Abstract
The breast and ovarian cancer suppressor BRCA2 controls the enzyme RAD51 during homologous DNA recombination (HDR) to preserve genome stability. BRCA2 binds to RAD51 through 8 conserved BRC repeat motifs dispersed in an 1127-residue region (BRCA2[BRC1–8]). Here, we show that BRCA2[BRC1–8] exerts opposing effects on the binding of RAD51 to single-stranded (ss) versus double-stranded (ds) DNA substrates, enhancing strand exchange. BRCA2[BRC1–8] alters the electrophoretic mobility of RAD51 bound to an ssDNA substrate, accompanied by an increase in ssDNA-bound protein assemblies, revealed by electron microscopy. Single-molecule fluorescence spectroscopy shows that BRCA2[BRC1–8] promotes RAD51 loading onto ssDNA. In contrast, BRCA2[BRC1–8] has a different effect on RAD51 assembly on dsDNA; it suppresses and slows this process. When homologous ssDNA and dsDNA are both present, BRCA2[BRC1–8] stimulates strand exchange, with delayed RAD51 loading onto dsDNA accompanying the appearance of joint molecules representing recombination products. Collectively, our findings suggest that BRCA2[BRC1–8] targets RAD51 to ssDNA while inhibiting dsDNA binding and that these contrasting activities together bolster one another to stimulate HDR. Our work provides fresh insight into the mechanism of HDR in humans, and its regulation by the BRCA2 tumor suppressor.
Keywords: DNA recombination, electron microscopy, single-molecule fluorescence spectroscopy, tumor suppressor
Homologous DNA recombination (HDR) is essential in somatic cells not only for the error-free repair of DNA double-strand breaks (DSBs), but also for the restoration of DNA replication forks that stall at lesions in the template strand (1). The central event in HDR is the synapsis of a single-stranded (ss)DNA molecule with homologous duplex DNA, which initiates the strand exchange that leads to recombination. Recombinase molecules, conserved from RecA in prokarya to RAD51 in eukaryal cells, mediate strand exchange via distinct reactions grouped into the presynaptic, synaptic, and postsynaptic phases (2).
The breast cancer suppressor protein, BRCA2, an ∼384-kDa molecule of 3418 residues, is essential in vivo for RAD51-mediated HDR (3). Several regions of BRCA2 may contribute to this role (4). Human BRCA2 interacts with RAD51 through 8 copies of the BRC repeat motifs of ≈40 residues each, embedded in a 1127-residue fragment (BRCA2[BRC1–8]) within exon 11 (5), as well as through an unrelated carboxyl-terminal motif in exon 27 (6). The BRC repeats appear to be the primary locus for the BRCA2-RAD51 interaction; unlike the C-terminal motif, their sequence is evolutionarily conserved in simple eukaryotes like fungi, as well as in plants (7). In addition, exon 17 of human BRCA2 encodes an ≈800 residue DNA-binding domain (BRCA2DBD) containing 3 oligonucleotide-binding (OB) folds, ssDNA-binding modules also found in the abundant ssDNA-binding protein, replication protein A (RPA) (8). In a fungal BRCA2 homolog, the OB-fold-containing, ssDNA-binding domain displaces RPA from DNA substrates in vitro to enable nucleation of the RAD51 filament by the single BRC repeat found in this organism (9). Moreover, fragments containing 2 to 4 of the 8 human BRC repeats fused to the ssDNA-binding BRCA2[DBD] domain can partially complement HDR in BRCA2-deficient cells in vivo (10) and promote RAD51-dependent strand exchange in vitro (11). Together, these observations suggest that the BRCA2[DBD] cooperates with the BRC repeats to target RAD51 to DNA substrates, acting as a recombination mediator.
The precise role in HDR played by the multi-BRC repeat region of human BRCA2 is not yet clear. Indeed, the BRC repeats are reported to have activities that could potentially inhibit (12–14) as well as stimulate (11, 15, 16) RAD51 function, under different experimental conditions. We have previously purified the exon 11 encoded 1,127-residue domain (BRCA2[BRC1–8]) from human BRCA2 containing all 8 BRC repeats within their natural context, and shown that BRCA2[BRC1–8] suffices to promote RAD51-mediated strand exchange, independent of the ssDNA-binding domain BRCA2[DBD] (16). Here, we report results that suggest a mechanism underlying the role of BRCA2[BRC1–8] in stimulating HDR. We find that BRCA2[BRC1–8] promotes RAD51 loading on ssDNA, while suppressing RAD51 assembly on dsDNA, and propose that both these opposing effects cooperate to enhance the efficiency of RAD51-mediated HDR. Our findings provide additional evidence in support of a recently proposed model (17) reported while this paper was under review.
Results and Discussion
Electrophoretic Mobility of Complexes Between BRCA2[BRC1–8], RAD51, and ssDNA.
We have previously shown, using a well-established in vitro assay for HDR, that BRCA2[BRC1–8] promotes strand exchange between circular φX174 ssDNA and a homologous linear duplex DNA (16). To identify potential mechanisms underlying this stimulatory activity, we first examined the effect of BRCA2[BRC1–8] on RAD51 binding to ssDNA. We generated a 2.5-kb resected double-stranded (ds)DNA substrate from φX174 RF1 DNA by λ exonuclease digestion according to the schematic shown in Fig. S1A. When radiolabeled at its 5′ end, the great majority of the resected dsDNA preparation (Fig. S1B, lane 1) binds in an electrophoretic mobility shift assay (EMSA) to the ssDNA binding protein, RPA in a concentration-dependent manner (Fig. S1B, lanes 2–4), in contrast to radiolabeled dsDNA (Fig. S1C).
