Abstract
Large databases of high-resolution structural MR images are being assembled to quantitatively examine the relationships between brain anatomy, disease progression, treatment regimens, and genetic influences upon brain structure. Quantifying brain structures in such large databases cannot be practically accomplished by expert neuroanatomists using hand-tracing. Rather, this research will depend upon automated methods that reliably and accurately segment and quantify dozens of brain regions. At present, there is little guidance available to help clinical research groups in choosing such tools. Thus, our goal was to compare the performance of two popular and fully automated tools, FSL/FIRST and FreeSurfer, to expert hand tracing in the measurement of the hippocampus and amygdala. Volumes derived from each automated measurement were compared to hand tracing for percent volume overlap, percent volume difference, across-sample correlation, and 3-D group-level shape analysis. In addition, sample size estimates for conducting between-group studies were computed for a range of effect sizes. Compared to hand tracing, hippocampal measurements with FreeSurfer exhibited greater volume overlap, smaller volume difference, and higher correlation than FIRST, and sample size estimates with FreeSurfer were closer to hand tracing. Amygdala measurement with FreeSurfer was also more highly correlated to hand tracing than FIRST, but exhibited a greater volume difference than FIRST. Both techniques had comparable volume overlap and similar sample size estimates. Compared to hand tracing, a 3-D shape analysis of the hippocampus showed FreeSurfer was more accurate than FIRST, particularly in the head and tail. However, FIRST more accurately represented the amygdala shape than FreeSurfer, which inflated its anterior and posterior surfaces.
Introduction
Rapid advances in magnetic resonance imaging (MRI) have enabled noninvasive exploration of the human brain with increasing level of detail. Quantitative MRI studies have revealed differences in the volume of particular brain structures in several neuropsychiatric conditions including depression (Videbech and Ravnkilde, 2004), posttraumatic stress disorder (Bremner et al., 1995, 1997), schizophrenia (Turetsky et al., 1995; Vita et al., 2006), and Alzheimer's disease (Apostolova et al., 2006). Large databases of MR images are being assembled to enable researchers to examine subtle relationships between quantitative brain anatomy, disease progression, treatment regimens, risk, and other factors for various diseases. This research has been recently extended to an investigation of genetic influences on brain structure. The advent of technologies capable of assaying many thousands of gene variations on single chip has resulted in demand for ever larger data sets.
These research programs rely upon accurate segmentation and quantification of brain regions from high-resolution MR images. Until recently, manual tracing of brain regions by experts in neuroanatomy has been the accepted standard. However, as the size of the MRI datasets has increased, the time and cost required for the labor intensive process of manual tracing has become prohibitive. An experienced researcher may require two hours to trace a single structure such as the hippocampus, and more than a week to trace all of the major structures of the brain. Differences in criteria among experts can lead to systematically different volume estimates of some brain regions, and so the highest consistency and sensitivity is achieved when a single individual traces the entire dataset. However, the criteria used by even a trained expert can subtly drift during the course of a long study. For these reasons, automated procedures for segmenting and quantifying the brain have attracted considerable interest. Automated methods provide consistent results with repeated iterations on a given dataset. Improvements in the segmentation algorithms can be accommodated with relative ease even on large data sets by re-analysis with updated software.
Despite the availability of several sophisticated automated and semi-automated segmentation algorithms, there have been relatively few published comparisons of automated segmentation and hand tracing of brain structures (Barnes et al., 2008; Jatzko et al., 2006; Powell et al., 2008). Thus, there is little guidance in the literature for clinical research groups embarking upon a large-scale quantitative study of human brain anatomy. Here, we assessed the performance of two popular fully automated segmentation and quantification software tools available in the public domain and developed by leading neuroimaging analysis groups. FreeSurfer [Martinos Center for Biomedical Imaging, Harvard-MIT, Boston USA] performs subcortical and cortical segmentation and assigns a neuroanatomical label to each voxel based on probabilistic information automatically estimated from a large training set of expert measurements (Fischl et al., 2002). FSL/FIRST [FMRIB Integrated Registration and Segmentation Tool, Oxford University, Oxford UK] performs subcortical segmentation using Bayesian shape and appearance models (technical report at http://www.fmrib.ox.ac.uk/fsl/first/index.html). Fischl et al. (2002) reported performance characteristics for FreeSurfer in 2002; however, since that report there has been steady improvement in the quality (contrast to noise ratio) of high-resolution structural MR image acquisition at 3 T and improvements in the FreeSurfer software. The FIRST software was released in 2007 and performance metrics have not been published. Image quality improvements are sure benefit to both automated and manual brain segmentation although automated algorithms are likely to incur greater benefit because an expert rater is better able to estimate boundaries by employing multiple heuristics when insufficient contrast or other feature information is available.
Our first goal was to compare the accuracy of automated segmentation of the hippocampus and amygdala by FreeSurfer and FIRST as compared to manual tracing. These brain structures were chosen because of their relevance to a variety of neuropsychiatric disorders, and because their segmentation is technically challenging. Comparative performance was measured with four metrics (i) percent volume overlap (ii) percent volume difference (iii) correlation with manual tracing across the sample, and (iv) group-level 3-D shape analysis. These are well accepted approaches for comparing performance of competing segmentation methodologies (Fischl et al., 2002).
Our second goal was to provide sample size estimates for each of the methods to study differences in hippocampal or amygdala volume between two groups of subjects (e.g., healthy control and clinical populations). Many disorders that are associated with region-specific volume differences are also accompanied by an increased variability in volume. Thus, comparison of clinical group to a healthy group requires measurements that reliably capture the true variance represented in each group. The introduction of additional variance from inaccurate segmentation leads to a corresponding reduction in power to detect differences. We estimated the number of subjects required to adequately power a study for a range of anticipated effect sizes using the variance derived from hand tracing as the standard.
Differences in hippocampal and amygdala volumes have been reported in the literature for various neuropsychiatric disorders and the magnitude of the difference depends on the disorder in question. For instance, change in hippocampal volume range from 25% for epilepsy (Theodore et al., 1999) to 10% for PTSD (Bremner et al., 1995, 1997), while amygdala volumes showed reduction of 32% in dissociative identity disorder (Vermetten et al., 2006) and 17% in pedophilia (Schiltz et al., 2007). As a final confirmatory step in sample size estimation, we used FreeSurfer and FIRST to measure an existing dataset within our lab to examine hippocampal and amygdala volume differences associated with major depression (Campbell et al., 2004; Kronmuller et al., 2008; Videbech and Ravnkilde 2004).
Methods
Subject data
We collected high-resolution structural MR images at the Duke-UNC Brain Imaging and Analysis Center from 20 participants without history of neurologic disorder or head trauma that were enrolled at the Durham Veterans Affairs (VA) Medical Center during 2006–2008. Participants provided written informed consent to participate in procedures approved by the Institutional Review Boards at Duke University and the Durham VA Medical Center. High resolution T1-weighted images with 1 mm isometric voxels were acquired on a General Electric 3 T EXCITE system using the Array Spatial Sensitivity Encoding Technique (ASSET) with fast spoiled gradient-recall (FSPGR). Image parameters were optimized for contrast between white matter, gray matter, and CSF (TR/TE/flip angle=7.484 ms/2.984 ms/12°, 256 mm FOV, 1 mm slice, 166 slices, 256×256 matrix, 1 Nex).
Manual tracing of the hippocampus and amygdala
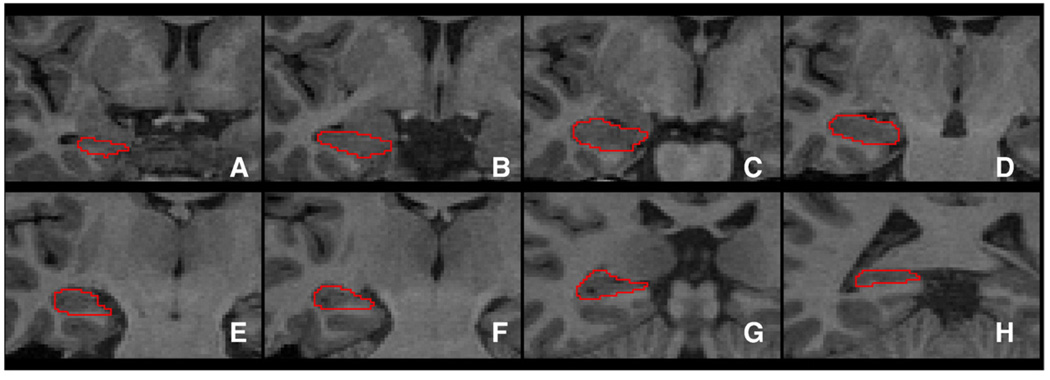
A detailed description of the manual segmentation protocol of hippocampus and amygdala are presented in Appendix A. Manual tracing in a sample subject is illustrated for eight representative coronal slices of the hippocampus in Fig. 1 and the amygdala in Fig. 2. Manual tracing of the hippocampus was performed by a single expert rater (YX) with experience performing over 500 hippocampal tracings (Lewis, 2005; Provenzale et al., 2008). Intrarater reliability for YX and a second expert rater, DVL, were measured for hippocampal volume measurement in a separate dataset using the intraclass correlation coefficient (ICC). Hippocampal volumes measured by DVL and YX had interrater ICCs of 0.89 for both sides together, 0.88 for the left side and 0.90 for the right side. The ICCs ranged from 0.87 to 0.97 for intrarater reliability of hippocampal volume readings. These indicate excellent agreement between and within these two observers. Manual tracing of the hippocampus was performed using ITK-SNAP v1.4.1 (Yushkevich et al., 2006) in native space and orientation on contiguous coronal slices proceeding from the most posterior to most anterior slice.
Fig. 1.
A selection of manually segmented hippocampal slices of a representative subject.
