Abstract
Gammaherpesviruses, including the human pathogens Epstein-Barr virus and Kaposi’s sarcoma–associated herpesvirus, are causative agents of lymphomas and other malignancies. The structural characterization of these viruses has been limited due to difficulties in obtaining adequate amount of virion particles. Here we report the first three-dimensional structural characterization of a whole gammaherpesvirus virion by an emerging integrated approach of the technique of cryo-electron tomography combined with single-particle cryo-electron microscopy, using murine gammaherpesvirus-68 (MHV-68) as a model system. We found that the MHV-68 virion consists of distinctive envelope and tegument compartments, and a highly conserved nucleocapsid. Two layers of tegument are identified: an inner tegument layer tethered to the underlying capsid and an outer, flexible tegument layer conforming to the overlying, pleomorphic envelope, consistent with the sequential viral tegumentation process inside host cells. Surprisingly, comparison of the MHV-68 virion and capsid reconstructions shows that the interactions between the capsid and inner tegument proteins are completely different from those observed in alpha and betaherpesviruses. These observations support the notion that inner layer tegument across different subfamilies of herpesviruses have evolved significantly to confer specific characteristics related to viral-host interactions, in contrast to a highly conserved capsid for genome encapsidation and protection.
Keywords: cryo-electron tomography, cryo-electron microscopy, tumor herpesvirus, murine gammaherpesvirus-68, tegument
Introduction
Gammaherpesviruses are a group of DNA tumor viruses that form a subfamily of the Herpesviridae (Chang et al. 1994; Damania et al. 2000; Ganem 1997; Roizman and Pellett 2001). The two known human members of the subfamily, Epstein-Barr virus (EBV) and Kaposi’s sarcoma–associated herpesvirus (KSHV), are significant pathogens associated with lymphomas and other malignancies (Carbone 2003; Chang et al. 1994; Damania et al. 2000; Damania et al. 1999; Mueller 1999). Along with herpes simplex virus-1 (HSV-1) -- the prototypic alphaherpesvirus that causes cold sores, and human cytomegalovirus (HCMV) -- a betaherpesvirus that is a leading cause of birth defects, gammaherpesviruses have a characteristic multilayered architecture. An infectious virion contains a double-stranded (ds) DNA genome, an icosahedral capsid shell, a thick, proteinaceous tegument compartment, and a lipid bilayer envelope spiked with glycoproteins (Liu and Zhou 2006; Rixon 1993; Steven and Spear 1997). The tegument, which surrounds the capsid and interacts with the envelope, is the least understood component, yet plays important roles in multiple aspects of the viral life cycle, including capsid transport to the nucleus, virion assembly and egress (Mettenleiter 2002; Steven and Spear 1997). The proximity of the tegument proteins to the surface of the viral capsid suggests that they may also be involved in modulating the interactions between viral particles and the host cellular proteins upon entry into the cell (Castillo and Kowalik 2002; Morrison et al. 1998).
The three-dimensional (3D) structures of the tegument and envelope have been difficult to characterize because of their asymmetry and fluidity. In gammaherpesviruses, they have been even more elusive due to additional difficulties in growing both EBV and KSHV in cell culture. KSHV, for example, tends to undergo latent replication in culture, and requires chemical treatment to induce lytic replication (Renne et al. 1996). These have made it difficult to study gammaherpesviruses using structural techniques such as single-particle cryo-electron microscopy (cryoEM), which requires highly concentrated samples.
We became interested in murine gammaherpesvirus-68 (MHV-68) as an alternative model for studying gammaherpesvirus structure, and in particular, the structural organization of the tegument compartment and its interaction with the underlying capsid. MHV-68 replicates to high titer in cell culture, and has been used widely to study gammaherpesvirus pathogenesis and infection (Ebrahimi et al. 2003; Virgin et al. 1997). Mass spectrometry has identified several virion-associated MHV-68 proteins -- including the five basic capsid proteins, and several tegument proteins and envelope glycoproteins -- that are homologous to proteins in EBV and KSHV (Bortz et al. 2003).
Cryo-electron tomography (cryoET) is an emerging structural tool for examining macromolecule complexes with heterogeneous, pleomorphic or dynamic characteristics (Baumeister 2002; McEwen and Marko 2001; Wagenknecht et al. 2002). Rather than averaging images of many different particles, as is the case in single-particle cryoEM, cryoET reconstructs a 3D volume of a single, asymmetric particle by combining images of the same particle taken over a range of tilt angles. Here we took an integrative approach of cryoET and single-particle cryoEM to determine the 3D structural organization of the MHV-68 virion and a subnanometer resolution structure of the MHV-68 capsid. We showed that the gammaherpesvirus has a highly conserved capsid structure with other herpesviruses. However, its tegument compartment is organized in an outer layer and a distinctive inner layer interacting with the underlying capsid in a manner completely different from those in alpha and betaherpesviruses. These results suggest that, while maintaining a relatively unchanged capsid to perform its conserved role of DNA packaging, herpesviruses have evolved their tegument compartments significantly to impart specificity in virion morphogenesis and other characteristics unique among alpha, beta and gammaherpesviruses and their infection processes.
Results
MHV-68 capsid structure by single-particle cryoEM
MHV-68 capsids were purified from host cell nuclei prior to tegumentation and envelopment, and were imaged in a 200-kV cryoEM instrument for 3D reconstruction. The 3D structure of MHV-68 capsid (Fig. 1B) was reconstructed by averaging over 4,000 particle images (Fig. 1A). Each icosahedrally averaged capsid structure has a polyhedral shape and consists of 12 pentons and 150 hexons, which are clamped together by 320 connecting triplexes on a T=16 icosahedral lattice (Fig. 1B). The unique structural building block responsible for forming the icosahedron is comprised of 1 pentameric subunit, hexons, and triplexes (for nomenclature and color coding see Fig.1C, D). Furthermore, detailed examination of the capsid shell interior revealed complex interactions between the structural components that were mediated through rod-shaped densities corresponding to long α helices. Interestingly, similar interactions were observed in HSV-1 suggesting the conservation of these structural elements across herpesvirus families (Baker et al. 2005; Zhou et al. 2000). We concluded that not only the structural organization and molecular interactions, but also many of the secondary structure elements within the capsid proteins are conserved among the capsids of MHV-68 and other herpesviruses.