We examined the effect of BRCA2[BRC1–8] on RAD51 binding to the resected dsDNA substrate. The reaction conditions used here were identical to those we have previously used for strand exchange stimulated by BRCA2[BRC1–8] (16), except that linear dsDNA was omitted. Reactions included ATP and resected dsDNA preincubated with a low, subsaturating concentration of RPA, within a range previously reported to promote strand exchange (18, 19). RAD51 (3.3 μM) was incubated in a reaction mix either in the absence or presence of a range of indicated BRCA2[BRC1–8] concentrations. The RPA-bound, radiolabeled, resected dsDNA substrate (10 μM) was subsequently added to these preformed complexes. After incubation at 37 °C for 60 min, reaction products were resolved by agarose gel electrophoresis (Fig. 1).
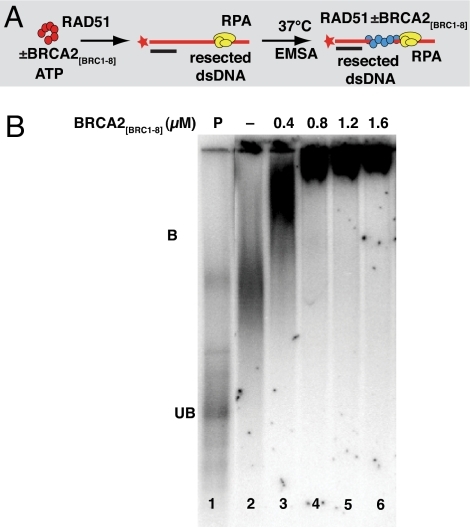
Fig. 1.
The electrophoretic mobility of RAD51 complexes on a resected dsDNA substrate is altered by BRCA2[BRC1–8]. (A) Schematic representation of the protocol and reagents used for EMSA to detect association between BRCA2[BRC1–8], RAD51, and resected dsDNA in the presence of RPA and ATP. (B) RAD51 forms complexes on a resected dsDNA substrate that are further retarded in an EMSA by increasing concentrations of BRCA2[BRC1–8], suggesting co-complex formation. Radiolabeled resected dsDNA (10 μM) was premixed with 0.07 μM RPA before adding 3.3 μM RAD51 in the absence (lane 1) or presence of increasing concentrations of BRCA2[BRC1–8] (0.4–1.6 μM; lanes 3–6) to reactions containing 1 mM ATP and incubated at 37 °C for 60 min. Protein-DNA complexes were resolved by gel electrophoresis and analyzed by phosphorimaging. In the presence of RAD51, the mobility of the resected dsDNA is resolved as a diffuse band (lane 2). The addition of increasing concentrations of BRCA2[BRC1–8] further retards this RAD51-resected dsDNA complex (lanes 3–6). The mobility of the radiolabeled resected dsDNA alone is indicated in lane 1 as unbound (UB). Note that lanes 2–6 in this figure and in Fig. S2 are isolated from the same original gel, and lane 1 has been duplicated.
RAD51 binds to the resected dsDNA substrate, retarding its electrophoretic mobility (Fig. 1B, compare lanes 1 and 2). Increasing concentrations of BRCA2[BRC1–8] affect this pattern (lanes 3–6), indicative of complex formation between BRCA2[BRC1–8], RAD51, and a resected dsDNA substrate that is predominantly single-stranded, with a short ds/ssDNA junction (Fig. S1A). Similar changes are observed when the nonhydrolyzable analog, AMP-PNP, is substituted for ATP (Fig. S2). Complex formation is evident with 0.4 μM BRCA2[BRC1–8] and plateaus between 1 to 2 μM. However, BRCA2[BRC1–8] alone at similar or higher concentrations does not detectably alter the mobility of the resected dsDNA substrate (Fig. S1D), suggesting that RAD51 mediates BRCA2[BRC1–8] binding to DNA in the complex.
BRCA2[BRC1–8] Increases ssDNA-Bound Protein Assemblies.
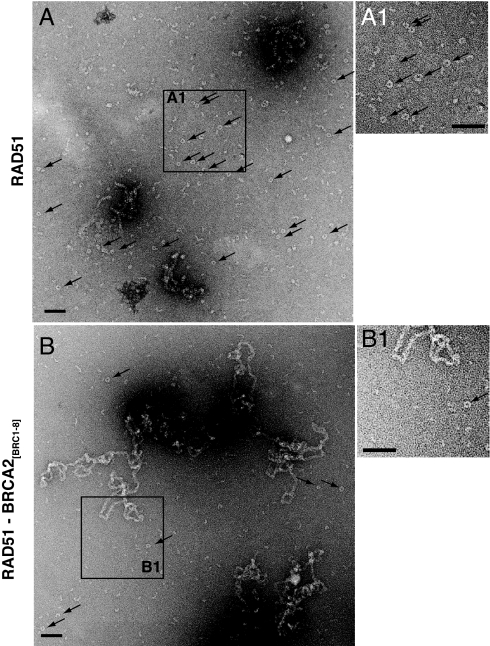
We examined by electron microscopy (EM) the complexes formed by ssDNA with RAD51 in the presence or absence of BRCA2[BRC1–8] (Fig. 2) under conditions similar to those used in Fig. 1. As before, 5 μM RAD51 and 15 μM ssDNA (φX174 circle) were mixed at the 1:3 ratio deemed optimal for assembly, and ATP was included. The absence of any ssDNA/dsDNA junction in the DNA substrate used for EM suggests such a region is not essential for the effects we observe. In the absence of BRCA2[BRC1–8], RAD51 is visualized predominantly in the form of oligomeric rings (arrows) apparently devoid of DNA, although DNA-bound assemblies are observed occasionally (Fig. 2A). The addition of 2 μM BRCA2[BRC1–8] markedly alters this pattern (Fig. 2B). There is an 11-fold reduction in the number of oligomeric RAD51 rings free of DNA, accompanied by an increase in ssDNA-bound protein assemblies, which become aggregated into large masses. Striations characteristic of ordered RAD51 filaments are not easily observed, due in part to the extensive aggregation of the assemblies, and/or the binding of the BRCA2 fragment to them, consistent with previous reports (20). Collectively, our EM findings suggest that BRCA2[BRC1–8] promotes the recruitment of RAD51 from DNA-free oligomeric rings to ssDNA-bound protein assemblies; this effect is accompanied by the changes in the electrophoretic mobility of the DNA-bound complexes shown in Fig. 1.
