Abstract
One of the most significant recent advances in health information systems has been the shift from paper to electronic documents. While research on automatic text and image processing has taken separate paths, there is a growing need for joint efforts, particularly for electronic health records and biomedical literature databases. This work aims at comparing text-based versus image-based access to multimodal medical documents using state-of-the-art methods of processing text and image components. A collection of 180 medical documents containing an image accompanied by a short text describing it was divided into training and test sets. Content-based image analysis and natural language processing techniques are applied individually and combined for multimodal document analysis. The evaluation consists of an indexing task and a retrieval task based on the “gold standard” codes manually assigned to corpus documents. The performance of text-based and image-based access, as well as combined document features, is compared. Image analysis proves more adequate for both the indexing and retrieval of the images. In the indexing task, multimodal analysis outperforms both independent image and text analysis. This experiment shows that text describing images can be usefully analyzed in the framework of a hybrid text/image retrieval system.
The shift from paper to electronic documents is one of the most significant advances in health information systems in recent years (Haux, 2006). While research on automatic text analysis and image processing has taken separate paths, there is a growing need for joint efforts. Modern electronic health documents such as electronic health records (EHR) or scholarly papers often intricately combine text and image media and need to be processed in a multimodal fashion (Lowe, Antipov, Hersh, & Smith, 1998). In recent years, several projects have considered alternatives to standard content-based image retrieval (CBIR; Smeulders, Worring, Santini, Gupta, & Jain, 2000) in order to accommodate the natural language processing (NLP) of documents where text and image co-occur (Müller, Michou, Bandon, & Geissbuhler, 2004). These efforts were also triggered by the habit users have of formulating text queries for information retrieval regardless of the medium of the documents they seek.
Practical Use Cases for Text/Image Retrieval
The need for multimodal (and more specifically text/image) retrieval may arise in various contexts. For example, consider the following scenarios:
Scholarly scenario: the need for a medical image to illustrate a particular situation, such as the presentation of a pathology in class for medical school professors or students. In this case, users would need to search a domain-specific database such as MEDLINE® or CISMeF1 (referencing curated content, as opposed to Google) by describing the desired image with natural language text.
Clinical scenario: the need for a second opinion when confronted with a new case for an intern or doctor. The clinician may have formed a hypothetical diagnosis and wish to search a database such as IRMA2 to consult similar cases and confirm his diagnosis. Currently, the search may be carried out using an image or code corresponding to a term in the IRMA terminology. Introducing a text query feature would make the system more approachable for users who are not familiar with the intricacies of the IRMA code. It would help them launch a search using text, knowing that they could later refine the search by selecting the most appropriate images returned by the text search.
In addition to these retrieval situations, it must be stressed that curated databases such as MEDLINE, CISMeF, or IRMA index the documents they reference using a controlled vocabulary. Therefore, automatic indexing tools that can efficiently help with the indexing effort are an asset to these projects.
Related Work in Text/Image Retrieval
Text/image retrieval belongs in the wider domain of information retrieval, where one of the most crucial issues is that of information representation. Specifically, what is the best way to represent queries and documents that will eventually need to be matched? Which features should be selected? Typical representation paradigms in text retrieval are text words (or sometimes sequences of characters), word stems, or keywords chosen in a controlled list. For image retrieval, Boujemaa, Fauqueur, and Gouet (2003) draw from text retrieval experience of complex query handling to refine the method of “query by visual example,” also referred to as the query-by-example (QBE) paradigm (Niblack et al., 1993). They develop a visual thesaurus composed of small images representative of color, texture, or objects. Using this thesaurus, the user can compose a query to indicate which elements should be in the images to be retrieved, such as “building AND blue region AND_NOT green region,” using three thesaurus images. Srihari, Zhang, and Rao (2000) show that multimodal queries (composed of text and image) improve the performance of retrieval in a multimodal database. In addition to providing the retrieval system with a more comprehensive information query, this method also takes advantage of the specific type of information that may be provided by each medium: information drawn from the text part of the query may be used to select a targeted image-analysis technique. For example, a text query referring to a person could trigger the use of a face-recognition component for image analysis, which would be useless in a search for a particular type of landscape. However, Byrne and Klein’s work on a database of text and image documents in the archeology and architectural history domain (Byrne & Klein, 2003) shows that combining NLP and CBIR approaches for image retrieval is not always successful. Rius (2006) explores a “keyword propagation” technique to provide automatic annotations for images. The hypothesis that images that have similar visual features should also share a similar textual description is tested through a retrieval task. Images in a news test collection are retrieved according to their visual content (using state-of-the-art CBIR features) and according to their textual description (using the statistical indexing system OKAPI; Robertson, Walker, & Beaulieu, 1998). The results show that there is no correlation between the two sets of documents retrieved for each test query. Rius concludes that the description techniques used on the images and accompanying text are not suitable to apply keyword propagation.
Specificities of Document Retrieval in the Medical Domain
As in other work on text-image retrieval in the general domain (e.g., Zhao & Grosky, 2002), Srihari et al. (2000) perform free-text indexing using methods such as latent semantic indexing (Deerwester, Dumais, Furnas, Landauer, & Harshman, 1990). In the biomedical domain, Shatkay, Chen, and Blostein (2006) also perform free-text indexing using Bayes classifiers in the Text REtrieval Conferences (TREC) Genomics track, and so do Ruiz and Srikanth (2004) using a vector-space model.
However, in the biomedical domain, indexing is more frequently performed using a predefined set of controlled, domain-specific terms. For example, in their experiments, Müller, Geissbuhler, and Ruch (2005) use Medical Subject Headings® (MeSH®). In Goldminer, a radiology image search engine, Kahn and Thao (2007) index the caption text of images with UMLS concepts in addition to free text.