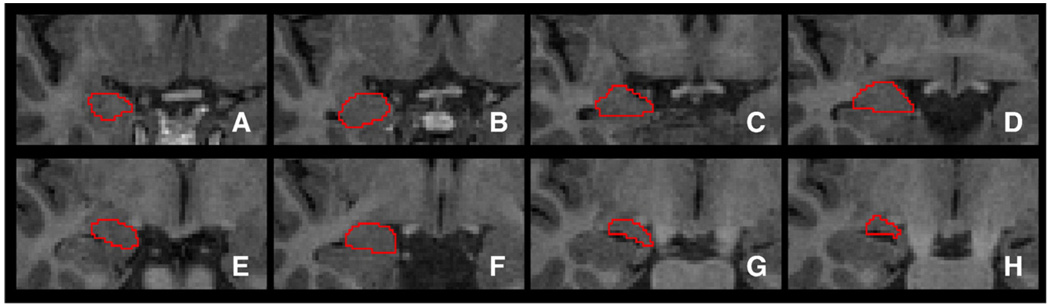
Fig. 2.
A selection of manually segmented amygdala slices of a representative subject.
Manual tracing of the amygdala was performed independently of the hippocampal tracing by rater YX with oversight by KSL. Tracing proceeded from posterior to anterior using coronally oriented slices.
The intraclass coefficients were calculated from a separate sample 47 subjects from a two-way mixed single measure coefficient, i.e. the raters, not selected at random, rated a random sample of subjects. Therefore, the raters are considered a fixed effect and the subjects are the random effect. The scans relevant to the ICC were collected on two different scanners, also different from the scanner used in the present study (see above). The first scanner was a GE 1.5 T with 3D, coronal, fast SPGR, TR/TE/flip angle=12 ms/5 ms/30°, full echo, 200 mm FOV, 1.5 mm slice, 124 slices, 256×192 matrix, 2 Nex, while the second scanner was a Siemens 1.5 T scanner with 3D, coronal, TR/TE/flip angle=12 ms/5 ms/20°, full echo, 20 cm FOV, 1.5 mm slice, 124 slices, 256×192 matrix, 2 averages. Therefore, it is likely that the higher resolution and enhanced contrast to noise resulting from improved scanner hardware and software in the present study would lead to higher ICC. The typical variation observed in automated segmentation of the amygdala with FIRST and FreeSurfer, and the hippocampus with FIRST was far greater than the variation associated with the quoted ICC values. Only the segmentation of the hippocampus with FreeSurfer yielded variation that was in the neighborhood of the ICC values.
Automated segmentation with FIRST and FreeSurfer
Automated segmentation of amygdala and hippocampus was performed using FIRST (FSL v4.0.1) which uses a Bayesian probabilistic approach. The shape and appearance models in FIRST are constructed froma library of manually segmented images. The manually generated labels are parameterized as surface meshes and then modeled as a point distribution. Using the learned models, FIRST searches through shape deformations that are linear combinations of the modes of variation to find the most probable shape instance given the observed intensities from the input image. Using T1 images with NIFTI headers in LAS orientation, the segmentation was performed with two-stage affine transformation to standard space of MNI 152 at 1mmresolution. The first stage was a standard 12 degrees of freedom registration to the template and the second stage applied 12 degrees of freedom registration using an MNI152 subcortical mask to exclude voxels outside the subcortical regions. Boundary voxels were thresholded at z=3, along with the recommended number of modes (iterations) for the hippocampus (30) and amygdala (50). The boundary voxel threshold is an important parameter that represents the z-score of the amount of noise in the boundary voxels. Thus a boundary voxel threshold of z=2 has less noise and therefore is stricter or more conservative than a boundary threshold of z=3. FIRST includes these boundary voxels as part of the segmented region. To our knowledge, there is little guidance from the FSL designers about the appropriate use or selection of a boundary threshold. Given that on average, FIRST overestimated the volume of both the hippocampus and amygdala relative to manual tracing, it seemed plausible that using a stricter boundary threshold might yield a volume that is more consistent with manual tracing. The main analysis was performed with threshold of z=3, but a secondary analyses with threshold of z=2 is included in Table 1. The only non-default option was the inclusion of a neck mask for improving the performance of the registration step with our T1 images. Overlap with neighboring structures was included to be consistent with our approach of independent manual tracing of hippocampus and amygdala. Segmented labels were automatically transformed back to native space by FIRST using the inverse transformation matrix derived in the initial registration step.
Table 1.
Comparison of automated measures to manual tracing
| Automated measure | Average volume±SD |
% Volume overlap | % Volume difference | Correlation with manual segmentation |
||
|---|---|---|---|---|---|---|
| Left | Right | Left | Right | |||
| Hippocampus | ||||||
| FreeSurfer | 4190±526.7 | 82%±1.5 | 82%±2.8 | 4%±2.1 | 5%±1.7 | r=0.82, y-intercept=496 |
| FSL-FIRST (threshold=2) | 4193±634.9 | 79%±3.6 | 80%±2.9 | 4%±2.4 | 4%±2.3 | r=0.66, y-intercept=1502 |
| FSL-FIRST (threshold=3) | 4843±743.4 | 78%±3.8 | 79%±2.8 | 6%±3.4 | 7%±2.3 | r=0.66, y-intercept=1480 |
| FSL-FIRST (version 4.1) | 4404±730.1 | 77%±5.9 | 80%±2.4 | 5%±3.2 | 5%±2.8 | r=0.66, y-intercept=1480 |
| Amygdala | ||||||
| FreeSurfer | 1945±266.5 | 75%±3.2 | 72%±4.0 | 7%±3.0 | 9%±2.7 | r=0.56, y-intercept=667 |
| FSL-FIRST (threshold=2) | 1469±243.8 | 73%±3.2 | 72%±4.5 | 3%±2.4 | 4%±3.2 | r=0.35, y-intercept=1024 |
| FSL-FIRST (threshold=3) | 1591±254.7 | 73%±3.2 | 73%±4.9 | 4%±3.1 | 5%±3.4 | r=0.24, y-intercept=1121 |
| FSL-FIRST (version 4.1) | 1526±295.2 | 70%±9.6 | 70%±6.6 | 5%±5.8 | 6%±4.9 | r=0.28, y-intercept=1125 |
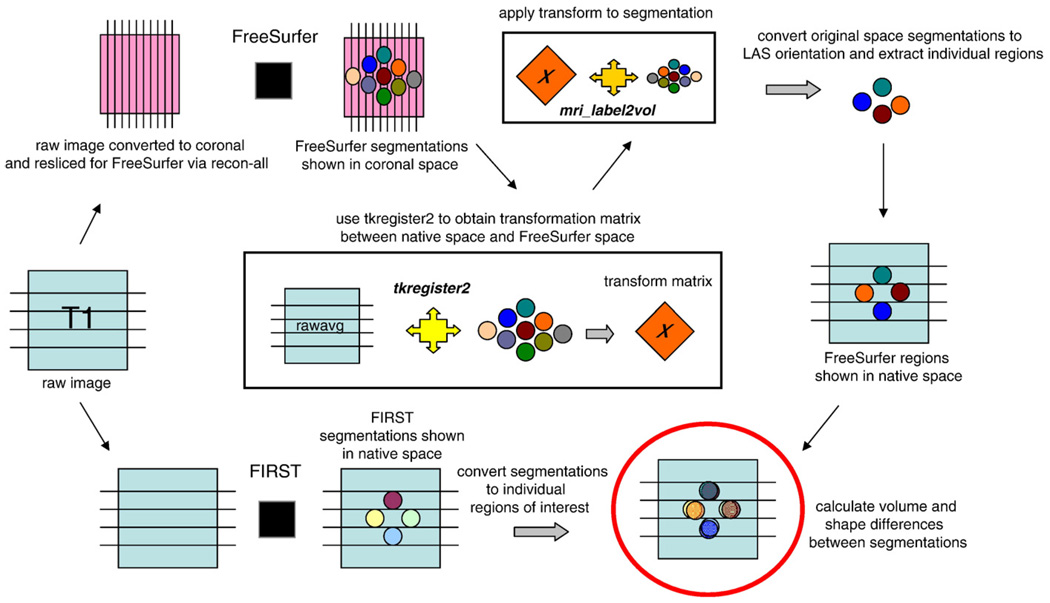
Automated segmentation and labeling of amygdala and hippocampus was also performed by FreeSurfer (v4.0.5) which utilizes an affine rigid linear transformation and combines information about voxel intensity relative to a probability distribution for tissue classes with information about the spatial relationship of the voxel to the location of neighboring structures obtained from a manually labeled atlas. Details of FreeSurfer subcortical segmentation are described in Fischl et al. (2002). Segmented labels were returned to native space using the FreeSurfer library function mri_label2vol and the transformation matrix generated by tkregister2 which minimizes the distortion introduced by interpolation. Individual areas were extracted from the large segmentation volume that contains all the regions of interest. The analysis pipeline of transformations for segmentation and reverse transformations to native space for volumetric comparisons are diagrammed in Fig. 3. (Analysis commands and scripts are detailed at http://fourier.biac.duke.edu/wiki/doku.php/mirecc:mireccanat)
Fig. 3.
Method for transforming brains from native space to standard space to perform segmentation with FreeSurfer and FIRST and then transform back to native space for volume and shape comparisons. The function tkregister2, available in the FreeSurfer library, performs transformation with minimal interpolation which typically leads to distortion. Therefore regions retain their original dimensions.
Analysis of automated segmentation performance
To validate the success of the automated segmentation procedure, we compared the results with manual tracing, the assumed reference standard, using a dataset of 20 brains. The automated segmentation methods were compared to manual tracing using the following criteria (i) percent volume overlap or Dice's coefficient as defined in Eq. 1 (ii) percent volume difference as defined in Eq. 2 (iii) correlation between automated measures and manual tracing, and (iv) 3-D shape difference analysis. Given two different labelings of a structure, L1 and L2, and a function V(L), which takes a label and returns its volume, the percent volume overlap is given by:
| (1) |
For identical labelings, O(L1,L2) achieves its maximum value of 100, with decreasing values indicating less perfect overlap. Note that the overlap between two different labelings will be reduced by slight shifts in the spatial location of one label with respect to another. Given that many neuroanatomical studies are only interested in quantifying volumetric changes, we quantified volume difference between two labelings that is insensitive to spatial shift.
| (2) |
For labels with identical volume, D(L1,L2) achieves its optimal value of zero, with increasing values indicating a greater volume difference between the two labelings. Note that greater values of D(L1,L2) lead directly to reduced statistical power to detect subtle volumetric changes in subcortical structures.
