Abstract
Purpose
The Cancer Vaccine Consortium of the Cancer Research Institute (CVC-CRI) conducted a multicenter HLA-peptide multimer proficiency panel (MPP) with a group of 27 laboratories to assess the performance of the assay.
Experimental design
Participants used commercially available HLA-peptide multimers and a well characterized common source of peripheral blood mononuclear cells (PBMC). The frequency of CD8+ T cells specific for two HLA-A2-restricted model antigens was measured by flow cytometry.
The panel design allowed for participants to use their preferred staining reagents and locally established protocols for both cell labeling, data acquisition and analysis.
Results
We observed significant differences in both the performance characteristics of the assay and the reported frequencies of specific T cells across laboratories. These results emphasize the need to identify the critical variables important for the observed variability to allow for harmonization of the technique across institutions.
Conclusions
Three key recommendations emerged that would likely reduce assay variability and thus move toward harmonizing of this assay. (1) Use of more than two colors for the staining (2) collect at least 100,000 CD8 T cells, and (3) use of a background control sample to appropriately set the analytical gates. We also provide more insight into the limitations of the assay and identified additional protocol steps that potentially impact the quality of data generated and therefore should serve as primary targets for systematic analysis in future panels. Finally, we propose initial guidelines for harmonizing assay performance which include the introduction of standard operating protocols to allow for adequate training of technical staff and auditing of test analysis procedures.
Electronic supplementary material
The online version of this article (doi:10.1007/s00262-009-0681-z) contains supplementary material, which is available to authorized users.
Keywords: HLA-peptide multimer, Tumor immunity, Flow cytometry, CTL
Introduction
A large number of clinical vaccination trials have been conducted in the last 15 years in cancer patients [1]. Only a few vaccine candidates, however, have advanced to phase III clinical trials, and none have been approved so far [2]. Recent insights into the mechanisms that regulate immune responses against tumors have led to the identification of a broad selection of compounds (antibodies, small-molecules, cytokines, TLR ligands) that have the potential to increase the efficacy of vaccine regimens [3]. However, the number of combinations that can be brought to advanced stages of clinical testing is limited due to the complex approval process for investigational new drugs. While it is not possible to predict clinical outcome of therapeutic tumor vaccination by results obtained from T cell immunomonitoring assays, development of validated surrogate immunological end points for tumor vaccine activity and ideally efficacy could dramatically accelerate their clinical development [4]. Consequently, the search for such surrogate immunological assays has become a high priority task for the field.
For cancer vaccines, this search has led to the continuous development of more powerful and sensitive assays to monitor immune responses in vaccinated patients [5, 6]. Indeed, improved immunization strategies and new monitoring techniques have led to a steady increase of the fraction of cancer patients with observed vaccine-related immune responses [7–10]. Nevertheless, clinical responses occur in a small fraction of patients, which may correlate with the presence of a specific immune response [11–13]. The apparent disconnect between immunomonitoring data and clinical events may be explained by several factors. Measuring a single immune system parameter in one tissue compartment, mostly IFN-gamma secreting T cells in the peripheral blood, as is the case for the majority of clinical trials thus far, might not be sufficient to capture an immunological signature correlating with the development of an appropriate anti-tumor response. There is more and more data suggesting that multiple functions of antigen-specific T cells rather than, or in addition to, T cell numbers correlate with clinical outcome, as described in studies of protective immune responses to microbial infections in mice and humans [14–16]. There is further evidence that classical T cell assays will have to be used to measure responses in the most appropriate tissue compartment [17, 18]. An additional important factor to consider for the observed lack of correlation between results from immunomonitoring and clinical events is the use of non standardized and non-validated immune monitoring assays. The use of such assays precludes the direct comparison of results obtained by labs across institutions [19] and also significantly impacts the ability to compare data obtained from different patients and time points within the same study. Thus, immune monitoring assays need to be standardized, validated (or at a minimum qualified) and auditable within laboratories before they can be appropriately applied to evaluate samples obtained from clinical trials. Finally, only qualified assays can be used to effectively guide the development of new drugs [20] or serve as surrogate clinical endpoints.
Approximately 3 years ago, two international associations (CVC-CRI and the Association for Immunotherapy of Cancer) initiated programs to address immune assay standardizations within the cancer scientific community [21]. A similar effort is ongoing in the field of infectious disease, mainly driven by HIV-researchers [22]. The primary goals of the CVC-CRI proficiency panel program are to allow for the harmonization of immune monitoring assays across institutions to a degree needed to represent stable biomarker assays and to provide for an external quality assurance resource for laboratories participating in the proficiency panel. The specific aims of the first CVC-CRI multimer proficiency panel presented in this manuscript were to (1) demonstrate the feasibility of such large international inter-laboratory testing project for HLA-peptide multimers, (2) identify protocol variables, reagent choices and strategies that are relevant in the formulation of harmonization guidelines for assay protocol optimization (3) provide each participating lab direct feed-back about their qualitative and quantitative performance in relation to the other members of the group (external-validation), (4) quantify the variation of results reported by such a large number of labs (inter-center variation), (5) determine the variation of results obtained within the same lab using the same protocol to quantify antigen-specific CD8+ T cells with the same samples doing the experiment at two different time points (intra-center variation).
Materials and methods
Panel design
The first CVC multimer panel was conducted with a group of 27 centers. Each lab had to determine the frequency of CD8+ T cells specific for two model antigens in cryopreserved peripheral blood mononuclear cell (PBMC) samples from five donors (D1–D5). All participants used either HLA-peptide tetramers or pentamers that were generously donated by Beckman Coulter (Fullerton, CA) and Proimmune (Oxford, UK), respectively. Two labs used both tetramers and pentamers and generated two separate data sets. Consequently, we were able to collect 29 complete data sets in a first step of testing. All participants were further offered the possibility to receive a second set of PBMC batches to allow for repetition of the experiments in a second step. Nine of the 27 labs made use of this offer and completed a second round of testing of the same PBMC samples with one lab contributing two separate data sets.
Participants and organizational setup
Participating laboratories were located in eight countries (Belgium, Canada, Germany, France, Sweden, Switzerland, UK, and USA). Each laboratory received an individual lab ID number. Panel leadership was provided by two scientific leaders experienced in MHC-peptide multimer staining, in collaboration with the CVC executive office. The Lausanne branch of the Ludwig Institute for Cancer Research (LICR-LB) performed extensive pre-testing of donor samples and selection of appropriate donors.
PBMCs and peptides
PBMCs from healthy donors were obtained from a commercial donor bank and processed under GMP conditions using established standard operating procedures at Cellular Technologies Limited (CTL), Cleveland, OH. PBMCs were frozen using a rate controlled freezer and transferred to the vapor phase of liquid nitrogen. Cell separation procedure and freezing of cells were conducted under validated conditions. It was demonstrated that functionality and viability were maintained throughout the procedure. Each vial of PBMCs contained enough cells to ensure a recovery of 10 million cells or more under CTL’s SOP. Cells were shipped to all participants for overnight delivery on sufficient dry ice for 48 h for centers within the USA. Centers located in Europe and Canada received cells shipped in dry shippers filled with sufficient liquid nitrogen to assure cell integrity for up to 7 days. Shipment of cells was performed by CTL under their existing SOPs.
