Abstract
The vaccinia virus E6R gene (VACVWR062) is conserved in all members of the poxvirus family and encodes a protein associated with the mature virion. We confirmed this association and provided evidence for an internal location. An inducible mutant that conditionally expresses E6 was constructed. In the absence of inducer, plaque formation and virus production were severely inhibited in several cell lines, whereas some replication occurred in others. This difference could be due to variation in the stringency of repression, since we could not isolate a stable deletion mutant even in the more “permissive” cells. Under non-permissive conditions, viral late proteins were synthesized but processing of cores proteins was inefficient, indicative of an assembly block. Transmission electron microscopy of sections of cells infected with the mutant in the absence of inducer revealed morphogenetic defects with crescents and empty immature virions adjacent to dense inclusions of viroplasm. Mature virions were infrequent and cores appeared to have lucent centers.
Introduction
Poxviruses are divided into chordopoxvirus and entomopoxvirus subfamilies, which infect vertebrate and insect hosts, respectively (Moss, 2007). Vaccinia virus (VACV), the prototype poxvirus, encodes nearly 200 proteins of which approximately 100 are conserved in all chordopoxviruses and about 50 of those can be identified in entompoxviruses as well (Upton et al., 2003). The genes encoding the majority of the highly conserved proteins are located in the central portion of the VACV genome and encode structural components of the virion and enzymes and factors needed for DNA replication and transcription (Condit et al., 2006). The less conserved genes, located at either end of the genome, encode a variety of proteins involved primarily in host interactions (Nazarian and McFadden, 2006). Although great progress has been made in characterizing VACV genes, the roles of some such as E6R (VACVWR062) remain unknown. E6, the protein encoded by the E6R open reading frame (ORF), is one of about 80 that were identified in VACV mature virions by mass spectrometry (Chung et al., 2006; Resch et al., 2007; Yoder et al., 2006). In a recent study (Kato et al., 2008), three temperature-sensitive mutants were mapped to the E6R open reading frame. We have taken a combined genetic and biochemical approach to characterize the E6 protein. We provide evidence that E6 is a component of the virion core and is essential for replication. When expression of E6 was repressed, virion morphogenesis was severely impaired at an early stage.
Results
Conservation of the E6R gene
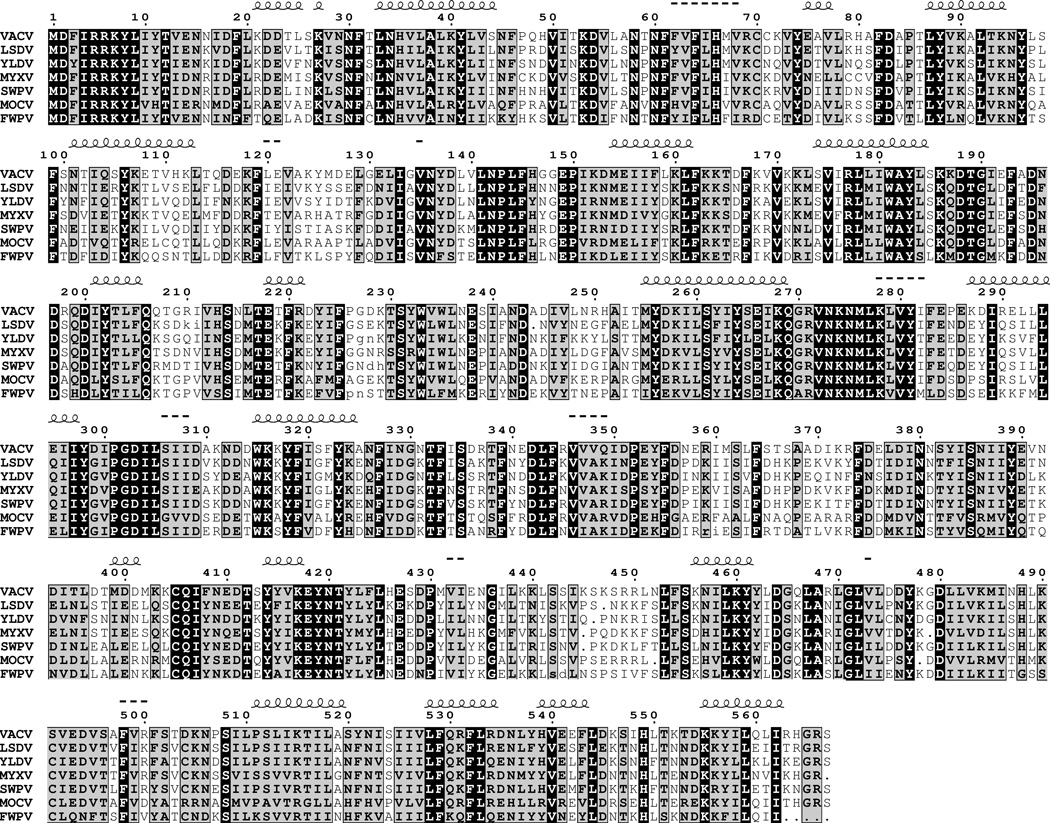
The VACV E6R ORF is predicted to encode a protein of 567 amino acids. A sequence alignment of E6 homologs from a representative of each chordopoxvirus genus is presented in Fig. 1. Among the orthopoxviruses, the sequence identity of E6 homologs is 98% or greater. For other chordopoxviruses, the identity with VACV E6 is between 44 and 63%. A putative homolog in the insect poxviruses Amsacta moorei entomopoxvirus and Melanoplus sanguinipes entomopoxvirus with approximately 18% sequence identity was detected using the position-specific iterative protein-protein basic local alignment search tool (psi-blastp) in the third iteration. To estimate the significance of the match, the Melanoplus sanguinipes entomopoxvirus sequence was shuffled 500 times and then each shuffled sequence was aligned with VACV E6. No shuffled sequence reached the score of the original alignments, allowing us to reject the null hypothesis of no homology (p < 0.002). The conclusion that E6 is conserved in all poxviruses agrees with Upton and coworkers (Upton et al., 2003). No homology was detected with non-poxvirus proteins, nor were any motifs suggestive of function or intracellular trafficking found. The nucleotides immediately upstream of the E6R ORF contain a variant TAAAAATG with extra As of the consensus late promoter motif TAAATG (Davison and Moss, 1989).
Fig. 1.
Multiple alignments of amino acid sequences of chordopoxvirus E6 homologs. Identical and conserved amino acids are indicated by black and gray backgrounds, respectively. Predicted coil-coil ( ) and β sheet (---) domains are indicated above the amino acid sequence. Abbreviations: VACV, vaccinia virus; LSDV, lumpy skin disease virus; YLDV, Yaba-like tumour virus; MYXV, myxoma virus; SWPV, swinepox virus; MOCV, molluscum contagiosum virus; FWPV, fowlpox virus.
) and β sheet (---) domains are indicated above the amino acid sequence. Abbreviations: VACV, vaccinia virus; LSDV, lumpy skin disease virus; YLDV, Yaba-like tumour virus; MYXV, myxoma virus; SWPV, swinepox virus; MOCV, molluscum contagiosum virus; FWPV, fowlpox virus.