The performance was compared using Multivariate Analysis of Variance (MANOVA) on two dependent variables, volume overlap and volume difference, with 3 factors: region (2 levels: amygdala, hippocampus), method (2 levels: FIRST, FreeSurfer), and hemisphere (2 levels: left, right).
Correlation of automated segmentation measures with manual tracing
To measure the ability of automated methods to capture the true variability in volume within a group, we computed Pearson correlation. We computed correlations between manual tracing and both the automated segmentation methods. A strong correlation yields small volumes for small structures and large volumes for large structures. Again, we assume that volumes calculated by manual tracing are closest to the true volumes for a given structure. The intercept of the fitted line provides information about systematic differences in volume estimates between measures.
Bland–Altman plots offer another approach to assessing agreement between measures. Bland–Altman analyses provide information about the interchangeability of two measures without assuming that either is the gold-standard and can be of value in clinical settings.
Assessing systematic shape bias between methods with 3D contour maps
We computed group-level difference maps between manual tracing and FreeSurfer and FIRST. Difference maps were created by SPHARM shape analysis tool v1.5 [UNC NeuroImage Analysis Laboratory (NIAL), Chapel Hill, NC]. The input — segmentations of the hippocampus and amygdala — were converted into a corresponding spherical harmonic description (SPHARM), and then sampled into triangulated surface meshes, and aligned. Differences between groups' surface characteristics were then computed using the Hotelling's T2 for two samples metric. The comparisons included (i) a significance map showing p-values corrected using permutation testing which provides a conservative (comparable to Bonferroni correction) and tractable approach for handling the multiple comparisons problem (Pantazis et al., 2004), (ii) a mean difference map thresholded by the distance between corresponding vertices on the mesh of the reference group and the comparison group, (iii) vectors superimposed on the difference maps that show the directionality between corresponding vertices, and (iv) ellipsoids superimposed on the difference map that show variance in the comparison group where the dimensions of the major axes (x, y, z) of the ellipsoid conveys the magnitude of variance (Styner et al., 2006). Difference maps and significance maps were computed for the hippocampus and amygdala between the reference group (manual tracing) and each of the comparison groups (FreeSurfer, FIRST).
Sample size estimation
Sample size estimates were computed for manual tracing, FreeSurfer, and FIRST segmentation of the amygdala and hippocampus to detect volumetric differences between a hypothetical reference group and a comparison group for a range of effect sizes (0.1 to 0.9) assuming a power level of 0.8, significance level of 0.05, and normal distributions in both groups. We estimated variance by computing the standard deviation while effect size was computed based on the standard deviation for manual tracing. The first step in the sample size estimation was to calculate adjusted volumes for each method by removing the influence of the actual mean of the group and retaining only the variance of the group. Thus, the adjusted volume was calculated by subtracting the mean volume of the method from each observation resulting in a fitted line with a zero-intercept. Using the adjusted volume, power was calculated based on a non-inferiority test of the difference of two means using the Power Analysis and Sample Size (PASS) software [NCSS: Kaysville, Utah USA] (Hintze, 2005). Note that since the empirically derived variances were different for each method, an effect size of 0.1 for manual tracing corresponded to a slightly different effect size for FreeSurfer and FIRST. Thus, the sample size requirements are provided for each method to detect an effect size of 0.1 or volume difference of 45.9 mm3 (ES×SDmanual=0.1×459), and so on for effect sizes up to 0.9 in increments of 0.1. For example, 45.9 mm3 corresponds to an effect size of 0.1 for manual tracing but an effect size of 0.092 for FreeSurfer based on a standard deviation of 501, and an effect size of 0.067 for FIRST based on a standard deviation of 688.
As a proof of concept, we examined hippocampal and amygdala volumes in an existing dataset of subjects with Major Depressive Disorder (MDD) to compare results from FreeSurfer and FIRST. The subject group was comprised of veterans who were enrolled in the Durham VA Medical Center between 2006 and 2008 using the scanning protocol described above. Participants were assessed for MDD using the Structured Clinical Interview for DSM-IV Axis I Disorders (SCID-I) (First et al., 2002). Nine participants with MDD, based on SCID diagnosis, were compared to 48 non-depressed individuals. To account for the unequal sample size in the control group, a subset of 10 non-depressed individuals were randomly selected for comparison to the nine participants with MDD (demographic characteristics described in Table 1).
Results
The major results of analyses are summarized in Table 1.
Performance of methods based on volume overlap
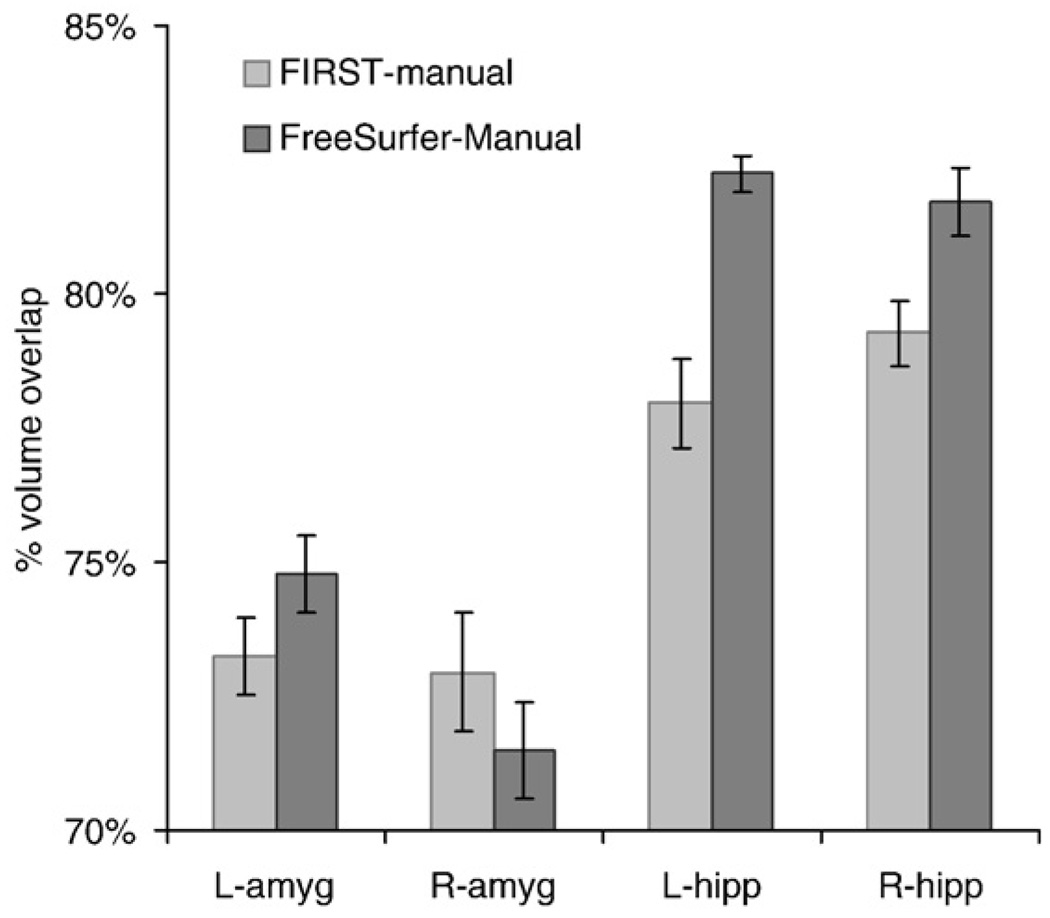
As assessed by volume overlap (Dice's coefficient), FreeSurfer was superior to FIRST for hippocampal segmentation but not for amygdala segmentation as seen in Fig. 4; significant interaction of region*method [F(1,19)=16.9; p<0.001]. The volume overlap for FreeSurfer and manual tracing was greater than for FIRST and manual tracing in the left and right hippocampus [t(40)=5.3, p<10−6]. There were no corresponding differences between methods for amygdala segmentation [t(40)=0.05; p>0.5]. Percent volume overlap was greater for the hippocampus than the amygdala regardless of the measurement method [t(40)=12.4, p<10−24].
Fig. 4.
Percent volume overlap (Dice's coefficient) between FreeSurfer segmentation and manual tracing is greater than the overlap between FIRST and manual tracing for the left and right hippocampus. In the amygdala, the overlap is not different between manual tracing and either of the two automated methods. Percent volume overlap is greater in the hippocampus than the amygdala regardless of the segmentation method.
