Abstract
A long-standing question in retrovirus biology is how RNA genomes are distributed among virions. In the studies presented in this report, we addressed this issue by directly examining HIV-1 RNAs in virions using a modified HIV-1 genome that contained recognition sites for BglG, an antitermination protein in the Escherichia coli bgl operon, which was coexpressed with a fragment of BglG RNA binding protein fused to a fluorescent protein. Our results demonstrate that the majority of virions (>90%) contain viral RNAs. We also coexpressed HIV-1 genomes containing binding sites for BglG or the bacteriophage MS2 coat protein along with 2 fluorescent protein-tagged RNA binding proteins. This method allows simultaneously labeling and discrimination of 2 different RNAs at single-RNA-detection sensitivity. Using this strategy, we obtained physical evidence that virions contain RNAs derived from different parental viruses (heterozygous virion) at ratios expected from a random distribution, and we found that this ratio can be altered by changing the dimerization sequences. Our studies of heterozygous virions also support a generally accepted but unproven assumption that most particles contain 1 dimer. This study provides answers to long-standing questions in HIV-1 biology and illustrates the power and sensitivity of the 2-RNA labeling method, which can also be adapted to analyze various issues of RNA biogenesis including the detection of different RNAs in live cell imaging.
Keywords: Bgl, MS2, dimerization, DIS, fluorescent protein
Like all known retroviruses, HIV-1 virions contain dimeric full-length viral RNAs (1). Each copy of RNA contains the full genetic information needed for viral replication in host cells. These RNAs serve as templates for the synthesis of viral DNA carried out by the virally encoded reverse transcriptase. Viral DNA is then integrated into the host cell genome to become a provirus. Progeny virions, which are formed by association of genomes transcribed from the provirus with viral proteins including uncleaved structural protein (Gag) precursors, followed by budding from the cell and processing of the proteins to yield mature virions, are characterized by a somewhat irregular conical core shell composed of the CA (p24) protein. Virions derived from cells containing more than 1 provirus can contain 2 copies of RNA from the same provirus (homozygous) or RNAs derived from different proviruses (heterozygous). Because portions of both copies of RNA can be used as templates for DNA synthesis, a recombinant can be formed during reverse transcription. When the 2 copies of RNA encode different genetic information, a recombinant with a phenotype different from either parent can be generated (1).
A long standing question in HIV-1 and retrovirus biology is the content of the viral genetic material in the particles. Although it is known that viral RNAs are enriched within virions, HIV-1, like most retroviruses also packages other RNAs, including transcripts from the producer cell genome that lack known packaging signals (2). Furthermore, full-length viral genomic RNA is not required for assembly, budding, and release of virus like particles (3). Most previous studies examined RNA content from viral populations; comparison of the RNA content of mutant particles with that of wild-type particles was used to define the level of deficiency in RNA packaging (4). In these studies, the proportion of particles in the “wild-type” population that actually contained the viral RNA genome was not known. Indeed, it is possible that a large portion of the HIV-1 particles may contain no viral genomes at all.
Retroviral particles lack the highly regular structures found in many other viruses; hence, the numbers of Gag and RNA genome molecules in HIV-1 virions have been difficult to estimate. Previous biochemical analyses of mature HIV-1 particles estimated 1,000 to 10,000 copies of viral RNA for 1 pg of capsid (p24) or ∼25 million capsid molecules (5). The number of Gag molecules in immature HIV-1 particles was estimated to be 1,400 to 2,400 (6, 7). Because each Gag polyprotein yields 1 processed, mature capsid protein; taken together, these numbers imply that each virion contains, on average, somewhere between 0.05 and 1 genomes. Based on the assumption that all viral RNAs are dimeric, we calculated that 2.5% to 50% of the particles have viral RNAs. This calculation yields a wide range in the estimated HIV-1 RNA encapsidation efficiency; additionally, with limitations on assay sensitivity, variation introduced in correlating different measurements, and gaps in our current knowledge, the accuracy of even this crude estimate is questionable.