E6 is a component of the virion core
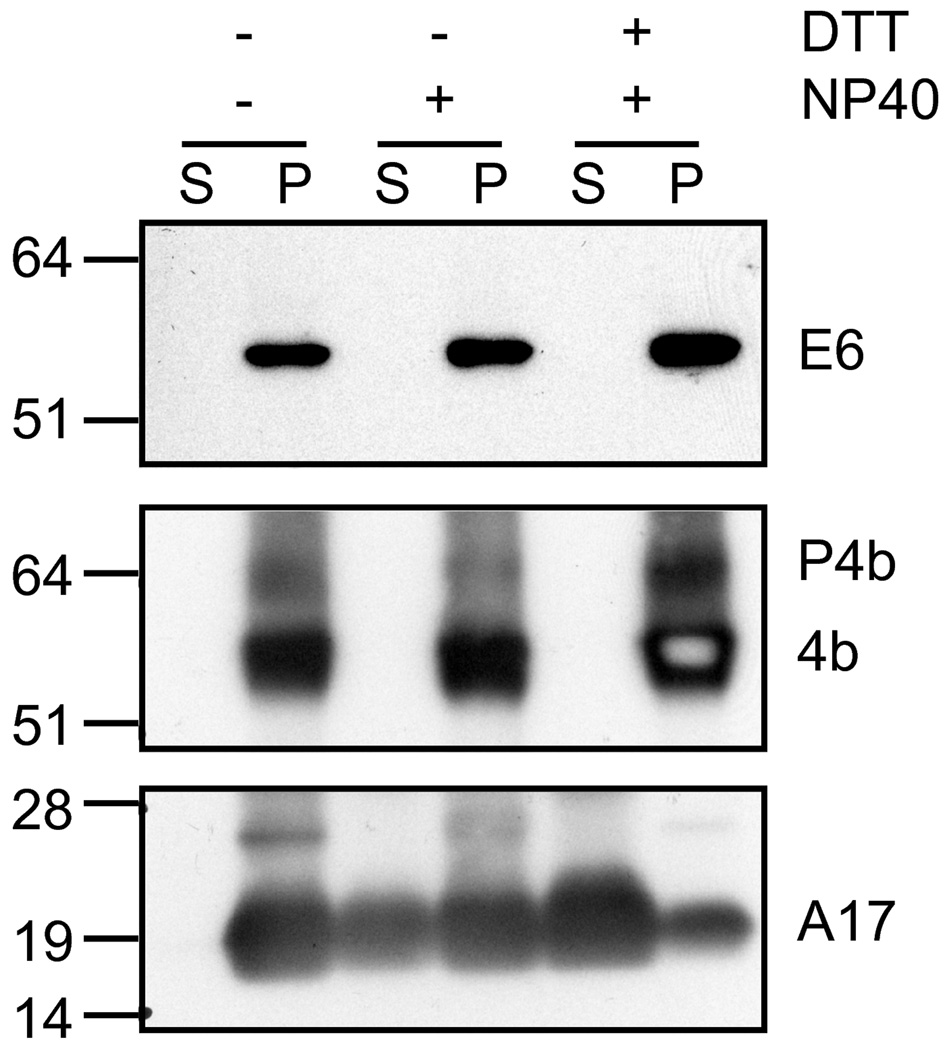
In order to detect the E6 protein, rabbits were immunized with a conjugated peptide derived from amino acids 439 to 452 of the predicted E6 amino acid sequence. Using the rabbit antiserum, a protein of approximately 60 kDa was detected by Western blotting of infected cell lysates at late times after infection. The presence of E6 in purified MVs was confirmed by Western blotting. The protein remained in the insoluble pellet fraction after addition of NP40 and dithiothreitol, similar to the core protein A3 and distinct from the membrane protein A17 (Fig. 2). These data suggested that E6 is an internal component of MVs.
Fig. 2.
Western blots of proteins extracted from purified virions. MVs were purified from VACV-infected cells by sucrose gradient sedimentation and treated with NP40 detergent and dithithreitol (DTT) reducing agent as indicated by plus and minus signs. Proteins were resolved by polyacrylamide gel electrophoresis, transferred to a membrane and detected by chemiluminescence following incubation with primary antibodies to E6, P4b/4b precursor and mature core proteins, and the A17 MV membrane protein followed by a secondary antibody conjugated to horse radish peroxidase. Abbreviations: S, soluble supernatant; P, insoluble pellet. Numbers on the left indicate the kDa and position of marker proteins.
Construction and infectivity of a recombinant VACV with an inducible E6R gene
The conservation of the E6R gene made it likely that it would be essential for virus replication. Therefore, we were not surprised by the failure to isolate a stable mutant, in which the E6R gene was replaced with a gene encoding GFP, by homologous recombination in BS-C-1 cells. These results suggested that the E6 gene is either essential or that deletion mutants are too enfeebled for plaque isolation.
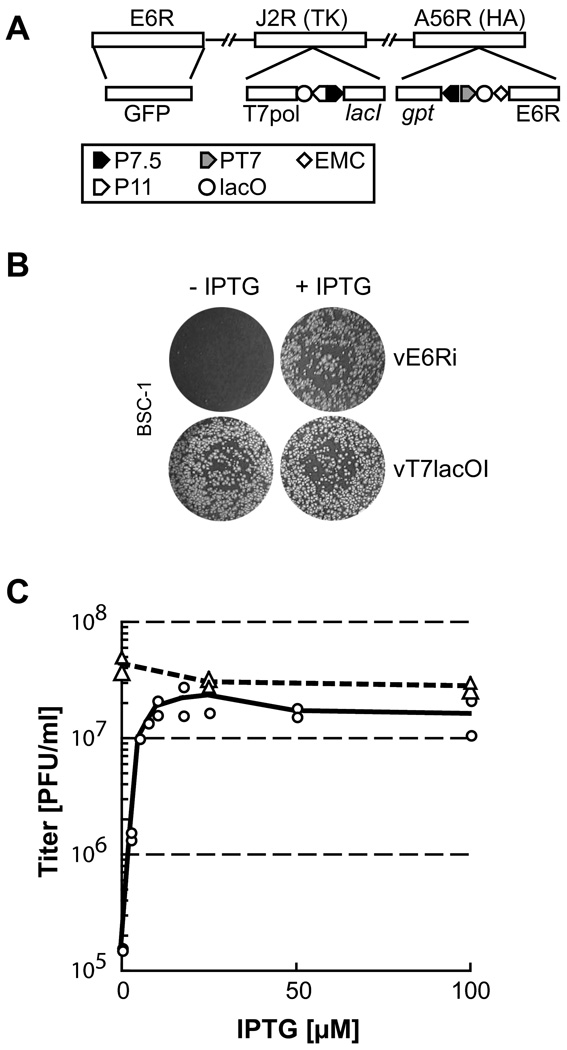
Our next approach to discerning the role of E6 was to make a mutant that conditionally expresses the protein. An inducible system based on the Eschericia coli lac operon was used (Ward et al., 1995; Zhang and Moss, 1991a). In brief, a copy of the E6R gene regulated by a bacteriophage T7 promoter was inserted into the A56R (hemagglutinin) locus of the recombinant vT7lacOI, which encoded the T7 RNA polymerase regulated by the late p11 VACV promoter and the E. coli lac repressor regulated by a VACV early/late promoter. Next, the original E6R gene was deleted and replaced by the gene encoding GFP (Fig. 3A). Plaques containing recombinant virus were recognized by their fluorescence and the virus vE6Ri was clonally purified in BS-C-1 cells by repeated plaque isolation in the presence of inducer isopropyl-β-D-thiogalactopyranoside (IPTG). Normal size plaques formed on a BS-C-1 monolayer in the presence but not absence of IPTG (Fig. 3B). By contrast, the parental vT7lacOI virus formed similar size plaques in the presence or absence of inducer. The IPTG requirement of vE6Ri to form plaques could arise from decreased virus replication or spread. One-step growth experiments indicated that IPTG was required for replication. Virus replication in BS-C-1 cells increased up to 150-fold in proportion with IPTG concentration and the maximum yield was obtained at approximately 25 µM (Fig. 3C).
