Abstract
MicroRNAs (miRNAs) play important roles in directing the differentiation of cells down a variety of cell lineage pathways. The planarian Schmidtea mediterranea can regenerate all lost body tissue after amputation due to a population of pluripotent somatic stem cells called neoblasts, and is therefore an excellent model organism to study the roles of miRNAs in stem cell function. Here, we use a combination of deep sequencing and bioinformatics to discover 66 new miRNAs in S. mediterranea. We also identify 21 miRNAs that are specifically expressed in either sexual or asexual animals. Finally, we identified five miRNAs whose expression is sensitive to γ-irradiation, suggesting they are expressed in neoblasts or early neoblast progeny. Together, these results increase the known repertoire of S. mediterranea miRNAs and identify numerous regulated miRNAs that may play important roles in regeneration, homeostasis, neoblast function, and reproduction.
Keywords: planarians, microRNAs, stem cells, deep sequencing, bioinformatics
INTRODUCTION
Planarians are an excellent and increasingly popular model system for studying stem cell biology, as they have the ability to completely replace tissue lost after injury (Newmark and Sanchez Alvarado 2002). This remarkable regenerative capacity is made possible by the presence of a population of pluripotent somatic stem cells called neoblasts that can give rise to all cell types in the animal, including germ cells (Baguñà et al. 1989; Wang et al. 2007), and comprise ∼30% of the entire organism. In addition to being required for regeneration, neoblasts are also essential for homeostasis where they replace cells lost during normal tissue turnover.
The planarian species that is predominantly studied and particularly amenable to molecular biology analysis is Schmidtea mediterranea (Sanchez Alvarado 2006). Two strains of S. mediterranea have been identified that differ significantly in their mode of reproduction (Newmark and Sanchez Alvarado 2002). One strain undergoes sexual reproduction as cross-fertilizing hermaphrodites and develops both testes and ovaries. In contrast, the second strain never develops reproductive organs but rather reproduces by asexual fission. The only known genetic difference between these two strains is a chromosomal translocation (Baguñà et al. 1999), although the precise loci involved are unknown.
MicroRNAs (miRNAs) are small (∼22 nucleotides [nt]) noncoding RNAs that regulate gene expression by base pairing to mRNAs where they direct either mRNA cleavage or translation repression (Eulalio et al. 2008). miRNAs are generated from longer primary transcripts called pri-miRNAs through a series of processing events (Kim 2005). In the first step of miRNA synthesis, the stem–loop structure containing the miRNA is cleaved from the pri-miRNA by the RNaseIII enzyme Drosha and its cofactor DGCR8 to generate a pre-miRNA. The pre-miRNA is then exported from the nucleus to the cytoplasm by exportin5 where it is further processed by a second RNaseIII enzyme Dicer and its cofactor TRBP. The mature miRNA is then loaded onto an Argonaute protein (Ago1, Ago2, Ago3, or Ago4 in humans) where it then interacts with and regulates the mRNA target.
We previously described the cloning and characterization of 63 miRNAs from S. mediterranea (Palakodeti et al. 2006). As these miRNAs were identified using traditional cloning and sequencing methods, these efforts were not exhaustive and it is likely that additional miRNAs remained to be identified in S. mediterranea. Here we use deep sequencing to further explore the diversity of S. mediterranea miRNAs and their expression patterns. Specifically, we expand the miRNA repertoire of S. mediterranea and identify 66 new miRNA encoding loci. We also identify 21 miRNAs that are expressed specifically in the sexual or asexual strains and five miRNAs that decrease upon irradiation. The regulated miRNAs are particularly intriguing as they may play important roles in the development of reproductive organs, the control of reproductive fission, and regulating the function of neoblasts or their progeny during regeneration and homeostasis.