Performance of methods based on volume difference
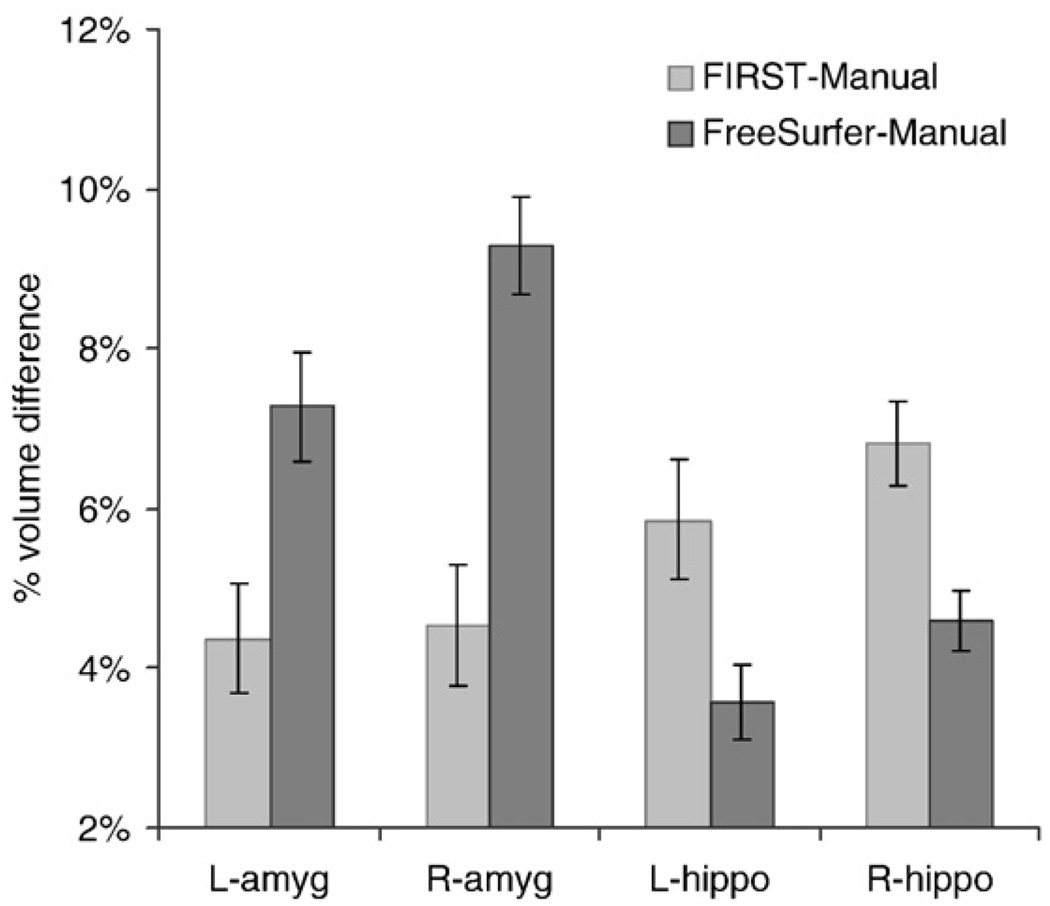
When comparing percent volume differences between automatic and manual measurements, FreeSurfer was superior to FIRST for hippocampal segmentation, but FIRST was superior to FreeSurfer for amygdala segmentation as seen in Fig. 5 (significant interaction of region*method [F(1,19)=125.6; p<0.0001]). Note that lower values of percent volume difference indicate superior performance. The volume difference for FreeSurfer was less than FIRST in the hippocampus [t(40)=4.1, p<10−4]. On the other hand, volume difference for FIRST was less than FreeSurfer in the left and right amygdala [t(40)=5.5, p<10−6]. Volume difference was smaller in the hippocampus than the amygdala; main effect of region [t(40)=2.3, p<0.02].
Fig. 5.
In the left and right hippocampus, the percent volume difference relative to manual tracing, is smaller with FreeSurfer than FIRST. In the left and right amygdala, the volume difference with manual tracing, is smaller with FIRST than FreeSurfer. There was a smaller FreeSurfer-Manual volume difference in the L-amygdala than the R-amygdala. No other laterality differences were detected.
A comparison of volume differences between automated and manual segmentation showed greater FreeSurfer-Manual volume difference in the L-amygdala than the R-amygdala [t(40)=2.2, p<0.05]. However, no FIRST-Manual volume difference was detected between the left and right amygdala [t(40)=0.16, p>0.8]. Comparing the left and right hippocampus, we failed to detect a Freesurfer-Manual volume difference [t(40)=1.1, p=0.3], or a FIRST-Manual volume difference [t(38)=1.7, p=0.1].
Correlation of automated segmentation methods
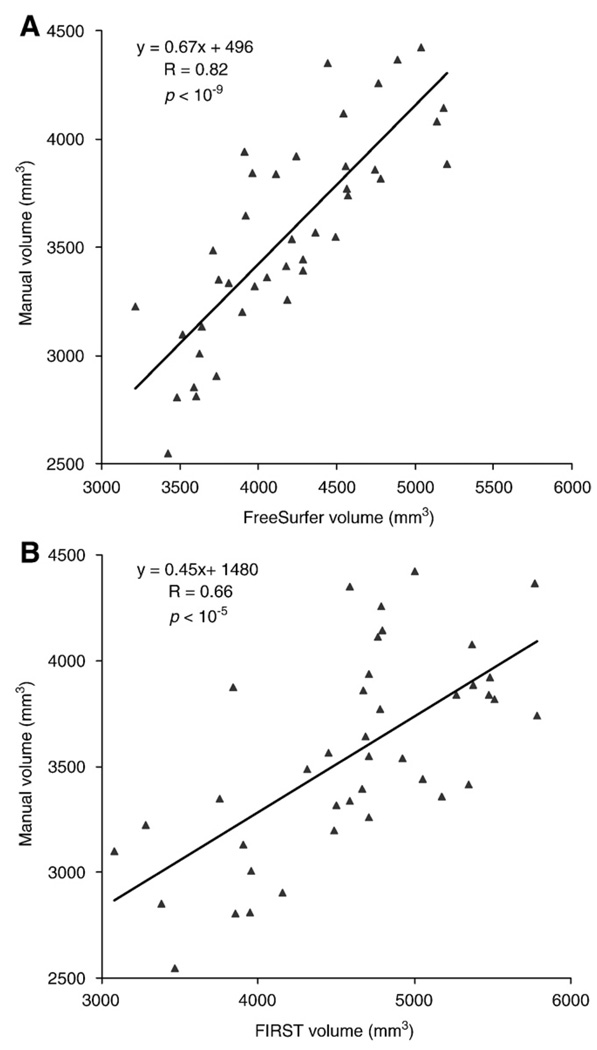
The correlation of hippocampal volume between FreeSurfer and manual tracing (R=0.82, p<10−9) was higher than the correlation between FIRST and manual tracing (R=0.66, p<10−5) (Fig. 6). Both automated methods yielded larger hippocampal volumes relative to manual segmentation.
Fig. 6.
(A) Hippocampal volume derived from FreeSurfer segmentation is highly correlated with manual tracing (R=0.82; p<10−9). (B) Hippocampal volume derived from FSL/FIRST segmentation is correlated with manual tracing (R=0.66; p<10−5).
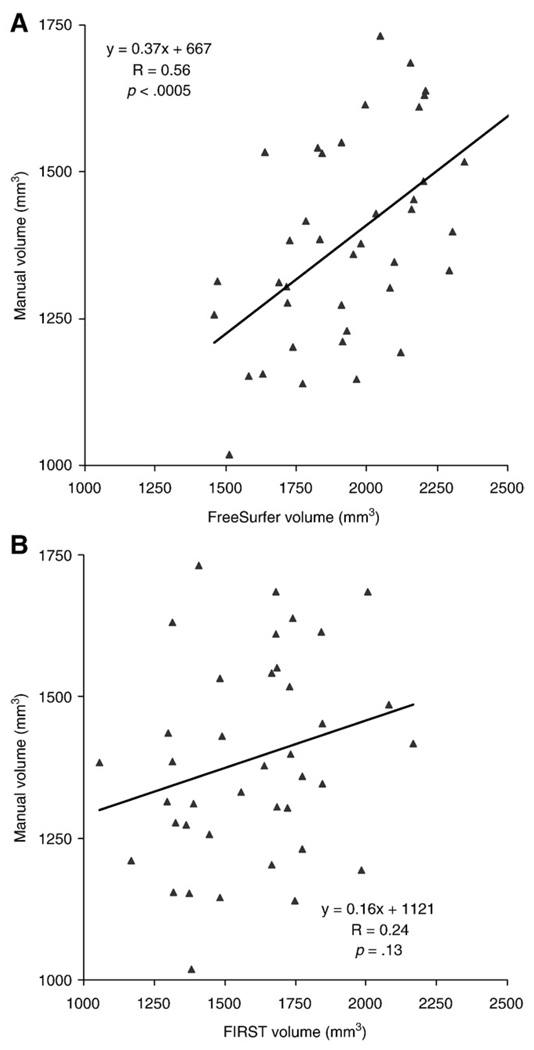
The correlation of amygdala volume between FreeSurfer and manual tracing (R=0.56, p<0.0005) was higher than the correlation between FIRST and manual tracing (R=0.24, p>0.13) (Fig. 7). Both automated methods yielded larger amygdala volumes relative to manual segmentation.
Fig. 7.
(A) Amygdala volume derived from FreeSurfer is correlated with manual tracing (R=0.56; p<0.0005). (B) Amygdala volume derived from FSL/FIRST was poorly correlated with manual tracing (R=0.24; p>0.13).
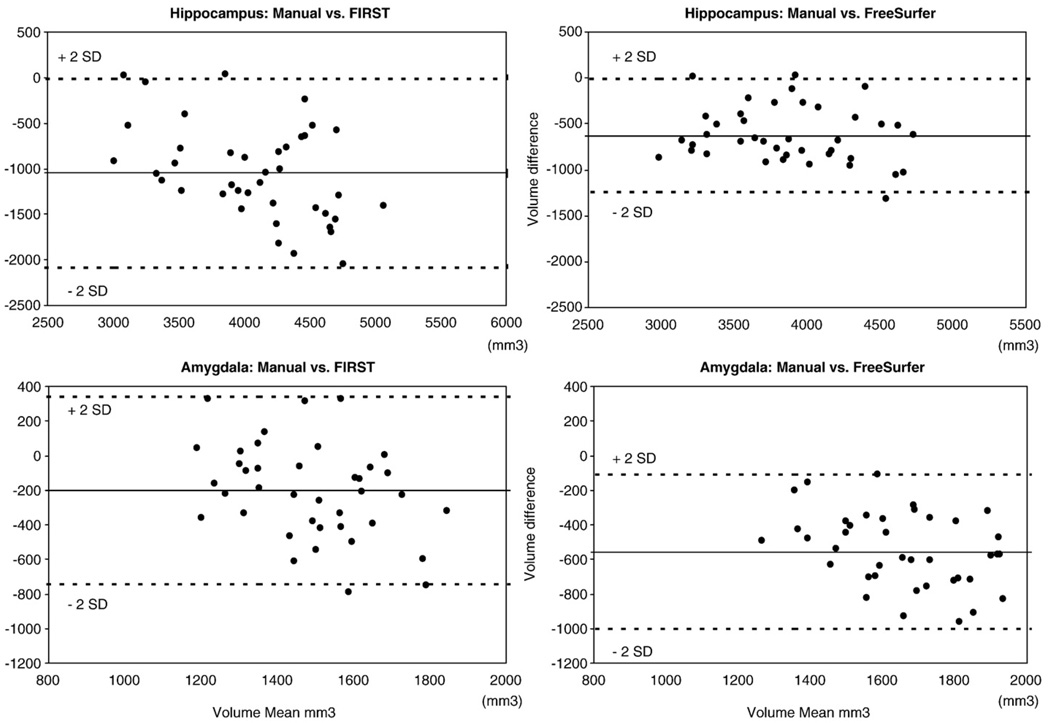
The Bland–Altman plots (see Fig. 8) confirm that both automated methods generated systematically larger volumes than manual tracing. We also examined the extent to which the automated measures performed poorly on the same images by comparing subjects whose volume differences approached or exceeded 2 SDs. Across the four plots, there were five data points showing an overestimation of automated volume compared to manual volume. However, these five data points represented images from five unique subjects, confirming that the automated techniques were not giving their worst performance on the same images. Thus, we find no evidence that gross anatomic anomalies that were unduly influencing results.