Fig. 2.
EM visualization of RAD51-ssDNA assemblies in the presence or absence of BRCA2[BRC1–8]. RAD51 (5 μM) was incubated with 15 μM φX174 circular ssDNA either in the absence (A) or presence (B) of 2 μM BRCA2[BRC1–8] in a reaction mix containing ATP for 15 min at 37 °C. After staining with 1% uranyl acetate, the grids were analyzed for protein-DNA complexes. In the absence of BRCA2[BRC1–8], visualization by EM reveals few DNA-bound assemblies of RAD51, but a large number of oligomeric RAD51 rings (black arrows) apparently free of DNA (compare A1 with B1). BRCA2[BRC1–8] markedly reduces the number of oligomeric RAD51 rings. This is accompanied by an increase in ssDNA-bound protein assemblies that appear as extensively aggregated masses. (Scale bar, 100 nm.) Quantitation (see SI Methods) to compare the ratios of areas occupied by rings, with areas enclosing filaments, revealed an 11-fold reduction.
Single Molecule Fluorescence Spectroscopy Reveals Increased RAD51-ssDNA Binding Promoted by BRCA2[BRC1–8].
We used single-molecule fluorescence spectroscopy to directly measure the effect of BRCA2[BRC1–8] on ssDNA binding by RAD51. Fluorescently labeled RAD51 (RAD51-Atto647N; 3.3 μM) and a fluorescently labeled, synthetic ssDNA oligomer (Alexa488-ssDNA; 10 μM) without any ssDNA/dsDNA junctions were incubated with ATP in the presence or absence of 1.2 μM BRCA2[BRC1–8]. The solution was then extensively diluted and analyzed by a single-molecule fluorescence spectroscopy technique, two-color coincidence detection (TCCD) (21) (Fig. 3A). In TCCD, fluorescent molecules diffusing in solution are excited as they pass through the overlapping subfemtoliter focal volumes generated by focusing a red and a blue laser through an objective of high numerical aperture. Complexes of RAD51 with ssDNA give rise to coincident bursts of blue fluorescence from Alexa488-ssDNA and far-red fluorescence from RAD51-Atto647N, allowing any complexes present in solution to be identified and analyzed.
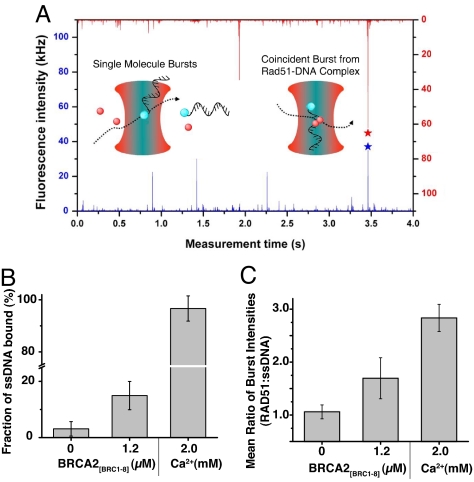
Fig. 3.
Effect of BRCA2[BRC1–8] on RAD51-ssDNA association measured by TCCD. (A) Single-molecule bursts from RAD51-Atto647N (red) and Alexa488-ss/dsDNA (blue). Coincident bursts in both channels indicate the presence of a RAD51-DNA complex diffusing through the probe volume. RAD51-Atto647N (3.3 μM) alone or in the presence of either 1.2 μM BRCA2[BRC1–8] or 2 mM Ca2+ ions were incubated with 10 μM Alexa488-ssDNA to measure (B) the fraction of Alexa488-ssDNA fluorescent bursts that show a coincident RAD51-Atto647N burst (corrected for detector efficiency) and (C) the mean ratio of intensities observed in coincident RAD51-Atto647N and Alexa488-ssDNA bursts. Calcium ions have been shown to stabilize RAD51 filaments on DNA (25) and were used as a positive control in the TCCD experiments.
We found that the frequency of coincident bursts from RAD51-ssDNA complexes decreased significantly after the first 6 min postdilution and became negligible after 30 min. This dissociation time agrees well with previous reports (22), and to optimize sampling of coincident bursts, the experiment was repeated 8 times, and data collection was carried out over the first 6 min after dilution only. The frequency of these coincident bursts was measured and compared with the overall frequency of bursts originating from Alexa488-ssDNA to determine the fraction of DNA bound to RAD51-Atto647N (Fig. 3B).