Biochemical and electron microscopy studies revealed that the RNA genomes in HIV-1 particles are noncovalently linked dimers that can be separated into monomers upon denaturation. Although it is clear that RNAs inside the virions are dimers, whether RNA dimerization initiated in the producer cell or the viral particle is still being debated. In 1 study, monomeric RNAs were detected in samples isolated from newly harvested viruses, leading to the hypothesis that 2 monomeric RNAs were packaged (8). Based on genetic studies, which showed that recombination rates can be regulated by the identity of the dimerization initiation signal (DIS) sequence, we hypothesized that RNA dimerization occurs before encapsidation (9, 10). The DIS is a 6-nt palindromic sequence located at the 5′ untranslated region (UTR) of HIV-1 RNA. Our current understanding is that 2 RNAs initiate dimerization by forming base pairs using the palindromic sequence of DIS (11). Previously, we showed that the genetic recombination rate of 2 HIV-1 variants decreased significantly when 1 contained the subtype B DIS (GCGCGC) and the other contained the subtype C DIS (GTGCAC) (9). We hypothesized that HIV-1 Gag packaged dimeric RNA and imperfect base-pairing of the DIS decreased heterodimer formation, thereby lowering the frequencies of heterozygous virions and the observed recombination rate. In contrast, the recombination rates increased significantly when 1 HIV-1 variant contained GGGGGG and the other contained CCCCCC in their DIS. We hypothesized that RNAs from these 2 viruses preferred to form heterodimers, thereby elevating the frequencies of heterozygous virions and the recombination rate (10). Additionally, how RNA genomes are distributed in virions generated from dually infected cells is another biologically important question that has not been directly answered. RNA derived from different proviruses may copackage together efficiently or there may be biases to preferentially copackage RNA from the same virus. Results from genetic assays can provide insights to RNA copackaging; however, these approaches are not without caveats as the results are indirect.
Although most studies have been performed using populations of viruses, much can be learned by analyzing individual HIV-1 particles. The coat protein of bacteriophage MS2 fused to a fluorescent protein has been used to study transport of murine leukemia virus RNAs and the dynamics of HIV-1 transcription using genomes modified to contain the specific binding site (12, 13). In this report, we developed an analogous system using the Escerichia coli BglG RNA-binding protein fused to a fluorescent protein to label virion RNA at single-RNA-detection sensitivity. We investigated the RNA content of HIV-1 particles using genomes containing MS2 or Bgl recognition sites and found that >90% of HIV-1 particles contain genome RNA. By combining the MS2 and the Bgl labeling methods, heterozygous particles were detected and were shown to form at a frequency similar to that predicted from random RNA assortment. The proportions of heterozygous virions can be altered by changing the identity of the DIS sequences, providing direct support for the model that HIV-1 Gag packages dimeric RNA and not 2 monomeric RNA molecules. These studies provide answers to several long standing questions in HIV-1 biology as well as an approach to study retrovirus replication mechanisms.
Results
Labeling of Specific RNA Reveals that Most HIV-1 Particles Contain Viral Genomic RNA.
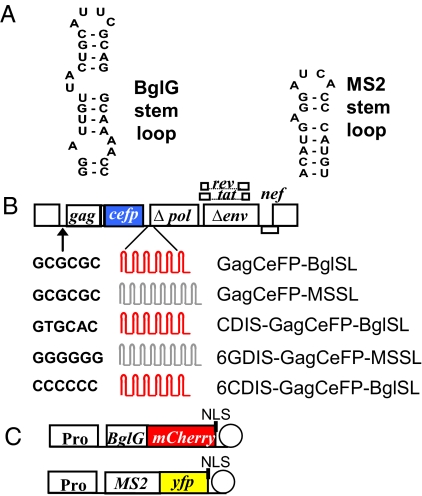
We developed an RNA-labeling system based on a regulatory element of the E. coli bgl operon, in which the antitermination protein, BglG, binds to a specific stem-loop (BglSL; Fig. 1A) to prevent early termination of RNA transcripts (14). For this purpose, 18 copies of the BglG stem-loops were inserted into the pol gene of a modified HIV-1 genome to detect full-length RNA, and the N-terminal portion of the BglG antitermination protein gene, encoding the RNA binding domain, was fused with the red fluorescent protein gene, mCherry (Fig. 1 B and C). To verify that the RNA signals detected were in viral particles, we also tagged the HIV-1 gag gene, which encodes the structural Gag protein, with cerulean fluorescent protein gene (cefp). When coexpressed with untagged Gag, such fusion proteins have been shown to coassemble into pseudovirions, which have normal morphology indistinguishable from particles generated from Gag alone and which bud from the cell surface with efficiency equivalent to wild-type virions (15, 16).
Fig. 1.
A bgl operon-based system for labeling RNA. (A) Sequence of the stem-loop used by the BglG antitermination protein and a modified stem-loop that can be bound by MS2 coat protein. (B) General structure of modified HIV-1 genomes used in the study. The DIS sequence is GCGCGC unless otherwise specified; 18 copies of BglG stem-loops (in red) or 24 copies of MS2 stem-loops (in gray) were inserted into the HIV-1 genome, although fewer copies are illustrated. For each HIV-1 genome illustrated in the figure, a partner genome lacking the cefp gene was also generated. In all of the transfection experiments, a mixture of plasmids containing the specified HIV-1 genome and its partner genome was used. (C) General structure of plasmids for expression of RNA-binding proteins. The circle represents the termination signal from the β-globin 3′ UTR polyadenylation site. Pro, promoter from the gene encoding the large subunit of RNA polymerase II; NLS, nuclear localization signal.