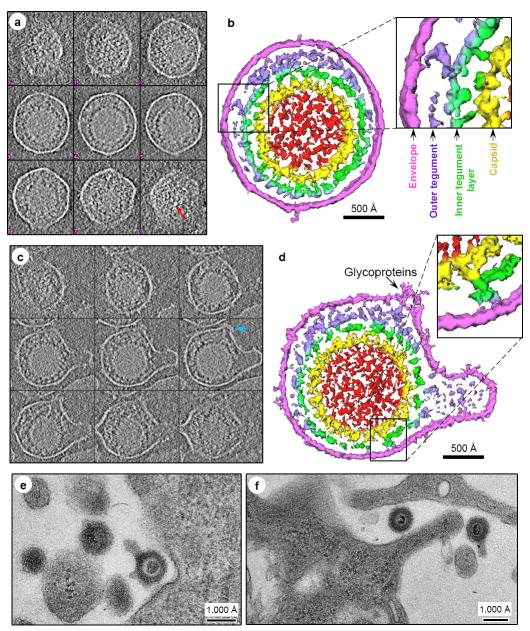
Figure 1. 3D structure of MHV-68 capsid by single-particle cryoEM.
(A) CryoEM digital image of ice-embedded MHV-68 capsids showing particle distribution. (B) Shaded surface representation of the MHV-68 capsid reconstruction as viewed along a threefold axis and colored according to radius. Three pentons (P), three structurally unique hexons (Hp, He, Hc for P, E and C hexons, respectively), and six structurally unique triplexes (Ta, Tb, Tc, Td, Te, and Tf) are labeled. (C) Front and (D) rear views of color coded unique components of the capsid comprising an asymmetric unit, including 2 and1/2 hexons, 1 pentameric subunit, and 5 and 1/3 triplexes. The full penton and E hexon are included for clarity. Color coding: yellow: penton; red: hexon c; light-blue: hexon p; dark green: hexon e; magenta: triplex a; purple: triplex b; orange: triplex c; light green: triplex d; blue: triplex e; beige: triplex f. (E) The interior of the capsid shell reveals interactions between capsomeric subunits mediated through sausage-shaped helices (arrow).
MHV-68 virion structure by cryoET
In order to reveal the structural organization of the pleomorphic components in MHV-68 virions, we performed 3D reconstruction of the infectious virions by cryoET. A total of 7 tomograms containing 20 MHV-68 particles were reconstructed. Figure 2A shows representative original image frames from one of these cryoET tilt series. Because particular care was taken during cryoET imaging to choose the thinnest possible vitreous ice while keeping the virions fully embedded, sufficient image contrast was obtained for image alignment without the use of gold fiducial markers. The resolutions of these tomograms were estimated to be around 6-8 nm based on geometric parameters of the tilt series (Radermacher 1992). The good quality of our reconstructions is reflected by the resolution of individual capsomers in the tomograms (Fig. 2B-D, see also Supplementary Video S1). The naked capsid visible in the field was used to validate the tomographic reconstruction by comparing it with the capsid reconstruction obtained by single-particle cryoEM averaging (Fig. 1B). The typical capsid morphology can be clearly seen in the 15-Å-thick slices from the tomogram (Fig. 2C). Also, the T=16 icosahedral lattice was clearly resolved, allowing the identification of pentons and hexons without ambiguity (Fig. 2D). Therefore, our tomograms obtained using the “marker-free” alignment approach (Winkler and Taylor 2006) were of sufficient quality to reliably discern structural features of the virion particles.
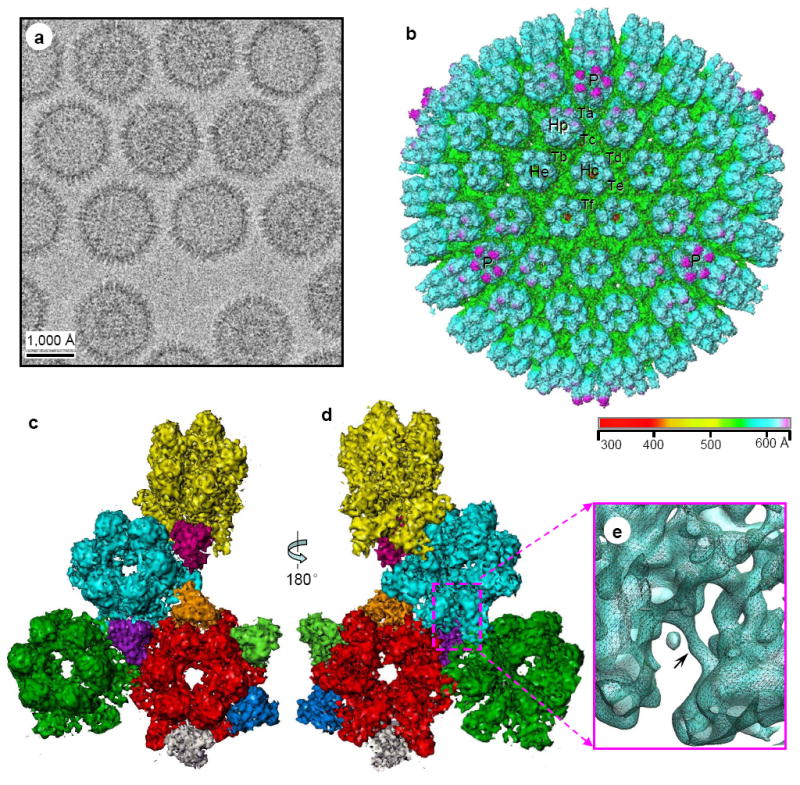
Figure 2. CryoET tilt series and quality assessment of the tomograms (see Supplementary Movie 1).
(A) Representative image frames from a typical cryoET tilt series of ice-embedded MHV-68 particles. This tilt series is chosen because a naked capsid (white arrowhead) is present in the field for validation of 3D reconstruction. A small portion of the carbon support film (indicated by black arrow) of the holey grid was included to reduce specimen charging during imaging. (B) A 15-Å thick slice taken from the 3D volume reconstructed from the tilt series in (A). The naked capsid is indicated by the arrowhead. (C) Density distribution in representative serial slices (15-Å thick each) extracted from the naked capsid indicated in (B). Note that individual capsomers are clearly revealed. (D) Individual capsomers, resembling those in the single-particle cryoEM reconstruction (Fig. 1B and Fig. 4B), are visible in the close-up of the slice boxed in (C). Hexons (H) and pentons (P) are indicated in the bottom panel. Following accepted convention, protein densities in the 3D reconstructions are displayed as white and background as dark, which is opposite to that used for displaying raw image data.