RESULTS AND DISCUSSION
Deep sequencing of planarian miRNAs
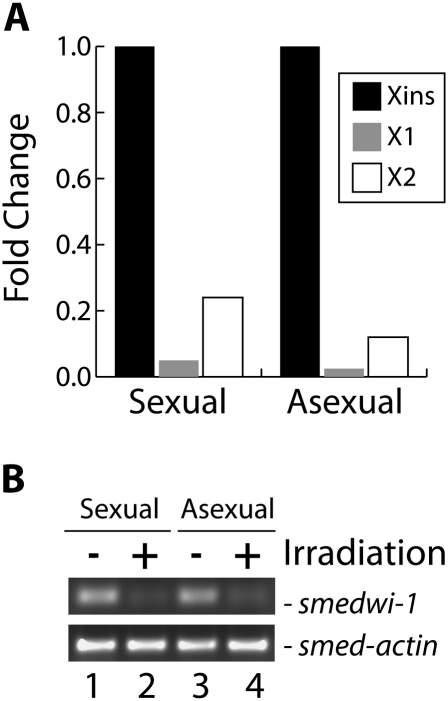
To identify additional S. mediterranea miRNAs we prepared four small RNA libraries from RNA isolated from nonirradiated sexual and asexual animals and from sexual and asexual animals four days after γ-irradiation. FACS analysis confirmed that the γ-irradiation treatment significantly depleted the animals of both dividing (X1) and nondividing (X2) neoblasts (Fig. 1A). Furthermore, RT-PCR of the four RNA samples demonstrated that the neoblast marker smedwi-1 (Reddien et al. 2005) was present at significantly lower levels in the irradiated samples than in the nonirradiated samples, while smed-actin was unaffected by irradiation (Fig. 1B). As piRNAs are significantly more abundant than miRNAs in S. mediterranea (Palakodeti et al. 2008), we first gel purified RNAs between 18 nt and 28 nt in length to enrich the libraries for miRNAs. Each library was sequenced in one lane of an Illumina Genome Analyzer II for 36 cycles. We obtained 11,059,352 and 11,826,579 individual reads from the nonirradiated sexual and asexual libraries, respectively, and an additional 9,343,092 and 10,703,240 individual reads from the irradiated sexual and asexual libraries, respectively. A total of 19,635,184 of these reads were between 18 and 24 nt in length.
FIGURE 1.
Sample quality control. (A) FACS analysis to monitor the neoblast population in untreated or γ-irradiated animals. The fold change of the X1, X2, and Xins populations in the irradiated sample compared with the untreated sample is graphed. The fold change of the Xins population was arbitrarily set to a value of 1.0 and the values for X1 and X2 adjusted accordingly. (B) Analysis of smedwi-1 mRNA level. The level of smedwi-1 mRNA, a neoblast marker (Reddien et al. 2005), was analyzed using semiquantitative RT-PCR. As a control, a parallel experiment was performed for smed-actin mRNA.
Detailed analysis of the 18–24 nt reads revealed that they map to all 63 previously identified miRNA precursors. However, our data suggests that three of the previously reported miRNAs are not actual miRNAs. First, although we obtained 9949 and 21,604 reads that perfectly aligned to the mir-749 and mir-751 pre-miRNAs, respectively, the boundaries of the reads on the predicted pre-miRNA structure are inconsistent with that predicted for Drosha and Dicer processing events (data not shown). Second, mir-753 appears to correspond to a repetitive element as it is present >520 times in the draft assembly of the S. mediterranea genome. Thus, our current data suggest that the previously reported miRNAs mir-749, mir-751, and mir-753 (Palakodeti et al. 2006) are not authentic miRNA encoding loci in S. mediterranea.
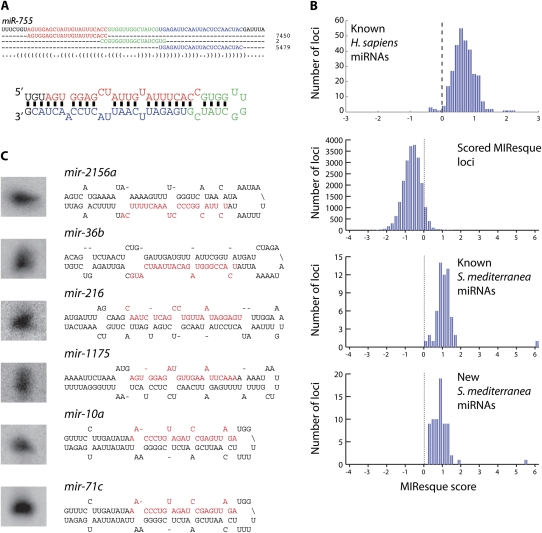
The sequencing data has also allowed us to more accurately annotate the known miRNAs (Fig. 2A). For example, in our initial study we obtained two sequences of different lengths for mir-755 leading to the annotation of the mir-755 mature strand as either 18 or 20 nt in length (Palakodeti et al. 2006). We have now obtained 27,268 reads that map perfectly to the mir-755 pre-miRNA and analysis of the most abundant reads that map to the 5′ or 3′ stems (Fig. 2A) suggests that the mature strand is actually 22 nt. Thus, the deep sequencing has dramatically improved the annotation of the known S. mediterranea miRNAs.