Fig. 3.
Structure and replication of a conditional lethal E6 mutant. (A) Diagram of selected regions of the vE6Ri genome. Upper rectangles labeled E6R, J2R, and A56R are ORFs used as sites of insertion of recombinant DNA. TK and HA, are abbreviations for thymidine kinase and hemagglutinin, respectively. Lower rectangles show inserted DNA encoding GFP; bacteriophage T7 RNA polymerase (T7pol) regulated by the late P11 promoter (P11) and lac operator (lacO); E. coli lac repressor (lacI) regulated by the VACV p7.5 early/late promoter (P7.5); E. coli xanthine-guanine phosphoribosyltransferase (gpt) regulated by P7.5; and E6R regulated by the T7 promoter (PT7), lacO, and encephalomyocarditis (EMC) cap-independent leader. (B) Plaque formation. BS-C-1 monolayers were infected with vE6Ri or the parent virus vT7lacOI in the presence (+) or absence (−) of IPTG and incubated in normal growth medium supplemented with 1% methyl cellulose as a thickening agent. After 48 h, the cells were stained with crystal violet. (C) IPTG-dependence of vE6Ri replication. BS-C-1 Cells were infected with 10 PFU per cell of vE6Ri (circles) or vT7lacOI (triangles) in the presence of indicated concentrations of IPTG. At 24 h after infection, cells were harvested, lysed by three freeze-thaw cycles and sonication. The virus titer was determined by plaque formation on BS-C-1 monolayers.
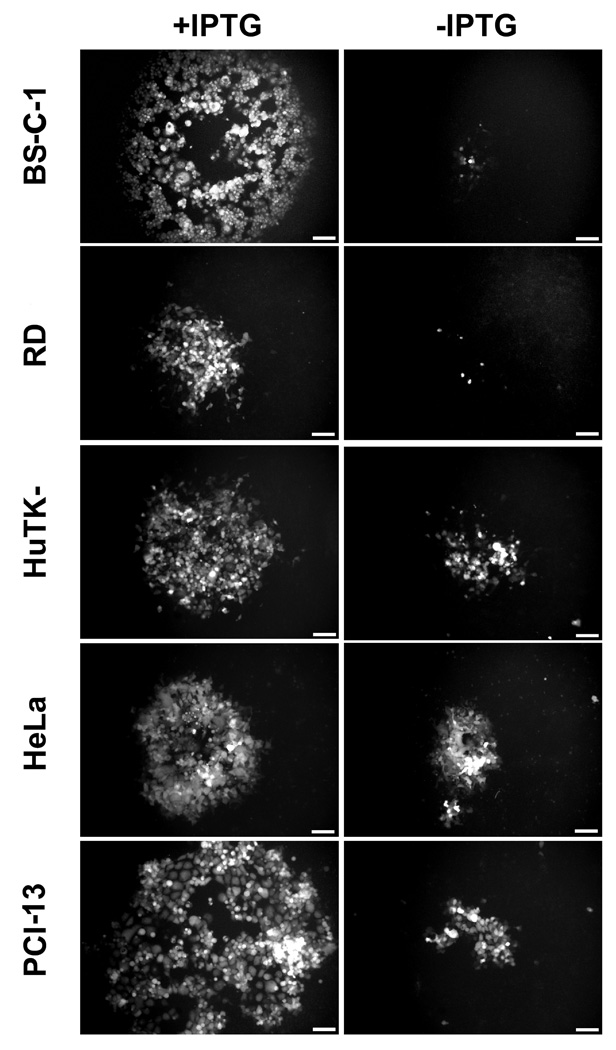
We tested a variety of cell lines to see if there were host differences in the dependence on IPTG for replication of vE6Ri. Wild-type plaques visualized by green fluorescence formed in the presence of IPTG in all cells tested, whereas in the absence of IPTG the plaques were pinpoint in BS-C-1, RD, BHK-21, 3T3 and RK13 cells and somewhat larger in HuTK-, HeLa and PCI-13 (Fig. 4 and data not shown). In view of the possibility that the latter cell lines would be more permissive for an E6R deletion mutant, we tried another isolation strategy. The E6R gene was deleted from the VACV genome in a bacterial artificial chromosome (Domi and Moss, 2002; Domi and Moss, 2005). Attempts to rescue infectious VACV lacking E6R in PCI-13 or HuTK− cells were unsuccessful, however, whereas rescue of a VACV bacterial artificial chromosome containing intact E6R was accomplished in parallel transfections (data not shown). It seems likely, therefore, that the difference in IPTG dependence resulted from less stringent repression of E6 in some cells rather than a true host-range effect. However, our antibody was not sufficiently sensitive and free of cross-reactivity to detect low expression.
Fig. 4.
Host cell effect on plaque formation. Monolayers of BS-C-1, RD, HuTK-, HeLa and PCI-13 cells were infected with vE6Ri in the presence (+) or absence (−) of 25µM IPTG. After 24 h, the cells were examined under a fluorescence microscope and images of individual typical plaques are shown. Scale bars correspond to 0.1 mm.