To illustrate key findings of the structural organization of the pleomorphic tegument and envelope of the MHV-68 virion, serial slices (15-Å thick each, 180 Å apart from each other) through the 3D volumes of two representative virions are shown as gray level displays (Fig. 3A and C), along with shaded surface representations of central slabs of the tomogram revealing molecular interactions (Fig. 3B, D). There is a substantial level of structural variability and pleomorphism in the structural organization of the tegument and envelope layers in the two virions although the structures of the capsids enclosed inside are indistinguishable (c.f.Fig. 3A vs. 3C and Fig. 3B vs. 3D). For example, the capsid is nearly centrally located in the first particle (Fig. 3B) but eccentrically located in the second particle (Fig. 3D). The envelope of the virions is clearly a continuous layer surrounding the tegument and sparsely spiked with densities of 100-150 Å in height, which we attributed to viral glycoproteins. The spiky glycoprotein densities are not evenly distributed on the envelope, and most notably, seem to cluster around regions with membrane distortion or protrusions, or envelope “blebs” (Fig. 3D). Although comparisons of only two particles are shown here, the pleomorphic nature of the tegument and envelope structures and the clustering of spiky glycoproteins are a general observation among all our tomograms.
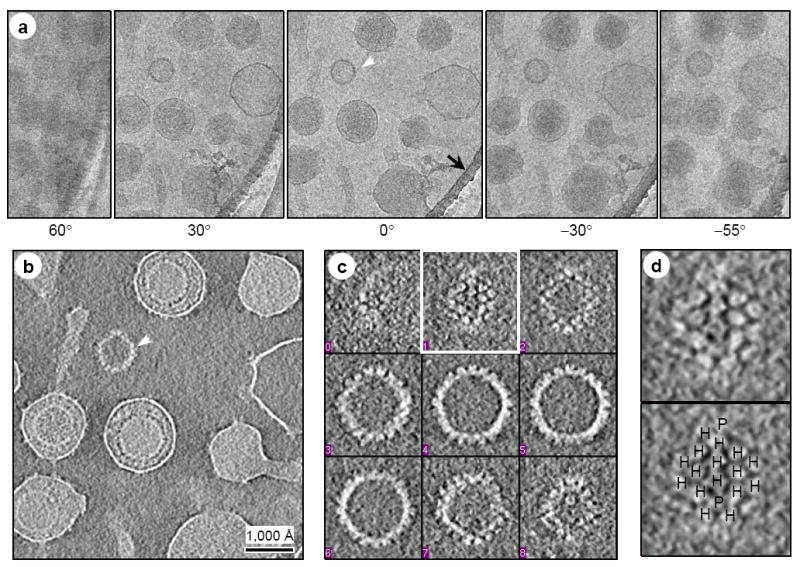
Figure 3. CryoET reconstructions of MHV-68 virions.
(A, C) Serial, evenly-spaced slices were selected from two representative MHV-68 virion tomograms, one in which the capsid is centrally located (A) and another in which the tegument and membrane are distorted (C). Red arrow in (A) indicates visible capsomers and blue arrow in (C) indicates a cluster of glycoprotein densities. (B, D) Shaded surface representations of central slabs (150 Å thick) of the virions segmented from the tomograms shown in (A, C), with close-ups showing tegument-capsid and tegument-envelope interactions. Color coded according to distance from the capsid center: red, internal densities in capsid; yellow, capsid shell; green, inner layer tegument; violet, flexible/variable outer layer tegument that conforms to shape of viral envelope; magenta, viral envelope. (E, F) EM images of thin sections of 3T3 cell infected with MHV-68 virions. During attachment and entry, both the circular shaped virions and virions with envelope protrusions of variable numbers and lengths are observed under the physiological infection conditions.
The tegument is organized into two distinctive layers. The inner layer (green in Fig. 3) forms a network of density enclosing the capsid, and interacts directly with the capsid through linker densities. The outer tegument (violet in Fig. 3) occupies the space between the inner layer and the envelope, and is much more loosely organized, as it conforms to the irregular shape of the viral envelope. Among most of the virions we examined, the outer tegument layer does not encircle the entire underlying inner tegument sphere completely, permitting the inner layer tegument layer to interact directly with the viral envelope (Fig. 3D).
Unique tegument/capsid interactions in gammaherpesvirus
It was previously shown that the capsid-proximal tegument proteins interact with the underlying capsid in an icosahedrally ordered manner in both alpha- (Zhou et al. 1999) and beta-herpesvirus (Chen et al. 1999; Trus et al. 1999). To see whether the inner tegument proteins of gammaherpesvirus also interact with the capsid in such manner, we performed single-particle cryoEM reconstruction by averaging 576 particle images of MHV-68 virions and compared it with the MHV-68 capsid reconstruction at the same resolution (Fig. 4A-C). Because the 3D reconstructions were performed by averaging a large number of particles and by imposing icosahedral symmetry, any structural components that are variably attached to the capsid and do not conform to the imposed symmetry would be reduced in signal. Indeed, the shaded surface representation of the virion reconstruction (Fig. 4B, top, left hemisphere) is characterized by spotty, disconnected low tegument densities surrounding the capsid. When displayed at a higher density contour level (Fig. 4B, top, right hemisphere), the spotty densities disappeared, revealing a structure that is identical to that of the naked capsid (Fig. 4B, bottom). Central slices of the reconstruction confirmed that the capsid in the virion is surrounded by a thick, yet low density layer corresponding to the overlying tegument (Fig. 4C, top). The weakness of the density of the tegument layer indicates that the tegument proteins do not conform to icosahedral symmetry, or only fill up a limited number of available binding sites on the capsid.
Figure 4. Different inner-layer-tegument/capsid interactions in gammaherpesvirus from those in alpha and betaherpesviruses by single-particle cryoEM averaging.