FIGURE 2.
Deep sequencing of planarian miRNAs. (A) Alignment of sequence reads to pre-miR-755. The most frequently obtained mature, loop, and miRNA* strand sequences are highlighted in red, blue, and green, respectively, and the number of total reads obtained for each sequence are shown on the right. The location of these sequences on the secondary structure of the pre-miRNA is depicted below the alignment. (B) Histograms of the MIResque scores for known human miRNAs, all S. mediterranea loci containing clusters of deep sequence reads, known S. mediterranea miRNAs, and new S. mediterranea miRNAs identified in this study. The dotted line indicates the threshold at which a genomic region is considered to be a candidate miRNA locus. (D) Northern blot validation of MIResque predicted miRNAs. The structure for each pre-miRNA is shown on the right.
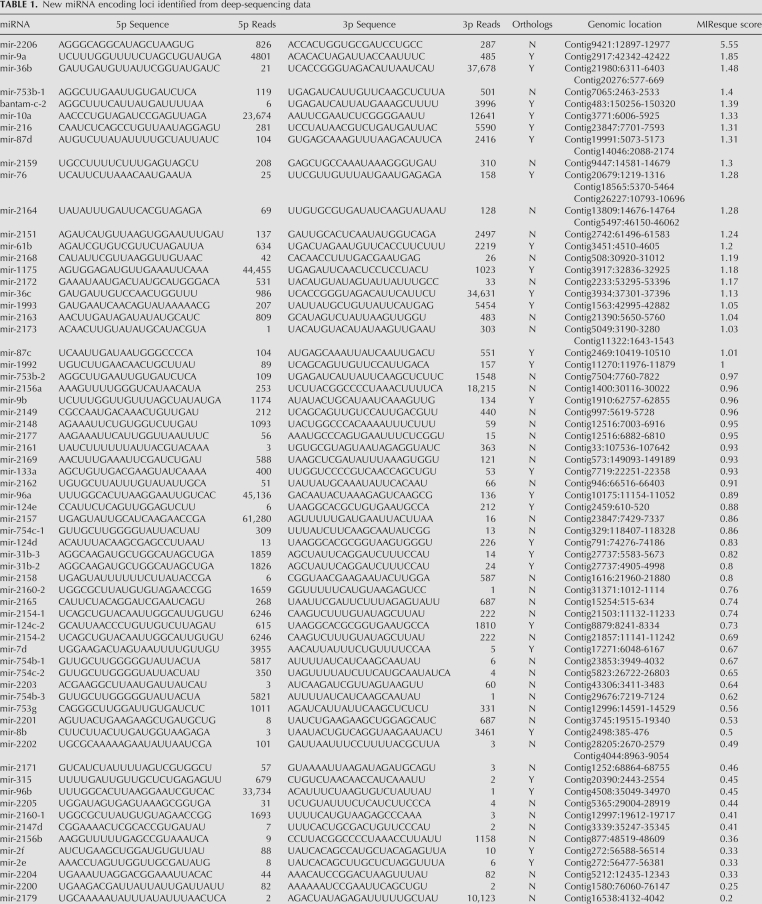
We have also utilized an algorithm called MIResque (GW Yeo and S Aigner, unpubl.) to identify new miRNA encoding loci from the deep-sequencing data (Table 1). This algorithm analyzes sequence reads that have been mapped to the genome, searches for clusters of reads, performs RNA folding on the sequence encompassed by the reads, and then assigns a score to the potential miRNA hairpin. The assigned score is derived from a regularized least-squares classification algorithm designed to score unknown miRNA hairpins given a positive set of known miRNA hairpins generated by deep sequencing. The classification strategy encompasses highly characteristic structural and sequence features of miRNAs and their precursors, such as duplex quality, average length of the mature miRNAs, and heterogeneity of the ends of the reads. MIResque utilized a data set of deep-sequencing reads aligned to a subset of known human miRNA hairpins as a training set. After training, MIResque analyzed the remainder of the known human miRNA hairpins and scored 98% (297/304) of these above the threshold assigned for a new miRNA (Fig. 2B). Interestingly, some of the poorly scored human miRNAs are actually not miRNAs, but rather ribosomal RNA (GW Yeo and S Aigner, unpubl.).