We used this system to examine the proportion of HIV-1 particles that contain HIV-1 RNA. Constructs expressing MS2 or BglG RNA-binding protein fusions were cotransfected with plasmids containing modified HIV-1 genomes; in each experiment, we used a pair of HIV-1 genomes that were identical, except that 1 encoded the wild-type Gag protein and the other encoded the GagCeFP fusion protein. The resulting viruses were harvested and examined by fluorescence microscopy; representative images are shown in Fig. 2. Because HIV-1 Gag was tagged with CeFP, the channel detecting CeFP signals identified all viral particles (Fig. 2 Left). When the HIV-1 genome containing the BglG stem-loops was coexpressed with the BglG-mCherry fusion protein, abundant mCherry signals were detected (Fig. 2A Middle). To assess the relationship between the RNA (mCherry) and the viral particles (CeFP), we merged the images detected in the 2 channels and shifted the mCherry image 4 pixels to the right (Fig. 2A Right). As shown in this image, most of the mCherry and CeFP signals overlapped, indicating that the RNA signals detected were indeed within the viral particles. Analyses of thousands of particles from different experiments revealed that the large majority of virus particles (>93%) contain HIV-1 RNAs (Table 1, line 1). To ensure that the mCherry signals were the result of specific detection of BglG RNA stem-loops in the viral RNA, we performed experiments with a similar HIV-1 genome that contained 24 copies of stem-loops from bacteriophage MS2 in the pol gene (17) (GagCeFP-MSSL) instead of the BglG stem-loops. As shown in a representative image (Fig. 2B Right), we did not detect mCherry signals when the HIV-1 genome did not contain BglG stem-loops, indicating that the positive mCherry signals were due to specific interactions between the BglG protein and the BglG stem-loops. These results also showed that the BglG protein did not interact with MS2 RNA stem-loops with high affinity, as we did not observe significant signals with the HIV-1 genome containing the 24 copies of MS2 stem-loops (Table 1, line 2).
Fig. 2.
Detection of HIV-1 RNA with the BglG or MS2 system. Representative images of virions labeled with Gag-CeFP as detected by the BglG (A) or MS2 (C) system. Channels detecting CeFP or mCherry signals are indicated (Top). In the “Merged and shifted” panels, images of the CeFP signals and mCherry (A) or YFP (C) signals were merged, and the mCherry or YFP image was shifted to the right by 4 pixels to allow easy identification of colocalization. (B) Representative images of virions containing the MS2 stem-loops produced by cells expressing the BglG fusion protein. (D) Representative images of virions containing the BglG stem-loops produced by cells expressing the MS2 fusion protein.
Table 1.
Proportions of virions expressing each fluorescent marker
| Cells transfected with | No. CeFP particles analyzed | % CeFP+ + YFP+ | % CeFP+ + mCherry+ | % CeFP+ + YFP+ + mCherry+ | RNA labeling efficiency* |
|---|---|---|---|---|---|
| 1. Bgl-mCherry + GagCeFP-BglSL | |||||
| Exp. a | 6092 | 0.00 | 93.4 | 0.05 | 93.5 |
| Exp. b | 2861 | 0.00 | 93.8 | 0.07 | 93.9 |
| 2. Bgl-mCherry + GagCeFP-MSSL | |||||
| Exp. a | 2136 | 0.37 | 0.00 | 0.00 | 0.37 |
| Exp. b | 2042 | 0.05 | 0.10 | 0.00 | 0.15 |
| 3. MS2-YFP + GagCeFP-MSSL | 1902 | 94.9 | 0.00 | 0.00 | 94.9 |
| 4. MS2-YFP + GagCeFP-BglSL | |||||
| Exp. a | 3872 | 0.46 | 0.00 | 0.00 | 0.46 |
| Exp. b | 2826 | 0.21 | 0.00 | 0.00 | 0.21 |
| 5. MS2-YFP + Bgl-mCherry + GagCeFP-MSSL + GagCeFP-BglSL | |||||
| Exp. a | 1753 | 19.7 | 30.2 | 44.0 | 94.0 |
| Exp. b | 3220 | 24.6 | 22.8 | 46.3 | 93.7 |
| 6. MS2-YFP + Bgl-mCherry + GagCeFP-MSSL + CDIS-GagCeFP-BglSL | |||||
| Exp. a | 1341 | 39.1 | 34.4 | 21.0 | 94.5 |
| Exp. b | 2003 | 46.8 | 27.8 | 19.3 | 93.9 |
| 7. MS2-YFP + Bgl-mCherry + 6GDIS-GagCeFP-MSSL + 6CDIS-GagCeFP-BglSL | |||||
| Exp. a | 498 | 5.2 | 17.1 | 69.5 | 91.8 |
| Exp. b | 1294 | 6.3 | 15.2 | 69.6 | 91.1 |
*RNA labeling efficiency is the sum of the 3 previous columns, i.e., %CeFP+ + YFP+, %CeFP+ + mCherry+, and %CeFP+ +YFP+ + mCherry+.