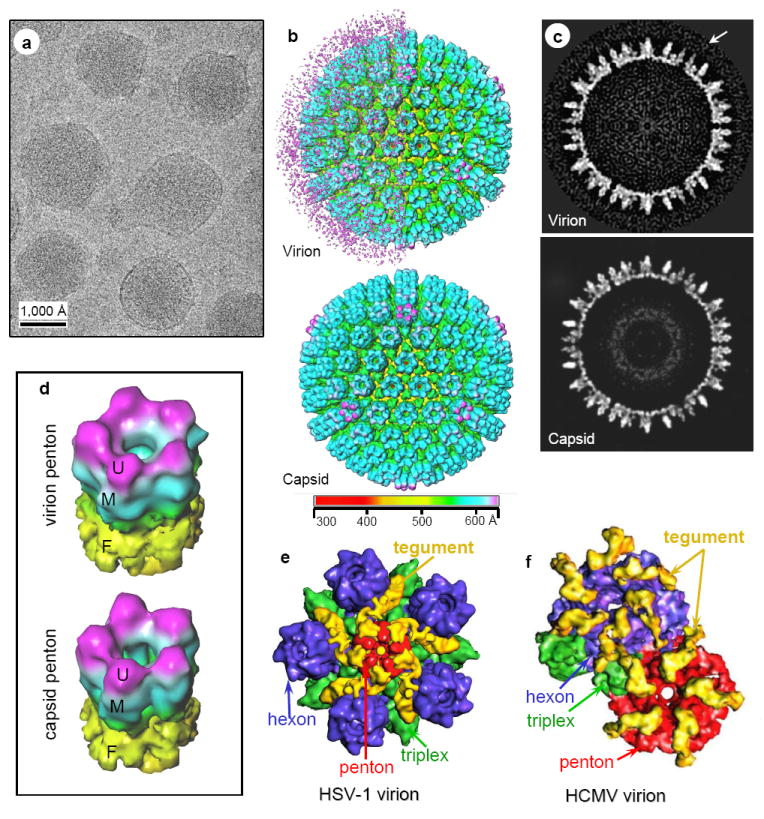
(A) CryoEM image of MHV-68 virions. (B) Shaded surface representations of MHV-68 reconstructions (30 Å resolutions) viewed along a threefold axis and colored according to radius. Top, Virion reconstruction with left hemisphere displayed at low threshold to show the layer of unordered structure surrounding the capsid. Right hemisphere is displayed at slightly higher density threshold, thus only revealing the enclosed capsid. Bottom, capsid reconstruction. (C) Central slices from the virion (top) and the capsid (bottom) reconstructions, taken perpendicular to a threefold axis. Arrow indicates tegument density layer surrounding capsid surface in the virion. (D) Tilted views of pentons computationally extracted from the virion (top) and capsid (bottom) reconstructions. U, upper domain; M, middle domain; F, floor domain. (E) Close-up view of a computationally extracted portion of the HSV-1 virion reconstruction. The tegument densities, colored in yellow, make contact with the upper domains of penton, and its adjacent triplexes, but not the upper domains of any hexons. (F) Close-up view of a computationally isolated portion of the HCMV virion reconstruction. The tegument densities interact with the upper domains of all penton, hexon and triplex by attaching one end to the upper domain of a hexon (or penton), and anchoring the other end on the upper domain of the adjacent triplex. Color codes in (E) and (F): red, penton; blue, hexon; green, triplex; yellow, tegument.
Because the capsid-proximal tegument proteins of HSV-1 and HCMV are known to attach to the pentons symmetrically, we computationally extracted pentons from our single-particle cryoEM reconstructions of MHV-68 virion and capsid for more detailed comparison (Fig. 4D). The virion and capsid pentons were nearly identical in shape and size, and there are no extra densities attached to the virion pentons. This observation is stark in contrast to the prominent tegument densities attached to the pentons of HSV-1 (Zhou et al. 1999) and HCMV (Chen et al. 1999), as exemplified by Fig. 4E and 4F, respectively. Furthermore, the MHV-68 pentons appear identical to those of other gammaherpesvirus capsids (Trus et al. 2001; Wu et al. 2000; Yu et al. 2003). Therefore, the pattern of interactions between the tegument layer and its underlying capsid in MHV-68 completely differs from those in alpha and betaherpesviruses, despite their highly conserved capsids.
Discussion
Our results showed that the MHV-68 capsid structure is highly similar to those reported for HSV-1 (Zhou et al. 2000) and cytomegalovirus (Butcher et al. 1998; Chen et al. 1999; Trus et al. 1999; Yu et al. 2005), members of the alphaherpesvirus and betaherpesvirus subfamilies, respectively. This is consistent with mass spectrometry results showing that the MHV-68 capsid consists of proteins that are highly homologous to capsid proteins of other herpesviruses (Bortz et al. 2003). Using cryoET, we further visualized the tegument organization: an inner layer tethered to the capsid and an outer flexible/variable layer with proteins that conform to the pleomorphic shape of the envelope. This is the first reported gammaherpesvirus reconstruction that resolved structural features beyond the capsid.
The presence of the inner and outer tegument layers seems to be a conserved feature across members of the herpesvirus family; however, the tegument compartments across alpha, beta and gammaherpesviruses differ in their patterns of interactions with the underlying capsid. In the alphaherpesvirus HSV-1, the inner tegument proteins are icosahedrally arranged and appear as convoluted, ribbon-like structures that anchor one end at the upper domain of the pentons and extend the other end to neighboring triplexes (Fig. 4E) (Zhou et al. 1999). The bulk of the tegument density is centered on the pentons, and makes only minimal contact with neighboring P hexons. The inner tegument protein has a different structure in the betaherpesvirus HCMV, forming a thin, net-like shell that surrounds the entire capsid (Fig. 4F) (Chen et al. 1999). In particular, the filamentous tegument densities interact with the upper domain of all the pentons, hexons and triplexes. However, the gammaherpesvirus MHV-68 inner tegument layer differs from those in HSV-1 and HCMV in that it does not make symmetric contacts with the capsid proteins. Nevertheless, the existence of inner tegument among all herpesvirus subfamilies suggests a structural role of the inner layer tegument in capsid integrity maintenance. The net-like enclosure formed by the inner tegument proteins may provide additional strength to constrain the packed DNA which has a tendency to remain in an energetically favorable unbent state. Indeed, betaherpesviruses, which contain the largest DNA genome among all herpesviruses, have the most extensive inner tegument/capsid interactions.