Synthesis and processing of viral late proteins
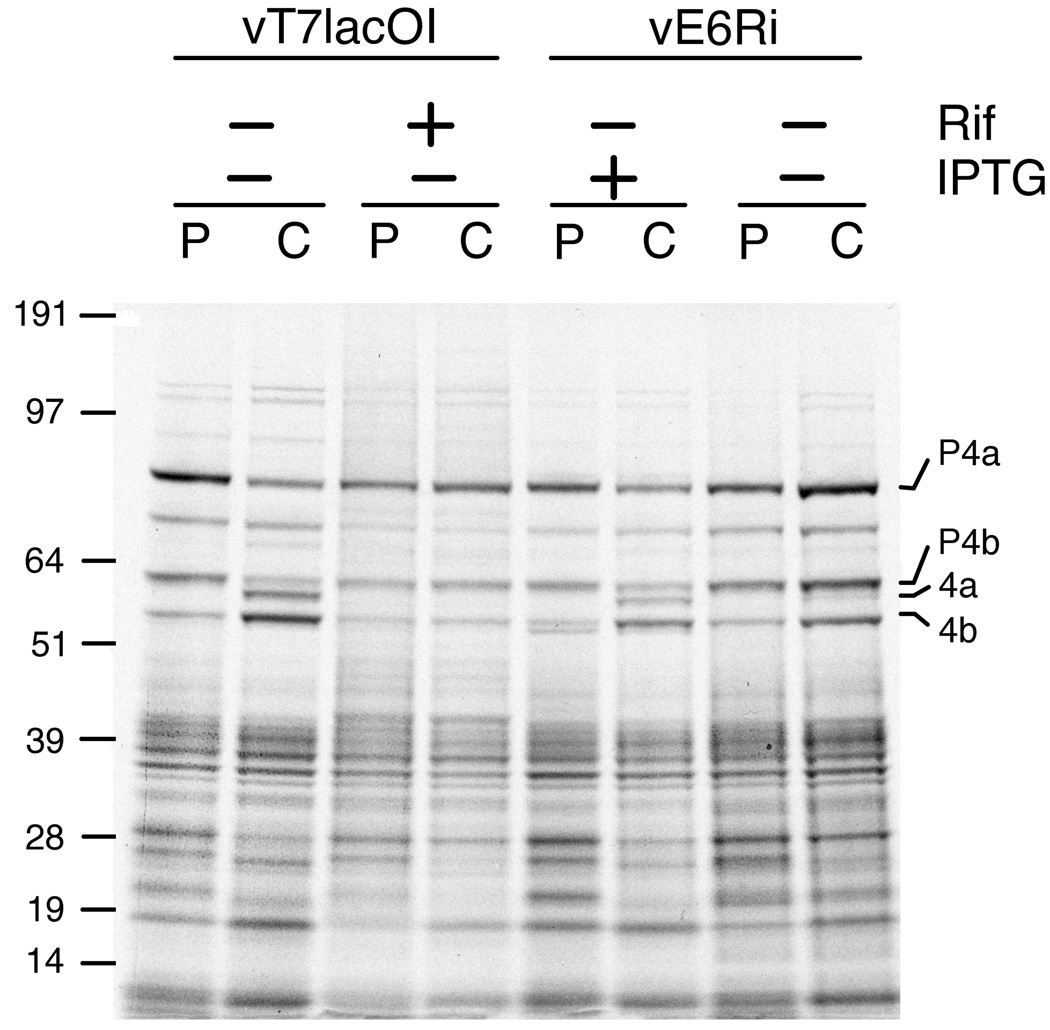
Further experiments were carried out in BS-C-1 or BHK-21 cells because of their stringent repression of replication. At 9 h after infecting BS-C-1 cells with vE6Ri or the parental vT7lacOI in the presence or absence of IPTG or the drug rifampicin, lysates prepared from pulse-labeled cells were analyzed by SDS polyacrylamide gel electrophoresis and autoradiography. The well-characterized pattern of radioactivity indicated that viral late protein synthesis proceeded similarly in the presence or absence of IPTG (Fig. 5). Several viral proteins undergo proteolytic processing during virus maturation, and this can be prevented by the drug rifampicin (Katz and Moss, 1970; Moss and Rosenblum, 1973). Following a chase with non-radioactive amino acids, the pattern of radioactivity indicated diminished processing of the precursors of the 4A (A10) and 4B (A3) proteins, although this was not as severe as with rifampicin (Fig. 5).
Fig. 5.
Effect of E6 repression on synthesis and processing of viral late proteins. BS-C-1 cells were infected with vE6Ri or vT7lacOI in the presence or absence of 25 µM IPTG or 100 µg/ml of rifampicin (Rif). At 9 h after infection, the cells were pulse-labeled (P) with [35S]methionine and [35S]cysteine for 30 min in methionine- and cysteine-deficient medium. The cells were then washed and incubated in regular medium for 15 h as a chase (C). Cell lysates were prepared and the proteins were separated on a SDS 4 to 12% polyacrylamide gradient gel. An autoradiogram is shown. The positions of the core proteins 4a and 4b and their precursors P4a and P4b are indicated on the right. The masses in kDa and positions of marker proteins are shown on the left.
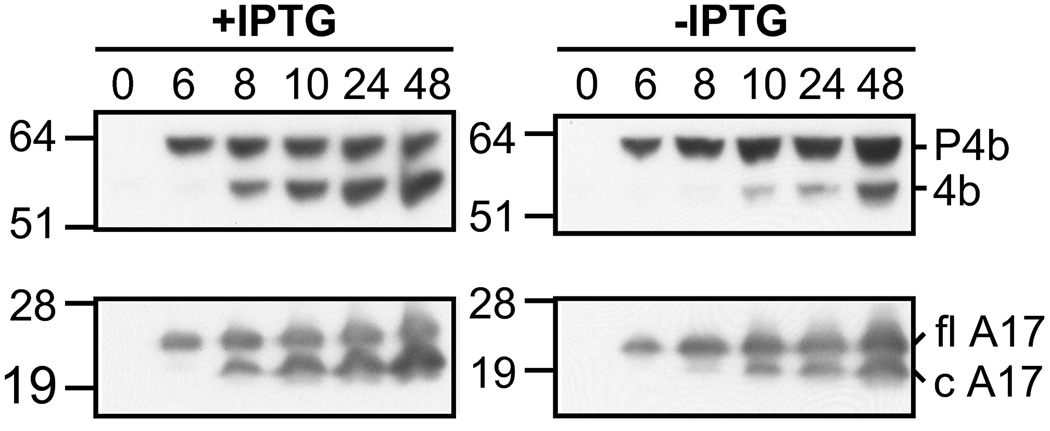
Western blotting was used to confirm the defect in protein processing. Cells were infected with vE6Ri in the presence or absence of IPTG and samples removed at intervals between 0 and 48 h. Following gel electrophoresis, the proteins were transferred to a membrane and probed with specific antibodies to the 4B core and A17 membrane proteins. In the presence of IPTG, processing of the 4B and A17 precursors were detected at 8 h after infection and by 24 h the processed proteins exceeded the amount of precursor (Fig. 6). In the absence of IPTG, processing of the 4B precursor was undetectable at 8 h and detectable only as faint bands at 12 and 24 h but increased by 48 h. Processing of the A17 membrane protein, which is not inhibited by rifampicin (Betakova et al., 1999b; Rodriguez et al., 1993), was only mildly affected by E6 repression (Fig. 6). These data suggested a defect at a post-membrane formation stage in virion morphogenesis.
Fig. 6.