Fig. 8.
Bland–Altman mean difference plots for hippocampal and amygdala volumes.
Shape analysis of segmentations
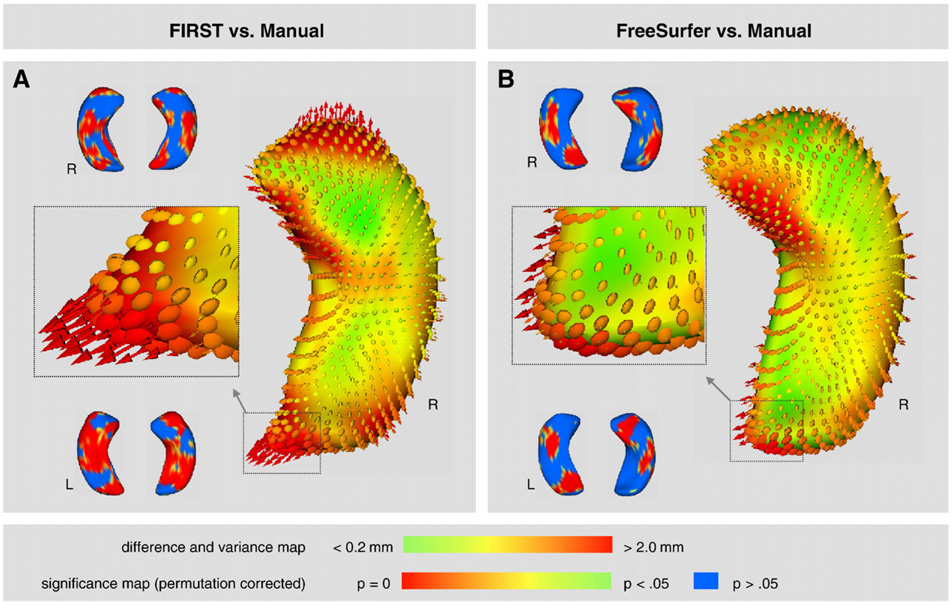
Group averaged 3D shape renderings generated for shape analyses demonstrate that FreeSurfer had better overall performance than FIRST in segmenting the hippocampus, particularly in the head and tail regions that are especially challenging. Difference maps (with variance) and significance maps for the hippocampus show shape differences between FIRST and manual tracing (Fig. 9A), and shape differences between FreeSurfer and manual tracing (Fig. 9B). The difference map for FIRST reveals that the head and tail had the largest shape difference and greater variance indicating that much of the inflated volume estimates of FIRST originate from extended surface estimations in the head and tail regions. The difference map for FreeSurfer shows the anterior-medial surface had prominent shape difference and some increased variance indicating that this region was the major source the inflated volume estimates from FreeSurfer when compared to hand tracing. Though ellipsoids indicate increased variance in the tail region, the difference maps indicate that the mean difference between surfaces is relatively small (suggested by the green color of the tail section). Significance maps (permutation corrected, p<0.05) confirm prominent shape differences between FIRST and manual tracing as well as between FreeSurfer and manual tracing. The latter comparison revealed less widespread shape differences providing additional evidence that FreeSurfer performed favorably relative to FIRST in the hippocampus.
Fig. 9.
Shape analysis of the hippocampus in 3-D where difference maps show the distance between segmentation contours thresholded from 0.2mmto 2.0 mm. Vectors indicate the directionality between the corresponding mesh vertices of the two segmentation methods. Ellipsoids indicate the [x, y, z] components of variance introduced by the automated segmentation. Significance maps (permutation corrected, p<0.05) highlight shape differences between automated segmentation and manual tracing. (A) FIRST compared to manual tracing, and (B) FreeSurfer compared to manual tracing.
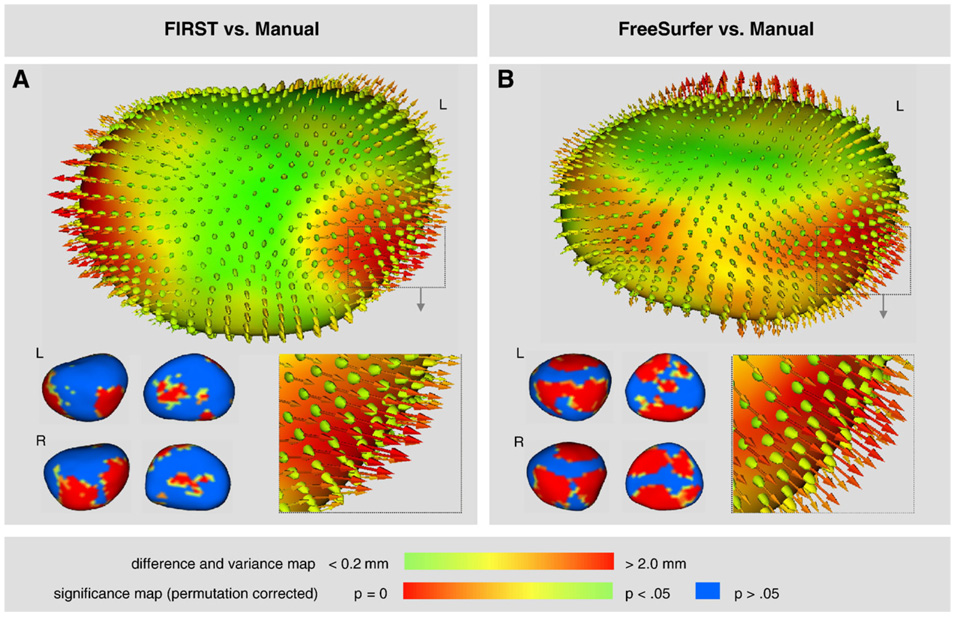
Shape analysis results suggest FIRST had better overall performance than FreeSurfer in segmenting the amygdala. Difference maps and significance maps for the amygdala show shape differences between FIRST and Manual tracing (Fig. 10A), and shape differences between FreeSurfer and Manual tracing (Fig. 10B). The maps suggest a general increase in volume in the anterior and posterior surfaces generated by FreeSurfer that is less pronounced with FIRST. This finding is consistent with the larger FreeSurfer-Manual volume difference (8.3%) than FIRST-Manual volume difference (4.5%) as represented in Fig. 5. Notably, the ellipsoids for both methods reflect the greater overall variance for the amygdala compared to the hippocampus. This is consistent with the relative variances seen in Fig. 6 and Fig. 7. Significance maps (permutation corrected, p<0.05) confirm prominent shape differences between FreeSurfer and manual tracing as well as between FIRST and manual tracing. The latter comparison revealed less widespread shape differences providing additional evidence that FIRST performed favorably relative to FreeSurfer in the amygdala.
Fig. 10.
Shape analysis of the amygdala in 3-D where difference maps show the distance between segmentation contours thresholded from 0.2 mm to 2.0 mm. Vectors indicate the directionality between the corresponding mesh vertices of the two segmentation methods. Ellipsoids indicate the [x, y, z] components of variance introduced by the automated segmentation. Significance maps (permutation corrected, p<0.05) highlight shape differences between automated segmentation and manual tracing. (A) FIRST compared to manual tracing, and (B) FreeSurfer compared to manual tracing.
Sample size estimation
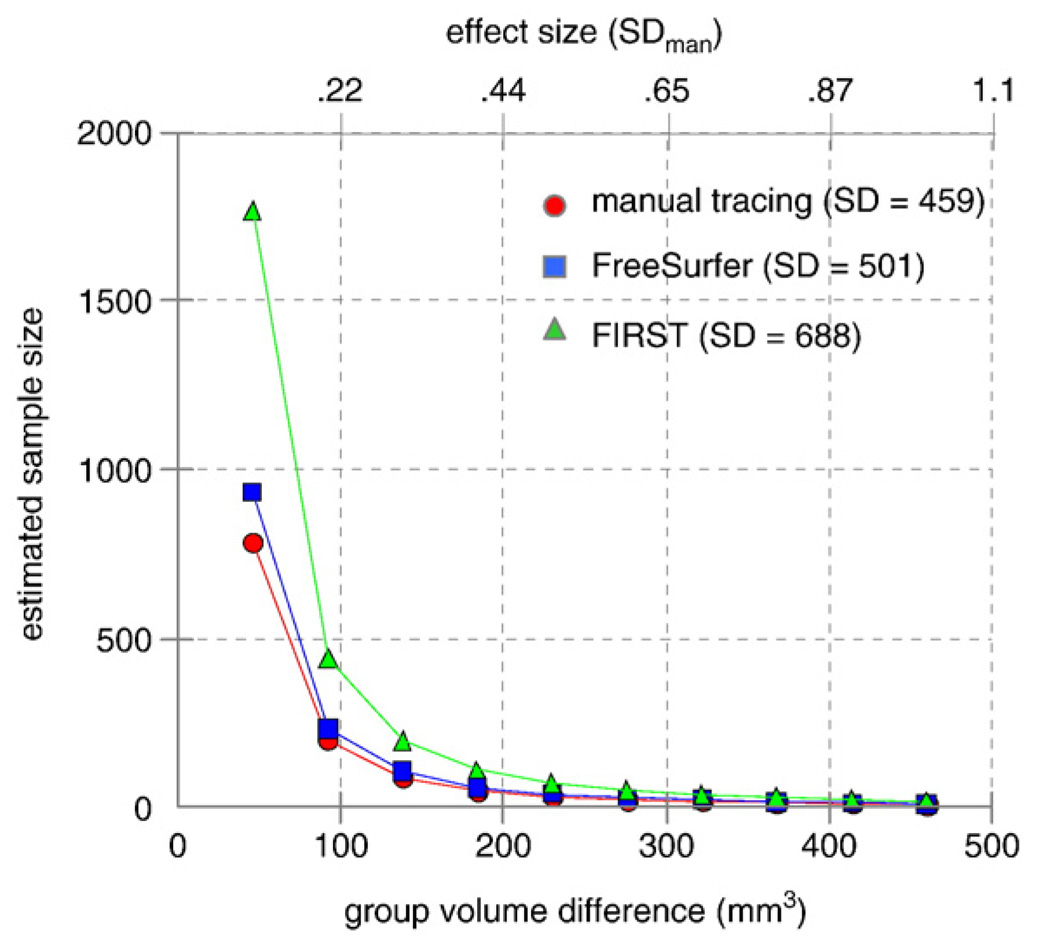
Based on the additional variance introduced by FreeSurfer and FIRST methods relative to manual tracing of the hippocampus, FreeSurfer requires a relatively small increase in sample size over hand tracing for the entire range of effect sizes. On the other, hand a substantial increase in sample size is required if FIRST is used for measurement compared to hand tracing. For example, for an effect size of 0.9, which reflects a change in volume of 414 mm3 (ES×SD=0.9×459) or about 12% of typical hippocampal volume (414/3560), would require a per group sample size of n=12 for manual tracing, n=14 for FreeSurfer, and n=24 for FIRST. Sample size estimates for a range of effect sizes (power=0.8; alpha=0.05) are shown in Fig. 11 for each method.