CryoET data on the HSV-1 virion have shown the polar packaging of the outer layer tegument, and that the capsid appears to be eccentrically located within the tegument (Grunewald et al. 2003). In majority of MHV-68 viral particles, the outer tegument seems to be more loosely organized, allowing the capsid and its associated inner tegument layer to “float” inside the envelope (Fig. 3B, D). It is possible that the outer tegument proteins unique to MHV-68 may contribute to the differences in fluidity and polarity of the packed outer tegument proteins.
Our finding of the existence of a distinct inner layer tegument has important biological implications in herpesvirus assembly and egress. The existence of an inner tegument protein layer across different herpesviruses is consistent with a common tegumentation and egress pathway (Mettenleiter 2002) (Fig. 5). Herpesviruses are distinctive among all viruses for their large number of structural and functional components packaged within the envelope. In MHV-68, at least 10 proteins have been demonstrated to be packaged in the tegument compartment, and this number is still growing (Bortz et al. 2003; Virgin et al. 1997). It remains unknown how and with what structural integrity these proteins are specifically recruited and assembled in the tegument compartment. It is now well recognized that tegumentation of herpesvirus capsid involves complex protein-protein interactions in both the nucleus and cytoplasm, and is further convoluted by the repeated steps of envelopment, de-envelopment and re-envelopment process (Mettenleiter 2002). As depicted in Fig. 5, it is likely that some components of the capsid-associated inner tegument proteins are assembled inside the host nucleus at or near the nuclear lamina prior to budding through the inner nuclear membrane in the early stage of virion maturation. Our observation of the outer tegument layer conforming to the pleomorphic shape of viral envelope implies that the outer tegument proteins might be packaged into maturing virion particles through interactions with the viral glycoproteins decorated on the trans Golgi membrane (Chi et al. 2005; Farnsworth et al. 2007; Fuchs et al. 2002). As the first to be released into the cellular environment upon infection, outer layer proteins are probably essential viral functional proteins during the early stages of infection.
Figure 5. Proposed MHV-68 tegumentation pathway.
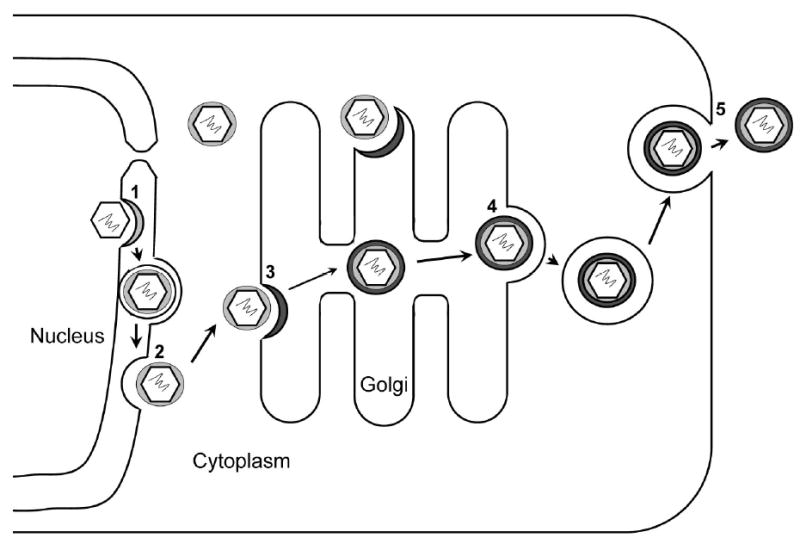
1. Acquirement of inner layer tegument proteins and envelopment by budding through the inner nuclear membrane. 2. De-envelopment at the outer nuclear membrane and release of partially tegumented capsids. 3. Acquisition of the outer tegument proteins and re-envelopment at the trans-Golgi apparatus. 4. Release of the virion-containing vesicles at the cis-Golgi. 5. Virion-containing vesicles fuse with cell membrane and virions are released to the extra-cellular space by exocytosis.
We also observed protrusions of various sizes and shapes, which we attribute to glycoprotein spikes, decorating the surface of the viral envelope. Their distribution seems to be random, although some envelope protruding regions contained a higher density of spikes (Fig. 3C, D). Although we cannot completely rule out the possibility that the pseudopod-like membrane extensions are simply artifacts introduced by the purification process, it is noteworthy that we tried several different purification procedures, including ficoll gradient centrifugation, skipping the initial pelleting step, and double sucrose cushion. Under all these conditions, the envelope blebs were a consistent observation. On the contrary, when the same procedures were used to isolate HCMV virions, the envelope blebs were very rarely present. Interestingly, we have also observed these envelope protrusions in infection experiments, where we captured images of virion particles during the attachment and entry process. These virions have a variety of shapes, including circular, as well as irregular with envelope protruding structures (Fig. 3E, F). Finally, observations similar to the glycoprotein clustering of Fig. 3D had been reported for HSV-1 by immunogold electron microscopy (Stannard et al. 1987).
At the present resolution of our cryoET reconstructions, it was not possible to discern whether there are several distinct types of glycoproteins on the viral envelope, similar to the gB, gC and gD proteins of HSV-1 (Grunewald et al. 2003; Stannard et al. 1987). Sequence analysis and mass spectrometry studies had identified at least five glycoproteins associated with MHV-68 virion, gB (Bortz et al. 2003), gH, and glycoprotein 150 (gp150) with apparent molecular weights of 105kDa, 83kDa, and 150kDa, respectively, as well as glycoproteins encoded by the ORF27 and ORF28 genes (Bortz et al. 2003; May et al. 2005; May et al. 2005). The size of the spike densities indicates that the spikes are probably homo- or hetero-oligomers of the above proteins. However, the direct identification and demonstration of their oligomeric states await future higher resolution cryoET reconstructions, perhaps together with antibody labeling (Lo et al. 2003; Yu et al. 2005).
Material and Methods
Capsid and virion purification
Baby Hamster Kidney (BHK-21) cells were infected with MHV-68 at a multiplicity of infection (MOI) of either 0.01 (for virions) or 5 (for capsids). We isolated MHV-68 virions and B-capsids from the extracellular media or the nuclei of infected BHK-21 cells, respectively. When the cultured cells exhibited 90% cytopathic effect, they were pelleted by centrifuging at 1,000 g for 10 min at 4°C.