We are particularly interested in controlled indexing in the context of our work with IRMA, which uses specific codes to describe images (see below) and CISMeF, which uses MeSH descriptors to describe documents. In fact, it is important to stress the descriptive aspect of a set of controlled terms assigned to a document. In addition to enabling document retrieval in a database, controlled terms may also be viewed as a way to provide users with a conceptual summary of documents’ subject matter. In this respect, it is important to consider which controlled vocabulary should be used to allow for a full description of the documents. Text/image experiments are sometimes limited to classifying images according to their modality (e.g., graph, radiography, etc.; see Rafkind, Lee, Chang, & Yu, 2006), leaving out content information.
Availability of Text-Image Collections for Experiments
Our scenario for text/image retrieval involves scholarly documents such as lecture notes or scientific articles. In both cases, the multimodal documents are often composed of paragraph text, images, and caption text. The hypothesis can be made that caption text and paragraph text mentioning an image should provide a natural language utterance directly related to the image. Rowe and Gugliemo (1993) advocate the use of NLP techniques to extract semantic triplets (i.e., a set of two concepts and the relation between them) from the caption text, which constitutes higher-level indexing than isolated keywords. The captions used in their experiments were produced specifically for image indexing in an image-retrieval system. It would be interesting to assess the viability of this type of high-level semantic indexing on independently annotated images in order to avoid any bias in the writing style of the caption.
In the case where one contemplates the use of captions already included in existing documents (i.e., in which the captions were not made specifically to accommodate image retrieval), the task of extracting images, captions, and the links between them constitutes a full research problem in itself (Chang, Kayed, Girgis, & Shaalan, 2006; Christiansen, Lee, & Chang, 2007). Assuming this material is available (a collection of images accompanied by their captions) Srihari et al. (2000, p. 246) rightly point out that even though the captions may contain descriptive information on the image, “objects mentioned in the caption are not always in the picture.” In fact, image captions—and paragraph text mentioning an image—constitute “secondary documents” (Briet, 1951), and while using them, one must keep in mind that they do not provide direct access to the image content. Pastra and Wilks (2004) have specifically pointed out the lack of text/image multimodal resources, especially those linking images with text naming the objects they represent along with gold-standard annotations.
Contribution of Our Work
In summary, there are limitations in both text and image analysis when retrieving documents from multimodal medical records. On the one hand, although NLP allows the mapping of free text to controlled vocabularies such as MeSH, a natural language description of medical images can be lacking to formulate information queries. On the other hand, while CBIR techniques enable adequate matching between images, the retrieval is lacking when appropriate query images are not available. These observations indicate a need to link image and text analysis in the framework of multimodal document retrieval. The goal of the work presented in this article is twofold. First, we aim at analyzing the potential of text and image approaches individually. We specifically intend to assess how appropriate paragraph text and caption text are for the description of medical images. Second, we want to assess the performance yielded by the combination of image and text analysis.
Materials and Method
Corpus and Reference Annotations
As mentioned above, extracting images and related text (such as an image caption or a paragraph discussing the image) is in fact a research problem in itself (Chang et al., 2006). For this reason, previous experiments sometimes used all the text that appears in the document containing the image (Srihari et al., 2000; Ruiz & Srikanth, 2004). The annotation of a corpus by domain experts is also a costly task. For this reason, evaluation campaigns such as CLEF focus on information-retrieval tasks and have annotators evaluate the relevance of an automatically selected subset of corpus documents to the evaluation queries (Hersh et al., 2006). For our experiments, we used a corpus of medical images accompanied by a caption and/or paragraph text. The documents were selected from institutional and educational health resources referenced in the CISMeF catalog and automatically collected from the Internet using a supervised wrapper system (Florea, 2006).
The corpus contains 180 radiographs with a text caption and/or paragraph of text mentioning the image. The text (caption or paragraph) in the corpus is in French. It generally contains one or more of the following elements on or about the image:
imaging modality
imaging angle
body part
biosystem (body system, such as “respiratory system”)
disease or pathology/diagnosis hypothesis
patient-related data (e.g., name, gender, or age)
physician-related data (e.g., name, service, or hospital)
Although in some cases, caption and paragraph contained almost the same text, in general, the paragraphs were significantly longer than captions (Table 1).
TABLE 1.
Length of caption and paragraph text in the corpora.
| Corpus | Type of text | Document count N | Word count |
|---|---|---|---|
| Training | Paragraph only | 13 | 26 ± 14 |
| Caption only | 85 | 13 ± 16 | |
| Test | Paragraph | 81a | 44 ± 42 |
| Caption | 81a | 17 ± 20 | |
| Test and Training | Paragraph | 94 | 42 ± 40 |
| Caption | 166 | 15 ± 18 |
In the test corpus, one radiograph showed two hands and had to be split in two images (one for each hand) to make IRMA indexing possible. For text processing, however, the same caption and paragraph text had to be used for both images.
We divided the corpus in two sets: a training set comprising 98 X-ray images with either only a caption (N = 85) or only a paragraph (N = 13), and a test set comprising 82 X-ray images with both caption and paragraph. To allow for a comparison of indexing and retrieval performance when caption versus paragraph text was used, it was decided to reserve the documents for which both types of text were available for the test set. In the training set, we identified 5 pairs of documents with identical or near-identical images. However, the texts associated with the images showed some variation, so we decided to keep them as such.
Images in the whole corpus have been annotated by an experienced radiologist with respect to the image type and content. Based on the framework for image retrieval in medical applications (IRMA; Lehmann et al., 2004), this information is conveyed using the IRMA code for medical-image description (Lehmann, Shubert, Keysers, Kohnen, & Wein, 2003). The IRMA code includes 4 fields to encode information related to the image:
modality (e.g., X-ray, MRI; 415 codes in total)
direction (e.g., frontal, sagittal; 115 codes in total)
body part (e.g., abdomen, thorax; 238 codes in total)
biosystem (e.g., digestive system, respiratory system; 355 codes in total).