Fig. 11.
FreeSurfer segmentation had the power to detect differences in hippocampal volume between groups for a range of effect sizes (power=0.8; alpha=0.05) with negligible increase in sample size relative to manual tracing. On the other hand, FIRST segmentation required a modest increase in sample size particularly to detect relatively small effects.
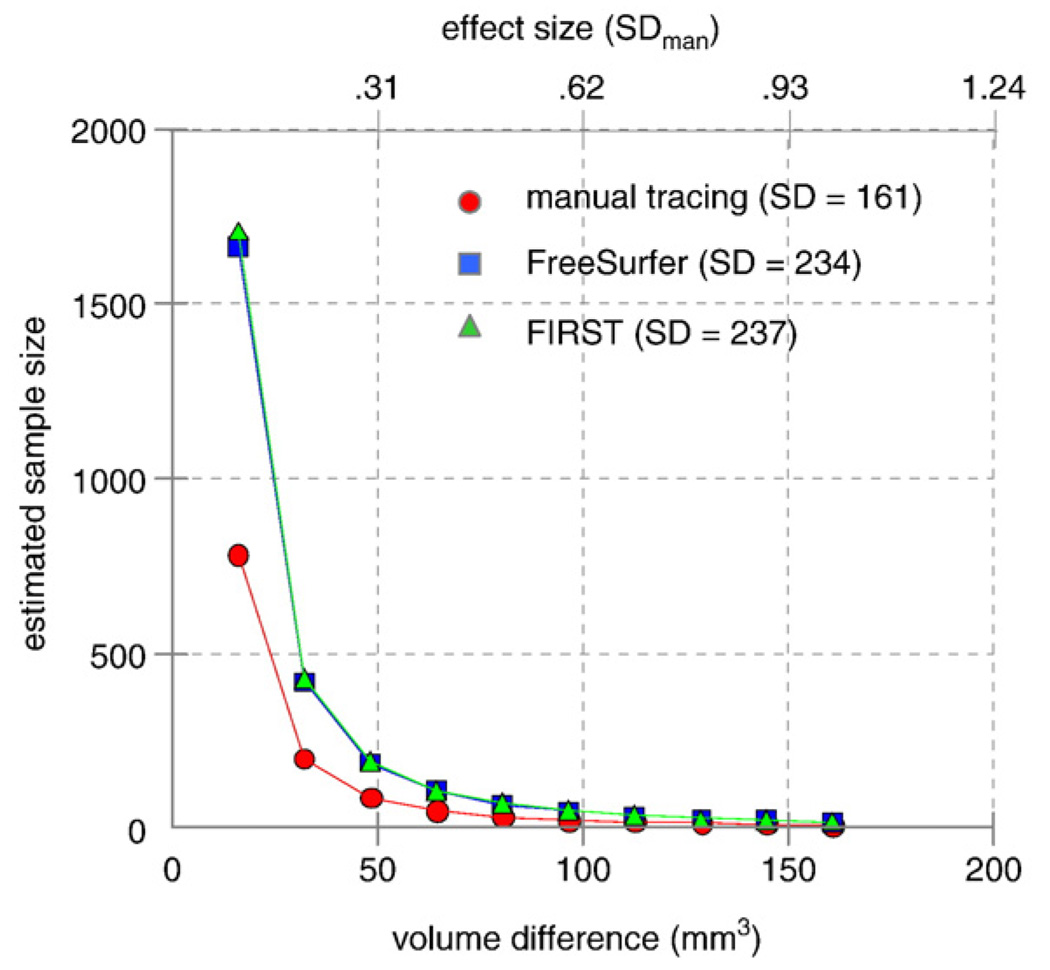
The sample size estimates for amygdala measurements show that both FreeSurfer and FIRST required considerably larger numbers of subjects relative to manual tracing. For example, for an effect size of 0.9, which reflects a change in volume of 145 mm3 (ES×SD=0.9×161) or about 10% of typical amygdala volume (145/1389), would require a per group sample size of n=12 for manual tracing, n=23 for FreeSurfer, and n=24 for FIRST. Sample size estimates for a range of effect sizes are shown in Fig. 12 for each method.
Fig. 12.
FreeSurfer and FIRST segmentation had roughly equal power in detecting differences in amygdala volume between groups for a range of effect sizes (power=0.8; alpha=0.05 but, both methods required a modest increase in sample size relative to manual tracing, particularly to detect relatively small effects. Note that sample size estimates were derived solely from standard deviation with correlation having no role in the estimation process.
It is important to note that standard deviation was the determining factor in estimation of sample size. Both automated segmentation methods introduced additional variance in measures of total volume relative to manual segmentation. For hippocampal measures, FreeSurfer (SD=501) and manual tracing (SD=459) introduced less variance than FIRST (SD=688). For amygdala measures, approximately equal variance was introduced by FreeSurfer (SD=234) and FIRST (SD=237) but, both were greater than manual (SD=161).
Utility of methods in major depression
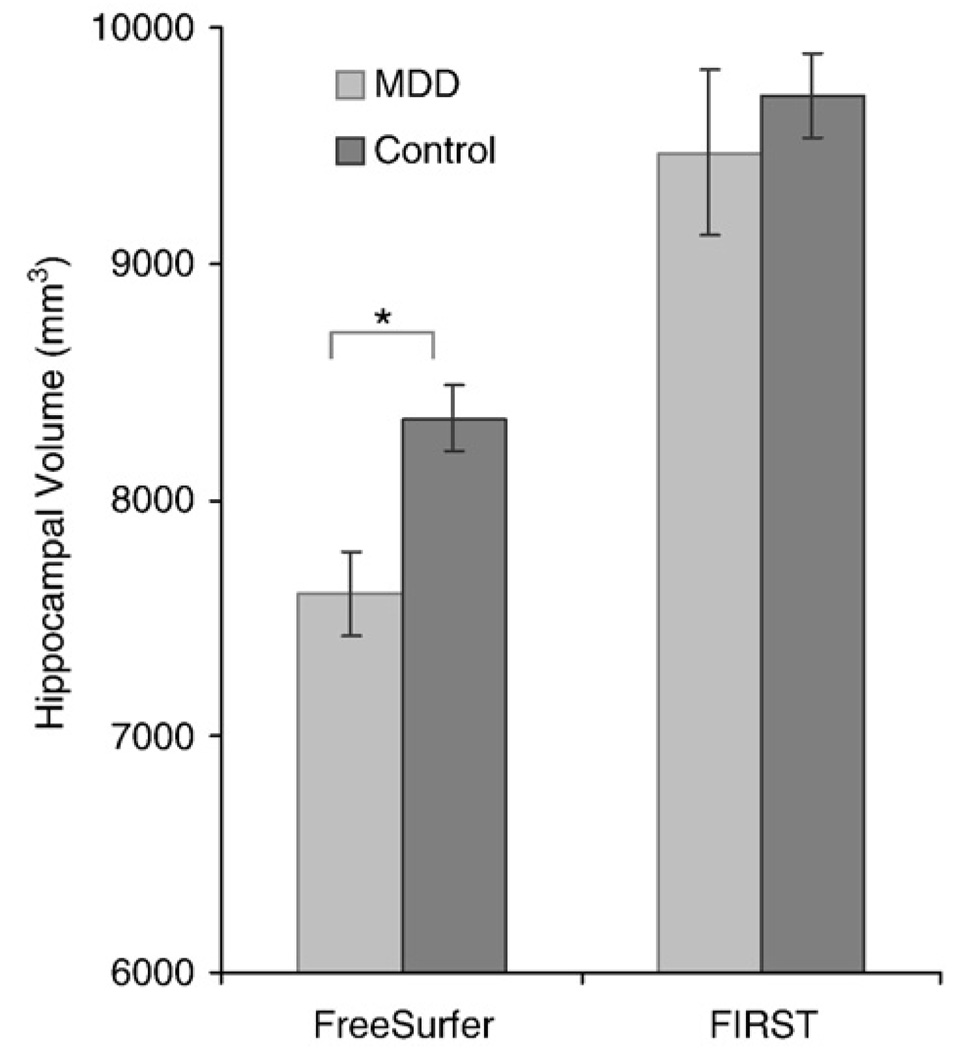
Analysis of hippocampal and amygdala volumes in MDD showed that hippocampal volume, measured by FreeSurfer, was reduced in depressed patients relative to controls (t(55)=2.22, p<0.04) but not when these same hippocampi were measured by FIRST (t(55)=0.54, p>0.59) (see Fig. 13). The difference in volume between the MDD and control groups as measured by FreeSurfer was about 9%, consistent with published studies (Videbech and Ravnkilde, 2004). Given the large disparity in sample size between MDD and control groups, we randomly selected 10 participants from the control group to serve as secondary reference group (see Table 2). Consistent results were obtained showing lower hippocampal volumes associated with depression using FreeSurfer (t(17)=2.14, p<0.05) but not FIRST (t(17)=0.57, p>0.57). The MDD and control groups did not differ in total cerebral volume (t(17)=0.05, p>0.96). Neither method showed differences in amygdala volume associated with depression.