Supernatants were collected for virion isolation by first centrifugation through a 20% sucrose cushion at 83,000 g for 1 h. The resultant pellet was resuspended and overlaid on a 15–35% potassium tartrate and 30–0% glycerol gradient, and centrifuged at 250,000 g for 20 min. The visible bands were collected and checked for virions using negative stain transmission electron microscopy. The virion-containing band was resuspended in phosphate buffered saline (PBS) and concentrated by centrifuging at 80,000 g for 60 min. In our latter experiments, ficoll gradient (5-20%) was also used to reduce possible osmotic effects due to sucrose. In this case, clear bands were visible in the gradient after centrifugation at 15,500 g for 2 h, which was followed by a resuspending and concentrating step at 28,500 g for 2 h.
For capsids, the cell pellets were washed with PBS and centrifuged at 1,000 g for 10 min at 4°C. Nuclei were released from the cells by resuspending the cell pellet in 50 ml of PBS containing 1% NP-40, incubating on ice for 5 min, and collected by centrifuging at 3,000 g for 5 min at 4°C. The nuclei in the pellet were then broken by resuspending the nuclear pellet in 7 ml of NTE (500 mM NaCl, 10 mM Tris, 1 mM EDTA pH 7.5) containing 1% NP-40 and passaging through a 23-gauge syringe (Becton-Dickinson) for 20 times. The resulting solution, containing free capsids, was centrifuged at 3,000 g for 5 min, overlaid on 5 ml of a double-layered sucrose cushion (4 ml of 41% sucrose in NTE over 1 ml of 60% sucrose in PBS and 1% NP-40). The capsids were centrifuged through the top layer at 98,000 g for 70 min at 4°C. The band located at the interface of the double-layered cushion containing capsids was collected. The capsids were then concentrated using Centricon YM-100 filter tubes (Millipore). The capsids were further purified by centrifuging through a 22–50% sucrose density gradient with 1% NP-40 at 62,000 g for 50 min at 4°C. Three bands were visible and contained A-, B-, and C-capsids, respectively. The middle, B-capsid band was collected and pelleted by centrifugation at 75,000 g for 60 min and resuspended in 20 μl PBS for cryoEM imaging.
MHV-68 infection experiment and thin section imaging
3T3 cells were infected with MHV-68 viral stock at a MOI of 150 in cold room so that cell membrane is immobilized. After 2 hours, unbound viruses were removed by centrifugation at 500-1000g for 2 minutes. The pellet contained cells with attached viral particles and was washed for three times in PBS, and fixed with 3% glutaraldehyde. The fixed cell pellet was then embedded in LX-112 resin and cut to 100nm thin sections for imaging.
Cryo-electron microscopy
We used established single-particle cryoEM procedures (Booth et al. 2004) to image the MHV-68 virions and naked capsids. Briefly, 3 μl of purified sample was applied to 400 mesh, 3.5/1.0 μm holey grids (Quantifoil, Germany) and quickly frozen in liquid ethane, so that viral particles were suspended in a thin layer of vitreous ice across the holes of the supporting film.
Focal-pair image frames of naked capsids were acquired using a 200-kV JEOL 2010F electron microscope equipped with a field emission gun and a Gatan US4000 16 megapixel charge-coupled device (CCD) camera (Booth et al. 2004). Digital cryoEM images were recorded at an effective magnification of 83,100× on the camera (60,000× on the film plane), corresponding to an effective pixel size of 1.805 Å/pixel at the specimen level, with an electron dosage of approximately 12 electrons/Å2/micrograph. Focal-pair images were recorded and preprocessed for translation and rotation correction using the JAMES semiautomatic data collection program (Booth et al. 2004). The electron beam was under-focused, with a 1.5-μm difference between the close-to-focus (~1 μm under focus) and far-from-focus (~2.5 μm under focus) images. For reconstruction, all images were averaged by combining adjacent pixels to yield a final sampling size of 3.61 Å/pixel on the specimen scale. Micrographs of intact virions were taken as focal pairs at 30,000× magnification, with a dosage of ~9 electrons/Å2/micrograph in a JEOL JEM1200 cryo-electron microscope operated at 100 kV with a cold stage at −167°C. Selected micrographs were digitized with a SCAI microdensitometer (Carl Zeiss, Inc., Englewood, Colo.) using a step size equivalent to 4.67 Å/pixel.
We processed the cryoEM images and carried out 3D reconstructions using the IMIRS software package (Liang et al. 2002; Zhou and Chiu 2003). 3D visualization was done using Iris Explorer (NAG, Downers Grove, IL) with custom-designed modules. The effective resolution of the map was estimated based on the criterion that the Fourier shell cross-correlation coefficient between two independent reconstructions must be larger than 0.5.
Cryo-electron tomography
CryoET tilt series were obtained at 300kV in a CM300 cryoEM instrument (FEI, Hillsboro, OR) equipped with a 4 megapixel CCD camera (TVIPS, Gauting, Germany). Holey grids (Quantifoil, Germany) with larger grid squares (200 mesh, 3.5/1.0 μm) were used to prepare frozen hydrated MHV-68 virion samples for tomography data collection with a specimen temperature of about -172°C. Tilt angles ranging from −60° to 60° were chosen according to the cosine rule (Saxton et al. 1984), which uses decreasing angle increments for large tilting angles. The image data was recorded on CCD using the TVIPS EMMENU tomography program, with an electron dosage of ~1 electron/Å2/frame, and a total dosage of 60-80 electron/Å2 for the whole tilt series. The magnification used was 37,500×, giving a CCD sampling pixel size of 6.4 Å/pixel. The underfocus value of the zero-tilt image was set to 6 μm.
Data processing for tomographic reconstruction was carried out using the “marker-free” program package ProjAlign (Winkler and Taylor 2006). A distinctive feature of this package is that no gold fiducial markers are needed for data processing. Briefly, a preliminary tomogram was obtained by merging the aligned images in the series after low-pass filtering, using the goniometer readings as the tilt angles. Translational and rotational alignment, and tilt axis refinement were subsequently performed using the preliminary tomogram. Several rounds of refinement of the alignment and area matching were carried out until no significant improvement in the alignment parameters and the 3D volume was detectable. The final 3D map was then computed by weighted back projection algorithm (Radermacher 1992). 3D tomographic reconstructions were low-pass filtered to 60 Å and segmented using either Amira (Mercury Computer Systems, Inc., Chelmsford, MA) or Iris Explorer (NAG, Downers Grove, IL) with custom-designed modules.