Figure 1 shows a sample image from the test corpus, with its accompanying caption and paragraph text. The IRMA code selected by the radiologist for this image is “1221-115-700-400,” which corresponds to a “plain analog radiography overview” taken in the “AP, coronal, upright” direction representing the “abdomen” within the “gastrointestinal system.” The annotations supplied by the radiologist for the entire corpus were considered as the gold standard (ground truth) for all experiments. The corpus is freely available for research purposes by request to the authors.
Figure 1.
Sample document in the test corpus. An English translation of the text is provided by the authors for illustration purposes: Caption: “François, 1 1/2 month: Abdomen without preparation.” Paragraph: “Based on the child’s age and the fact that he experienced projectile vomiting, a diagnosis of pyloric stenosis can be made. An emergency abdomen without preparation is ordered for François; no evidence of gastric dilatation is visible.”
Processing Multimodal Medical Documents
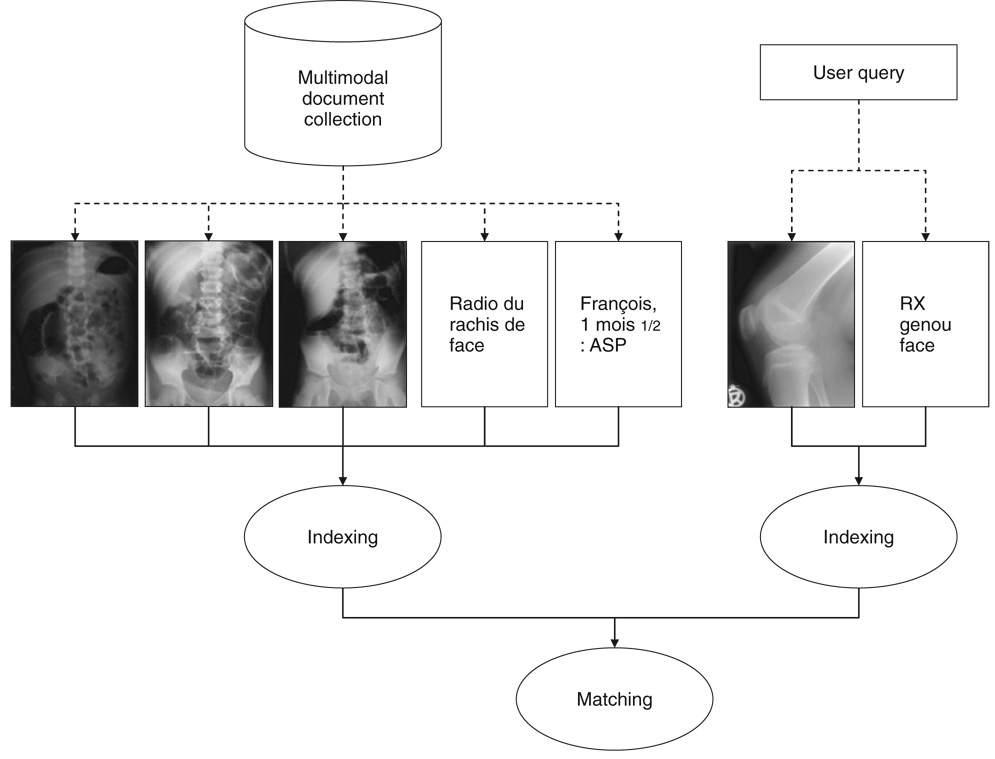
To perform retrieval in a collection of multimodal medical documents, it is necessary to devise a representation scheme that allows the description of both text and image components. The critical issue is the compatibility between image and text representations, so that the system can take advantage of image description using text and also interpret both text and image queries for retrieval. Using adequate controlled terms for the indexing of text and image components can facilitate the document/query matching phase. For example, IRMA codes may be used for the representation of medical images, ICD-9 codes may be used for clinical text, MeSH terms for bibliographical text, and so on. Furthermore, relationships between the concepts of these various controlled vocabularies can support the conjoint use of more than one vocabulary for representing multimodal documents. Figure 2 illustrates the general workflow of such a retrieval system. As detailed in the next section, our work focused on image representation using content-based analysis and two types of descriptive texts. The indexing relied on IRMA codes, as did the queries used in the retrieval experiments.
Figure 2.
Processing multimodal biomedical documents for information retrieval.
Experiments
The automatic systems for image annotation are compared to the gold standard obtained from our radiologist. Since we are working with radiographs only, the modality is already known. Furthermore, since the biosystem information is of minor relevance for document retrieval, we decided to focus our efforts on the direction of projection and the body part that is displayed in the image. Two experiments are performed on the data: an indexing task and a retrieval task.