Effect of E6 repression on the synthesis and processing of representative core and membrane proteins determined by Western blotting. BS-C-1 cells were infected with vE6Ri in the absence (−) or presence (+) of 25 µM IPTG. At indicated times between 0 and 48 h, lysates were prepared and the proteins resolved by gel electrophoresis as described in the legend to Fig. 5. The proteins were transferred to a membrane and probed with rabbit polyclonal antiserum to the 4b core protein or A17 membrane protein followed by anti-rabbit immunoglobulin G conjugated to horseradish peroxidase. The proteins were visualized by chemiluminescence. The positions of precursor 4b (P4b) and processed 4b and of full length (fl) A17 and C-terminal processed (c) A17 are indicated on the right. The masses in kDa and positions of marker proteins are shown on the left.
Virus morphogenesis
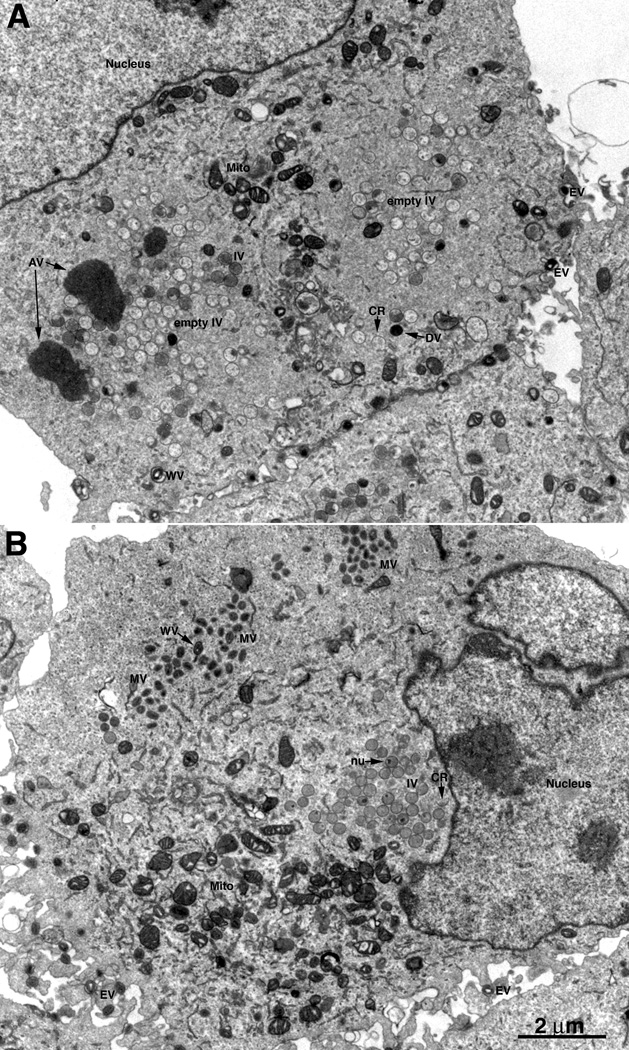
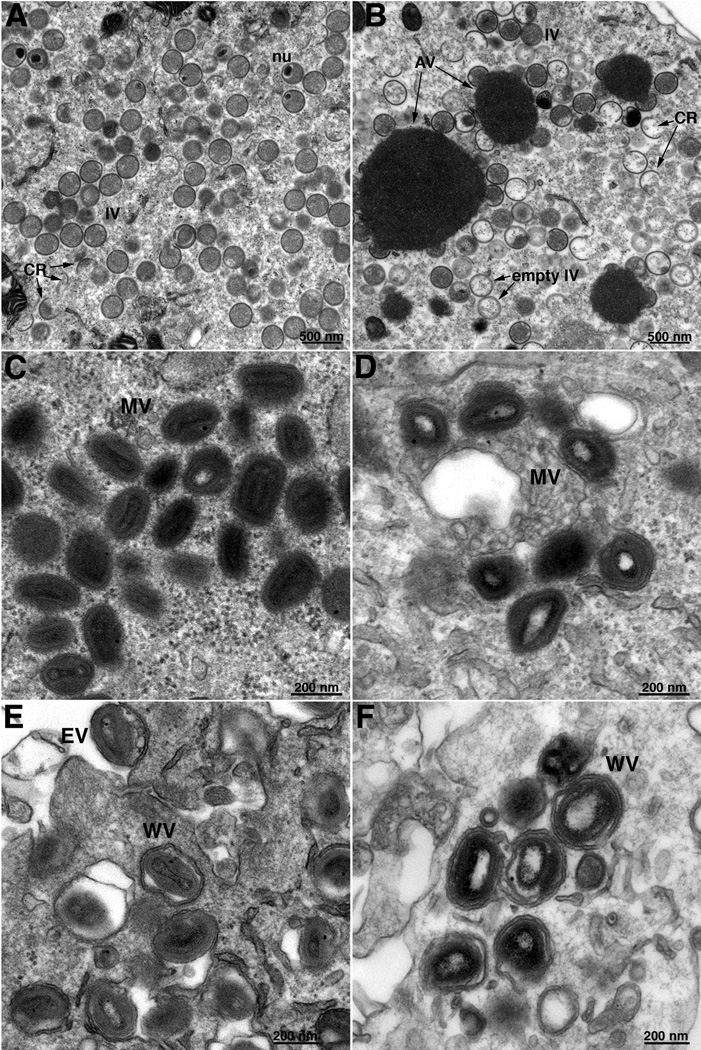
Thin sections of BS-C-1 cells infected with vE6Ri in the absence and presence of IPTG were examined by transmission electron microscopy. In the presence of IPTG, circular IVs with granular material, MVs with well-defined cores, and membrane wrapped virions were abundant (Fig. 7B). In the absence of IPTG, many crescents and IVs with and without internal granular material were adjacent to dense inclusions not seen in the presence of inducer and some round, dense virions were also present (Fig. 7A). Brick-shaped MVs and WVs were infrequent and the cores usually had a lucent appearance (Fig. 7A). The differences between cells infected in the presence (A, C, E) and absence (B, D, F) of IPTG were more clearly seen in the higher power images in Fig. 8. Similar images were also found on examination of BHK-21 cells infected with vE6Ri in the absence of IPTG (not shown).
Fig. 7.
Low magnification transmission electron microscopic images of cells infected with vE6Ri. BS-C-1 cells were infected with vE6Ri in absence (A) or presence (B) of 25 µM IPTG and fixed after 24 h. Magnification is indicated by the bar at the bottom of panel B. Abbreviations: AV, aggregated viroplasm; CR, crescent, IV, immature virion, MV, mature virion; WV, wrapped virion, EV, enveloped virion; mito, mitochondrion.
Fig. 8.
High magnification electron microscopic images of cells infected with vE6Ri. Infection was carried out in presence (A, C, E) or absence (B, D, F) of IPTG as described in the legend to Fig. 7. Fields containing mostly IVs (A, B), MVs (C, D) and EVs (E, F). Abbreviations are the same as in the legend to Fig. 7. Magnification is indicated in bar near lower right corner of each image.