To further examine possible cross-reactivity of the BglG system with the existing MS2 system (17, 18), we also cotransfected modified HIV-1 genomes containing BglG stem-loops or MS2 stem-loops with MS2-YFP, a plasmid expressing the MS2 coat protein fused with the yellow fluorescent protein gene (yfp). Representative results are shown in Fig. 2 C and D. Similar to the results obtained with the BglG system, we found that when MS2-YFP was coexpressed with an HIV-1 genome containing MS2 stem-loops, most of the viral particles displayed YFP signals (≈95%; Fig. 2C and Table 1, line 3), confirming the observation that most HIV-1 virions contain viral RNA. The MS2 protein-RNA interaction was also specific, because particles generated by HIV-1 containing BglG RNA stem-loops did not generate significant YFP signals, indicating that the MS2 protein did not bind to BglG RNA stem-loops with high specificity (Fig. 2D and Table 1, line 4). Therefore, the BglG and MS2 systems have distinct specificities and did not share significant cross-reactivity under our experimental conditions.
Simultaneous Detection of 2 Different RNA Species Shows Efficient Heterozygous Particle Formation with a Frequency Similar to that Predicted from RNA Random Assortment.
HIV-1 RNA genomes are present as dimers in mature virions. The copackaged RNAs can be from different proviruses (i.e., in heterozygous virions) or from the same provirus (i.e., in homozygous virions). To date, the relative frequencies of the 2 types of virions have been inferred only from indirect measurements, such as recombination frequency (19–21). To address this issue directly, we used the BglG and MS2 systems to examine RNA copackaged in viral particles at single-RNA-detection sensitivity by coexpressing HIV-1 genomes containing either BglG or MS2 stem-loops along with plasmids that express RNA-binding proteins. Representative results are shown in Fig. 3A. As before, Gag was tagged with CeFP, MS2 coat protein was tagged with YFP, and BglG protein was tagged with mCherry. Thus, the channel detecting CeFP fluorescence identified viral particles, whereas the channel detecting mCherry or YFP fluorescence identified RNA containing BglG or MS2 stem-loops, respectively. For comparison, we also show images with all of the signals merged (Fig. 3, merged and shifted): The image from the YFP channel was shifted 4 pixels to the right, and the image from the mCherry channel was shifted 8 pixels to the right. For clarity, we have marked 6 CeFP-positive virions (Fig. 3A, 1 to 6): Particles 1, 3, 4, and 6 had positive YFP signals, whereas particles 2, 3, 4, and 5 had positive mCherry signals. Thus, particles 1 and 6 exhibited CeFP and YFP signals, indicating that they contained HIV-1 RNA with MS2 stem-loops. Particles 2 and 5 were positive for CeFP and mCherry signals, indicating that they contained HIV-1 RNA with BglG stem-loops. Particles 3 and 4 were positive with CeFP, YFP, and mCherry signals, indicating that they contained HIV-1 RNA with MS2 stem-loops and HIV-1 RNA with BglG stem-loops, i.e., were heterozygous virions. We analyzed ≈5,000 particles and found that heterozygous particles comprised ≈45% of the viral population (Table 1, line 5) or ≈48% of the particles with RNA signals. RNAs containing MS2 or BglG stem-loops were incorporated into particles at similar levels. If the copackaging of these 2 RNA species was random, we expected 50% of the virions to be heterozygous (analogous to a Hardy-Weinberg equilibrium), which is very close to the observed ≈45% frequency of heterozygous particles detected in our system. This finding provides physical evidence of HIV-1 particles containing 2 different RNAs as well as of efficient copackaging of RNAs derived from different HIV-1 genomes.
Fig. 3.