Semantic indexing task
The semantic indexing task consists of producing a single IRMA code as an annotation for a medical image. Here, a single IRMA code is assigned automatically to each of the images from the test corpus. For example, in the case of the sample image shown in Figure 1, the correct code is “1121-115-700-400.” In the IRMA taxonomy, each nonzero digit in a particular section of the code conveys additional precision. For example, the anatomy code 710 (upper abdomen) is more precise than 700 (abdomen), since the two codes are hierarchically related. For this reason, we considered the number of digits in common in our evaluation. For example, if the system produces the code 910 where the reference advocates 914, we consider that there is a two-digit match. This evaluation is also intended to account for cases where the text-analysis method may not be able to retrieve an exact match, such as the case of “left foot” (914) being mapped to “foot” (910) instead of the more precise code (914). As explained below, our text-analysis system is based on MeSH indexing, and MeSH does not distinguish between the concepts “foot” and “left foot.” For the semantic indexing task, correctness in terms of the number of correct digits in each code is computed. Here, the precision p corresponds to the number of codes correctly assigned to the images over the total number of codes retrieved; recall r corresponds to the number of codes correctly assigned to the images over the number of codes that were expected. In addition, we calculate a balanced F-measure f (Hripcsak & Rothschild, 2005):
Retrieval task
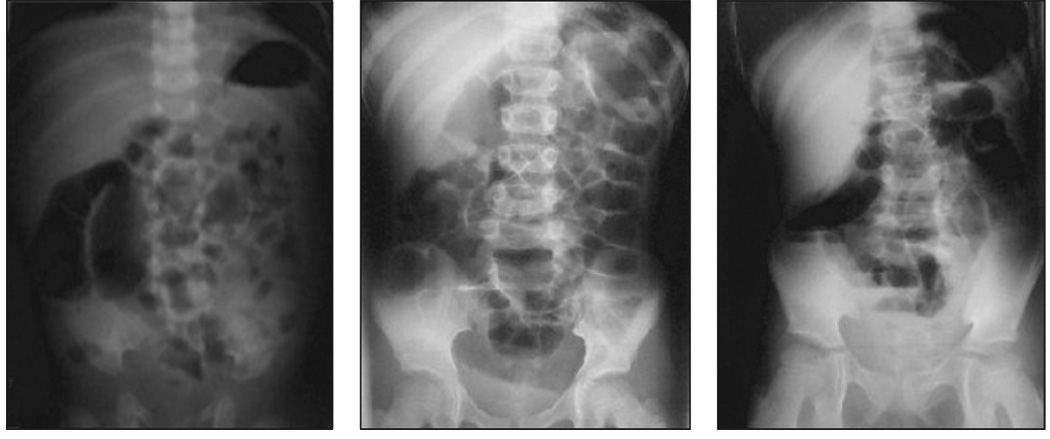
The retrieval task consists of matching images from the test collection to a query in the form of an IRMA code. Figure 3 shows the set of images matching a sample query.
Figure 3.
Set of images in the test corpus matching the query “1121-115-700-400”.
This experiment is performed for each individual code that is present in the test corpus. For this task, the documents in the test corpus are annotated with IRMA codes to which the query will be matched. Contrary to the semantic indexing task, more than one code may be assigned to each image. For example, if an image is assigned the two codes 800 (pelvis) and 950 (upper leg), it may be retrieved by queries containing either code.
Precision p corresponds to the number of images correctly retrieved for a given query over the total number of images retrieved. Recall r corresponds to the number of images correctly retrieved for a given query over the number of images that were expected for the query. Again, we also calculate the balanced F-measure f.
Methods for Automatic Annotation
In this section, we describe the methods we used for automatic annotation based on text analysis, image analysis, and the combination of both. Figure 4 illustrates the different text and image analyses detailed in this section.
Figure 4.
Text and image analysis performed to assign IRMA codes to a test document. Text analysis (solid line and box) involved applying a dictionary and using the training collection to retrieve the 5 nearest neighbors (5-NN) for the text portion of a query document. The image analysis (dashed lines and boxes) used either the training collection or the IRMA database to retrieve the k nearest neighbors (k-NN) based on content-based image retrieval (CBIR) features.
Text analysis: Natural language processing
In these experiments, the text analysis was performed at two levels: First, dictionaries containing a list of IRMA terms and corresponding terms or expressions that can be used to refer to them in natural language were applied to the text data to directly retrieve IRMA index terms. For example, according to this dictionary, if the words “abdomen,” “abdominal,” or “belly” appear in the text, they will be mapped to the IRMA term “abdomen” with code 700. In a second step, these IRMA indexing terms were used as a normalized description of the documents. Hence, for each document in the test set, the nearest-neighbor documents were retrieved from the training set in order to infer the final indexing.
An existing MeSH dictionary was used to extract body-part information from the caption text (Névéol, Douyère, Rogozan, & Darmoni, 2007). Previous work by the IRMA team produced a mapping table between MeSH and IRMA codes, wherever possible. The links in this table were established based on the English versions of MeSH and IRMA terminologies. To increase the coverage of the mapping from MeSH terms to IRMA codes, the links between a given MeSH term and a given IRMA code were propagated to the descendants of the MeSH terms, provided these descendants had no existing link to IRMA. This mapping was possible because the MeSH hierarchy that contains anatomy terms contains no cycles (Mougin & Bodenreider, 2005). For example, the MeSH term “toes” was mapped to the IRMA code 911—“toe.” “hallux” is a descendant of “toes” in the MeSH hierarchy and was not mapped to any IRMA code. By propagating the relation between “toes” and “toe,” “hallux” was also mapped to “toe,” as shown in Figure 5.
Figure 5.
Sample IRMA-MeSH equivalences made into dictionary entries (using MeSH hierarchy).
The dictionary we used for extracting body-part codes from the captions contained entries related to all the MeSH terms that could be mapped to IRMA codes.
Some adaptations were also necessary to accommodate the retrieval of terms that were more specific than what is currently available in the MeSH (Figure 6). For example, there are two different IRMA codes for “right foot” and “left foot,” while both terms can be mapped to the MeSH term “foot.” In practice, this meant that our text system was not able to distinguish between the concepts “right foot” and “left foot.” Such phrases yielded the term “foot” and resulted in the assignment of the code 910.
Figure 6.
Sample IRMA-MeSH equivalences made into dictionary entries (adapting to IRMA specificity).
However, MeSH does not include terms describing image direction. A set of image-direction terms in French was developed in order to extract the corresponding IRMA codes from text using regular expressions. Relevant image direction terms were sought from the training data. Specifically, the caption or paragraph texts accompanying images in the training corpus was manually analyzed (by the first author: AN) in order to infer which expression in the text referred to the image angle. These findings were presented to a radiologist (JND, Rouen University Hospital) for validation. This led to discussions in which the radiologist was often able to offer additional expressions that may be used by health professionals to describe imaging angles. For example, as shown in Table 2, the expression “de face” (coronal) corresponds to IRMA image-direction code 100.