TABLE 1.
New miRNA encoding loci identified from deep-sequencing data
To identify as many new planarian miRNAs as possible, we pooled the sequence data obtained from all four samples and mapped them to the draft assembly of the S. mediterranea genome while allowing for up to two mismatches. Of the 60 previously identified S. mediterranea miRNA loci with sequence reads mapping to both the 5′ and 3′ stems, 57 (95%) scored above the threshold (Fig. 2B). Interestingly, three miRNAs—let-7a, mir-1c, and mir-754a—were not scored by MIResque. First, mir-754a did not receive a score because we only obtained reads that mapped to the mature strand. Second, mir-1c was not scored because numerous reads were obtained whose 5′ ends mapped to the beginning of the loop and the 3′ ends mapped in the middle of the mature strand, which is inconsistent with Dicer processing. Finally, let-7a was not scored because the pre-miRNA sequence is not present in the draft assembly of the S. mediterranea genome even though several overlapping sequence reads containing the let-7a pre-miRNA generated during sequencing of the genome are present in the NCBI trace archives. Nonetheless, as we obtained 352, 54,437, and 654,138 reads for mir-754a, mir-1c, and let-7a, respectively, we believe that these are authentic miRNAs.
MIResque identified 200 hairpins that scored above the miRNA threshold and had a total of at least 50 reads that mapped to the pre-miRNA with up to two mismatches. As MIResque was trained on human miRNAs, not planarian miRNAs, we manually analyzed the predicted miRNA hairpins to assess whether they were actual miRNAs. This analysis eliminated 77 hairpins that clearly did not encode miRNAs, but received high scores due to the number of mapped reads and secondary structure. The discarded hairpins typically corresponded to highly expressed piRNA clusters that fortuitously resembled miRNA hairpins and for which 18–25 nt sequences were obtained due to partial degradation of piRNAs (data not shown). As 57 of the remaining 123 MIResque hairpins corresponded to previously known miRNAs, MIResque identified 66 new pre-miRNA hairpins corresponding to 72 loci in the S. mediterranea genome (Table 1). We have validated the expression of seven of the new MIResque predicted miRNAs by Northern blots (Figs. 2C, 3C). These results suggest that most, if not all of the miRNA encoding loci predicted by MIResque are in fact authentic. When combined with the 60 previously identified miRNAs, we have now identified a total of 126 distinct pre-miRNA hairpins corresponding to 138 loci in the S. mediterranea genome (Supplemental Table 1). Thirty-one of the new miRNA loci encode miRNAs that belong to miRNA families present in other species (e.g., mir-124d, mir-36b, etc.), while 41 of these loci encode miRNAs that have only been identified in S. mediterranea. Thus, S. mediterranea contains at least 48 miRNAs that are specifically found in planarians and 78 miRNAs that are members of evolutionarily conserved miRNA families.
FIGURE 3.
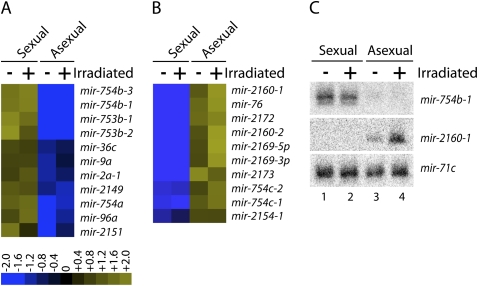
Strain-specific miRNA Expression. (A) MicroRNAs that are expressed at least twofold higher in sexual animals than in asexual animals were identified, and the expression ratios were log2 transformed and depicted in heat map representation. (B) MicroRNAs that are expressed at least twofold higher in asexual animals than in sexual animals were identified, and the expression ratios were log2 transformed and are depicted in heat map representation. (C) Northern blots for mir-754b-1 and mir-2160-1 were performed with total RNA isolated from intact (−) and irradiated (+) sexual and asexual animals. The microRNA mir-71c was used as a probe to control for equal RNA loading.