In the presence of BRCA2[BRC1–8], the fraction of ssDNA bound to RAD51 was observed to be 15 ± 5%, significantly higher than in its absence (3 ± 2%). For coincident bursts from the RAD51-ssDNA complex, the ratio of the fluorescence intensity of RAD51-Atto647N and Alexa488-ssDNA in the presence of BRCA2[BRC1–8] was found to be 1.7 ± 0.4. This was higher than in the absence of BRCA2[BRC1–8], where the ratio was 1.1 ± 0.1 (Fig. 3C). This indicates that the number of RAD51 molecules bound to ssDNA is on average higher in the presence of BRCA2[BRC1–8], but absolute stoichiometries cannot be inferred due to nonuniform fluorescent labeling of RAD51.
Taken together, these observations indicate that BRCA2[BRC1–8] promotes the binding of RAD51 to ssDNA substrates (Fig. 3), leading to the formation of protein-DNA complexes (Fig. 1) in which RAD51 is recruited from DNA-free oligomeric rings into DNA-bound protein assemblies (Fig. 2). This prompted us to investigate the influence of BRCA2[BRC1–8] on the interaction between RAD51 and dsDNA (Fig. 4).
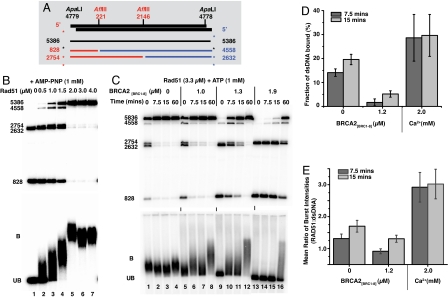
Fig. 4.
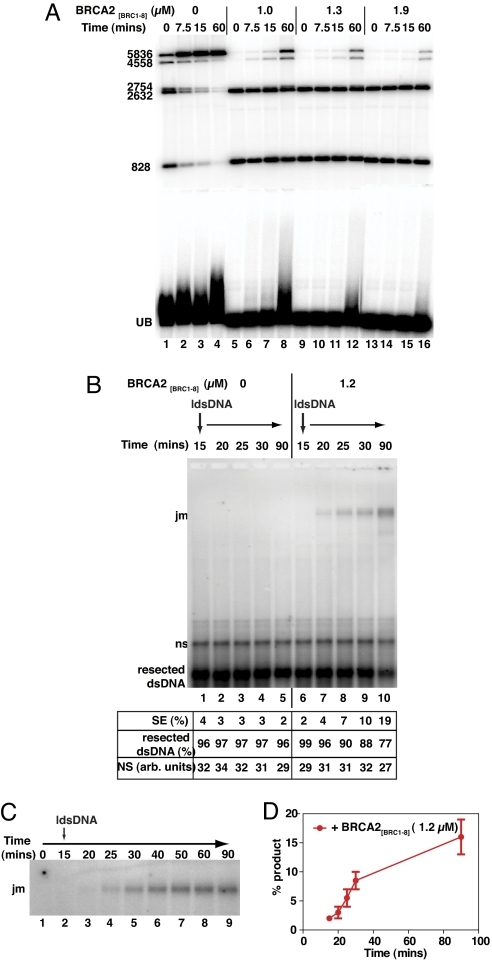
BRCA2[BRC1–8] delays RAD51 assembly on dsDNA. (A) Schematic of the AflIII restriction enzyme digestion pattern of radiolabeled ApaLI-linearized ΦX174 dsDNA. (B) Radiolabeled dsDNA (7 μM) was incubated either in the absence (lane 1) or presence of increasing concentrations of RAD51 (0.5–4.0 μM; lanes 2–7) with AMP-PNP at 37 °C for 60 min. Half of the reaction was removed and mixed with gel loading buffer for EMSA, while the other half was treated with 10 U AflIII at 37 °C for 15 min before being deproteinized. The AflIII digestion pattern (Upper) was then directly compared to ternary complex formation (Lower). Migration of the ternary complex was progressively retarded as a function of RAD51 concentration and correlated with a decrease in the ability of AflIII to digest the dsDNA (compare lane 1 with lanes 2–7). Note that only the radiolabeled end-fragments are visible in lane 1 (828 and 2632 bp); the central fragment is unlabeled and can only be seen in a partial digest. The radiolabeled linear dsDNA alone (lane 1) is marked UB. (C) BRCA2[BRC1–8] delays RAD51 assembly on dsDNA. Reactions were assembled and analyzed as above, except that ATP was used instead of AMP-PNP and BRCA2[BRC1–8] was added at the indicated concentrations (lanes 5–16). Whereas 3.3 μM RAD51 assembles rapidly and completely on dsDNA, conferring protection from restriction digestion, the addition of 1–1.9 μM BRCA2[BRC1–8] retards assembly in a concentration-dependent manner. Single-molecule fluorescence data, in which 3.3 μM RAD51-Atto647N alone or in the presence of either 1.2 μM BRCA2[BRC1–8] or 2 mM Ca2+ ions were incubated with 10 μM Alexa488-dsDNA for 7.5 min (dark) and 15 min (light) to measure (D) the fraction of Alexa488-dsDNA fluorescent bursts that show a coincident RAD51-Atto647N burst (corrected for detector efficiency) and (E) the mean ratio of intensities observed in coincident RAD51-Atto647N and Alexa488-dsDNA bursts.
Inhibitory Effects of BRCA2[BRC1–8] on RAD51 Binding to dsDNA.