Detection of the homozygous and heterozygous HIV-1 particles at single-RNA-molecule sensitivity. All particles were produced by cells transfected with Bgl-mCherry and MS2-YFP. (A) Images of particles generated from coexpression of 2 HIV-1 genomes, 1 containing MS2 stem-loops and the other containing BglG stem-loops. Both genomes contained GCGCGC in the dimerization initialization signal (DIS). (B) Particles generated by coexpression of an HIV-1 genome with GCGCGC in the DIS and an HIV-1 genome with GTGCAC in the DIS. (C) Particles generated by coexpression of an HIV-1 genome with GGGGGG in the DIS and CCCCCC in the DIS. Abbreviations are the same as in Fig. 2.
Base-Pairing of the DIS Sequences Is a Critical Step for Selecting Copackaged Viral RNA and Determines the Proportion of Heterozygous Particles in the Virus Population.
Based on previous recombination studies, we hypothesized that the mismatch between the DIS sequences from subtype B and subtype C viruses decreased the base-pairing of these 2 RNAs and reduced the formation of heterozygous particles (9). RNA containing GGGGGG and RNA containing CCCCCC in the DIS were inferred to form heterodimers more efficiently than homodimers, thereby increasing the proportion of heterozygous virions in the viral population (10). Using the aforementioned RNA-labeling system, we examined whether similar alterations to the DIS sequences could also change the copackaging of HIV-1 genomes and affect the proportion of heterozygous virions in the viral population.
For this purpose, we examined virions from cells expressing 1 HIV-1 genome containing the subtype B DIS (GCGCGC) and MS2 stem-loops and another HIV-1 genome containing the subtype C DIS (GTGCAC) and BglG stem-loops. Cells were cotransfected with these vectors along with plasmids expressing the RNA-binding proteins; the resulting particles were harvested and examined by fluorescence microscopy. As shown in Fig. 3B and Table 1, line 6, we observed many virus particles with YFP or mCherry signals; however, the percentage of particles containing both YFP and mCherry signals was only 20% of the viral population, approximately half of the value obtained from viruses with 2 subtype B DIS sequences. We also examined particles generated from cotransfection of a viral genome containing GGGGGG in the DIS and MS2 stem-loops and a viral genome containing CCCCCC in the DIS and BglG stem-loops (Fig. 3C and Table 1, line 7). We observed that indeed most of the particles had both YFP and mCherry. These results provide physical evidence that the proportion of HIV-1 heterozygous virions can be altered by changing the DIS sequence. They also verify the accuracy and sensitivity of the dual-RNA-labeling system. In all experiments presented in Table 1, 91%–95% of the viral particles had RNA signals, including those that had mostly heterozygous virions, and hence the detection of RNA was at the single-RNA-molecule level. These unique findings demonstrate the simultaneous detection of 2 RNA species, each at single-RNA-molecule sensitivity. These results also illustrate the feasibility of using the dual-labeling system to study various experimental questions on RNA biogenesis.
Discussion
Our understanding of viral replication has been shaped by analyzing populations of viruses using various physical and genetic techniques. Studies at the single-virion level can provide insights that these traditional approaches cannot reveal. Using either the MS2 or the BglG-based RNA-labeling system, we found that >93% of the HIV-1 particles contained full-length viral genomes, as spliced HIV-1 RNA lacked RNA binding protein recognition sites inserted in the pol gene. Our results provide quantification of the percentage of retroviral particles that contains the viral RNA genome and show that HIV-1 RNA encapsidation is a remarkably efficient process, >90% in all cases. This result was unexpected, given the previous rough estimates of the genome RNA content of virions and the high efficiency of virion formation that occurs in the absence of genome RNA. A recent study indicated that, under optimal conditions, 1 in 8 to 1 in 20 virions can initiate an infection event, despite earlier estimates that a large portion of HIV-1 particles is noninfectious (22). Our results indicate that most of viral particles have full-length viral RNAs and a low efficiency of genomic RNA packaging is not a major contributor to the formation of noninfectious virions.