PBMCs were pretested at the LICR-LB for reactivity against the HLA-A2-restricted Influenza-M158–66 (antigen “A1” = GILGFVFTL) [23] and Melan-A/Mart-126–35A27L (antigen “A2” = ELAGIGILTV) [24]. PBMCs from five donors were selected for no, low, medium and strong responses against the two tested antigens. Pre-testing was performed by generating three independent data sets using HLA-peptide tetramers (Beckman Coulter Immunomics), HLA-peptide pentamers (ProImmune) and HLA-peptide tetramers manufactured at the LICR-LB for research use. For the proficiency panel testing, MHC-peptide tetramers (Beckman Coulter) and pentamers (Proimmune) specific for both model antigens were provided in sufficient amount to perform 50 stainings per center and had to be applied as 10 μl volume per staining. All cells and reagents sent to participants in both panels were obtained from the same batches.
HLA-peptide multimer staining
One of the main features of the first CVC multimer panel was that all participants (in both steps) used their preferred reagents and locally established protocols to determine the percentage of antigen-specific CD8+ T cells. The only requirement was to use the HLA-peptide multimers provided by the respective commercial sources. Moreover, the organizers avoided offering recommendations, providing protocols, or asking for any other measure of harmonization.
Statistical analysis
The following parameters were calculated for both the overall panel and the individual participant’s performance, using the lab-specific reported percentage of multimer-positive CD8-positive cells: the mean, standard deviation, and coefficient of variation (CV), the median, 25th and 75th percentiles, minimum, and maximum percentage of CD8-positive cells for each donor and antigen.
Due to the extreme variation of the reported values we applied two data filters to remove a large proportion of non meaningful results from the final analysis. The first filter was based on the coefficient of variation (CV) from the replicates within a lab for a given donor and antigen and used to eliminate outlier replicate values from the final analysis. Data sets with either replicate that had a CV greater than 75 or only one measurement for a given donor and antigen combination did not pass this filter. The second filter was based on the dot plots generated from the lab’s analysis of the flow cytometer results. An independent evaluator examined all the dot plots and assigned a score based on the clustering of positive events. A score of 0 was given when there was no clustering, a score of 1 for ambiguous results, and a score of 2 when there was a clearly clustered population of positive events in the upper right quadrant. Labs with replicates that had a total score of less than 4 did not pass this filter. Therefore, there had to be at least one staining with clear clustering from a given replicate to pass the filter. Labs that passed both filters were considered to have detected a response for that donor and antigen. The association between detected response rate and number of counted CD8+ T cells was tested using the Chi square test.
The questionnaire responses outlining the various protocols used by all the labs were summarized using frequency tabulations. The following parameters were calculated to summarize the overall recovery and viability for each of the thawed PBMC donor samples: mean, median, minimum, and maximum.
For laboratories that participated in the second step, the individual laboratory results from the second step were presented in the same format as in the first step. Within an individual laboratory, the results from the two steps were compared by calculating the percent difference from the first step, the standard deviation of the means from both steps and the corresponding coefficient of variation (CV). The average CV across the ten donor antigen combinations was computed as a general indication of how similar the results were in the same laboratory at two different time points.
Results
All 27 participating centers received the necessary PBMC samples, commercially available HLA-peptide multimers and instructions, and were able to perform the requested stainings. A total number of 29 expected questionnaires, report forms and dot plots were successfully collected for analysis via a web-based database specifically designed to administer and process large amount of data from immune monitoring assays. The organizers also received and analyzed ten complete data sets from the second step. After each step individualized reports were provided to all participating labs.
Human auditing process of final results
Before final analysis, all collected data sets were screened for obvious failures and inconsistencies. These included incorrect gating by some participants, uneven quality of electronic compensation of flow cytometry based event acquisition (Figs. 1a, b, 2b) and aberrant reported values from two data sets that did not reflect the number of events shown in the corresponding dot plots. The latter originated in a systematic error when entering the event counts for all four quadrants into the spread sheet (not shown). Together, these findings emphasize the importance of systematic auditing of assay results.
Fig. 1.
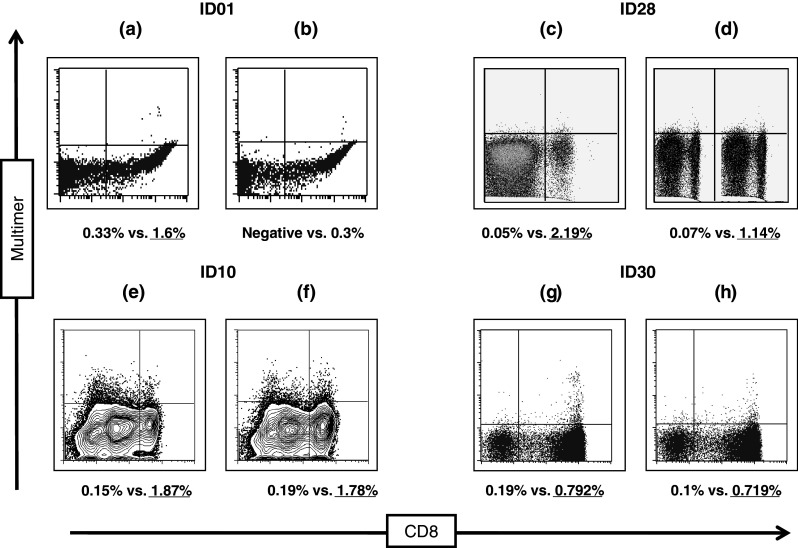
The figure shows eight selected examples (a–h) of dot plots where gating led to reporting of increased number of events in the upper right quadrant. All dot plots show the CD8-staining on the x-axis and the staining with the HLA-peptide multimer on the y-axis. Dot plots were chosen from laboratories ID01, ID10, ID28 and ID30. Under each dot plot the expected versus the reported (underlined) frequency of multimer-positive CD8 cells is indicated
Fig. 2.