We previously described 6 miRNA clusters that contain between two to four miRNAs (Palakodeti et al. 2006). Analysis of the genomic coordinates of the newly identified miRNAs revealed six additional miRNAs clusters—mir-2148/mir-2177, mir-mir-31b-2/mir-31b-3, mir-747/mir-2158, mir-36a/mir-36c, mir-216/mir-2157, and mir-mir-2e/mir-2f—where the miRNAs are located between 35 and 617 nt apart (Table 1). Thus, S. mediterranea contains at least 12 miRNA clusters encoding a total of 28 miRNAs.
Strain-specific miRNA expression
We next examined whether the expression of any miRNAs differed between the sexual and asexual strains. To do this, we compared the relative abundance of each miRNA between the sexual and asexual libraries. While most miRNAs were expressed at similar levels in each strain, we identified 21 miRNAs whose expression was significantly restricted to either the sexual or asexual strains (Fig. 3). Eleven of these miRNAs are expressed at least two- to threefold higher in sexual animals than in asexual animals (Fig. 3A), while the other 10 miRNAs are expressed at least threefold higher in asexual animals than in the sexual animals (Fig. 3B).
To validate these findings, we performed Northern blots using probes complementary to mir-754b-1 and mir-2160-1 which our sequence data indicated were significantly enriched in sexual and asexual animals, respectively. Specifically, mir-754b-1 was sequenced 3228 times in nonirradiated sexual animals, but only 23 times in nonirradiated asexual animals (Supplemental Table 2). In contrast, mir-2160-1 was sequenced 740 times in nonirradiated asexual animals, but only 36 times in nonirradiated sexual animals (Supplemental Table 2). Consistent with our sequence data, the Northern blot experiments show that mir-754b-1 and mir-2160-1 are indeed expressed in a strain-specific manner, whereas mir-71c is expressed equally in all samples (Fig. 3C). Interestingly, the Northern blots also showed that mir-2160-1 is expressed higher in irradiated animals than in nonirradiated animals (Fig. 3C, lanes 3,4), again consistent with our sequencing data. Thus, these data demonstrate that a subset of miRNAs are expressed in a strain-specific manner.
Interestingly, the majority of the strain-specific miRNAs are planarian-specific: only five of the 21 strain-specific miRNAs are members of miRNA families that exist in other organisms. Moreover, only one miRNA, mir-76, belonging to a known miRNA family is expressed specifically in the asexual strain. Thus, asexual animals express nine planarian-specific miRNAs at much higher levels than sexual animals. This raises the intriguing possibility that these miRNAs participate in either repressing development of the reproductive tissues or controlling reproductive fission, which only occurs in asexual animals. Conversely, it is likely that the miRNAs that are expressed specifically in the sexual animals enhance the development of testes and ovaries or repress fission.
Irradiation sensitive miRNAs
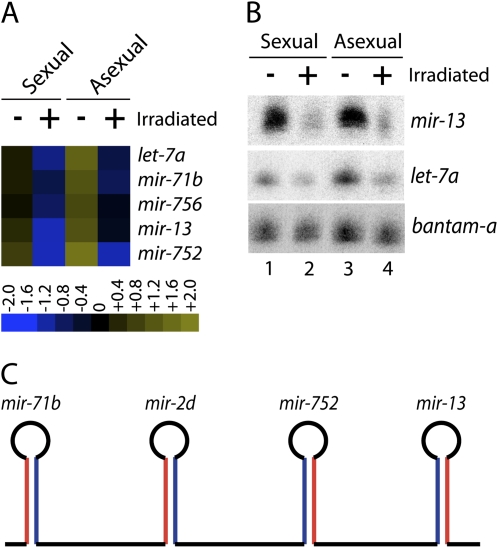
To identify miRNAs that may be expressed in neoblasts or neoblast progeny and potentially play a role in stem cell function we examined whether the expression of any miRNAs was reduced upon irradiation. This analysis identified 5 miRNAs—let-7a, mir-71b, mir-756, mir-13, and mir-752—that were reduced at least twofold upon irradiation (Fig. 4). For example, let-7a was sequenced 266,326 times from nonirradiated asexual animals, but only 92,535 times from irradiated asexual animals (Supplemental Table 2). Similarly, mir-13 was sequenced 2554 and 1257 times from nonirradiated and irradiated asexual animals, respectively. Consistent with the sequence results, Northern blot analysis confirmed that the levels of let-7a and mir-13 decrease upon irradiation, while the levels of a control miRNA, bantam-a, were similar between all samples (Fig. 4B). Intriguingly, at least one other member of the let-7 family appears to be expressed in irradiation sensitive cells. Specifically, our sequence data indicated that let-7b levels decrease 1.5-fold upon irradiation (Supplemental Table 2) and Northern blot experiments confirm this observation (data not shown). These results demonstrate that the expression of these miRNAs is sensitive to irradiation in both sexual and asexual animals. Together, this suggests that these miRNAs are expressed in either neoblasts or their progeny and are strong candidates for miRNAs that are involved in some aspect of stem cell function. It is important to note that the sequencing and Northern blot experiments were all conducted 4 d after irradiation. Thus, it is quite likely that the levels of other miRNAs will decrease at later time points.