Discussion
Using a specific antibody for Western blotting, we confirmed the association of E6 with purified MVs, which had previously been determined by mass spectrometry (Chung et al., 2006; Resch et al., 2007; Yoder et al., 2006). Our inability to extract the protein in a soluble form by incubating purified MVs with NP40 and reducing agent suggested that E6 is an internal component of the MV. Such insolubility was the major criterion used to assign 47 other VACV proteins to the core or lateral body (Condit et al., 2006). Of these internal proteins, 19 are like E6 in having no known or predicted enzymatic function and are presumed to have structural roles. Our attempts to isolate a viable E6 deletion mutant failed, leading us to construct a recombinant virus in which the E. coli lac operator regulated E6 expression. In the absence of inducer, plaque formation and virus yield were severely inhibited in several cell lines including monkey BS-C-1, rabbit RK13, hamster BHK-21, mouse 3T3 and human RD cells. However, in the absence of inducer replication occurred to varying degrees in human HuTK-, HeLa and PCI-13 cells. Initially, we considered that E6 might be a host range factor. However, we were unable to rescue a VACV bacterial artificial chromosome with an E6 deletion in either HuTK- or PCI-13 cells. It seems likely that low amounts of E6 allow limited replication and that the repression is incomplete in the more permissive cell lines. However, our antibody was not sufficiently sensitive or free of non-specific binding to test this possibility. Therefore, we proceeded to use BS-C-1 and BHK-21 cells to study the replication defect.
Amino acid labeling studies indicated that E6 was not required for late gene expression. However, the proteolytic processing of core proteins 4a and 4b was inhibited. By contrast, processing of the A17 membrane protein was much less affected. Although the I7 protease is responsible for cleavage of several core proteins and at least one membrane protein (Ansarah-Sobrinho and Moss, 2004; Byrd et al., 2003), the latter is less dependent on morphogenesis and is not blocked by rifampicin (Betakova et al., 1999b; Rodriguez et al., 1993), which prevents the association of the D13 scaffold with membranes to form crescent structures (Moss et al., 1969; Szajner et al., 2005; Zhang and Moss, 1992).
Transmission electron microscopy studies revealed severe defects in morphogenesis that corresponded with the protein processing data. Although crescent membranes and IVs were visualized, they were frequently adjacent to dense inclusions of viroplasm, which contain the core proteins. In addition, many of the IVs appeared empty because of the absence of enclosed viroplasm and MVs were sparse and many appeared defective. With regard to the inclusions, the phenotype resembled that obtained with the conditional lethal A15, A30, D2, D3, G7 and J1 mutants, which interact to form a complex (Chiu et al., 2005; Mercer and Traktman, 2005; Szajner et al., 2003; Szajner et al., 2004a; Szajner et al., 2004b; Szajner et al., 2001). However, with the latter mutants, the IVs appear more uniformly empty and MVs do not form, whereas many IVs did contain viroplasm in cells infected with the E6 mutant and there were abnormal MVs. Abnormal MVs that appear more spherical or have internal defects are found with mutations of some other non-enzymatic core proteins (Heljasvaara et al., 2001; Kato et al., 2004; Klemperer et al., 1997; Williams et al., 1999; Zhang and Moss, 1991b).
Kato and coworkers (Kato et al., 2008) recently mapped three temperature sensitive mutants, including Dts41 (#6606) and Dts80 (#9021) from the S. Dales collection, to the E6R gene. Both Dts41 and Dts80 were described as having defects in morphogenesis at the non-permissive temperature (Dales et al., 1978). Electron micrographs of cells infected with these mutants were not published; instead the mutants were placed in morphogenetic classes and only images of other representatives of these classes were shown. The morphogenetic classes go from A to Q, in order of increasing development e.g. A has no virus structures and Q has normal looking IVs and MVs. Of these, the representative image shown of an F class mutant seems to most resemble vE6Ri in the absence of IPTG, as both contain masses of viroplasm and empty IVs. Dts80, however, was placed in the H class and the representative image for this group has no empty IVs, although masses of viroplasm are present. The Dales H class is defined as a composite of classes D–F so there may be some variation in the individual mutants placed in this class and the phenotype of Dts80 may have additional similarities or differences relative to vE6Ri. Dts80 was also subclassified as Q, indicating some normal-looking virions. Dts41 was classified as having a P phenotype, which is defined as normal-looking immature particles and mature particles with cores that are predominantly lucent at the center. Although we did find mature particles with lucent centers in cells infected with vE6Ri in the absence of IPTG, the more obvious defects were at early stages. It is certainly possible that temperature sensitive and inducible null mutants will exhibit phenotypic differences. However, direct comparisons using the same strains of virus, cell lines and microscopic procedures are needed to make unambiguous conclusions.
Materials and methods
Cell lines and virus strains
BS-C-1 (African green monkey kidney cells; ATCC:CCL-26), RD (human rhabdomyosarcoma cells; ATCC:CCL-136), HuTK− 143 (non-producer human cells induced by murine sarcoma virus; (Rhim et al., 1975), HeLa (human adenocarcinoma celss; ATCC:CCL-2), and PCI-13 (human pharyngeal carcinoma cells) cells were maintained under standard culture conditions. The Western Reserve (WR) strain of VACV and the vT7lacOI recombinant (Alexander et al., 1992) were propagated as described previously. vE6Ri was propagated in the presence of 25 µM IPTG.
Antibodies
Rabbit antiserum was raised against a peptide derived from the predicted E6 amino acids 439 to 452 (KKLSSIKSKSRRLN) plus a C-terminal cysteine added for coupling to keyhole limpet hemocyanin (Covance Research Products, Denver, PA). Anti-A17-N (Betakova et al., 1999a) and anti-A3 (P4b/4b) (Resch et al., 2005) rabbit antisera were described previously.
Plasmid and recombinant VACV construction
To construct pVOTE-E6R, the E6R ORF was amplified by PCR from genomic DNA using the oligonucleotide primers 5’-GTA TTA CCT GCA TAC CAT GGA TTT TAT TCG TAG AAA GT-3’ and 5’-CTA TGA TCT ACC GTG TCT TAT CAA TTG AAG TAT-3’, and the PCR product was inserted within the NcoI-SmaI sites of pVOTE.1 downstream of an encephalomyocarditis virus leader sequence (Ward et al., 1995). This plasmid was used to introduce the repressible copy of E6R into the hemagglutinin (A56R) locus of vT7lacOI by homologous recombination and mycophenolic acid selection as described previously (Ward et al., 1995), resulting in the intermediate virus vE6R/E6Ri. The presence of the inducible E6R copy was verified by PCR amplification and sequence analysis. The endogenous E6R ORF was replaced with the GFP marker gene by homologous recombination with a linear DNA fragment containing the GFP sequence fused to E6R-flanking sequences. This fragment was generated in three steps. First, three independent PCR amplifications were carried out to amplify the flanking regions and the GFP sequence. The upstream flanking region was amplified with the oligonucleotide primers 5’-TCT TTA TCA AAC AGA TGG TAG AAC AAT GTG TGA CAT CCC-3’ and 5’-TTG CTC ACC ATT TTT ATA GAA GAG AGT TAT CTA TGA TAC-3’. The GFP sequence was amplified with oligonucleotide primers 5’-TCT CTT CTA TAA AAA TGG TGA GCA AGG GCG AGG AGC TG-3’ and 5’-ATA TTT GGT CTG TTT TAT CTA GAT CCG GTG GAT CCC GGG C-3’. The lower flanking region was amplified by using the oligonucleotide primers 5’-GGA TCT AGA TAA AAC AGA CCA AAT ATA TTA TTA ATA ATT T-3’ and 5’-AGA GTT GTT CTA GCA TCC ATT CAT TGT CTT TAC ATC CGT T-3’. The separate products were combined using their terminal overlaps in a final PCR reaction using the outer forward and reverse primers. The final PCR product was transfected into cells infected with vE6R/E6Ri at 0.5 PFU per cell using Lipofectamine 2000 (Invitrogen). Recombinant viruses expressing GFP were isolated by five rounds of plaque purification using an inverted fluorescence microscope. The correct site of recombination was verified by PCR analysis.
