeric heterochromatin. Mol Cell. 2007;26:593–602. doi: 10.1016/j.molcel.2007.05.004. [DOI] [PubMed] [Google Scholar]
- Rehwinkel J, Behm-Ansmant I, Gatfield D, Izaurralde E. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA. 2005;11:1640–1647. doi: 10.1261/rna.2191905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider MD, Najand N, Chaker S, Pare JM, Haskins J, Hughes SC, Hobman TC, Locke J, Simmonds AJ. Gawky is a component of cytoplasmic mRNA processing bodies required for early Drosophila development. J Cell Biol. 2006;174:349–358. doi: 10.1083/jcb.200512103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takimoto K, Wakiyama M, Yokoyama S. Mammalian GW182 contains multiple Argonaute-binding sites and functions in microRNA-mediated translational repression. RNA. 2009;15:1078–1089. doi: 10.1261/rna.1363109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till S, Lejeune E, Thermann R, Bortfeld M, Hothorn M, Enderle D, Heinrich C, Hentze MW, Ladurner AG. A conserved motif in Argonaute-interacting proteins mediates functional interactions through the Argonaute PIWI domain. Nat Struct Mol Biol. 2007;14:897–903. doi: 10.1038/nsmb1302. [DOI] [PubMed] [Google Scholar]
- Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ding L, Cheung TH, Dong M-Q, Chen J, Sewell AK, Liu X, Yates JR, Han M. Systematic identification of C. elegans miRISC proteins, miRNAs, and mRNA targets by their interactions with GW182 Proteins AIN-1 and AIN-2. Mol Cell. 2007;28:598–613. doi: 10.1016/j.molcel.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipprich JT, Bhattacharyya S, Mathys H, Filipowicz W. Importance of the C-terminal domain of the human GW182 protein TNRC6C for translational repression. RNA. 2009;15:781–793. doi: 10.1261/rna.1448009. [DOI] [PMC free article] [PubMed] [Google Scholar]