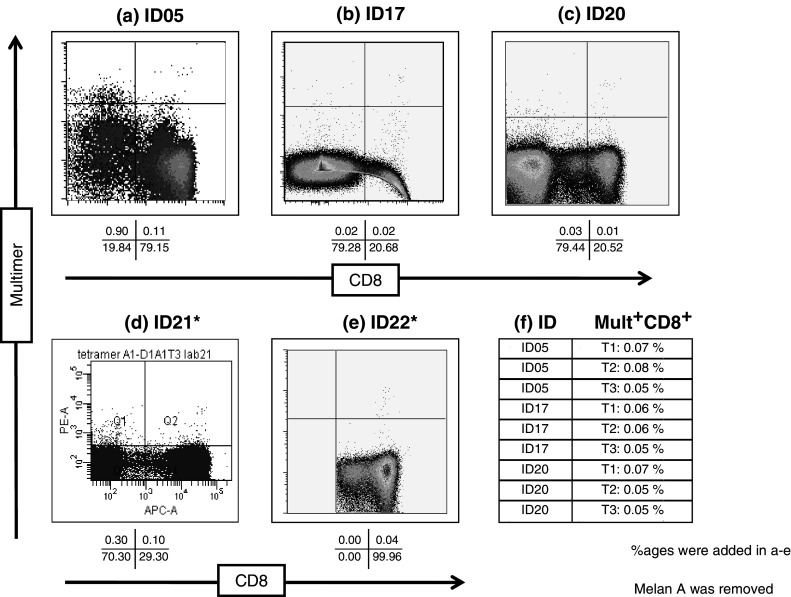
Figures a–e show representative dot plots from centers ID05, 17, 20, 21 and 22 reported for staining samples from donor1 with the Influenza-M1 multimer. *Center ID21 set the analytical gate in such a way that multimer-negative cells are shown in the upper right quadrant. **Center ID22 used an atypical gating strategy in which CD8-negative cells were removed at an earlier step of the analysis. The inserted table (f) shows reported values for non-specific binding of the Melan-A/Mart-1-specific multimer in samples from triplicate analysis (T1, T2 and T3) of donor 1 performed by centers ID05, 17 and 20
Overview of assay protocols currently in use at the international level
Each participant provided detailed information about experimental protocol and reagents used. It became clear that multimer labeling is currently performed using a broad variety reagents and procedures. Supplementary table 1 which is available online shows the distribution of labs for 11 variables with the potential to influence the sensitivity of the multimer labeling assay (multimer source, use of DNAse during thawing, counting method, type of flow cytometer used, staining performed in tubes or plates, conjugate staining order, number of fluorochromes, method for dead cell exclusion, anti-CD3 staining, use of a dump channel and antibodies used for co-staining) and provides a comprehensive overview of the protocols that were applied. When looking at a distinct subgroup of labs sharing one variable, it became clear that the expression of the other ten variables was still randomly distributed within the subgroups.
Inter- and intra-center variation
For our group of 27 laboratories we found an unexpectedly high variation among the reported 29 datasets for eight of the ten different donor antigen combinations with CVs ranging from 47 to 158 (Table 1). This was the case even for the three highest responses (Influenza in D2/D4/D5 with corresponding CVs of 47.2/93.7/57.1). The even higher variation found in donor 1 result from the fact that no or extremely low number of antigen-specific T cells were present in this donor. The liberal design of this panel provides a measure of the variation of results that may be representative of current immune monitoring of antigen-specific CD8+ T cell responses using the multimer-based assay.
Table 1.
Percentage of CD8-specific multimer binding based on the mean of the triplicates
| Antigen | Donor | Median | 25th Percentile | 75th Percentile | Mean | SD | CV | Min | Max |
|---|---|---|---|---|---|---|---|---|---|
| Influenza-M1 | D1 | 0.07 | 0.04 | 0.15 | 0.32 | 0.67 | 210.58a | 0.00 | 2.87 |
| D2 | 0.67 | 0.50 | 0.89 | 0.73 | 0.34 | 47.16 | 0.24 | 2.05 | |
| D3 | 0.16 | 0.11 | 0.32 | 0.29 | 0.35 | 122.95 | 0.00 | 1.53 | |
| D4 | 0.19 | 0.17 | 0.27 | 0.28 | 0.26 | 93.66 | 0.07 | 1.44 | |
| D5 | 0.35 | 0.31 | 0.41 | 0.44 | 0.25 | 57.05 | 0.21 | 1.21 | |
| Melan-A/Mart-1 | D1 | 0.07 | 0.03 | 0.19 | 0.23 | 0.48 | 207.48b | 0.00 | 2.53 |
| D2 | 0.10 | 0.03 | 0.17 | 0.17 | 0.27 | 156.14 | 0.00 | 1.30 | |
| D3 | 0.12 | 0.06 | 0.23 | 0.25 | 0.40 | 157.76 | 0.02 | 2.09 | |
| D4 | 0.10 | 0.04 | 0.15 | 0.17 | 0.23 | 135.44 | 0.01 | 1.06 | |
| D5 | 0.08 | 0.03 | 0.17 | 0.18 | 0.26 | 142.63 | 0.01 | 1.06 |
The table shows the overall results (median, 25th, 75th percentile, mean, standard deviation, coefficient of variation, minimum and maximum) for all ten antigen-donor combinations reported by the whole group
aDonor 1 was considered to have a very low response against Influenza-M1 which was below the limit of quantification
bDonor 1 was considered to have no detectable response against Melan-A/Mart-1
It is well established that the validation process of any diagnostic test, including cellular assays, should include the prior determination of accuracy, specificity, sensitivity, reliability, linearity and range determination as well as important precision parameters such as the intra-assay and inter-assay variation of results [25]. Nine labs repeated the panel at a separate time point. As one center generated two separate data sets with HLA-peptide tetramers as well as pentamers the whole group submitted ten complete datasets. In order to quantify the intra-lab variation we calculated the mean CD8+ specific T cell binding at each time and compared these means by determining the absolute and percentage-wise difference between the results from both steps (not shown) as well as the coefficient of variation of both results (details are shown in Supplementary table 2). Five participating centers reported pairs of results that were very close to each other (average intra-center CV < 30) for most of the eight depicted antigen-donor combinations showing that reproducibility within an acceptable range can be obtained for a broad range of detected antigen-specific T cell responses. The other five laboratories reported results for step 2 that were quite different from step 1 (average intra-center CV > 30). Consequently, intra-center variation was unexpectedly large for these labs.
Limitations of the multimer assay
A small group of five centers were able to detect a small but distinct population of Influenza-M1-specific CD8+ cells in donor D1 and reported an average value of 0.11% multimer-positive CD8+ T cells in this donor. Representative dot plots are shown in Fig. 2a–e. On the other hand none of the participating centers was able to detect Melan-A/Mart-1-specific CD8+ T cells in the same donor. We made use of the reported data from this donor to get more insights into the technical limitations of the technology. Our aim was to determine if the reported mean value of 0.11% Influenza-M1-specific CD8 T cells could indeed be considered as being above the normal variation of the background usually found in an average lab. ID21 set the analytical gates in such a way that a high number of clearly multimer-negative events fell into the upper right quadrant (Fig. 2d, Supplementary figures 1 and 2). ID22 used an atypical gating strategy which removed dead and CD8-negative cells and reported an extremely low background of non-specific multimer staining (Supplementary figure 2a). Three centers (ID05, ID17 and ID20) used a classical gating strategy and reported values for the non-specific binding of the Melan-A/Mart-1 multimer that were similar to results obtained by a large number of other participants and therefore appropriately represent the background staining normally found in a number of commonly used protocols (Supplementary figure 2a). These three centers reported a mean of 0.06% multimer-positive CD8+ T cells using the Melan-A/Mart-1 multimer in donor 1 with a standard deviation of 0.01% (Fig. 2f). Based on these results we roughly estimated a limit of detection (LOD) of 0.09% which resulted from adding three times the standard deviation to the mean. In addition we estimated a limit of quantification (LOQ) of 0.16% which resulted from adding ten times the standard deviation to the mean.