Supplementary Material
Acknowledgments
We thank Pierrette Lo for assistance in manuscript preparation. This research was supported by National Institutes of Health grants (CA94809 and AI069015 to ZHZ, CA091791/DE015752 to RS and GM30598 to KAT). WD is partially supported by a training fellowship from the W.M. Keck Foundation to the Gulf Coast Consortia. We acknowledge the use of cryoEM facility at the National Center for Macromolecular Imaging (NIH P41RR02250 to Wah Chiu).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baker ML, Jiang W, Rixon FJ, Chiu W. Common ancestry of herpesviruses and tailed DNA bacteriophages. J Virol. 2005;79:14967–14970. doi: 10.1128/JVI.79.23.14967-14970.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumeister W. Electron tomography: towards visualizing the molecular organization of the cytoplasm. Curr Opin Struct Biol. 2002;12:679–684. doi: 10.1016/s0959-440x(02)00378-0. [DOI] [PubMed] [Google Scholar]
- Booth CR, Jiang W, Baker ML, Zhou ZH, Ludtke SJ, Chiu W. A 9 angstroms single particle reconstruction from CCD captured images on a 200 kV electron cryomicroscope. J Struct Biol. 2004;147:116–127. doi: 10.1016/j.jsb.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Bortz E, Whitelegge JP, Jia Q, Zhou ZH, Stewart JP, Wu TT, Sun R. Identification of proteins associated with murine gammaherpesvirus 68 virions. J Virol. 2003;77:13425–13432. doi: 10.1128/JVI.77.24.13425-13432.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher SJ, Aitken J, Mitchell J, Gowen B, Dargan DJ. Structure of the human cytomegalovirus B capsid by electron cryomicroscopy and image reconstruction. J Struct Biol. 1998;124:70–76. doi: 10.1006/jsbi.1998.4055. [DOI] [PubMed] [Google Scholar]
- Carbone A. Emerging pathways in the development of AIDS-related lymphomas. Lancet Oncol. 2003;4:22–29. doi: 10.1016/s1470-2045(03)00957-4. [DOI] [PubMed] [Google Scholar]
- Castillo JP, Kowalik TF. Human cytomegalovirus immediate early proteins and cell growth control. Gene. 2002;290:19–34. doi: 10.1016/s0378-1119(02)00566-8. [DOI] [PubMed] [Google Scholar]
- Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- Chen DH, Jiang H, Lee M, Liu F, Zhou ZH. Three-dimensional visualization of tegument/capsid interactions in the intact human cytomegalovirus. Virology. 1999;260:10–16. doi: 10.1006/viro.1999.9791. [DOI] [PubMed] [Google Scholar]
- Chi JH, Harley CA, Mukhopadhyay A, Wilson DW. The cytoplasmic tail of herpes simplex virus envelope glycoprotein D binds to the tegument protein VP22 and to capsids. J Gen Virol. 2005;86:253–261. doi: 10.1099/vir.0.80444-0. [DOI] [PubMed] [Google Scholar]
- Damania B, Choi JK, Jung JU. Signaling activities of gammaherpesvirus membrane proteins. J Virol. 2000;74:1593–1601. doi: 10.1128/jvi.74.4.1593-1601.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damania B, Lee H, Jung JU. Primate herpesviral oncogenes. Mol Cells. 1999;9:345–349. [PubMed] [Google Scholar]
- Ebrahimi B, Dutia BM, Roberts KL, Garcia-Ramirez JJ, Dickinson P, Stewart JP, Ghazal P, Roy DJ, Nash AA. Transcriptome profile of murine gammaherpesvirus-68 lytic infection. J Gen Virol. 2003;84:99–109. doi: 10.1099/vir.0.18639-0. [DOI] [PubMed] [Google Scholar]
- Farnsworth A, Wisner TW, Johnson DC. Cytoplasmic residues of herpes simplex virus glycoprotein gE required for secondary envelopment and binding of tegument proteins VP22 and UL11 to gE and gD. J Virol. 2007;81:319–331. doi: 10.1128/JVI.01842-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs W, Klupp BG, Granzow H, Hengartner C, Brack A, Mundt A, Enquist LW, Mettenleiter TC. Physical interaction between envelope glycoproteins E and M of pseudorabies virus and the major tegument protein UL49. J Virol. 2002;76:8208–8217. doi: 10.1128/JVI.76.16.8208-8217.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem D. KSHV and Kaposi’s sarcoma: the end of the beginning? Cell. 1997;91:157–160. doi: 10.1016/s0092-8674(00)80398-0. [DOI] [PubMed] [Google Scholar]
- Grunewald K, Desai P, Winkler DC, Heymann JB, Belnap DM, Baumeister W, Steven AC. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science. 2003;302:1396–1398. doi: 10.1126/science.1090284. [DOI] [PubMed] [Google Scholar]
- Liang Y, Ke EY, Zhou ZH. IMIRS: a high-resolution 3D reconstruction package integrated with a relational image database. J Struct Biol. 2002;137:292–304. doi: 10.1016/s1047-8477(02)00014-x. [DOI] [PubMed] [Google Scholar]
- Liu F, Zhou ZH. In: Comparative virion structures of human herpesviruses Human Herpesviruses: Biology, Therapy and Immunoprophylaxis. Arvin A, Campadelli-Fiume G, Moore P, et al., editors. Cambridge, UK: Cambridge University Press; 2006. pp. 27–42. [Google Scholar]
- Lo P, Yu X, Atanasov I, Chandran B, Zhou ZH. Three-Dimensional Localization of pORF65 in Kaposi’s Sarcoma-Associated Herpesvirus Capsid. J Virol. 2003;77:4291–4297. doi: 10.1128/JVI.77.7.4291-4297.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May JS, Colaco S, Stevenson PG. Glycoprotein M is an essential lytic replication protein of the murine gammaherpesvirus 68. J Virol. 2005;79:3459–3467. doi: 10.1128/JVI.79.6.3459-3467.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May JS, Walker J, Colaco S, Stevenson PG. The murine gammaherpesvirus 68 ORF27 gene product contributes to intercellular viral spread. J Virol. 2005;79:5059–5068. doi: 10.1128/JVI.79.8.5059-5068.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen BF, Marko M. The emergence of electron tomography as an important tool for investigating cellular ultrastructure. J Histochem Cytochem. 2001;49:553–564. doi: 10.1177/002215540104900502. [DOI] [PubMed] [Google Scholar]