We used a radiolabeled linear dsDNA molecule that contains 2 internal AflIII restriction sites (Fig. 4A) to elucidate the temporal pattern of RAD51 loading over time, combining analysis of ternary complex formation by EMSA with restriction enzyme protection (23). The effect of a range of RAD51 concentrations on mobility shift was first compared with sensitivity to enzymatic cleavage (Fig. 4B). Radiolabeled dsDNA (7 μM) was incubated either in the absence (Fig. 4B, lane 1) or presence of various concentrations of RAD51 (Fig. 4B, lanes 2–7) for 60 min, before AflIII restriction digestion (Fig. 4B Upper) or EMSA (Fig. 4B Lower). As expected, the extent of RAD51-dsDNA interaction revealed by EMSA is inversely correlated with the sensitivity of the dsDNA substrate to enzymatic cleavage (Fig. 4B, lanes 2–7). RAD51 concentrations between 0.5–4 μM, which induce increasing EMSA shifts, also progressively render the dsDNA immune to AflIII cleavage (Fig. 4B, lanes 2–5). These phenomena plateau at RAD51 concentrations between 2–4 μM, consistent with the reported optimum molar ratio for RAD51 binding (16). Addition of BRCA2[BRC1–8] suppresses and slows RAD51 loading onto radiolabeled dsDNA in a dose-dependent manner over time, as monitored by protection from enzyme digestion (Fig. 4C). Whereas 3.3 μM RAD51 binds rapidly to dsDNA in the absence of BRCA2[BRC1–8], conferring near-complete protection from digestion (0 min, lane 1), the addition of 1–1.9 μM BRCA2[BRC1–8] detectably inhibits this process (compare lanes 5–16). When 1.9 μM BRCA2[BRC1–8] is present (lanes 13–16), RAD51 binding to dsDNA is significantly suppressed and delayed, approaching completion by only ≈60 min. These findings are in marked contrast to the effect of BRCA2[BRC1–8] on the binding of RAD51 to ssDNA substrates.
We used single-molecule fluorescence spectroscopy to further investigate the suppression of RAD51-dsDNA binding by BRCA2[BRC1–8]. A fluorescently labeled dsDNA oligomer (Alexa488-dsDNA; 10 μM) was incubated with 3.3 μM RAD51-Atto647N and ATP in the presence or absence of 1.2 μM BRCA2[BRC1–8]; aliquots were analyzed by TCCD. After 7.5 min incubation, the fraction of dsDNA bound to RAD51-Atto647N in the presence of BRCA2[BRC1–8] (Fig. 4D) was observed to be 1.7 ± 1.4%, significantly lower than in its absence (14 ± 2%). Furthermore, for the coincident bursts observed from dsDNA-RAD51 complexes, the ratio of the fluorescence intensity of RAD51-Atto647N and Alexa488-dsDNA (Fig. 4E) was found to be lower in the presence of BRCA2[BRC1–8] (0.9 ± 0.1) than in its absence (1.3 ± 0.1). This suggests that the number of RAD51 molecules bound to dsDNA is on average lower in the presence of BRCA2[BRC1–8]. After 15 min incubation, both the fraction of dsDNA bound and ratio of fluorescence intensities have increased in the presence and absence of BRCA2[BRC1–8], confirming that the effect of BRCA2[BRC1–8] is to suppress and delay the binding of Rad51 to dsDNA (Fig. 4 D and E).
BRCA2[BRC1–8] Regulates Recombination Between Resected ssDNA and dsDNA.
We reasoned that these opposing effects of BRCA2[BRC1–8] on RAD51 loading might preferentially slow down dsDNA capture by RAD51 in reactions where ssDNA and homologous, linear dsDNA substrates are both present. To test this hypothesis, we used the EMSA and enzyme-protection assays to monitor the impact of BRCA2[BRC1–8] on linear dsDNA binding during a reconstituted strand exchange reaction containing RAD51, RPA, and resected dsDNA, followed by homologous, radiolabeled linear dsDNA over 60 min (Fig. 5). Reaction conditions mirrored those we have previously found to be optimal for the stimulation of RAD51-mediated strand exchange by BRCA2[BRC1–8] (16). In these conditions, a narrow, ∼1–2 μM, concentration range of BRCA2[BRC1–8] stimulates strand exchange by 3.3 μM RAD51; titrations were therefore performed in this range. Only the radiolabeled dsDNA substrate is visible in EMSA, and it is vulnerable to digestion with AflIII unless coated by RAD51, enabling visualization of the temporal progression and extent of RAD51 binding to dsDNA.
Fig. 5.
BRCA2[BRC1–8] stimulates RAD51-mediated strand exchange while delaying RAD51-dsDNA assembly. Strand exchange reactions between 10 μM resected dsDNA and 7 μM homologous dsDNA mediated by 3.3 μM RAD51 were assembled in the presence or absence of BRCA2[BRC1–8]. (A) The dsDNA substrate was radiolabeled to allow RAD51 assembly on dsDNA to be monitored via protection from AflIII digestion as described in Fig. 4. (B and C) The resected dsDNA substrate was labeled to allow the detection of joint molecule products of recombination. Without BRCA2[BRC1–8] (A, lanes 1–4), rapid protection of the dsDNA by RAD51 is evident between 0–7.5 min; and joint molecule (jm) formation is not detectable (B, lanes 1–5). By contrast, the addition of 1–1.9 μM BRCA2[BRC1–8] to the strand exchange reactions markedly delays RAD51 assembly on dsDNA (A, lanes 5–16). Delayed RAD51 assembly on dsDNA by 1.3 μM BRCA2[BRC1–8] between 15–60 min (A, lanes 9–12) is accompanied by the stimulation of jm formation (B, lanes 6–10). (B) A DNA species co-purifying with the resected dsDNA substrate (Fig. S1B) is marked “ns,” whose relative intensity is unaltered between the lanes, indicating it does not interfere with the reaction. The bottom panel compares the formation of jm products in the presence or absence of BRCA2[BRC1–8] as a percentage of the resected dsDNA substrate, with the intensity of the ns band expressed in arbitrary units. (C) Shows an extended time-course analysis for a strand-exchange reaction carried out as in B, in the presence of BRCA2[BRC1–8]. (D) Shows the quantitation of jm formation from the data in B and C. Error bars indicate SEM.