Individual laboratory performance
A total of nine positive responses could have been detected in the five donors (for all details see Table 2) from which 1 was considered to be below the estimated limit of quantification (defined as <0.16%), 2 were low (defined as <0.2%), 5 were moderate (defined as ≥0.2 and ≤0.5%) and 1 was high (defined as >0.5%). Table 1 indicates the median, 25th, 75th percentile and mean, for each of the ten measurements. The report from the first step of the panel displayed the percentages for antigen-specific CD8+ cells from all 29 protocols for all ten antigen-donor combinations (Supplementary figures 1 and 2).
Table 2.
Expected percentage of antigen-specific CD8 T cells within the distributed samples
| Antigen | Donor | Frequency | Response |
|---|---|---|---|
| Influenza-M1 | D1 | 0.11 | >LOD and <LOQ |
| D2 | 0.76 | High | |
| D3 | 0.26 | Moderate | |
| D4 | 0.28 | Moderate | |
| D5 | 0.39 | Moderate | |
| Melan-A/Mart-1 | D1 | 0.00 | Negative |
| D2 | 0.22 | Moderate | |
| D3 | 0.31 | Moderate | |
| D4 | 0.19 | Low | |
| D5 | 0.16 | Low |
The table shows that a total of eight positive responses could have been detected in the five donors (D1–D5) from which one was above the LOD value but below the LOQ value, two were considered as being low (defined as <0.2%), five were moderate (defined as ≥0.2 and ≤0.5%) and one was high (defined as >0.5%)
To define if a participant had successfully detected a response, two acceptance criteria were introduced and applied. On one hand, replicates were excluded from further analysis when they had a high variation (defined as CV > 75). A total of 24 replicates (8.2%) were found to have unacceptably high variation and were therefore rejected (details are shown in Supplementary table 3). On the other hand, replicates that did not show clear and clustered populations of events in the upper right quadrants indicating CD8+ multimer+ cells, as assessed by an independent evaluator, were also excluded (details are shown in Supplementary table 4). Due to this rule a total of 101 replicates (34%) were discarded. Based on this definition of response, the whole group was able to detect 66% of all possibly detectable responses, with some labs not able to detect any responses and only a few detecting all nine responses (Table 3). Donor D1 with Melan-A/Mart-1 was a negative control donor. No lab reported a false positive response for this antigen-donor combination. Supplementary figure 3, which is also available as electronic supplementary material, shows selected results from the optical evaluation to allow the reader to discern the amount of clustering that was considered to be clearly negative (0), ambiguous result (1) or clearly positive (2). The figure also makes clear that optical evaluation of dot plots is a very subjective approach as an objective definition of a “clustered population” does not exist. Consequently, interpretation of results by optical evaluation should be regarded with caution, especially when very small populations are judged. However, one independent evaluator examined and rated all of the dot plots and applied the same criterion to all dot plots. Hence, there was a uniform criterion applied to all dot plots even though a subjective definition was used.
Table 3.
Number of labs that detected each possible number of accepted responses
| No. of detected responses | No. of labs | Percentage |
|---|---|---|
| 0 | 2 | 7 |
| 1 | 1 | 3 |
| 3 | 3 | 10 |
| 4 | 1 | 3 |
| 5 | 2 | 7 |
| 6 | 5 | 17 |
| 7 | 5 | 17 |
| 8 | 7 | 24 |
| 9 | 3 | 10 |
The table shows the summary of the distribution of the number of times a lab was considered of having detected a response
Subgroup-analysis
One aim of the CVC multimer panel was to deduce recommendations for initial harmonization guidelines of the assay. One clear finding from the panel was that the number of CD8+ cells counted per replicate critically determines the chance that an immune response is detected. The majority of results were based on collection of 30,000–100,000 CD8 positive (CD8+) events. In the subgroup in which less than 10,000 CD8+ cells were collected, only 13% detected a response (Table 4). In contrast, 87% of replicates with more than 100,000 accumulated CD8+ cells detected a response correctly. There was a statistically significant difference (P = 0.001, Chi square test) when comparing the response detection rates in triplicates with at least 100,000 CD8+ cells counted (39/45 = 87%) versus less than 100,000 CD8+ cells counted (133/218 = 61%).
Table 4.
Number of CD8-positive cells counted and proportion of responses detected
| Number of CD8-positive cells | Number of triplicates | Number of responses | Responses (%) |
|---|---|---|---|
| <10,000 | 32 | 4 | 13 |
| 10–30,000 | 40 | 24 | 60 |
| 30–100,000 | 144 | 105 | 73 |
| >100,000 | 45 | 39 | 87 |
| Number of CD8-positive cells | Number of triplicates | Number of low responses | Low responses (%) |
|---|---|---|---|
| <10,000 | 15 | 0 | 0 |
| 10–30,000 | 18 | 4 | 22 |
| 30–100,000 | 63 | 34 | 54 |
| >100,000 | 20 | 16 | 80 |
The table shows the average number of CD8-positive cells counted for a given replicate and the proportion of responses detected within the specified counting range (first column). The mean number of CD8+ T cells counted per replicate was calculated and all tests were attributed to one of four groups (<10,000/10–30,000/30–100,000/>100,000). The table shows the total number of responses and the percentage of responses detected by labs in each of the four groups. The upper half of the table shows the results from all ten donor antigen combinations. The lower half of the table displays the results of testing for the four lowest responses in the distributed samples (Melan-A/Mart-1 response for donors D2, D4 or D5 and Influenza-M1 response for donor D1)
We then focused only on the results obtained for the four lowest responses (Melan-A/Mart-1 response for donors D2, D4 or D5 and Influenza-M1 response for donor D1). The lower part of Table 4 shows that 12% (4/33) of the lower responses were detected by triplicates with less than 30,000 positive CD8+ cells counted, whereas 80% were detected by triplicates with more than 100,000 CD8+ cells counted. There was a statistically significant difference (P = 0.001, Chi square test) when comparing the response detection rates in triplicates with at least 100,000 CD8+ cells counted (16/20 = 80%) versus less than 100,000 CD8+ cells counted (38/96 = 40%).
A second source of variation was related to electronic gate setting during data analysis. Most of the labs that reported out of range high values of specific multimer binding did so because they set the gates too low thereby increasing the number of dots on the upper right quadrant (Fig. 1). Thus, a method that helps setting the gates correctly would lead to a reduction of the inter-lab variation.
Although the majority of labs (66%) were not able to detect eight or nine responses, ten of the participating labs were able to do so. Analysis of the protocols used by these ten high performing labs indicated that comparable results could be obtained with protocols that differed widely from each other (not shown). For instance, high performing labs were found among those that (1) reported low or high cell viability after thawing, (2) used tetramers as well as pentamers, (3) used or did not use DNAse for thawing, (4) used manual or automated cell counting methods, (5) stained in tubes or in plates, (6) first stained with the multimer or simultaneously stained with multimer and antibodies for co-staining, (7) used three or four fluorochromes, (8) used or did not use dead cell staining, (9) used or did not use anti-CD3 staining, (10) used or did not use a dump channel, or (11) reported low, medium or high values for non-specific multimer binding. The only subgroup that did not include any of the ten high performing labs was the one that only used two fluorochromes (anti-CD8 vs. HLA-peptide multimer) for the staining (two laboratories).