Western blot analysis
Proteins of MV particles, purified by sucrose gradient centrifugation (Earl and Moss, 1998), or infected cell lysates were resolved by SDS-polyacrylamide gel electrophoresis on 4 to 12% gradient SDS-polyacrylamide gels with 3-(N-morpholino)propanesulfonic acid running buffer (Invitrogen) and electrophoretically transferred to nitrocellulose membranes. Nonspecific binding sites were blocked with 5% nonfat dried milk or 5% bovine serum albumin in Tris-buffered saline. The membranes were probed with rabbit polyclonal antiserum, followed by anti-rabbit immunoglobulin G conjugated to horseradish peroxidase. Bound antibody was detected by chemiluminescence with commercial reagents (Pierce, Rockford, IL).
Pulse-labeling of proteins with [35S]methionine and [35S]cysteine
BS-C-1 cells were incubated with 5 PFU of virus per cell for 1 h at 37°C. After adsorption, cells were washed three times and incubated at 37°C in Eagle modified essential medium supplemented with 2.5% fetal bovine serum in the presence or absence of 25 µM IPTG. At 30 min before labeling, the medium was replaced with cysteine- and methionine-free medium, and cells were incubated for 30 min at 37°C, followed by the addition of 100 µCi of a mixture of [35S]methionine and [35S]cysteine per ml of deficient medium. After 30 min, some cells were harvested and others were washed with regular medium and incubated for an additional 15 h. Harvested cells were washed once with cold phosphate-buffered saline and whole-cell lysates were prepared in 1X sample buffer (Invitrogen). Samples were separated on an SDS 4 to 12% polyacrylamide gradient gel with 3-(N-morpholino)propanesulfonic acid running buffer (Invitrogen).
Transmission electron microscopy
BS-C-1 cells were grown in 60-mm-diameter dishes and infected with 3 PFU per cell of vE6Ri in the presence or absence of 25 µM IPTG. At 24 h after infection, cells were fixed and prepared for transmission electron microscopy as described previously (Senkevich et al., 2008).
Acknowledgments
We thank Arban Domi for materials and advice regarding use of the vaccinia virus bacterial artificial chromosome and Norman Cooper for technical assistance with cell lines.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexander WA, Moss B, Fuerst TR. Regulated expression of foreign genes in vaccinia virus under the control of bacteriophage T7 RNA polymerase and the Escherichia coli lac repressor. J. Virol. 1992;66:2934–2942. doi: 10.1128/jvi.66.5.2934-2942.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansarah-Sobrinho C, Moss B. Role of the I7 protein in proteolytic processing of vaccinia virus membrane and core components. J. Virol. 2004;78:6335–6343. doi: 10.1128/JVI.78.12.6335-6343.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betakova T, Wolffe EJ, Moss B. Membrane topology of the vaccinia virus A17L envelope protein. Virology. 1999a;261:347–356. doi: 10.1006/viro.1999.9870. [DOI] [PubMed] [Google Scholar]
- Betakova T, Wolffe EJ, Moss B. Regulation of vaccinia virus morphogenesis: phosphorylation of the A14L and A17L membrane proteins and C-terminal truncation of the A17L protein are dependent on the F10L protein kinase. J. Virol. 1999b;73:3534–3543. doi: 10.1128/jvi.73.5.3534-3543.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd CM, Bolken TC, Hruby DE. Molecular dissection of the vaccinia virus I7L core protein proteinase. J. Virol. 2003;77:11279–11283. doi: 10.1128/JVI.77.20.11279-11283.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu WL, Szajner P, Moss B, Chang W. Effects of a temperature sensitivity mutation in the J1R protein component of a complex required for vaccinia virus assembly. J Virol. 2005;79:8046–8056. doi: 10.1128/JVI.79.13.8046-8056.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung CS, Chen CH, Ho MY, Huang CY, Liao CL, Chang W. Vaccinia virus proteome: Identification of proteins in vaccinia virus intracellular mature virion particles. J. Virol. 2006;80:2127–2140. doi: 10.1128/JVI.80.5.2127-2140.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condit RC, Moussatche N, Traktman P. In a nutshell: structure and assembly of the vaccinia virion. Adv. Virus Res. 2006;66:31–124. doi: 10.1016/S0065-3527(06)66002-8. [DOI] [PubMed] [Google Scholar]
- Dales S, Milovanovitch V, Pogo BGT, Weintraub SB, Huima T, Wilton S, McFadden G. Biogenesis of vaccinia: isolation of conditional lethal mutants and electron microscopic characterization of their phenotypically expressed defects. Virology. 1978;84:403–428. doi: 10.1016/0042-6822(78)90258-1. [DOI] [PubMed] [Google Scholar]
- Davison AJ, Moss B. The structure of vaccinia virus late promoters. J. Mol. Biol. 1989;210:771–784. doi: 10.1016/0022-2836(89)90108-3. [DOI] [PubMed] [Google Scholar]
- Domi A, Moss B. Cloning the vaccinia virus genome as a bacterial artificial chromosome in Escherichia coli and recovery of infectious virus in mammalian cells. Proc. Natl. Acad. Sci. USA. 2002;99:12415–12420. doi: 10.1073/pnas.192420599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domi A, Moss B. Engineering of a vaccinia virus bacterial artificial chromosome in Escherichia coli by bacteriophage lambda-based recombination. Nature Methods. 2005;2:95–97. doi: 10.1038/nmeth734. [DOI] [PubMed] [Google Scholar]
- Earl PL, Moss B. Characterization of recombinant vaccinia viruses and their products. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current Protocols in Molecular Biology. Vol. 2. New York: Greene Publishing Associates & Wiley Interscience; 1998. pp. 16.18.1–16.18.11. [Google Scholar]
- Heljasvaara R, Rodriguez D, Risco C, Carrascosa JL, Esteban M, Rodriguez JR. The major core protein P4a (A10L gene) of vaccinia virus is essential for correct assembly of viral DNA into the nucleoprotein complex to form immature viral particles. J. Virol. 2001;75:5778–5795. doi: 10.1128/JVI.75.13.5778-5795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato SEM, Moussatche N, D'Costa SM, Bainbridge TW, Prins C, Strahl AL, Shatzer AN, Brinker AJ, Kay NE, Condit RC. Marker rescue mapping of the combined Condit/Dales collection of temperature-sensitive vaccinia virus mutants. Virology. 2008;375:213–222. doi: 10.1016/j.virol.2008.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato SEM, Strahl AL, Moussatche N, Condit RC. Temperature-sensitive mutants in the vaccinia virus 4b virion structural protein assemble malformed, transcriptionally inactive intracellular mature virions. Virology. 2004;330:127–146. doi: 10.1016/j.virol.2004.08.038. [DOI] [PubMed] [Google Scholar]