TABLE 2.
Sample expressions used to retrieve image directions.
| Text | IRMA | Code |
|---|---|---|
| de face | coronal, unspecified | 100 |
| AP | coronal, anteroposterior | 120 |
| AP AND debout | coronal, anteroposterior, upright | 125 |
With this method, the use of the training data was limited to a contribution to augmenting existing dictionaries of biomedical terms.As a first step of the text analysis, the indexing resulting from direct dictionary lookup was considered as the final indexing for the images.
In a second step, the dictionary was used to index the text contained in both the training and test sets3 so that the training set was used to retrieve the 5 nearest neighbors (5-NN) for each test document.4 Similarity between the documents was computed using a cosine measure between vectors of IRMA codes. The final indexing consisted of the majority vote between the neighbors.
Content-based image analysis
The methodology for content-based annotation of images is adopted from the IRMA framework according to the experiments previously performed in the image challenge of the Cross Language Evaluation Forum (CLEF) (Müller et al., in press) and on dental radiographs (Deselaers, Müller, Clough, Ney, & Lehmann, 2007). Each image is represented by a global signature that is built from numerical features that are extracted from the pixel gray-scale values of the entire image. In particular, we applied a combination of the Tamura texture measures (TTM; Tamura, Mori, & Yamawaki, 1978), the cross-correlation function (CCF), and the image-distortion model (IDM; Keysers, Dahmen, Ney, Wein, & Lehmann, 2003).
Unlike approaches that are based on co-occurrence matrices (Haralick, Shanmugam, & Dinstein, 1973), TTM directly captures coarseness, directionality, and contrast. To take size variations into account, the analysis of local averages, properties of image gradients, and local variance of pixel intensities are aggregated into one histogram per image and compared using the Jensen-Shannon divergence. The CCF models differences in the global positioning of the patients within the imaging device. The IDM is a correlation-based feature that also models local deformations. This is important to cope with interindividual variations of living tissue as well as intraindividual differences resulting from pathologies, growth, or aging. Because TTM represents each image by 384 numerical features, the X-ray images are ultimately represented by 1,408 or 640 feature values, while IDM and CCF are based on downsized images of 32 × 32 or 16 × 16 pixels, respectively. The vector of feature values is also referred to as global signature (Deserno, Antani, & Long, 2007).
For indexing and retrieval, the signature of the query image is compared to those of a database of 10,000 reference images that have been annotated by experienced radiologists (ground truth, as used for the CLEF campaign). In particular, each classifier results in a distance d (r, q) between the query and reference feature vectors,q and r, respectively. Let C denote a class and the set of according reference vectors r. A parallel combination of N classifiers yields
| (1) |
where denotes the normalized distance, and the weights each classifier contributes to the final result.
Based on the training set of 98 images, optimal weights λTTM, λCCF, and λIDM are determined to be 0.30, 0.45, and 0.25, respectively. Neighbor images are then retrieved from the IRMA collection (N = 10,000). For comparison, the λTTM, λCCF, and λIDM that were used in the previous CLEF experiments were 0.40, 0.18, and 0.42, respectively.
In order to optimize the results for both indexing and retrieval, the confidence in the code assignments must be assessed. During the training phase, a threshold of required similarity smin is computed. This corresponds to the lowest similarity obtained for a correctly assigned image. After a temporary majority vote is obtained from the nearest neighbors retrieved, the score of the best neighbor with the temporary code is compared to the threshold. If the score is above the threshold, the temporary code is rejected and no code is assigned for the image considered. In our experiments, this method successfully rejected erroneous codes but did not reject any valid codes.
Multimodal document analysis
Multimodal document analysis consists of a combination of the results obtained from both NLP of caption text and CBIR. Based on the results of the text analysis, only the caption text was used. The indexing candidates resulting from the top 5 neighbors extracted with image analysis were also considered to index images in a multimodal fashion.
For the indexing task, the majority votes for imaging-angle and body-part code were selected as the final indexing codes. When using “equal vote,” i.e., given the same weight for both image and text code candidates, little change was observed. This was to be expected since text analysis provides an average of 1.3 candidates (each occurring once) versus 2.2 (with a total of 5 occurrences) for the image analysis. For further testing, “balanced vote” was applied. A weight of 2 was given to the indexing angle codes, a weight of 3 was given to the body-part codes, and a weight of 1 was given to each occurrence of candidates obtained through image analysis.
For the retrieval task, we first considered the final code assigned to the images (one code per image). Then, we also considered a pooling of all candidate codes (considered equally).
Results
A total of 22 different codes were assigned to the images in our test corpus by the radiologist who reviewed them and assigned the gold-standard codes (ground truth). In the tables below, bolded figures show the best performance obtained for the indexing and retrieval tasks.
Semantic Indexing Task
Table 3 presents the performance of text analysis for the assignment of IRMA imaging-angle and body-part codes. For text analysis, we show the results obtained when caption text and paragraph text was processed (column 4 and 5) as well as when the codes were used to retrieve the 5 nearest-neighbor documents in the training set to infer final indexing codes (column 3).
TABLE 3.