In the absence of BRCA2[BRC1–8], RAD51 alone rapidly loads onto dsDNA even when ssDNA is present, engendering significant protection to AflIII digestion within 7.5–15 min (Fig. 5A, lanes 1–4). Nevertheless, this is slower and less complete than when linear dsDNA alone is present (compare with Fig. 4C, lanes 1–4), suggesting that ssDNA competes with dsDNA for a fraction of the bound RAD51. However, the presence of BRCA2[BRC1–8] markedly retards the progression of RAD51 loading on dsDNA in a dose-dependent manner over time (Fig. 5A, lanes 5–16). When 1–1.9 μM BRCA2[BRC1–8] are added to the reaction, little loading of RAD51 onto dsDNA is evident even after 60 min, with incomplete protection from digestion even at this time point (compare lanes 4, 8, 12, and 16). These findings suggest that BRCA2[BRC1–8] significantly suppresses the otherwise avid loading of RAD51 onto dsDNA substrates during strand exchange reactions even when ssDNA is present; this is likely to reflect the net outcome of its opposing activities in promoting ssDNA binding by RAD51, while retarding dsDNA loading. These opposing activities could conceivably bolster one another to promote stepwise progression through HDR, suppressing the premature capture of RAD51 by dsDNA and targeting it to ssDNA instead.
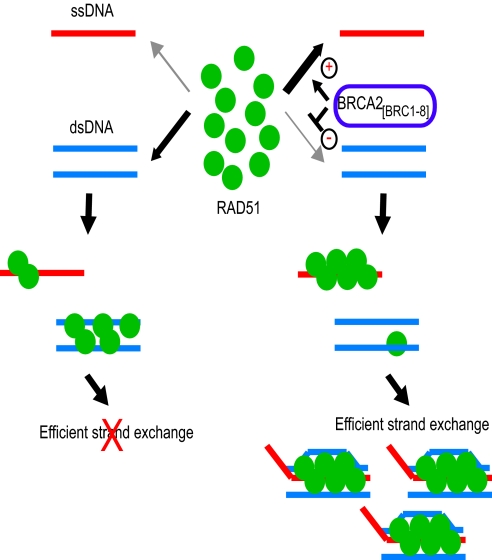
To investigate this notion further, we examined the temporal relationship between the appearance of joint molecules signifying recombination products (Fig. 5 B and C) and the observed extent and timing of dsDNA protection from enzyme digestion (Fig. 5A). Strand exchange reactions were assembled as in Fig. 5A, except that radiolabeled ssDNA was used to enable detection of joint molecules. Also, reactions were performed in the absence or presence of 1.2 μM BRCA2[BRC1–8], a concentration effective in slowing dsDNA loading by RAD51 (Figs. 4C and 5A), as well as in stimulating RAD51-mediated strand exchange under these conditions (16). In the absence of BRCA2[BRC1–8], the generation of recombination products even after 90 min is low (Fig. 5B, lanes 1–5). It is accompanied by extensive and early RAD51 loading onto dsDNA marked by substantial protection from enzyme digestion some 7.5 min after initiation of the reaction (compare with Fig. 5A, lanes 1–4). In contrast, when BRCA2[BRC1–8] is present, joint molecule formation is visible some 20 min after initiation (Fig. 5B, lanes 7–10) with increasing product formation over time (Fig. 5C). The appearance of joint molecule products in the presence of BRCA2[BRC1–8] correlates with the delayed onset and extent of dsDNA binding by RAD51 marked by the observed changes in enzyme protection (Fig. 5A, lanes 9–12). These findings suggest a model in which BRCA2 promotes sequential completion of the presynaptic and synaptic steps of HDR (Fig. 6) by differentially regulating RAD51 loading onto ssDNA versus dsDNA, favoring ssDNA binding, before capture of the incoming duplex followed by strand exchange.
Fig. 6.
Opposing effects of BRCA2[BRC1–8] on RAD51 binding to ssDNA versus dsDNA together bolster one another to stimulate strand exchange. A hypothetical model for the regulation of RAD51-mediated HDR by BRCA2[BRC1–8] is depicted. It refers to events that occur after the resection of a DSB to generate a 3′-tailed ssDNA substrate, but does not show the putative role of the C-terminal ssDNA-binding domain of BRCA2 in displacing RPA, and guiding RAD51 to its substrate near ssDNA/dsDNA junctions (9). When BRCA2[BRC1–8] is absent (left-hand side), ssDNA and dsDNA compete for RAD51 binding. Strand exchange is inefficient because premature, stable RAD51 assembly on dsDNA is futile. BRCA2[BRC1–8] favors the preferential assembly of RAD51 onto ssDNA because it promotes RAD51-ssDNA binding while suppressing RAD51-dsDNA binding (right-hand side). These opposing effects reinforce one another to guide stepwise progression from the presynaptic to the synaptic step of strand exchange.
Regulation of RAD51-Mediated Strand Exchange by BRCA2.