- Mettenleiter TC. Herpesvirus assembly and egress. J Virol. 2002;76:1537–1547. doi: 10.1128/JVI.76.4.1537-1547.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison EE, Stevenson AJ, Wang YF, Meredith DM. Differences in the intracellular localization and fate of herpes simplex virus tegument proteins early in the infection of Vero cells. J Gen Virol. 1998;79:2517–2528. doi: 10.1099/0022-1317-79-10-2517. [DOI] [PubMed] [Google Scholar]
- Mueller N. Overview of the epidemiology of malignancy in immune deficiency. J Acquir Immune Defic Syndr. 1999;21:S5–10. [PubMed] [Google Scholar]
- Radermacher M. Weighted back-projection methods. In: Frank J, editor. Electron Tomography - Three-dimensional imaging with the Transmission Electron Microscopy. New York: Plenum Press; 1992. pp. 91–115. [Google Scholar]
- Renne R, Zhong W, Herndier B, McGrath M, Abbey N, Kedes D, Ganem D. Lytic growth of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat Med. 1996;2:342–346. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- Rixon FJ. Structure and assembly of herpesviruses. Seminars in Virology. 1993;4:135–144. [Google Scholar]
- Roizman B, Pellett PE. In: Herpesviridae: A Brief Introduction Fields Virology. Fields BN, Knipe DM, Howley PM, et al., editors. Vol. 2. Philadelphia: Lippincott Williams & Wilkins; 2001. pp. 2381–2398. [Google Scholar]
- Saxton WO, Baumeister W, Hahn M. Three-dimensional reconstruction of imperfect two-dimensional crystals. Ultramicroscopy. 1984;13:57–70. doi: 10.1016/0304-3991(84)90057-3. [DOI] [PubMed] [Google Scholar]
- Stannard LM, Fuller AO, Spear PG. Herpes simplex virus glycoproteins associated with different morphological entities projecting from the virion envelope. Journal of General Virology. 1987;68:715–725. doi: 10.1099/0022-1317-68-3-715. [DOI] [PubMed] [Google Scholar]
- Steven AC, Spear PG. In: Herpesvirus capsid assembly and envelopment Structural Biology of Viruses. Chiu W, Burnett RM, Garcea R, editors. New York: Oxford University Press; 1997. pp. 312–351. [Google Scholar]
- Trus BL, Gibson W, Cheng N, Steven AC. Capsid structure of simian cytomegalovirus from cryoelectron microscopy: evidence for tegument attachment sites. J Virol. 1999;73:2181–2192. doi: 10.1128/jvi.73.3.2181-2192.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trus BL, Heymann JB, Nealon K, Cheng N, Newcomb WW, Brown JC, Kedes DH, Steven AC. Capsid structure of Kaposi’s sarcoma-associated herpesvirus, a gammaherpesvirus, compared to those of an alphaherpesvirus, herpes simplex virus type 1, and a betaherpesvirus, cytomegalovirus. J Virol. 2001;75:2879–2890. doi: 10.1128/JVI.75.6.2879-2890.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgin HW, Latreille P, Wamsley P, Hallsworth K, Weck KE, Dal Canto AJ, Speck SH. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J Virol. 1997;71:5894–5904. doi: 10.1128/jvi.71.8.5894-5904.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagenknecht T, Hsieh CE, Rath BK, Fleischer S, Marko M. Electron tomography of frozen-hydrated isolated triad junctions. Biophys J. 2002;83:2491–2501. doi: 10.1016/S0006-3495(02)75260-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler H, Taylor KA. Accurate marker-free alignment with simultaneous geometry determination and reconstruction of tilt series in electron tomography. Ultramicroscopy. 2006;106:240–254. doi: 10.1016/j.ultramic.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Wu L, Lo P, Yu X, Stoops JK, Forghani B, Zhou ZH. Three-dimensional structure of the human herpesvirus 8 capsid. J Virol. 2000;74:9646–9654. doi: 10.1128/jvi.74.20.9646-9654.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Shah S, Atanasov I, Lo P, Liu F, Britt WJ, Zhou ZH. Three-dimensional localization of the smallest capsid protein in the human cytomegalovirus capsid. J Virol. 2005;79:1327–1332. doi: 10.1128/JVI.79.2.1327-1332.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Trang P, Shah S, Atanasov I, Kim YH, Bai Y, Zhou ZH, Liu F. Dissecting human cytomegalovirus gene function and capsid maturation by ribozyme targeting and electron cryomicroscopy. Proc Natl Acad Sci U S A. 2005;102:7103–7108. doi: 10.1073/pnas.0408826102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu XK, O’Connor CM, Atanasov I, Damania B, Kedes DH, Zhou ZH. Three-dimensional structures of the A, B, and C capsids of rhesus monkey rhadinovirus: insights into gammaherpesvirus capsid assembly, maturation, and DNA packaging. J Virol. 2003;77:13182–13193. doi: 10.1128/JVI.77.24.13182-13193.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZH, Chen DH, Jakana J, Rixon FJ, Chiu W. Visualization of tegument-capsid interactions and DNA in intact herpes simplex virus type 1 virions. J Virol. 1999;73:3210–3218. doi: 10.1128/jvi.73.4.3210-3218.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZH, Chiu W. Determination of icosahedral virus structures by electron cryomicroscopy at subnanometer resolution. Advances in Protein Chemistry. 2003;64:93–124. doi: 10.1016/s0065-3233(03)01003-9. [DOI] [PubMed] [Google Scholar]
- Zhou ZH, Dougherty M, Jakana J, He J, Rixon FJ, Chiu W. Seeing the herpesvirus capsid at 8.5 Å. Science. 2000;288:877–880. doi: 10.1126/science.288.5467.877. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.