Many aspects of the molecular mechanisms of HIV-1 RNA packaging remain unknown; for example, does HIV-1 Gag recruit monomeric or dimeric RNA? How are RNAs derived from different proviruses sorted into virions? Lastly, what is the limit of RNA packaging capacity? To address these issues, we examined the frequency of heterozygous and homozygous virions in the viral population. This analysis was made possible by lack of cross-reactivity between the BglG and MS2 systems, allowing these 2 binding proteins to be used simultaneously to detect 2 RNA species. Previously, our understanding of heterozygous virion formation was mainly inferred from genetic studies examining recombination. Here, we provide direct physical evidence that 1 particle contains RNA derived from 2 HIV-1 genomes and that genetically distinct RNAs segregate according to a random distribution. Our study also demonstrates that changing the DIS sequences affects the proportion of the heterozygous virions, which is in general agreement with our genetic recombination studies (9, 10). Compared with 2 subtype B viruses with GCGCGC in their DIS sequences, when 1 subtype B virus had GCGCGC in its DIS and 1 virus had GTGCAC in its DIS, we observed decrease in both recombination rate (4-fold) and proportion of heterozygous virions (2.3-fold). Similarly, when 1 virus had GGGGGG in its DIS and 1 virus had CCCCCC in its DIS, we observed increase in recombination rate (1.9-fold) and the proportion of heterozygous virions (1.6-fold). Therefore, results from the 2 systems lead to the same conclusions. It is possible that virions containing 2 RNAs with imperfectly matched DIS sequences have a slightly lower infectivity causing an elevated change in the recombination assay. Because altering DIS sequences can change the proportion of heterozygous virions, these results also imply that HIV-1 Gag mainly packages a dimeric RNA rather than 2 monomeric RNAs. If 2 unassociated monomeric RNAs were packaged into virions, changing a few nucleotides in the DIS sequences would not have affected the frequency of heterozygous virions. Interestingly, we still observed heterozygous virions when the 2 HIV-1 genomes contained imperfectly matched DIS sequences; hence it remains possible that, at a low frequency, HIV-1 Gag packages 2 monomers. Together, these results also imply that association of 2 RNA molecules to form dimers must occur at a location in the cell that is well separated from the site of RNA synthesis.
Although it is known that HIV-1 RNAs exist as dimers in particles, there is no direct evidence indicating how many dimers are packaged into a particle. A generally accepted but unproven assumption is that 1 dimer is encapsidated in each virion. Our heterozygous particle analyses also provide evidence to support this view. We observed that 45% of the particles had RNAs from 2 parental viruses; considering that ≈94% of the particles had RNA signals, ≈48% of the particles with detectable RNA signals were heterozygous particles. This value is very close to the 50% frequency predicted from random assortment. If HIV-1 packages more than 1 dimer, we would have observed far more particles with 2 different RNA signals. For example, if 2 dimers were packaged, we would have observed that 87.5% of the virions had both mCherry and YFP signals (assuming random RNA assortment). A mathematically plausible alternative explanation is that all RNAs form homodimers and 2 dimers are packaged into a particle by random assortment; this scenario will also predict 50% particles with mCherry and YFP signals. However, if RNA packaging indeed operates by such mechanism, changing the DIS sequences should not be able to alter the frequency of heterozygous virions. Therefore, our results strongly support the view that most virions package 1 dimer.
The experiments described in this report used 2 different labeling systems to analyze RNA in viral particles. Furthermore, we have demonstrated that the detection sensitivity for each label is at single-RNA-molecule sensitivity. This combination of systems can be applied to study various experimental questions related to RNA biogenesis and transport. Using a second RNA labeling system (BglG) that does not have cross-reactivity to the previously established RNA labeling system (MS2) allows simultaneous detection of 2 RNA species and comparison of their behavior in the same cell or the same experiment. The ability to perform such a comparative study significantly increases the tools available for imaging analyses to study biological questions.
Despite the worldwide epidemic, a preventive vaccine against HIV-1 is not likely to be available in the near future, and generation and transmission of drug-resistant viruses threaten the effectiveness of existing antiviral drug treatments. A major reason for the difficulty of generating a preventive vaccine and effective treatments is the high genetic diversity of HIV-1 populations (23). Among the key components HIV-1 uses to generate high genetic diversity are the high viral load and frequent recombination. By directly examining the RNA content of the viral particles, we have shown that RNA packaging during particle assembly is a surprisingly efficient process and that heterozygous viruses are generated efficiently, providing the basis for frequent reassortment of variant viral sequences. These results provide answers to long-standing questions in retrovirus biology and and provide insights into the mechanisms that generate high genetic variation of the HIV-1 population.
Methods
Plasmid Construction.