Performance of text in the indexing task. Precision p, recall r, and balanced F-measure f are given as percentages.
| Text |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 5-NN |
Caption |
Paragraph |
||||||||
| Digit | p | r | f | p | r | f | p | r | f | |
| Imaging angle | ||||||||||
| 1 | 97.40 | 91.46 | 94.34 | 93.33 | 34.15 | 50.00 | 66.67 | 9.76 | 17.02 | |
| 2 | 84.42 | 79.27 | 81.76 | 0.00 | 0.00 | 0.00 | 16.67 | 2.44 | 4.26 | |
| 3 | 77.92 | 73.17 | 75.47 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| Body part | ||||||||||
| 1 | 51.95 | 48.78 | 50.31 | 94.03 | 76.83 | 84.56 | 67.92 | 43.90 | 53.33 | |
| 2 | 38.96 | 36.59 | 37.74 | 59.70 | 48.78 | 53.69 | 37.74 | 24.39 | 29.63 | |
| 3 | 31.17 | 29.27 | 30.19 | 35.82 | 29.27 | 32.21 | 16.98 | 10.98 | 13.33 | |
Table 4 presents the performance of text analysis for the assignment of IRMA imaging-angle and body-part codes. We show the results obtained when CLEF weights are used and 1 nearest neighbor is retrieved from the training set (column 3), as well as when weights obtained from the training set are used to retrieve the 5 nearest-neighbor documents in the IRMA database (column 4).
TABLE 4.
Performance of image analysis in the indexing task. Precision p, recall r, and balanced F-measure f are given as percentages.
| Image |
|||||||
|---|---|---|---|---|---|---|---|
|
1-NN |
5-NN |
||||||
| Digit | p | r | f | p | r | f | |
| Imaging angle | |||||||
| 1 | 98.72 | 93.90 | 96.25 | 93.75 | 91.46 | 92.59 | |
| 2 | 78.21 | 74.39 | 76.25 | 88.75 | 86.59 | 87.65 | |
| 3 | 70.51 | 67.07 | 68.75 | 86.25 | 84.15 | 85.19 | |
| Body part | |||||||
| 1 | 66.67 | 63.41 | 65.00 | 90.00 | 87.80 | 88.89 | |
| 2 | 61.54 | 58.54 | 60.00 | 85.00 | 82.93 | 83.95 | |
| 3 | 56.41 | 53.66 | 55.00 | 81.25 | 79.27 | 80.25 | |
Table 5 presents the performance of combined image (5-NN) and text analysis (caption) for the assignment of IRMA imaging-angle and body-part codes.
TABLE 5.
Performance of combined text and image analysis in the indexing task. Precision p, recall r, and F-measure f are given as percentages.
| Text/Image |
|||||||
|---|---|---|---|---|---|---|---|
| Equal vote |
Balanced |
||||||
| Digit | p | r | f | p | r | f | |
| Imaging angle | |||||||
| 1 | 93.90 | 93.90 | 93.90 | 97.40 | 91.46 | 94.34 | |
| 2 | 91.46 | 91.46 | 91.46 | 96.10 | 90.24 | 93.08 | |
| 3 | 85.37 | 85.37 | 85.37 | 92.21 | 86.59 | 89.31 | |
| Body part | |||||||
| 1 | 87.80 | 87.80 | 87.80 | 98.70 | 92.68 | 95.60 | |
| 2 | 82.93 | 82.93 | 82.93 | 85.71 | 80.49 | 83.02 | |
| 3 | 79.27 | 79.27 | 79.27 | 79.22 | 74.39 | 76.73 | |
It can be observed that for all methods, performance decreases with the number of correct digits assigned. However, the gap is wider for text analysis than image analysis.
Retrieval Task
Table 6 presents the performance of image and text analysis for image retrieval, after IRMA codes have been assigned using the methods described previously. For text analysis, we show the results obtained when caption text was processed and the 5 nearest neighbors retrieved from the training set.
TABLE 6.
Performance of text and image analysis in the retrieval task. Precision p, recall r, and F-measure f are given as percentage.
| Retrieved | Correct | Precision | Recall | F-measure | |
|---|---|---|---|---|---|
| Image (5-NN) | |||||
| Final code only | 70 | 63 | 90.00 | 76.83 | 82.89 |
| All codes | 139 | 73 | 52.52 | 89.02 | 66.06 |
| Text/Image | |||||
| Balanced Final code | 71 | 64 | 90.14 | 78.05 | 83.66 |
| Weighted Final code | 135 | 66 | 48.89 | 80.49 | 60.83 |
| All codes | 371 | 76 | 20.49 | 92.68 | 33.55 |
| Text (Captions) | |||||
| Final code only | 49 | 29 | 59.18 | 35.37 | 44.27 |
| All codes | 76 | 36 | 47.37 | 43.90 | 45.57 |
| Text (5-NN) | |||||
| Final code only | 65 | 12 | 18.46 | 14.63 | 16.32 |
| All codes | 202 | 47 | 23.27 | 57.32 | 33.10 |
Discussion
Comparison With Other Research
In previous text-image retrieval experiments using the Casimage database, Müller et al. (2004) found that the best results were obtained when image analysis was given a much higher weight than text analysis (80%).
While using a portion of the corpus built by Florea (2006) for the indexing of images according to 6 modalities and 19 body parts, our experiments were focused on the indexing of images according to the 115 imaging angles and 238 anatomy codes listed in the IRMA terminology. There is a difference in the indexing terms used (more specifically, the terms themselves, the aspect of image description they represent, and the scale of the indexing sets). Moreover, our choice of selecting images for which both paragraph and caption text was available enabled us to compare the benefit of using either type of text for image indexing. The drawback was that we had to use a smaller corpus.
Compared to the image-CLEF experiments reported in (Müller et al., 2004) our corpus is also significantly smaller (82 images vs. several thousand), but has the advantage of having been annotated manually by a radiologist. The reference used for the Casimage database resulted from a pool of automatic annotations produced by the systems competing in CLEF. In fact, our reference annotations can be considered as a gold standard according to the criteria described by Lehmann (2002).