FIGURE 4.
Irradiation-sensitive miRNA Expression. (A) MicroRNAs whose levels are reduced at least twofold after irradiation were identified and the expression ratios were log2 transformed and depicted in heat map representation. (B) Northern blots for mir-13 and let-7a were performed with total RNA isolated from intact (−) and irradiated (+) sexual and asexual animals. The microRNA bantam-a was used as a probe to control for equal RNA loading. (C) Schematic depiction of the organization of the mir-71b/mir-2d/mir-752/mir-13 miRNA cluster (red, mature strand; blue * strand).
Interestingly, mir-13, mir-71b, and mir-752 are contained in a cluster that also contains mir-2d. Within this cluster, mir-2d is located between mir-752 and mir-71b (Fig. 4C). Surprisingly, while the expression of mir-13, mir-71b, and mir-752 decreases at least twofold upon irradiation, the levels of mir-2d does not change (1.08-fold decrease). This suggests that the expression of the miRNAs in this cluster may be individually regulated at a post-transcriptional step.
The observation that let-7a, and to a lesser extent let-7b, is expressed in irradiation sensitive cells (most likely neoblasts) is intriguing, as let-7 has been linked to cellular proliferation and differentiation in many organisms. In Caenorhabditis elegans, let-7 is required for the terminal differentiation of seam cells—in let-7 mutants, the seam cells fail to exit the cell cycle and instead continue to proliferate (Reinhart et al. 2000). Similarly, vertebrate let-7 has been shown to be involved in the differentiation of embryonic and cancer stem cells. For example, let-7 expression is low in both cancer and embryonic stem cells and becomes upregulated upon differentiation (Johnson et al. 2005; Lee et al. 2005; Yu et al. 2007). Finally, expression of the let-7 family has been found to change significantly during regeneration in the newt (Tsonis et al. 2007). Specifically, let-7 expression decreases during the initiation of lens and inner ear hair cell regeneration that involves dedifferentiation. Thus, in most organisms, let-7 is expressed in differentiated cells, not in stem cells. While our data links let-7a expression to neoblast function, it is not yet clear how let-7a functions in planarians. One possibility is that rather than being involved in inducing differentiation, in planarians let-7a could be expressed in neoblasts where it functions to prevent differentiation. In this case, neoblast differentiation would require let-7a down-regulation. It is worth noting that we have not yet identified a miRNA that perfectly matches the canonical let-7a miRNA, which is 100% conserved in all other metazoans (Pasquinelli et al. 2000); smed-let-7a differs by 5 nt from the cannonical let-7a (Palakodeti et al. 2006). On the other hand, let-7a could function similarly in planarians and other organisms where it acts to induce differentiation. In this regard, we imagine that let-7a would either be transiently expressed in the neoblasts or the early neoblast progeny (Eisenhoffer et al. 2008) to initiate differentiation, but that its expression would decrease soon after differentiation began. This scenario would explain the absence of let-7a expression in both irradiated (Fig. 4) and smedwi-2(RNAi) (data not shown) animals. Nonetheless, our results strongly suggest that let-7a plays an important role in some aspect of neoblast function.
MATERIALS AND METHODS
Planarian cultures
The hermaphroditic sexual (Zayas et al. 2005) and asexual (Sanchez Alvarado et al. 2002) strains of S. mediterranea used for these experiments were provided by P. Newmark (University of Illinois) and A. Sánchez-Alvarado (University of Utah). The animals were maintained essentially as described (Cebria and Newmark 2005) at 22°C in ddH2O supplemented with 1.6 mM NaCl, 1.0 mM CaCl2, 1.0 mM MgSO4, 0.1 mM MgCl2, 0.1 mM KCl, and 1.2 mM NaHCO3, and fed homogenized organic beef liver. All animals were starved 1 wk prior to experiments.