To identify additional variables that may influence the sensitivity of the assay, we used the results from the questionnaires to define 2–3 subgroups for 11 protocol variables. We then compared the ability to successfully detect one of the nine responses for all different subgroups of labs. Supplementary table 5 summarizes the lab characteristics and reports the number of detected responses and the average proportion of detected responses for each subgroup. Responses were more likely to be detected if labs had more than 75% viability (vs. less than 75%), used pentamers (vs. tetramers), used DNAse (vs. no DNAse), used a manual counting method (vs. machine), did multimer and then co-staining (vs. simultaneously), used three or four colors (vs. two colors), did not do CD3 staining (vs. did CD3 staining) or used a dump channel (vs. no dump channel). It is important to stress that the labs within each subgroup applied protocols that differed widely from each other. This means that differences found between some of the subgroups in our panel do not formally prove that the variable that was selected to define the subgroup was the defining factor that influenced the performance of those labs. Although we were not able to deduce additional recommendations from this analysis we classify the subgroups that showed the highest differences as interesting targets for more systematic analysis in future proficiency panels.
Discussion
The specific aims of the first CVC-CRI multimer proficiency panel presented in this manuscript were to establish the foundations for developing a robust and validated MHC multimer assay which can be harmonized across immune monitoring laboratories by: (1) demonstrating the feasibility of such large international inter-laboratory testing project for HLA-peptide multimers, (2) identifying protocol variables, reagent choices and strategies that are relevant for assay protocol optimization (3) providing each participating lab direct feed-back about their qualitative and quantitative performance in relation to the other members of the group, (4) quantifying the variation of results reported by such a large number of labs (inter-center variation), and (5) determining the variation of results obtained within the same lab using the same protocol with the same samples doing the experiment at two different time points (intra-center variation).
The results from this panel demonstrated the feasibility of such a large inter-laboratory testing project for MHC-peptide multimer staining. Importantly, they reveal the wide variety of different protocols and strategies used with multimers, which appears representative of the most commonly used protocols worldwide. This unique situation allowed us to determine the average amount of non-specific multimer binding normally observed in a broad range of commonly applied protocols (Fig. 2f, Supplementary figure 2a). Based on these data we estimated an LOD and LOQ. Such performance-related thresholds determined by data sets obtained in large-scale quality assurance programs are valuable tools to more objectively rank the performance of participating labs and might be used for assay certification purposes in the future.
The results obtained in this CVC-CRI multimer panel allowed, for the first time, the quantification of the actual variation in results between different labs across the United States and Europe. The high inter- and intra-center variation were unexpected and stand in contrast to a report from a smaller European group [26]. Although the performance of cellular immunoassays tested in single centers or within selected groups that either used one common protocol [27] or went through intensified validation and harmonization steps appears quite robust [28, 29], it is important to point out that the performance of this assay in a non-selected group of labs is clearly not robust and results are subject to a large degree of variation. The results generated by this large international panel can now serve the scientific community as a benchmark for future similar projects. Every newly introduced measure to harmonize the technique should lead to a reduction of the variation found in this panel.
Our results emphasize the urgent need for harmonization of the multimer labeling technique. Despite this conclusion, it is important to state that the presented data should not be used to justify the non-critical discarding of data obtained by HLA-peptide multimer staining. Recently a systematic analysis of cellular based immunoassays clearly showed that ELIPOT, cytokine flow cytometry as well as HLA-peptide multimer staining can be used to precisely and stably quantify antigen-specific T cell responses within a broad range of frequencies within highly validated laboratories [30]. The fact that a third of the labs involved in this proficiency panel were able to detect eight or nine of all responses included in the distributed samples and that five out of ten labs were able to (qualitatively and quantitatively) reproduce their results in a second step throughout a broad range of T cell responses of different frequencies demonstrate that staining with HLA-peptide multimers is a sensitive technique that can be reliably used to quantify antigen-specific T cells in clinical trials. At the same time, these observations pose major challenges for assay harmonization as it is clear that two-thirds of the labs involved had difficulty in accurately and reproducibly measuring specific T cell frequencies.
Initial harmonization guidelines to improve assay performance
Based on the results of this multimer panel, we outline initial assay harmonization guidelines addressing four areas of immunomonitoring by HLA-peptide multimers (Table 5). These guidelines include basic recommendations (a) for the staining procedure, (b) the analysis of data, (c) the auditing of data obtained before they are released and (d) the qualification of staff members involved in the analysis. Use of these guidelines may substantially reduce assay variability while allowing individual laboratories to use their respective assay protocols.
Table 5.
MHC-peptide multimer harmonization guidelines to optimize assay performance
| (A) Establish lab SOP for MHC-peptide multimer staining |
| A1 Count at least 100,000 CD8 T cells per staining |
| A2 Introduce a background control to set gates |
| A3 Use more than two colors for staining |
| (B) Establish SOP for software analyses of stained samples, including |
| B1 Gating strategy |
| B2 Rules to set the gates |
| (C) Establish a human auditing process of all final results |
| C1 Are all dot plots correctly compensated? |
| C2 Have the gates been set correctly? |
| C3 Are the calculated frequencies of multimer-positive cells plausible? |
| (D) Only let well trained personnel (per lab SOP) conduct assay |
Standard operating procedure for HLA-peptide multimer staining
Three clear recommendations for the staining procedure can be deduced from the analysis of inter-laboratory result variation. These are to (1) use of a protocol that includes more than two colors for staining, (2) evaluate at least 100,000 CD8+ T cells and (3) use a background control to set the gates.
Although a simple two-color staining is adequate to characterize the HLA-peptide multimer binding capacity of T cell lines or clones, it is clear that such a simple protocol does not allow reliable quantification of the frequency of antigen-specific T cells in peripheral blood specimens which include non CD8 T cell subsets that either specifically or non-specifically bind specific detection reagents. Non-CD8 T cell specific staining becomes a major problem when low frequencies of antigen-specific T cells need to be visualized, which is the case for ex vivo detection of most virus-specific T cell responses that are regularly found to represent less than 1% of CD8+ T cells in chronic viral infections [31] or tumor-specific T cells that are present in even lower numbers [32, 33]. One clear recommendation from this panel is to use additional fluorochromes and establish a methodology which minimizes background; this conclusion is supported by an earlier report elaborating the relevant factors for staining with HLA-peptide multimers [34]. The use of additional fluorochromes is technically feasible and easily allows gating to remove dead cells, B cells, monocytes, CD4+ cells or NK cells to focus more specifically on the cell population of interest, thereby reducing the chance of displaying cells that all can contribute to non-relevant events.
The second recommendation to increase the number of counted cells confirms data from an inter-laboratory comparison of different labs conducted by the European CIMT Monitoring panel [35] demonstrating the high impact of this parameter.