The formation of a presynaptic filament of RAD51 on ssDNA is a prerequisite for HDR. However, several characteristics of RAD51 binding to ssDNA affect both its kinetics and efficiency. Recent studies suggest that when ATP is present, this process is driven predominantly by the nucleation of RAD51 onto ssDNA at multiple sites, because binding is not highly cooperative, and dissociative events are prominent (24, 25). Thus, RAD51-ssDNA assemblies may consist of multiple, short protein patches that incompletely cover the DNA and from which RAD51 dynamically dissociates. This is in contrast to the bacterial recombinase, RecA, for which less frequent nucleation events on ssDNA can nonetheless engender extensive filament formation through rapid, cooperative growth from the nucleation sites (26). Moreover, unlike RecA, human RAD51 can bind in vitro with similar affinity to both ssDNA and dsDNA (27–29). Therefore, in vitro strand exchange reactions have usually been carried out sequentially, with the addition of dsDNA to preformed complexes between ssDNA and RAD51 (30, 31).
In the nuclear milieu, where both ssDNA and dsDNA substrates are generated together in the vicinity of a DSB, these features of DNA binding by RAD51 could lead to the inefficient coupling of presynaptic filament formation on ssDNA, with subsequent reactions that involve the capture of dsDNA. Our results show that BRCA2[BRC1–8] promotes RAD51 binding to ssDNA, while slowing down its loading onto dsDNA, a competing process that would be inhibitory to strand exchange if performed prematurely. We reason that these opposing effects of BRCA2[BRC1–8] on RAD51-DNA binding are therefore likely to reinforce one another, working together to favor sequential progression through the steps involved in strand exchange (Fig. 6). Thus, the findings we report here identify previously unrecognized functions for the multi-BRC repeat region of human BRCA2 in the regulation of RAD51, which provide general insight into the mechanism of HDR in mammalian cells.
Materials and Methods
Protein Expression and Purification.
Human BRCA2[BRC1–8], RAD51, and RPA used in this study were prepared as previously described (16). See SI Methods for details.
EMSA, Strand Exchange, and Restriction Site Protection Assays.
These assays were assembled as previously described (16). Experimental details are described in the text, figure legends, and SI Methods.
Negative Staining EM.
RAD51 either in the absence or presence of BRCA2[BRC1–8] (2 μM) was assembled in a reaction mix containing 1 mM ATP. Preincubation for 5 min at 37 °C was followed by addition of φX174 circular ssDNA and further incubation at 37 °C for 15 min. The samples were diluted 1:10 in Buffer A (see EMSA Assay). Negatively stained EM grids were prepared according to standard procedures (see SI Methods). Quantitation of oligomeric RAD51 rings per unit area in images of RAD51-ssDNA and of BRCA2[BRC1–8]-RAD51-ssDNA is described in SI Methods.
Two-Color Coincidence Detection.
TCCD was carried out using Alexa488-labeled ssDNA or dsDNA substrates and Atto647N-labeled RAD51, whose preparation is described in SI. Samples, in Buffer B (40 mM Tris, pH 7.8, 1 mM MgCl2, 1 mM ATP), consisted of 3.3 μM RAD51-Atto647N either in the absence or presence of 1.2 μM BRCA2[BRC1–8], 10 μM Alexa488-ssDNA, or 10 μM Alexa488-dsDNA, were incubated at 37 °C. At indicated times, aliquots were removed and immediately flash-frozen to permit multiple samples to be analyzed. A control experiment was carried out to confirm that there was no difference in coincidence or relative burst intensities before and after flash-freezing. Samples were thawed individually and diluted in Buffer B to a final fluorophore concentration of 150 pM onto a cover-glass precoated with BSA for immediate analysis by TCCD (32). The procedure used for TCCD is further described in SI. Positive controls were carried out similarly, in the absence of BRCA2[BRC1–8] but with the addition of 2 mM calcium chloride to Buffer B.
Supplementary Material
Acknowledgments.
We thank Luca Pellegrini and Owen Davies (University of Cambridge, Cambridge, U.K.) for providing BRCA2[BRC1–8] protein, Angel Orte for assistance with TCCD, John Finch for assistance with EM, and Richard Henderson for comments on the manuscript. S.R.M. was supported by a studentship from the Engineering and Physical Sciences Research Council, and E.R. and J.S. were supported by studentships from the Medical Research Council, which also funded this work in A.R.V.'s laboratory.
Footnotes
Conflict of interest statement: The editor and A.V. recently (March 2009) co-authored a paper on a related topic.
This article contains supporting information online at www.pnas.org/cgi/content/full/0906208106/DCSupplemental.