Fig. 13.
FreeSurfer showed a 9% reduction in hippocampal volume for participants diagnosed with major depressive disorder (MDD) compared to a matched Control Group. However, FIRST did not show differences between MDD and Control groups.
Table 2.
Demographic and clinical characteristics of MDD and control groups
| Characteristic | MDD (n=9) | Control (n=10) | t/Chi square | p |
|---|---|---|---|---|
| Age (years), [std dev] | 37.2 [8.5] | 35.4 [11] | 0.40 | >0.69 |
| Gender, no. (%) of females | 3 (33) | 4 (40) | 0.90 | >0.76 |
| Handedness, no. (%) right-handed | 7 (78) | 9 (90) | 1.20 | >0.54 |
| Ethnicity, no. (%) of Caucasian subjects | 5 (56) | 3 (30) | 1.27 | >0.25 |
| Education (years), [std dev] | 13.6 [0.9] | 14.4 [1.2] | 1.76 | >0.09 |
Discussion
FreeSurfer was superior for segmenting the hippocampus by all of the objective measures we utilized, including volume overlap with manual tracing, volume difference with manual tracing, correlation to manual tracing, sample size estimation, and group-level 3-D shape analysis. However, assessment of amygdala segmentation was more equivocal. FreeSurfer and FIRST had comparable volume overlap and sample size estimates, while FIRST had smaller volume difference. FreeSurfer volumes correlated more strongly with manual tracing, suggesting higher validity, although for study of amygdala morphometry, FIRST may be preferable. The correlation of amygdala volumes between FIRST and hand tracing was not significant. Both methods overestimate the amygdala volume with larger overestimates by FIRST than FreeSurfer. These analyses were repeated using Version 4.1 of FIRST, released shortly after the initial submission of this paper, showed very similar results (summarized in Table 1).
We estimate that in group comparison studies of hippocampal volume, FreeSurfer can be used with only a marginal increase in the sample size relative to manual tracing. By comparison, the FSL/FIRST method performs less favorably with a substantially greater variance than the manual segmentation. FreeSurfer showed a 9% reduction in hippocampal volume for participants diagnosed with MDD, while FSL/FIRST did not show differences between MDD and control groups. These findings provide evidence for the utility of FreeSurfer in detecting differences in hippocampal volume associated with neuropsychiatric disorders. On the other hand, sample size estimates of amygdala segmentation show that both FreeSurfer and FIRST require appreciably greater numbers of subjects relative to manual tracing. It is interesting to observe that while the power calculation shows negligible change in sample size, FreeSurfer provides a volume measure of amygdala that varies among subjects similarly to hand tracing. Thus, in evaluating the automated methods, it is important to consider the combined merits of the entire constellation of performance metrics, as focusing on a specific metric might be misleading.
It is notable that both automatic methods provided larger volume estimates for both hippocampus and amygdala than hand tracing. Systematic differences in segmentation contours between automated and manual methods were expressed as group difference maps rendered in 3-D space. The hippocampal shape analysis showed that when compared to manual tracing, FreeSurfer performed better than FIRST, which tended to inflate surfaces in the head and tail regions. The amygdala shape analysis showed that when compared to manual tracing, FIRST performed better than FreeSurfer which tended to inflate generally over the entire amygdala but particularly in the anterior and posterior surfaces. The maps highlight significant discrepancies in the surface contours generated by the automated methods that may guide users interested in performing “post hoc editing” targeted at areas with systematic biases. It is possible that surface inaccuracies are introduced by conversion of the surface to an image and back again to a surface. Super-sampling the original surfaces for spherical harmonics is one approach to mitigating possible distortions, although these distortions are expected to be minimal.
We conclude that FreeSurfer is a reasonable substitute for manual tracing of the hippocampus and may be an acceptable substitute for the amygdala, depending on the scope of the study. Recent findings comparing hippocampal segmentation by FreeSurfer to manual tracing as well as other automated measures support this conclusion. There is evidence for accurate diagnostic classification of patients with temporal lobe epilepsy using FreeSurfer (McDonald et al., 2008) and greater agreement with manual tracing relative to automated segmentation of the hippocampus using the Individual Brain Atlases Statistical Parametric Mapping (IBASPM) (Tae et al., 2008). However, users should be aware that volume estimates of these regions are consistently inflated relative to manual tracing. FreeSurfer could be especially beneficial in studies where a large cohort of subjects is required based on an expected effect size that is small and/or the association with multiple or noisy outcome variables. The complexity can be further aggravated by outcome variables such as behavioral differences and the role of genetics. Longitudinal multi-site studies are particularly well suited for adoption of an automated approach such as FreeSurfer. In these studies, maintaining consistency across raters at centers that are geographically distant over long durations where staff transitions are commonplace may be especially challenging. The sample size estimates we provide may guide investigators in the design and feasibility of structural MRI studies.
Additional techniques have recently been proposed that may improve upon the volumetric output provided by FreeSurfer, including combining FreeSurfer with large deformation diffeomorphic metric mapping (LDDMM) (Khan et al., 2008). There is some evidence that combining these methods increases segmentation reliability and accuracy compared to using FreeSurfer alone for regions such as the caudate nucleus and putamen. However, the evidence for improvement of hippocampal estimates was less convincing, particularly for older adults. This suggests that combining the methods may be useful for some structures, but not all. Researchers may therefore need to weigh the potential benefit of combining the two methods against the limitation of added computational time required to implement LDDMM.
Limitations
One important source of variance that can influence the outcome of segmentation is associated with the MRI scanner. The possibility that scan–rescan differences may confound results was investigated in five subjects that were scanned 7–9 days apart using the same acquisition sequence and scanner. The scan–rescan segmentation by FreeSurfer (see Supplementary data, Figure S1) showed consistent results in the hippocampus and slightly less consistent results in the amygdala. The scan–rescan segmentation by FIRST (see Figure S1) showed less consistent results in the hippocampus and the amygdala. It is interesting to note however, that the rescan values are not identical, suggesting that small differences in image contrast, ostensibly in boundary voxels, can result in the segmentation algorithms computing non-trivial differences in volume. Although these results are preliminary, this issue warrants systematic investigation in a much larger scan–rescan sample. We expect more robust reproducibility with a larger sample that more accurately captures scanner associated variance. Nonetheless, this information highlights the importance of MRI scanner quality assurance and careful technique calibration.
While manual tracing was our reference standard, it is unknown how well manual tracing itself represents the true volume of the structures examined particularly because there are several difficulties inherent in this method. For example, manual labeling may lead to inconsistent labeling across slice directions (Fischl et al., 2002). Post-mortemvolume assessment may provide the closest possible estimate of true volume but is clearly impractical. Therefore, for this study we assume, in a manner consistent with the literature, that manual tracing is an accurate representation of the true boundaries, and that these boundaries can be discerned by objective means. Finally, it may also be the case that even post-mortem examination is limited, e.g. a transition layer where cells share features of both structures. In this case, conventions are established for defining boundaries based on anatomical landmarks or some other feature(s). These issues border on the larger issues of epistemology faced by all methods of observation and measurement in scientific inquiry. These limitations may be partially overcome in the future with the promise of high-field (7 T) structural MRI capable of revealing details at ~100 µm in-plane resolution. Finally, these findings are reflective of the MR acquisition sequence used in the present study and may not fully generalize to other scanners or sequences.
Conclusion
We found that FreeSurfer and FIRST are not equal when compared to manual tracing and we provide practical information for making decisions in the choice of segmentation tools depending on the scope of the study. Based on converging data from shape and volume measures, we conclude that FreeSurfer generally is preferred to FIRST for automated segmentation of the hippocampus, with results from the amygdala being less robust and more equivocal across automated methods.
Supplementary Material
Acknowledgments
We gratefully acknowledge assistance from Douglas Greve, Ph.D., David N. Kennedy, Ph.D., Joshua Bizzell, M.S., Jeff Hoerle, M.S., Nicolas Augier, B.S., Lihong Wang, Ph.D., and Srishti Seth, B.S.
Appendix A
Manual tracing of the hippocampus
Boundaries of the tail of the hippocampus
The most posterior hippocampal slice was measured at the point where portions of both the crus of the fornix and hippocampal tissue were clearly visible, with sufficient hippocampal tissue visible to outline an ROI even though the full extent of the crus may appear on a more anterior slice. We did not trace more posterior slices where the crus could not be discerned. The superior boundary of the tail is limited by but does not include the crus and, medially, the lateral extent of the splenium. More anteriorly, the upper surface abuts the lower margin of the thalamus, which is usually a lighter shade of gray (in T1 sequences) than the hippocampal tissue.
Boundaries of the body of the hippocampus
CSF defines the lateral and superior boundaries in the ventricle and ambient cistern, except near the tail where the superior surface is the interface between hippocampus and inferior thalamus. We did not include the fimbria outline in the tracing, and the boundary between fimbria and hippocampal tissue was defined by a smooth line following the curve of the alvear surface of the hippocampus. The inferior boundary was defined inferiorly by the interface of the subicular gray matter and the underlying white matter. The medial limit of subicular/presubicular gray was defined by a line drawn from the superior portion of the most medial extent of the underlying white matter across the subicular gray to the cistern. The point at which this line crossed the subicular gray to the cistern was estimated by imagining a horizontal line tangential to the surface of the subicular gray matter projecting to meet a vertical line along the surface of the parahippocampal gray. The line of crossing the subicular gray starts at the most medial and superior aspect of the underlying white matter aimed towards the intersection of the imaginary lines.