Small RNA cloning and sequencing
Libraries for the Illumina small RNA sequencing were constructed using the Illumina Small RNA Sample Prep Kit, essentially as described by the manufacturer. Briefly, small RNAs of 18–28 nt in length were gel-purified from 10 μg of total RNA isolated from sexual and asexual animals that were either untreated or irradiated. Eluted RNAs were successively ligated to 5′ and 3′ adaptors with RNA ligase and gel purified after each ligation step. RNAs were reverse transcribed and the resultant cDNA amplified by PCR for 16 cycles. PCR products were purified on a nondenaturing acrylamide gel and sequenced for 36 cycles on the Illumina Genome Analyzer II.
Small RNA genomic analysis
Sequences from the Illumina Genome Analyzer image files were extracted using Firecrest and Bustard. Sequences were trimmed for adaptor sequences and 18–25 base sequences were mapped to the supercontigs of the draft S. mediterranea genome assembly (version 3.1) with Bowtie (Langmead et al. 2009). For the MIResque analysis, the mapping was performed to identify reads that map to the genome with up to two mismatches. To compare the expression levels of all miRNAs, reads were only considered that mapped perfectly to the pre-miRNA sequences. MIResque is implemented as a combination of Perl scripts and Matlab code for assigning scores to candidate miRNA hairpins (GW Yeo and S Aigner, unpubl.).
γ-Irridiation and FACS analysis
Animals were γ-irradiated with a cesium source essentially as previously described (Palakodeti et al. 2008) except that the animals were exposed to one 30-Gy dose and the animals analyzed 4 d later. FACS analysis of the neoblast populations were carried out as previously described (Palakodeti et al. 2008) based on earlier protocols (Reddien et al. 2005; Hayashi et al. 2006). Planarians were cut into small pieces on ice in cold calcium, magnesium-free medium (CMF) (15 mM HEPES, 400 mg/L NaH2PO4, 800 mg/L NaCl, 1200 mg/L KCl, 800 mg/L NaHCO3, 240 mg/L glucose [pH 7.3]) and washed twice with CMF. The fragments were soaked in CMF containing 0.25% (w/v) Trypsin (GIBCO) and 1% BSA (Fisher Scientific) at 20°C for ∼60 min. The fragments were then completely dissociated into single cells by gentle pipetting. The cell mixture was sequentially filtered through 40 μm and 20 μm nylon filters (Millipore). Cells were collected by centrifugation, resuspended in fresh CMF supplemented with 1% BSA, 0.5 μg/mL Calcein AM (Sigma), and 10 μg/mL Hoescht 3342 (Sigma) and incubated at 20°C for 2 h. Finally, 2 μg/mL propidium iodide (Sigma) was added and the samples were kept on ice for analysis with a LSR II flow cytometer (Becton-Dickinson).
Northern blotting
Twenty miocrgrams of total RNA were resolved on a denaturing polyacrylamide gel, transferred to nitrocellulose, and hybridized as described previously (Palakodeti et al. 2006). Three 10-min post-hybridization washes were performed at room temperature in 2× SSC and 0.1% SDS. The membranes were then visualized with a Cyclone PhosphorImager (Perkin-Elmer).
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank the University of Connecticut Health Center Translational Genomics Core Facility for use of the Illumina Genome Analyzer. S.A. is a Damon Runyon Fellow supported by the Damon Runyon Cancer Research Foundation (DRG-1859-05). Genomic sequence data were produced by the Washington University Genome Sequencing Center in St. Louis. This work was generously supported by the Raymond and Beverly Sackler Fund for the Arts and Sciences (to B.R.G.). Y.L. prepared the small RNA libraries and performed FACS and deep sequencing; M.S., D.P., and Y.L. performed Northern blots; S.A. prepared small RNA libraries; M.L. and G.Y. performed the MIResque analysis; Y.L., D.P., and B.G. analyzed the data; Y.L. and B.G. wrote the paper with input from all authors; B.G. supervised the project.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1702009.