In summary, our experiments differ from the literature on medical-image annotations in the following respects:
We are using a small, manually annotated test corpus. Both caption and paragraph texts are available for each image in the test corpus.
We are indexing images with controlled index terms providing comprehensive coverage of imaging angles and body parts.
What We Learned From the Experiment
Image versus text analysis
The results of both indexing and retrieval using image analysis were generally better than those obtained with text analysis. However, in some cases, text analysis provided correct codes for the indexing task where image analysis did not. This suggests that there is room for general performance improvement when text and image analysis are combined.
For the text analysis, the results of both tasks (indexing and retrieval) indicate that the caption text is more suitable as a secondary source of information on images than paragraph text.
Moreover, results also show that, in our test corpus, body-part information is more readily available in the caption or paragraph texts than image-direction information. In fact, image-direction information seems to be implicit in many cases. For example, we learned from our expert radiologists that “intravenous urographies” (IVU) are always performed while the patient is in an “AP, coronal, supine” position (IRMA code 127); therefore this type of image-direction information would be redundant with the modality information and is not explicitly stated in the text. Our experiment of assigning codes through the retrieval of nearest-neighbor documents in the training set confirms this intuition, as this method performed much better for the retrieval of imaging-angle codes. However, it was less successful with the retrieval of body-part codes. The size of the training set cannot fully explain this phenomenon as CBIR performed better using the same training set.
In general the information in the text caption is also more subjective than the “unique” description provided by our expert radiologist with IRMA annotations. For example, for an image of a foot where a fracture can be seen on the hallux (big toe), a text caption may refer to the “hallux fracture,” which is the point of interest on the image, instead of the whole foot, which is actually shown on the image. Such a caption would lead the text-based system to extract the code 911 “toe” instead of 910 “foot” selected by the radiologist. Arguably, both codes could be considered suitable for the image. Furthermore, for two images in the test collection, the CBIR system recommended the code 413, “carpal bones,” where the radiologist provided 421, “left carpal joint.” After a discussion in which the test images and the images from the RMA collection that led to the recommendation of 413 were shown to the radiologist, he concluded that both codes could be considered suitable for the test images.
Results obtained with the image analysis are slightly better for the imaging angle than the body part. However, the disparity is not as significant as it is with text analysis. One of the strong points of the image-indexing method is that a code can be easily provided for each image, whereas the text analysis may be silent in some cases.
Multimodal analysis
The results of the multimodal analysis show that the combination of text and image analysis seems to be more beneficial for indexing than for retrieval; in our experiment, retrieval using image analysis alone produced very close results to retrieval using multimodal analysis.
This study could be useful for future work addressing the accommodation of multimodal queries, for instance, retrieving images with a text query.
Conclusion
We have shown that caption text seems to be more appropriate than paragraph text to describe the content of medical images. Furthermore, although image analysis performs better than text analysis for medical image description and retrieval, some of the results are complementary, and combining the techniques improves the description precision and the retrieval recall. However these results—especially text analysis—need to be confirmed by further experiments on a larger corpus.
Acknowledgments
This research was supported in part by an appointment of A. Névéol to the Lister Hill Center Fellows Program and an appointment of T.M. Deserno, né Lehmann, to the Lister Hill Center Visitor Program both sponsored by the National Library of Medicine and administered by the Oak Ridge Institute for Science and Education. The authors would like to thank Jean-Nicolas Dasher for his insight in creating the imaging-angle dictionary for text analysis, Filip Florea for supplying the corpus data used in this work, and Bastian Ott for supplying IRMA reference coding for the dataset.
Footnotes
French acronym for the Catalog of Online Medical Information in French (CHU de Rouen, 2008).
Image Retrieval in Medical Applications (Aachen University of Technology, 2008).
To avoid cases where no IRMA representation was available for the test documents, we used the representation yielded from the caption analysis, or, when it was not available, the representation yielded by the paragraph analysis.
Using controlled vocabulary (here, IRMA terms) for representing the documents yielded better results than directly using the plain text (either caption or paragraph) of the documents. This was also observed recently in a similar experiment assigning controlled terms to clinical text (Aronson et al., 2007).
Contributor Information
Aurélie Névéol, U.S. National Library of Medicine, National Institutes of Health, 8600 Rockville Pike, Bethesda, MD 20894. E-mail: neveola@nlm.nih.gov.
Thomas M. Deserno, U.S. National Library of Medicine, National Institutes of Health, 8600 Rockville Pike, Bethesda, MD 20894, and Department of Medical Informatics, Aachen University of Technology (RWTH), Pauwelsstrasse 30, 52057, Aachen, Germany. E-mail: tdeserno@mi.rwth-aachen.de
Stéfan J. Darmoni, CISMeF Group, Rouen University Hospital and GCSIS, LITIS EA 4051, Institute of BioMedical Research, University of Rouen, 1 rue de Germont, 76031 Rouen Cedex, France. E-mail: Stefan.Darmoni@chu-rouen.fr
Mark Oliver Güld, Department of Medical Informatics, Aachen University of Technology (RWTH), Pauwelsstrasse 30, 52057, Aachen, Germany. E-mail: mgueld@mi.rwth-aachen.de.
Alan R. Aronson, U.S. National Library of Medicine, National Institutes of Health, 8600 Rockville Pike, Bethesda, MD 20894. E-mail: alan@nlm.nih.org
References
- Aachen University of Technology Image retrieval in medical applications. 2008 Retrieved March 28, 2008, from http://www.irma-project.org/index_en.php.