- Katz E, Moss B. Formation of a vaccinia virus structural polypeptide from a higher molecular weight precursor: inhibition by rifampicin. Proc. Natl. Acad. Sci. USA. 1970;6:677–684. doi: 10.1073/pnas.66.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemperer N, Ward J, Evans E, Traktman P. The vaccinia virus I1 protein is essential for the assembly of mature virions. J. Virol. 1997;71:9285–9294. doi: 10.1128/jvi.71.12.9285-9294.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer J, Traktman P. Genetic and cell biological characterization of the vaccinia virus A30 and G7 phosphoproteins. J. Virol. 2005;79:7146–7161. doi: 10.1128/JVI.79.11.7146-7161.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss B. Poxviridae: the viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. Vol. 2. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 2905–2946. [Google Scholar]
- Moss B, Rosenblum EN. Protein cleavage and poxvirus morphogenesis: Tryptic peptide analysis of core precursors accumulated by blocking assembly with rifampicin. J. Mol. Biol. 1973;81:267–269. doi: 10.1016/0022-2836(73)90195-2. [DOI] [PubMed] [Google Scholar]
- Moss B, Rosenblum EN, Katz E, Grimley PM. Rifampicin: A specific inhibitor of vaccinia virus assembly. Nature. 1969;224:1280–1284. doi: 10.1038/2241280a0. [DOI] [PubMed] [Google Scholar]
- Nazarian SH, McFadden G. Immune evasion by poxviruses. Future Virology. 2006;1:123–132. [Google Scholar]
- Resch W, Hixson KK, Moore RJ, Lipton MS, Moss B. Protein composition of the vaccinia virus mature virion. Virology. 2007;358:233–247. doi: 10.1016/j.virol.2006.08.025. [DOI] [PubMed] [Google Scholar]
- Resch W, Weisberg AS, Moss B. Vaccinia virus nonstructural protein encoded by the A11R gene is required for formation of the virion membrane. J. Virol. 2005;79:6598–6609. doi: 10.1128/JVI.79.11.6598-6609.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhim JS, Cho HY, Huebner RJ. Non-producer human cells induced by murine sarcoma virus. Int. J. Cancer. 1975;15:23–29. doi: 10.1002/ijc.2910150104. [DOI] [PubMed] [Google Scholar]
- Rodriguez D, Rodriguez JR, Esteban M. The vaccinia virus 14–kilodalton fusion protein forms a stable complex with the processed protein encoded by the vaccinia virus A17L gene. J. Virol. 1993;67:3435–3440. doi: 10.1128/jvi.67.6.3435-3440.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senkevich TG, Wyatt LS, Weisberg AS, Koonin EV, Moss B. A conserved poxvirus NlpC/P60 superfamily protein contributes to vaccinia virus virulence in mice but not to replication in cell culture. Virology. 2008;374:506–514. doi: 10.1016/j.virol.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szajner P, Jaffe H, Weisberg AS, Moss B. Vaccinia virus G7L protein interacts with the A30L protein and is required for association of viral membranes with dense viroplasm to form immature virions. J. Virol. 2003;77:3418–3429. doi: 10.1128/JVI.77.6.3418-3429.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szajner P, Jaffe H, Weisberg AS, Moss B. A complex of seven vaccinia virus proteins conserved in all chordopoxviruses is required for the association of membranes and viroplasm to form immature virions. Virology. 2004a;330:447–459. doi: 10.1016/j.virol.2004.10.008. [DOI] [PubMed] [Google Scholar]
- Szajner P, Weisberg AS, Lebowitz J, Heuser J, Moss B. External scaffold of spherical immature poxvirus particles is made of protein trimers, forming a honeycomb lattice. J. Cell. Biol. 2005;170:971–981. doi: 10.1083/jcb.200504026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szajner P, Weisberg AS, Moss B. Physical and functional interactions between vaccinia virus F10 protein kinase and virion assembly proteins A30 and G7. J. Virol. 2004b;78:266–274. doi: 10.1128/JVI.78.1.266-274.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szajner P, Weisberg AS, Wolffe EJ, Moss B. Vaccinia virus A30L protein is required for association of viral membranes with dense viroplasm to form immature virions. J. Virol. 2001;75:5752–5761. doi: 10.1128/JVI.75.13.5752-5761.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upton C, Slack S, Hunter AL, Ehlers A, Roper RL. Poxvirus orthologous clusters: toward defining the minimum essential poxvirus genome. J. Virol. 2003;77:7590–7600. doi: 10.1128/JVI.77.13.7590-7600.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward GA, Stover CK, Moss B, Fuerst TR. Stringent chemical and thermal regulation of recombinant gene expression by vaccinia virus vectors in mammalian cells. Proc. Natl. Acad. Sci. USA. 1995;92:6773–6777. doi: 10.1073/pnas.92.15.6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams O, Wolffe EJ, Weisberg AS, Merchlinsky M. Vaccinia virus WR gene A5L is required for morphogenesis of mature virions. J. Virol. 1999;73:4590–4599. doi: 10.1128/jvi.73.6.4590-4599.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JD, Chen TS, Gagnier CR, Vemulapalli S, Maier CS, Hruby DE. Pox proteomics: mass spectrometry analysis and identification of Vaccinia virion proteins. Virol. J. 2006;3:10. doi: 10.1186/1743-422X-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Moss B. Inducer-dependent conditional-lethal mutant animal viruses. Proc. Natl. Acad. Sci. USA. 1991a;88:1511–1515. doi: 10.1073/pnas.88.4.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Moss B. Vaccinia virus morphogenesis is interrupted when expression of the gene encoding an 11-kilodalton phosphorylated protein is prevented by the Escherichia coli lac repressor. J. Virol. 1991b;65:6101–6110. doi: 10.1128/jvi.65.11.6101-6110.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Moss B. Immature viral envelope formation is interrupted at the same stage by lac operator-mediated repression of the vaccinia virus D13L gene and by the drug rifampicin. Virology. 1992;187:643–653. doi: 10.1016/0042-6822(92)90467-4. [DOI] [PubMed] [Google Scholar]