REFERENCES
- Baguñà J, Saló E, Auladell C. Regeneration and pattern formation in planarians III. Evidence that neoplasts are totipotent stem cells and the source of blastema cells. Development. 1989;107:77–86. [Google Scholar]
- Baguñà J, Carranza S, Pala M, Ribera C, Giribet G, Arnedo MA, Ribas M, Riutort M. From morphology and karyology to molecules. New methods for taxonomical identification of asexual populations of freshwater planarians. A tribute to Professor Mario Benazzi. Ital J Zool (Modena) 1999;66:207–214. [Google Scholar]
- Cebria F, Newmark P. Planarian homologs of netrin and netrin receptor are required for proper regeneration of the central nervous system and the maintenance of nervous system architecture. Development. 2005;132:3691–3703. doi: 10.1242/dev.01941. [DOI] [PubMed] [Google Scholar]
- Eisenhoffer G, Kang H, Alvarado A. Molecular analysis of stem cells and their descendants during cell turnover and regeneration in the planarian Schmidtea mediterranea. Cell Stem Cell. 2008;3:327–339. doi: 10.1016/j.stem.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Asami M, Higuchi S, Shibata N, Agata K. Isolation of planarian X-ray-sensitive stem cells by fluorescence-activated cell sorting. Dev Growth Differ. 2006;48:371–380. doi: 10.1111/j.1440-169X.2006.00876.x. [DOI] [PubMed] [Google Scholar]
- Johnson S, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert K, Brown D, Slack F. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Kim VN. MicroRNA biogenesis: Coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Kim H, Chung S, Kim K, Dutta A. Depletion of human micro-RNA miR-125b reveals that it is critical for the proliferation of differentiated cells but not for the down-regulation of putative targets during differentiation. J Biol Chem. 2005;280:16635–16641. doi: 10.1074/jbc.M412247200. [DOI] [PubMed] [Google Scholar]
- Newmark P, Sanchez Alvarado A. Not your father's planarian: A classic model enters the era of functional genomics. Nat Rev Genet. 2002;3:210–219. doi: 10.1038/nrg759. [DOI] [PubMed] [Google Scholar]
- Palakodeti D, Smielewska M, Graveley B. MicroRNAs from the planarian Schmidtea mediterranea: A model system for stem cell biology. RNA. 2006;12:1640–1649. doi: 10.1261/rna.117206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palakodeti D, Smielewska M, Lu Y-C, Yeo G, Graveley B. The PIWI proteins SMEDWI-2 and SMEDWI-3 are required for stem cell function and piRNA expression in planarians. RNA. 2008;14:1174–1186. doi: 10.1261/rna.1085008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli A, Reinhart B, Slack F, Martindale M, Kuroda M, Maller B, Hayward D, Ball E, Degnan B, Muller P, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- Reddien P, Oviedo N, Jennings J, Jenkin J, Sanchez Alvarado A. SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science. 2005;310:1327–1330. doi: 10.1126/science.1116110. [DOI] [PubMed] [Google Scholar]
- Reinhart B, Slack F, Basson M, Pasquinelli A, Bettinger J, Rougvie A, Horvitz H, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Sanchez Alvarado A. Planarian regeneration: Its end is its beginning. Cell. 2006;124:241–245. doi: 10.1016/j.cell.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Sanchez Alvarado A, Newmark P, Robb S, Juste R. The Schmidtea mediterranea database as a molecular resource for studying platyhelminthes, stem cells, and regeneration. Development. 2002;129:5659–5665. doi: 10.1242/dev.00167. [DOI] [PubMed] [Google Scholar]
- Tsonis P, Call M, Grogg M, Sartor M, Taylor R, Forge A, Fyffe R, Goldenberg R, Cowper-Sallari R, Tomlinson C. MicroRNAs and regeneration: Let-7 members as potential regulators of dedifferentiation in lens and inner ear hair cell regeneration of the adult newt. Biochem Biophys Res Commun. 2007;362:940–945. doi: 10.1016/j.bbrc.2007.08.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zayas R, Guo T, Newmark P. Nanos function is essential for development and regeneration of planarian germ cells. Proc Natl Acad Sci. 2007;104:5901–5906. doi: 10.1073/pnas.0609708104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, Huang Y, Hu X, Su F, Lieberman J, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- Zayas R, Hernandez A, Habermann B, Wang Y, Stary J, Newmark P. The planarian Schmidtea mediterranea as a model for epigenetic germ cell specification: Analysis of ESTs from the hermaphroditic strain. Proc Natl Acad Sci. 2005;102:18491–18496. doi: 10.1073/pnas.0509507102. [DOI] [PMC free article] [PubMed] [Google Scholar]