A strategy that helps to set gates correctly would prevent the errors found in this panel and may lead to greatly reduced inter-laboratory variation and optimized levels of detection and quantification as these critically depend on the observed background noise. In this regard, the use of a “negative” control staining to define the background multimer labeling remains controversial. Centers that already apply negative control stainings use one of the following three strategies. (1) Fluorescence minus one (“FMO”) stainings that include all antibodies/dead cell markers but no HLA-peptide multimer. (2) Negative control stainings with HLA-peptide multimer specific for well defined peptides from human immunodeficiency virus (HIV). (3) Commercially available preparations of irrelevant HLA-peptide multimers. An alternative strategy may be to search for new negative control peptides and to systematically test HLA-peptide multimers containing these putative candidates for their suitability as negative controls in patients with cancer, infectious and autoimmune diseases. All strategies that are commonly used to define background of HLA-peptide multimer staining have specific advantages and disadvantages and as yet they have not been systematically compared with each other. Regardless, it is clear that the use of a negative control for defining the background staining of HLA-peptide multimers should be applied as a guide for standardized gate setting. Valuable software solutions that apply mathematical algorithms to set gates independent of the operator’s subjective view are in development but are so far not available for broad use [36].
In addition to these three initial recommendations we identified primary targets for systematic analysis in future panels that have a high probability to influence test sensitivity (Supplementary table 5). The influence of these variables will be systematically addressed in future proficiency panels.
Training of staff members
Although the staining with MHC-peptide multimers seems, at first sight, to be a simple and straightforward procedure, our results have shown that even the use of one and the same batch of HLA-peptide multimers can lead to different results if applied in different staining protocols. The high values for intra-center variability detected in five from ten labs participating in a repetition of the experiment showed that even when a staining is repeated in the same lab by using the same protocol it does not automatically lead to similar results (Supplementary table 2). Each step, from the collection of cell material to reporting of results, adds sources for variation which separately contribute to the variation of results obtained either by different labs worldwide or from measurements by the same lab at different time points. It is thus necessary that staff members performing the measurements should be aware of the sources of variation within their responsibility and be trained in all measures that need to be taken to keep variation as low as possible. The regular and specific education of staff members and mechanisms to control their performance are well established and mandatory requirements for all laboratories working under rules defining a good manufacturing practice but have so far not been systematically applied for immunomonitoring of clinical samples. In a previous ELISPOT proficiency panel conducted by the CVC, it could be shown that establishing SOPs for assay validation and staff training clearly lead to an increased performance of participating labs [37]. We therefore propose to implement formalized assay specific education of all staff members involved in immune monitoring with HLA-peptide multimers before clinical material should be allowed to be processed.
In conclusion, our study reveals the variety of HLA-peptide multimer-based assay formats currently in use in the tumor immunology and immunotherapy community. Moreover, it has allowed us to measure the magnitude of both inter and intra-laboratory result variation. We have gained detailed insight into technical limitations of commonly applied protocols and provide estimates for expectable limits of detection and quantification. Finally, valuable recommendations could be derived that should guide the future assay harmonization efforts spearheaded by the CVC-CRI as well as other consortia active in this domain.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
C.M.B. was supported by a Dr. Mildred Scheel Grant from the Deutsche Krebshilfe (“German Cancer Aid”). The authors would like to thank Proimmune and Beckman Coulter for their generous offer to provide the necessary HLA-peptide multimers to all participants. The companies’ organizational support to reach our aim to harmonize immunoassays is very much appreciated. We are especially grateful that Dr. Jeremy Fry (Proimmune, Oxford, UK) and Dr. Stephen Haley (Beckman Coulter, Fullerton, CA) continuously supported the proficiency panel program. P.R. was supported in part by the EU FP6 grant “Cancerimmunotherapy”.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Collaborators of the CRI-CVC HLA-peptide multimer proficiency panel
(1) Ahmed RK, Swedish Institute for Infectious Disease Control and the Karolinska Institute, Sweden
(2) Bain C, Transgene SA, France
(3) Barnby-Porritt H, ProImmune Ltd, UK
(4) Baumgartner P, Ludwig Institute for Cancer Research, Lausanne Branch, Switzerland
(5) Bercovici N, IDM Pharma, France
(6) Brockstedt D, Anza Therapeutics, USA
(7) Cerundolo V, Weatherall Institute of Molecular Medicine, UK
(8) Channon JY, Dartmouth Medical School, USA
(9) Chikoti P, Oxford BioMedica PLC, UK
(10) Cox J, Henry M. Jackson Foundation, USA
(11) Currier JR, Henry M. Jackson Foundation, USA
(12) Curtsinger J, University of Minnesota Cancer Center Translational Cell Therapy Core Facility, USA
(13) Desmaretz C, Innogenetics NV, Belgium
(14) Gagnon D, University of Montreal (NIML), Canada
(15) Gearhart J, AERAS Global TB Vaccine Foundation, USA
(16) Gillanders W, Washington University School of Medicine, USA
(17) Hampl, J, Anza Therapeutics, USA
(18) Harrop R, Oxford BioMedica PLC, UK
(19) Hobeika A, Duke University Medical Center, Program in Molecular Therapeutics, USA
(20) Kaempgen E, University of Nuernberg/Erlangen, Germany
(21) Kaufman J, Rockefeller University, USA
(22) Labroquere K, IDM Pharma, France
(23) Landry C, University of Montreal (NIML), Canada
(24) Maeurer M, Swedish Institute for Infectious Disease Control and the Karolinska Institute, Sweden
(25) Matijevic M, MGI Pharma Biologics, USA
(26) Mueller S, AERAS Global TB Vaccine Foundation, USA
(27) Mohanakumar T, Washington University School of Medicine, USA
(28) Olson W, Human Immune Therapy Center, University of Virginia, USA
(29) Pohla H, Ludwig-Maximilians University of Munich, Germany
(30) Ramesh R, MGI Pharma Biologics, USA
(31) Rooke R, Transgene SA, France
(32) Samorski R, immatics biotechnologies GmbH, Germany
(33) Sékaly, R-P, University of Montreal (NIML), Canada
(34) Schullery D, University of Pennsylvania, USA
(35) Slingluff C, Human Immune Therapy Center, University of Virginia, USA
(36) Scheibenbogen C, Charité Berlin, Germany
(37) Schmitt-Händle M, University of Nuernberg/Erlangen, Germany
(38) Smith K, MannKind Corporation, USA
(39) Speiser D, Ludwig Institute for Cancer Research, Lausanne Branch, Switzerland
(40) Tarlton A, Weatherall Institute of Molecular Medicine, UK
(41) Trautmann L, University of Montreal (NIML), Canada
(42) Van Der Aa A, Innogenetics NV, Belgium
(43) Walter S, immatics biotechnologies GmbH, Germany
(44) Wolchok J, Memorial Sloan-Kettering Cancer Center, USA
(45) Yuan J, Memorial Sloan-Kettering Cancer Center, USA.
Contributor Information
Cedrik Michael Britten, Email: cebritten@web.de.
Pedro Romero, Phone: +41-21-3140198, Email: pedro.romero@licr.unil.ch.