References
- 1.Li X, Heyer WD. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 2008;18:99–113. doi: 10.1038/cr.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 3.Moynahan ME, Pierce AJ, Jasin M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol Cell. 2001;7:263–272. doi: 10.1016/s1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- 4.Thorslund T, West SC. BRCA2: A universal recombinase regulator. Oncogene. 2007;26:7720–7730. doi: 10.1038/sj.onc.1210870. [DOI] [PubMed] [Google Scholar]
- 5.Wong AK, Pero R, Ormonde PA, Tavtigian SV, Bartel PL. RAD51 interacts with the evolutionarily conserved BRC motifs in the human breast cancer susceptibility gene brca2. J Biol Chem. 1997;272:31941–31944. doi: 10.1074/jbc.272.51.31941. [DOI] [PubMed] [Google Scholar]
- 6.Sharan SK, et al. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature. 1997;386:804–810. doi: 10.1038/386804a0. [DOI] [PubMed] [Google Scholar]
- 7.Lo T, Pellegrini L, Venkitaraman AR, Blundell TL. Sequence fingerprints in BRCA2 and RAD51: Implications for DNA repair and cancer. DNA Repair (Amst) 2003;2:1015–1028. doi: 10.1016/s1568-7864(03)00097-1. [DOI] [PubMed] [Google Scholar]
- 8.Yang H, et al. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science. 2002;297:1837–1848. doi: 10.1126/science.297.5588.1837. [DOI] [PubMed] [Google Scholar]
- 9.Yang H, Li Q, Fan J, Holloman WK, Pavletich NP. The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA-ssDNA junction. Nature. 2005;433:653–657. doi: 10.1038/nature03234. [DOI] [PubMed] [Google Scholar]
- 10.Saeki H, et al. Suppression of the DNA repair defects of BRCA2-deficient cells with heterologous protein fusions. Proc Natl Acad Sci USA. 2006;103:8768–8773. doi: 10.1073/pnas.0600298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.San Filippo J, et al. Recombination mediator and Rad51 targeting activities of a human BRCA2 polypeptide. J Biol Chem. 2006;281:11649–11657. doi: 10.1074/jbc.M601249200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pellegrini L, et al. Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature. 2002;420:287–293. doi: 10.1038/nature01230. [DOI] [PubMed] [Google Scholar]
- 13.Davies AA, et al. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol Cell. 2001;7:273–282. doi: 10.1016/s1097-2765(01)00175-7. [DOI] [PubMed] [Google Scholar]
- 14.Chen CF, Chen PL, Zhong Q, Sharp ZD, Lee WH. Expression of BRC repeats in breast cancer cells disrupts the BRCA2-Rad51 complex and leads to radiation hypersensitivity and loss of G(2)/M checkpoint control. J Biol Chem. 1999;274:32931–32935. doi: 10.1074/jbc.274.46.32931. [DOI] [PubMed] [Google Scholar]
- 15.Galkin VE, et al. BRCA2 BRC motifs bind RAD51-DNA filaments. Proc Natl Acad Sci USA. 2005;102:8537–8542. doi: 10.1073/pnas.0407266102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shivji MK, et al. A region of human BRCA2 containing multiple BRC repeats promotes RAD51-mediated strand exchange. Nucleic Acids Res. 2006;34:4000–4011. doi: 10.1093/nar/gkl505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carreira A, et al. The BRC repeats of BRCA2 modulate the DNA binding selectivity of RAD51. Cell. 2009;136:1032–1043. doi: 10.1016/j.cell.2009.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sung P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science. 1994;265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- 19.Baumann P, Benson FE, West SC. Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell. 1996;87:757–766. doi: 10.1016/s0092-8674(00)81394-x. [DOI] [PubMed] [Google Scholar]
- 20.Petalcorin MI, Galkin VE, Yu X, Egelman EH, Boulton SJ. Stabilization of RAD-51-DNA filaments via an interaction domain in Caenorhabditis elegans BRCA2. Proc Natl Acad Sci USA. 2007;104:8299–8304. doi: 10.1073/pnas.0702805104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, Ying L, Green JJ, Balasubramanian S, Klenerman D. Ultrasensitive coincidence fluorescence detection of single DNA molecules. Anal Chem. 2003;75:1664–1670. doi: 10.1021/ac026367z. [DOI] [PubMed] [Google Scholar]
- 22.van der Heijden T, et al. Real-time assembly and disassembly of human RAD51 filaments on individual DNA molecules. Nucleic Acids Res. 2007;35:5646–5657. doi: 10.1093/nar/gkm629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chi P, Van Komen S, Sehorn MG, Sigurdsson S, Sung P. Roles of ATP binding and ATP hydrolysis in human Rad51 recombinase function. DNA Repair (Amst) 2006;5:381–391. doi: 10.1016/j.dnarep.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 24.Ristic D, et al. Human Rad51 filaments on double- and single-stranded DNA: Correlating regular and irregular forms with recombination function. Nucleic Acids Res. 2005;33:3292–3302. doi: 10.1093/nar/gki640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mine J, et al. Real-time measurements of the nucleation, growth and dissociation of single Rad51-DNA nucleoprotein filaments. Nucleic Acids Res. 2007;35:7171–7187. doi: 10.1093/nar/gkm752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galletto R, Amitani I, Baskin RJ, Kowalczykowski SC. Direct observation of individual RecA filaments assembling on single DNA molecules. Nature. 2006;443:875–878. doi: 10.1038/nature05197. [DOI] [PubMed] [Google Scholar]
- 27.Sung P, Robberson DL. DNA strand exchange mediated by a RAD51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell. 1995;82:453–461. doi: 10.1016/0092-8674(95)90434-4. [DOI] [PubMed] [Google Scholar]
- 28.Mazin AV, Zaitseva E, Sung P, Kowalczykowski SC. Tailed duplex DNA is the preferred substrate for Rad51 protein-mediated homologous pairing. EMBO J. 2000;19:1148–1156. doi: 10.1093/emboj/19.5.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benson FE, Stasiak A, West SC. Purification and characterization of the human Rad51 protein, an analogue of E. coli RecA. EMBO J. 1994;13:5764–5771. doi: 10.1002/j.1460-2075.1994.tb06914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sigurdsson S, Trujillo K, Song B, Stratton S, Sung P. Basis for avid homologous DNA strand exchange by human Rad51 and RPA. J Biol Chem. 2001;276:8798–8806. doi: 10.1074/jbc.M010011200. [DOI] [PubMed] [Google Scholar]
- 31.Liu Y, et al. Conformational changes modulate the activity of human RAD51 protein. J Mol Biol. 2004;337:817–827. doi: 10.1016/j.jmb.2004.02.022. [DOI] [PubMed] [Google Scholar]
- 32.Orte A, et al. Direct characterization of amyloidogenic oligomers by single-molecule fluorescence. Proc Natl Acad Sci USA. 2008;105:14424–14429. doi: 10.1073/pnas.0803086105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.