Plasmids pSL-MS2–24X, which contained 24 copies of the MS2 stem-loop, and pMS2-YFP were a generous gift from Dr. Robert Singer (Albert Einstein Medical College). Plasmid pBgl-mCherry was constructed by replacing the MS2 coat protein gene sequence of pMS2-YFP with the E. coli sequence encoding the N-terminal RNA binding domain of BglG protein and the yfp gene with the mCherry gene. Sequences containing BglG stem-loops were generated by annealing synthesized oligomeric nucleotides and inserting the resulting DNA fragments into a plasmid. DNA fragments containing the MS2 stem-loops (24 copies) or BglG stem-loops (18 copies) were inserted into the pol gene of a modified HIV-1 genome to generate GagCeFP-MSSL or GagCeFP-BglSL. This modified NL43-based HIV-1 genome contains the following mutations: A portion of the pol gene was deleted, and portions of vif, vpr, vpu, and env were deleted to inactivate these genes. However, this genome expresses functional Gag, Tat, and Rev and contains cis-acting elements important for viral replication including the LTRs, intact 5′ UTR, Rev-Response element, and intact 3′ UTR. Two related constructs, Gag-MSSL and Gag-BglSL, were also generated; these constructs had the same structures as GagCeFP-MSSL and GagCeFP-BglSL, respectively, except that they did not contain the cefp gene. Mutations were introduced into the DIS regions by replacing the DNA fragment containing subtype B DIS with those containing other DIS sequences. Molecular cloning was performed using standard molecular biology techniques; the general structures of all plasmids were verified by restriction enzyme mapping, and DNA sequences generated by PCR were confirmed by DNA sequencing to avoid inadvertent mutations.
Cell Culture, Transfection, and Viral Particle Preparation.
Human 293T cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% FCS, 50 U/mL penicillin, and 50 U/mL streptomycin. All cultured cells were maintained at 37 °C with 5% CO2. Transient transfection was performed with FuGENE HD (Roche) according to the manufacturer's recommendation. To generate virus particles, 293T cells were cotransfected with HIV-1 DNA containing or lacking cefp; for example, GagCeFP-BglSL and Gag-BglSL at an equal weight ratio. Virus particles were harvested 24 h posttransfection, clarified through a 0.45-μm-pore-size filter to remove cellular debris, and used immediately or stored at −80 °C before use. Polybrene was added to the culture supernatant, which was then placed in a glass-bottom dish and incubated for 2 h at 37 °C with 5% CO2 before image acquisition.
Microscopy, Image Acquisition, and Image Analyses.
Epifluorescence microscopy was performed with an inverted Nikon TE2000E2 microscope and a 100× 1.40 numerical aperture oil objective, using an X-Cite 120 system (EXFO Photonic Solution) for illumination. Digital images were acquired using a Hammamatsu ORCA-ER camera and Openlab software (Improvision) with the excitation and emission filter sets 427/10 nm and 472/30 nm for CeFP, 504/12 nm and 542/27 nm for YFP, and 577/25 nm and 632/60 nm for mCherry, respectively.
Virus particle and RNA signal analysis and colocalization were performed by using custom software developed with Matlab (MathWorks) and DIPimage (TU Delft). Fluorescence-tagged particles were automatically detected in the images using intensity-based thresholding. For each image, the histogram of the intensity distribution was calculated. The most frequent pixel intensity in the image was treated as the mean value (M) of the background intensities in the image, and the standard deviation of the background intensities (S) was calculated from the intensity distribution between zero intensity and the mean background intensity. The threshold intensity used to segment the images into background and signal regions was set to M + KS, where K was a constant between 5 to 7, depending on the channel used. Points in the segmented image that were intensity peaks (i.e., had greater intensity than all of their 8 nearest-neighbor pixels) were considered to represent virus particles. Particles detected in different channels were considered to be in the same virus if they were less than 3 pixels from each other. Merged images and pseudocolored images were generated using ImageJ software.
Acknowledgments.
We thank Anne Arthur for her expert editorial help; Drs. Steve Hughes and Eric Freed for suggestions, discussions, and critical reading of the manuscript; Delft University, The Netherlands, for providing DIPimage program; and Dr. Vitaly Boyko for assistance in setting up epifluorescence microscopy. This work was supported in part by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research, National Institutes of Health Grants GM38035 (to A.W.) and R37 CA089441 (to J.M.C.), and in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract no. HHSN261200800001E. J.M.C. was a Research Professor of the American Cancer Society, with support from the George Kirby Foundation.
Footnotes
The authors declare no conflict of interest.