Boundaries of head of hippocampus
The limit of the anterior superior hippocampal surface and lateral surface was often defined by ventricular CSF. The lateral inferior limit was the junction of gray hippocampal tissue and the collateral white matter. The medial inferior hippocampal boundary was defined to include the subicular gray matter and was defined by the junction of the subicular gray and the underlying white matter, with the medial extent of this junction defined as above in the description of the body. In more anterior slices of the head, the uncinate gyrus was identified as it turns superiorly to join the amygdala. The most superior limit of the uncinate gyrus was defined by a tangent line to the upper surface of the hippocampal head extended to cut across the uncinate gyrus medially.
Tracing proceeded to the most anterior slice, defined as follows. ITK-SNAP allows simultaneous views of coronal, axial and sagittal planes. With this feature, the anterior superior boundary of the hippocampal head in the sagittal plane was used to mark the most anterior slice of that structure in all three planes. Also, in the most anterior portions of the hippocampus, the upper boundary of the hippocampus was often defined by the alveus or the uncal recess of the temporal horn of the lateral ventricle if it was present.
Manual tracing of the amygdala
The most posterior slice
The most posterior slice of the amygdala was that slice in which the uncinate gyrus that connects the hippocampus to the overlying amygdala, first becomes visible while proceeding in an anterior direction. Proceeding anteriorly for several slices, the inferior border of the amygdala was bounded by the CSF in the uncal recess of the temporal horn of the lateral ventricle. The superior margin was bounded medially by the CSF in the entorhinal sulcus and laterally by a line extending from the optic tract to the most inferior portion of the circular sulcus. The lateral margin was bounded by the white matter of the temporal stem.
Mid-level slices
Proceeding anteriorly, the inferior border of the amygdala was indicated by the alvear white matter on the superior surface of the head of the hippocampus. The superior border was defined by the white matter of the junction of the internal capsule and cerebral peduncle, and the lateral border by the temporal stem. The medial border was bounded by CSF in the ambient cistern and entorhinal sulcus.
Anterior slices
The tracing was continued anteriorly to the slice just posterior to the crossing of the anterior commissure where an ovoid convexity, created by following the grey matter contours in the sagittal and axial planes, forms a closed surface in the coronal plane that is bounded anteriorly by the uncus.
Appendix B. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.neuroimage.2008.12.033.
Footnotes
This research was supported by the National Institutes of Health Grant K23 MH073091 (RAM), NS 43209 from NINDS (DVL, YX), the Department of Veterans Affairs, Mental Illness Research Education and Clinical Center Grant for Post-Deployment Mental Health (RAM, CSP, JPH, GM), and the Duke Silvio O. Conte Center for the Neuroscience of Depression (P50-MH60451).
References
- Apostolova LG, Dinov ID, Dutton RA, Hayashi KM, Toga AW, Cummings JL, Thompson PM. 3D comparison of hippocampal atrophy in amnestic mild cognitive impairment and Alzheimer's disease [Erratum appears in Brain. 2007 Sep;130(Pt 9):2474] Brain. 2006;129:2867–2873. doi: 10.1093/brain/awl274. [DOI] [PubMed] [Google Scholar]
- Barnes J, Foster J, Boyes RG, Pepple T, Moore EK, Schott JM, et al. A comparison of methods for the automated calculation of volumes and atrophy rates in the hippocampus. NeuroImage. 2008;40:1655–1671. doi: 10.1016/j.neuroimage.2008.01.012. [DOI] [PubMed] [Google Scholar]
- Bremner JD, Randall P, Scott TM, Bronen RA, Seibyl JP, Southwick SM, et al. MRI-based measurement of hippocampal volume in patients with combat-related posttraumatic stress disorder. Am. J. Psychiatry. 1995;152:973–981. doi: 10.1176/ajp.152.7.973. [See comment] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremner JD, Randall P, Vermetten E, Staib L, Bronen RA, Mazure C, et al. Magnetic resonance imaging-based measurement of hippocampal volume in posttraumatic stress disorder related to childhood physical and sexual abuse—a preliminary report. Biol. Psychiatry. 1997;41:23–32. doi: 10.1016/s0006-3223(96)00162-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell S, Marriott M, Nahmias C, MacQueen GM. Lower hippocampal volume in patients suffering from depression: a meta-analysis. Am. J. Psychiatry. 2004;161:598–607. doi: 10.1176/appi.ajp.161.4.598. [DOI] [PubMed] [Google Scholar]
- First MB, Spitzer RL, Gibbon M, Williams JB. Biometrics Research. New York: New York State Psychiatric Institute; 2002. Structured Clinical Interview for DSM-IV-TR Axis I Disorders, Research Version, Patient Edition With Psychotic Screen (SCID-I/P W/ PSY SCREEN) [Google Scholar]
- Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, et al. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33:341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- Hintze JL. PASS 2005 User's Guide. Kaysville, Utah: NCSS; 2005. [Google Scholar]
- Jatzko A, Rothenhofer S, Schmitt A, Gaser C, Demirakca T, Weber-Fahr W, et al. Hippocampal volume in chronic posttraumatic stress disorder (PTSD): MRI study using two different evaluation methods. J. Affect. Disord. 2006;94:121–126. doi: 10.1016/j.jad.2006.03.010. [DOI] [PubMed] [Google Scholar]
- Khan AR, Wang L, Beg MF. FreeSurfer-initiated fully-automated subcortical brain segmentation in MRI using large deformation diffeomorphic metric mapping. NeuroImage. 2008;41:735–746. doi: 10.1016/j.neuroimage.2008.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronmuller KT, Pantel J, Kohler S, Victor D, Giesel F, Magnotta VA, et al. Hippocampal volume and 2-year outcome in depression. Br. J. Psychiatry. 2008;192:472–473. doi: 10.1192/bjp.bp.107.040378. [DOI] [PubMed] [Google Scholar]
- Lewis DV. Losing neurons: selective vulnerability and mesial temporal sclerosis. Epilepsia. 2005;46 Suppl. 7:39–44. doi: 10.1111/j.1528-1167.2005.00306.x. [DOI] [PubMed] [Google Scholar]
- McDonald CR, Hagler DJ, Ahmadi ME, Tecoma E, Iragui V, Dale AM, Halgren E. Subcortical and cerebellar atrophy in mesial temporal lobe epilepsy revealed by automatic segmentation. Epilepsy Res. 2008;79:130–138. doi: 10.1016/j.eplepsyres.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantazis D, Leahy RM, Nichol TE, Styner M. Statistical surface-based morphometry using a non-parametric approach. Int Symposium on Biomedical Imaging (ISBI) 2004:1283–1286. [Google Scholar]
- Powell S, Magnotta VA, Johnson H, Jammalamadaka VK, Pierson R, Andreasen NC. Registration and machine learning-based automated segmentation of subcortical and cerebellar brain structures. NeuroImage. 2008;39:238–247. doi: 10.1016/j.neuroimage.2007.05.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provenzale JM, Barboriak DP, VanLandingham K, MacFall J, Delong D, Lewis DV. Hippocampal MRI signal hyperintensity after febrile status epilepticus is predictive of subsequent mesial temporal sclerosis. AJR Am. J. Roentgenol. 2008;190:976–983. doi: 10.2214/AJR.07.2407. [DOI] [PubMed] [Google Scholar]
- Schiltz K, Witzel J, Northoff G, Zierhut K, Gubka U, Fellmann H, et al. Brain pathology in pedophilic offenders: evidence of volume reduction in the right amygdala and related diencephalic structures. Arch. Gen. Psychiatry. 2007;64:737–746. doi: 10.1001/archpsyc.64.6.737. [DOI] [PubMed] [Google Scholar]
- Styner M, Oguz I, Xu S, Brechbuler C, Pantazis D, Gerig G. Statistical Shape Analysis of Brain Structures using SPHARM-PDM. Insight Journal DSpace. 2006 [PMC free article] [PubMed] [Google Scholar]
- Tae W, Kim S, Lee K, Nam EC, Kim K. Validation of hippocampal volumes measured using a manual method and two automated methods (FreeSurfer and IBASPM) in chronic major depressive disorder. Neuroradiology. 2008;50:569–581. doi: 10.1007/s00234-008-0383-9. [DOI] [PubMed] [Google Scholar]
- Theodore WH, Bhatia S, Hatta J, Fazilat S, DeCarli C, Bookheimer SY, Gaillard WD. Hippocampal atrophy, epilepsy duration, and febrile seizures in patients with partial seizures. Neurology. 1999;52:132–136. doi: 10.1212/wnl.52.1.132. [DOI] [PubMed] [Google Scholar]
- Turetsky B, Cowell PE, Gur RC, Grossman RI, Shtasel DL, Gur RE. Frontal and temporal lobe brain volumes in schizophrenia. Relationship to symptoms and clinical subtype. Arch. Gen. Psychiatry. 1995;52:1061–1070. doi: 10.1001/archpsyc.1995.03950240079013. [DOI] [PubMed] [Google Scholar]
- Vermetten E, Schmahl C, Lindner S, Loewenstein RJ, Bremner JD. Hippocampal and amygdalar volumes in dissociative identity disorder. Am. J. Psychiatry. 2006;163:630–636. doi: 10.1176/appi.ajp.163.4.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Videbech P, Ravnkilde B. Hippocampal volume and depression: a meta-analysis of MRI studies. Am. J. Psychiatry. 2004;161:1957–1966. doi: 10.1176/appi.ajp.161.11.1957. [DOI] [PubMed] [Google Scholar]
- Vita A, De Peri L, Silenzi C, Dieci M. Brain morphology in first-episode schizophrenia: a meta-analysis of quantitative magnetic resonance imaging studies. Schizophr. Res. 2006;82:75–88. doi: 10.1016/j.schres.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, Gerig G. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. NeuroImage. 2006;31:1116–1128. doi: 10.1016/j.neuroimage.2006.01.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.