References
- 1.Mocellin S, Mandruzzato S, Bronte V, Lise M, Nitti D. Part I: vaccines for solid tumours. Lancet Oncol. 2004;5:681–689. doi: 10.1016/S1470-2045(04)01610-9. [DOI] [PubMed] [Google Scholar]
- 2.Finke LH, Wentworth K, Blumenstein B, Rudoplph NS, Hoos A. Lessons from randomized phase III studies with active cancer immunotherapies—outcomes from the 2006 meeting of the Cancer Vaccine Consortium (CVC) Vaccine. 2007;25:B97–B109. doi: 10.1016/j.vaccine.2007.06.067. [DOI] [PubMed] [Google Scholar]
- 3.Cheever MA. Twelve immunotherapy drugs that could cure cancers. Immunol Rev. 2008;222:357–368. doi: 10.1111/j.1600-065X.2008.00604.x. [DOI] [PubMed] [Google Scholar]
- 4.Hoos A, Parmiani G, Hege K, Sznol M, Loibner H, Eggermont A, Urba W, Blumenstein B, Sacks N, Keilholz U, Nichol G, the Cancer Vaccine Clinical Trial Working Group A clinical development paradigm for cancer vaccines and related biologics. J Immunother. 2007;30:1–15. doi: 10.1097/01.cji.0000211341.88835.ae. [DOI] [PubMed] [Google Scholar]
- 5.Yee C, Greenberg P. Modulating T cell immunity to tumours: new strategies for monitoring T cell responses. Nat Rev Cancer. 2002;2:409–419. doi: 10.1038/nrc820. [DOI] [PubMed] [Google Scholar]
- 6.Appay V, Douek DC, Price DA. CD8+ T cell efficacy in vaccination and disease. Nat Med. 2008;14:623–628. doi: 10.1038/nm.f.1774. [DOI] [PubMed] [Google Scholar]
- 7.Connerotte T, Van PA, Godelaine D, Tartour E, Schuler-Thurner B, Lucas S, Thielemans K, Schuler G, Coulie PG. Functions of Anti-MAGE T cells induced in melanoma patients under different vaccination modalities. Cancer Res. 2008;68:3931–3940. doi: 10.1158/0008-5472.CAN-07-5898. [DOI] [PubMed] [Google Scholar]
- 8.Schmitt M, Schmitt A, Rojewski MT, Chen J, Giannopoulos K, Fei F, Yu Y, Gotz M, Heyduk M, Ritter G, Speiser DE, Gnjatic S, Guillaume P, Ringhoffer M, Schlenk RF, Liebisch P, Bunjes D, Shiku H, Dohner H, Greiner J. RHAMM-R3 peptide vaccination in patients with acute myeloid leukemia, myelodysplastic syndrome, and multiple myeloma elicits immunologic and clinical responses. Blood. 2008;111:1357–1365. doi: 10.1182/blood-2007-07-099366. [DOI] [PubMed] [Google Scholar]
- 9.Adams S, O’Neill DW, Nonaka D, Hardin E, Chiriboga L, Siu K, Cruz CM, Angiulli A, Angiulli F, Ritter E, Holman RM, Shapiro RL, Berman RS, Berner N, Shao Y, Manches O, Pan L, Venhaus RR, Hoffman EW, Jungbluth A, Gnjatic S, Old L, Pavlick AC, Bhardwaj N. Immunization of malignant melanoma patients with full-length NY-ESO-1 protein using TLR7 agonist imiquimod as vaccine adjuvant. J Immunol. 2008;181:776–784. doi: 10.4049/jimmunol.181.1.776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Speiser DE, Baumgaertner P, Voelter V, Devevre E, Barbey C, Rufer N, Romero P. Unmodified self antigen triggers human CD8 T cells with stronger tumor reactivity than altered antigen. Proc Natl Acad Sci USA. 2008;105:3849–3854. doi: 10.1073/pnas.0800080105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosenberg SA, Yang JC, Restifo NP. Cancer immunotherapy: moving beyond current vaccines. Nat Med. 2004;10:909–915. doi: 10.1038/nm1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Curigliano G, Spitaleri G, Pietri E, Rescigno M, de Braud F, Cardillo A, Munzone E, Rocca A, Bonizzi G, Brichard V, Orlando L, Goldhirsch A. Breast cancer vaccines: a clinical reality or fairy tale? Ann Oncol. 2006;17:750–762. doi: 10.1093/annonc/mdj083. [DOI] [PubMed] [Google Scholar]
- 13.Diefenbach CS, Gnjatic S, Sabbatini P, Aghajanian C, Hensley ML, Spriggs DR, Iasonos A, Lee H, Dupont B, Pezzulli S, Jungbluth AA, Old LJ, Dupont J. Safety and immunogenicity study of NY-ESO-1b peptide and montanide ISA-51 vaccination of patients with epithelial ovarian cancer in high-risk first remission. Clin Cancer Res. 2008;14:2740–2748. doi: 10.1158/1078-0432.CCR-07-4619. [DOI] [PubMed] [Google Scholar]
- 14.Darrah PA, Patel DT, De Luca PM, Lindsay RW, Davey DF, Flynn BJ, Hoff ST, Andersen P, Reed SG, Morris SL, Roederer M, Seder RA. Multifunctional TH1 cells define a correlate of vaccine-mediated protection against Leishmania major . Nat Med. 2007;13:843–850. doi: 10.1038/nm1592. [DOI] [PubMed] [Google Scholar]
- 15.Duvall MG, Precopio ML, Ambrozak DA, Jaye A, McMichael AJ, Whittle HC, Roederer M, Rowland-Jones SL, Koup RA. Polyfunctional T cell responses are a hallmark of HIV-2 infection. Eur J Immunol. 2008;38:350–363. doi: 10.1002/eji.200737768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Precopio ML, Betts MR, Parrino J, Price DA, Gostick E, Ambrozak DR, Asher TE, Douek DC, Harari A, Pantaleo G, Bailer R, Graham BS, Roederer M, Koup RA. Immunization with vaccinia virus induces polyfunctional and phenotypically distinctive CD8(+) T cell responses. J Exp Med. 2007;204:1405–1416. doi: 10.1084/jem.20062363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Vries I, Bernsen MR, Lesterhuis WJ, Scharenborg NM, Strijk SP, Gerritsen MJ, Ruiter DJ, Figdor CG, Punt CJ, Adema GJ. Immunomonitoring tumor-specific T cells in delayed-type hypersensitivity skin biopsies after dendritic cell vaccination correlates with clinical outcome. J Clin Oncol. 2005;23:5779–5787. doi: 10.1200/JCO.2005.06.478. [DOI] [PubMed] [Google Scholar]
- 18.Janossy G, Barry SM, Breen RA, Hardy GA, Lipman M, Kern F. The role of flow cytometry in the interferon-gamma-based diagnosis of active tuberculosis and its coinfection with HIV-1—a technically oriented review. Cytometry B Clin Cytom. 2008;74(Suppl 1):S141–S151. doi: 10.1002/cyto.b.20381. [DOI] [PubMed] [Google Scholar]
- 19.Keilholz U, Martus P, Scheibenbogen C. Immune monitoring of T cell responses in cancer vaccine development. Clin Cancer Res. 2006;12:2346s–2352s. doi: 10.1158/1078-0432.CCR-05-2540. [DOI] [PubMed] [Google Scholar]