References
- 1.Goff S. Retroviridae: The retroviruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. vol II. Philadelphia, PA: Lippincott, Williams & Wilkins; 2007. pp. 1999–2069. [Google Scholar]
- 2.Rulli SJ, Jr, et al. Selective and nonselective packaging of cellular RNAs in retrovirus particles. J Virol. 2007;81:6623–6631. doi: 10.1128/JVI.02833-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muriaux D, Mirro J, Harvin D, Rein A. RNA is a structural element in retrovirus particles. Proc Natl Acad Sci USA. 2001;98:5246–5251. doi: 10.1073/pnas.091000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gorelick RJ, Chabot DJ, Rein A, Henderson LE, Arthur LO. The two zinc fingers in the human immunodeficiency virus type 1 nucleocapsid protein are not functionally equivalent. J Virol. 1993;67:4027–4036. doi: 10.1128/jvi.67.7.4027-4036.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Piatak M, Jr, et al. High levels of HIV-1 in plasma during all stages of infection determined by competitive PCR. Science. 1993;259:1749–1754. doi: 10.1126/science.8096089. [DOI] [PubMed] [Google Scholar]
- 6.Zhu P, et al. Electron tomography analysis of envelope glycoprotein trimers on HIV and simian immunodeficiency virus virions. Proc Natl Acad Sci USA. 2003;100:15812–15817. doi: 10.1073/pnas.2634931100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carlson LA, et al. Three-dimensional analysis of budding sites and released virus suggests a revised model for HIV-1 morphogenesis. Cell Host Microbe. 2008;4:592–599. doi: 10.1016/j.chom.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song R, Kafaie J, Yang L, Laughrea M. HIV-1 viral RNA is selected in the form of monomers that dimerize in a three-step protease-dependent process; the DIS of stem-loop 1 initiates viral RNA dimerization. J Mol Biol. 2007;371:1084–1098. doi: 10.1016/j.jmb.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 9.Chin MP, Rhodes TD, Chen J, Fu W, Hu WS. Identification of a major restriction in HIV-1 intersubtype recombination. Proc Natl Acad Sci USA. 2005;102:9002–9007. doi: 10.1073/pnas.0502522102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moore MD, et al. Dimer initiation signal of human immunodeficiency virus type 1: Its role in partner selection during RNA copackaging and its effects on recombination. J Virol. 2007;81:4002–4011. doi: 10.1128/JVI.02589-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paillart JC, Shehu-Xhilaga M, Marquet R, Mak J. Dimerization of retroviral RNA genomes: An inseparable pair. Nat Rev. 2004;2:461–472. doi: 10.1038/nrmicro903. [DOI] [PubMed] [Google Scholar]
- 12.Basyuk E, et al. Retroviral genomic RNAs are transported to the plasma membrane by endosomal vesicles. Dev Cell. 2003;5:161–174. doi: 10.1016/s1534-5807(03)00188-6. [DOI] [PubMed] [Google Scholar]
- 13.Boireau S, et al. The transcriptional cycle of HIV-1 in real-time and live cells. J Cell Biol. 2007;179:291–304. doi: 10.1083/jcb.200706018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Houman F, Diaz-Torres MR, Wright A. Transcriptional antitermination in the bgl operon of E. coli is modulated by a specific RNA binding protein. Cell. 1990;62:1153–1163. doi: 10.1016/0092-8674(90)90392-r. [DOI] [PubMed] [Google Scholar]
- 15.Larson DR, Johnson MC, Webb WW, Vogt VM. Visualization of retrovirus budding with correlated light and electron microscopy. Proc Natl Acad Sci USA. 2005;102:15453–15458. doi: 10.1073/pnas.0504812102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jouvenet N, Bieniasz PD, Simon SM. Imaging the biogenesis of individual HIV-1 virions in live cells. Nature. 2008;454:236–240. doi: 10.1038/nature06998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fusco D, et al. Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Curr Biol. 2003;13:161–167. doi: 10.1016/s0960-9822(02)01436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bertrand E, et al. Localization of ASH1 mRNA particles in living yeast. Mol Cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 19.Rhodes T, Wargo H, Hu WS. High rates of human immunodeficiency virus type 1 recombination: Near-random segregation of markers one kilobase apart in one round of viral replication. J Virol. 2003;77:11193–11200. doi: 10.1128/JVI.77.20.11193-11200.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rhodes TD, Nikolaitchik O, Chen J, Powell D, Hu WS. Genetic recombination of human immunodeficiency virus type 1 in one round of viral replication: Effects of genetic distance, target cells, accessory genes, and lack of high negative interference in crossover events. J Virol. 2005;79:1666–1677. doi: 10.1128/JVI.79.3.1666-1677.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Onafuwa A, An W, Robson ND, Telesnitsky A. Human immunodeficiency virus type 1 genetic recombination is more frequent than that of Moloney murine leukemia virus despite similar template switching rates. J Virol. 2003;77:4577–4587. doi: 10.1128/JVI.77.8.4577-4587.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas JA, Ott DE, Gorelick RJ. Efficiency of human immunodeficiency virus type 1 postentry infection processes: Evidence against disproportionate numbers of defective virions. J Virol. 2007;81:4367–4370. doi: 10.1128/JVI.02357-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coffin JM. HIV population dynamics in vivo: Implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]