- 20.van der Burg SH. Therapeutic vaccines in cancer: moving from immunomonitoring to immunoguiding. Expert Rev Vaccines. 2008;7:1–5. doi: 10.1586/14760584.7.1.1. [DOI] [PubMed] [Google Scholar]
- 21.Britten CM, Janetzki S, van der Burg SH, Gouttefangeas C, Hoos A. Toward the harmonization of immune monitoring in clinical trials: quo vadis? Cancer Immunol Immunother. 2007;57:285–288. doi: 10.1007/s00262-007-0379-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cox JH, Ferrari G, Kalams SA, Lopaczynski W, Oden N, D’souza MP. Results of an ELISPOT proficiency panel conducted in 11 laboratories participating in international human immunodeficiency virus type 1 vaccine trials. AIDS Res Hum Retrovirus. 2005;21:68–81. doi: 10.1089/aid.2005.21.68. [DOI] [PubMed] [Google Scholar]
- 23.Bednarek MA, Sauma SY, Gammon MC, Porter G, Tamhankar S, Williamson AR, Zweerink HJ. The minimum peptide epitope from the influenza virus matrix protein. Extra and intracellular loading of HLA-A2. J Immunol. 1991;147:4047–4053. [PubMed] [Google Scholar]
- 24.Valmori D, Fonteneau JF, Lizana CM, Gervois N, Lienard D, Rimoldi D, Jongeneel V, Jotereau F, Cerottini JC, Romero P. Enhanced generation of specific tumor-reactive CTL in vitro by selected Melan-A/MART-1 immunodominant peptide analogues. J Immunol. 1998;160:1750–1758. [PubMed] [Google Scholar]
- 25.Walker EB, Disis ML. Monitoring immune responses in cancer patients receiving tumor vaccines. Int Rev Immunol. 2003;22:283–319. doi: 10.1080/08830180305226. [DOI] [PubMed] [Google Scholar]
- 26.Heijnen IA, Barnett D, Arroz MJ, Barry SM, Bonneville M, Brando B, D’hautcourt JL, Kern F, Totterman TH, Marijt EW, Bossy D, Preijers FW, Rothe G, Gratama JW. Enumeration of antigen-specific CD8+ T lymphocytes by single-platform, HLA tetramer-based flow cytometry: a European multicenter evaluation. Cytometry B Clin Cytom. 2004;62:1–13. doi: 10.1002/cyto.b.20028. [DOI] [PubMed] [Google Scholar]
- 27.Maecker HT, Rinfret A, D’Souza P, Darden J, Roig E, Landry C, Hayes P, Birungi J, Anzala O, Garcia M, Harari A, Frank I, Baydo R, Baker M, Holbrook J, Ottinger J, Lamoreaux L, Epling CL, Sinclair E, Suni MA, Punt K, Calarota S, El-Bahi S, Alter G, Maila H, Kuta E, Cox J, Gray C, Altfeld M, Nougarede N, Boyer J, Tussey L, Tobery T, Bredt B, Roederer M, Koup R, Maino VC, Weinhold K, Pantaleo G, Gilmour J, Horton H, Sekaly RP. Standardization of cytokine flow cytometry assays. BMC Immunol. 2005;24:6–13. doi: 10.1186/1471-2172-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horton H, Thomas EP, Stucky JA, Frank I, Moodie Z, Huang Y, Chiu YL, McElrath MJ, De Rosa SC. Optimization and validation of an 8-color intracellular cytokine staining (ICS) assay to quantify antigen-specific T cells induced by vaccination. J Immunol Methods. 2007;323:39–54. doi: 10.1016/j.jim.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Asai T, Storkus WJ, Whiteside TL. Evaluation of the modified ELISPOT assay for gamma interferon production in cancer patients receiving antitumor vaccines. Clin Diagn Lab Immunol. 2000;7:145–154. doi: 10.1128/cdli.7.2.145-154.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maecker HT, Hassler J, Payne JK, Summers A, Comatas K, Ghanayem M, Morse MA, Clay TM, Lyerly HK, Bhatia S, Ghanekar SA, Maino VC, Delarosa C, Disis ML. Precision and linearity targets for validation of an IFN gamma ELISPOT, cytokine flow cytometry, and tetramer assay using CMV peptides. BMC Immunol. 2008;9:9. doi: 10.1186/1471-2172-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ogg GS, Jin X, Bonhoeffer S, Dunbar PR, Nowak MA, Monard S, Segal JP, Cao Y, Rowland-Jones SL, Cerundolo V, Hurley A, Markowitz M, Ho DD, Nixon DF, McMichael AJ. Quantitation of HIV-1-specific cytotoxic T lymphocytes and plasma load of viral RNA. Science. 1998;279:2103–2106. doi: 10.1126/science.279.5359.2103. [DOI] [PubMed] [Google Scholar]
- 32.Boon T, Coulie PG, Van den Eynde BJ, van der Bruggen P. Human T cell responses against melanoma. Annu Rev Immunol. 2006;24:175–208. doi: 10.1146/annurev.immunol.24.021605.090733. [DOI] [PubMed] [Google Scholar]
- 33.Coulie PG, Somville M, Lehmann F, Hainaut P, Brasseur F, Devos R, Boon T. Precursor frequency analysis of human cytolytic T lymphocytes directed against autologous melanoma cells. Int J Cancer. 1992;50:289–297. doi: 10.1002/ijc.2910500220. [DOI] [PubMed] [Google Scholar]
- 34.Meidenbauer N, Hoffmann TK, Donnenberg AD. Direct visualization of antigen-specific T cells using peptide-MHC-class I tetrameric complexes. Methods. 2003;31:160–171. doi: 10.1016/S1046-2023(03)00126-9. [DOI] [PubMed] [Google Scholar]
- 35.Britten CM, Gouttefangeas C, Welters MJ, Pawelec G, Koch S, Ottensmeier C, Mander A, Walter S, Paschen A, Muller-Berghaus J, Haas I, Mackensen A, Kollgaard T, thor SP, Schmitt M, Giannopoulos K, Maier R, Veelken H, Bertinetti C, Konur A, Huber C, Stevanovic S, Wölfel T, van der Burg SH. The CIMT-monitoring panel: a two-step approach to harmonize the enumeration of antigen-specific CD8+ T lymphocytes by structural and functional assays. Cancer Immunol Immunother. 2008;57:289–302. doi: 10.1007/s00262-007-0378-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chan C, Feng F, Ottinger J, Foster D, West M, Kepler TB. Statistical mixture modeling for cell subtype identification in flow cytometry. Cytometry A. 2008;73:693–701. doi: 10.1002/cyto.a.20583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Janetzki S, Panageas KS, Ben-Porat L, Boyer J, Britten CM, Clay TM, Kalos M, Maecker HT, Romero P, Yuan J, Kast WM, Hoos A. Results and harmonization guidelines from two large-scale international Elispot proficiency panels conducted by the Cancer Vaccine Consortium (CVC/SVI) Cancer Immunol Immunother. 2008;57:303–315. doi: 10.1007/s00262-007-0380-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