- Aronson AR, Bodenreider O, Demner-Fushman D, Fung KW, Lee VK, Mork JG, et al. From indexing the biomedical literature to coding clinical text : Experience with MTI and machine learning approaches. In: Cohen K, Demner-Fushman D, Friedman C, Hirschman L, Pestian J, editors. Proceedings of the ACL 2007 Workshop on Biological, Translational, and Clinical Language Processing (BioNLP); 2007. pp. 105–112. [Google Scholar]
- Boujemaa N, Fauqueur J, Gouet V. IAPR International Conference on Image and Signal Processing (ICISP’ 2003), Agadir, Morocco. What's beyond query by example? 2003 June; Retrieved August 29, 2008, from http://www-rocq.inria.fr/~gouet/Recherche/Papiers/icisp03/pdf.
- Briet S. Qu’est-ce que la documentation? [What is documentation?] Paris: Éditions Documentaires et Industrielles; 1951. [English translation retrieved April 29, 2007, from http://martinetl.free.fr/suzanne_briet.htm and archived at http://www.webcitation.org/5OUOoaTjW] [Google Scholar]
- Byrne K, Klein E. Image retrieval using natural language and content-based techniques. In: de Vries AP, editor. Proceedings of the Fourth Dutch-Belgian Information Retrieval Workshop (DIR 2003); Amsterdam: Institute for Logic, Language, and Computation; 2003. pp. 57–62. [Google Scholar]
- Chang CH, Kayed M, Girgis MR, Shaalan K. A survey of Web information extraction systems. IEEE Transactions on Knowledge and Data Engineering. 2006;18:1411–1428. [Google Scholar]
- Christiansen A, Lee DJ, Chang Y. Finding relevant PDF medical journal articles by the content of their figures. In: Horii SC, Andriole KP, editors. Medical Imaging 2007: PACS and Imaging Informatics; Proceedings of the International Society for Optical Engineering (SPIE); 2007. pp. OK1–OK12. [Google Scholar]
- CHU de Rouen Catalogue et Index des Sites Médicaux Francophones [Catalog and index of online health information in French] 2008 Retrieved March 28, 2008, from http://www.cismef.org.
- Deerwester S, Dumais S, Furnas G, Landauer T, Harshman R. Indexing by latent semantic analysis. Journal of American Society for Information Science. 1990;6:391–407. [Google Scholar]
- Deselaers T, Müller H, Clough P, Ney H, Lehmann TM. The CLEF 2005 automatic medical image annotation task. International Journal of Computer Vision. 2007;74:51–58. [Google Scholar]
- Deserno TM, Antani S, Long LR. Exploring access to scientific literature using content-based image retrieval; Proceedings of the International Society for Optical Engineering (SPIE); 2007. pp. OL1–OL8. [Google Scholar]
- Florea FI. Combining textual and visual information for automatic annotation of online medical images (Technical Report) Rouen, France: Laboratoire LITIS; 2006. [Google Scholar]
- Haralick RM, Shanmugam K, Dinstein I. Textural features for image classification. IEEE Transactions on Systems, Man, and Cybernetics. 1973;3:610–621. [Google Scholar]
- Haux R. Health information systems: Past, present, and future. International Journal of Medical Informatics. 2006;75:268–281. doi: 10.1016/j.ijmedinf.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Hersh WR, Müller H, Jensen JR, Yang J, Gorman PN, Ruch P. Advancing biomedical image retrieval: Development and analysis of a test collection. Journal of the American Medical Informatics Association. 2006;13:488–496. doi: 10.1197/jamia.M2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hripcsak G, Rothschild AS. Agreement, the f-measure, and reliability in information retrieval. Journal of the American Medical Informatics Association. 2005;12:296–298. doi: 10.1197/jamia.M1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn CE, Thao C. GoldMiner: A radiology image search engine. American Journal of Roentgenology. 2007;188:1475–1478. doi: 10.2214/AJR.06.1740. [DOI] [PubMed] [Google Scholar]
- Keysers D, Dahmen J, Ney H, Wein BB, Lehmann TM. A statistical framework for model-based image retrieval in medical applications. Journal of Electronic Imaging. 2003;12:59–68. [Google Scholar]
- Lehmann TM. From plastic to gold: A unified classification scheme for reference standards in medical image processing; Proceedings of the International Society for Optical Engineering (SPIE); 2002. pp. 1819–1827. [Google Scholar]
- Lehmann TM, Güld MO, Thies C, Fischer B, Spitzer K, Keysers D, et al. Content-based image retrieval in medical applications. Methods of Information in Medicine. 2004;43:354–361. [PubMed] [Google Scholar]
- Lehmann TM, Schubert H, Keysers D, Kohnen M, Wein BB. The IRMA code for unique classification of medical images; Proceedings of the International Society for Optical Engineering (SPIE); 2003. pp. 109–117. [Google Scholar]
- Lowe HJ, Antipov I, Hersh W, Smith CA. Towards knowledge-based retrieval of medical images. The role of semantic indexing, image content representation, and knowledge-based retrieval; Proceedings of the American Medical Informatics Association (AMIA) Symposium; 1998. pp. 882–886. [PMC free article] [PubMed] [Google Scholar]
- Mougin F, Bodenreider O. Approaches to eliminating cycles in the UMLS Metathesaurus: Naïve vs. formal; Proceedings of the American Medical Informatics Association (AMIA) Fall Symposium; 2005. pp. 550–554. [PMC free article] [PubMed] [Google Scholar]
- Müller H, Deselaers T, Deserno TM, Clough P, Kim E, Hersch W. Overview of the ImageCLEFmed 2006 medical retrieval and annotation tasks. In: Peters C, Clough P, Gey F, Karlgren J, Magnini B, Oard DW, de Rijke M, Stempfhuber M, editors. Evaluation of Multilingual and Multi-modal Information Retrieval – 7th Workshop of the Cross-Language Evaluation Forum, CLEF 2006. Lecture Notes in Computer Science, vol. 4730. New York: Springer Verlag; 2007. pp. 595–608. [Google Scholar]
- Müller H, Geissbuhler A, Ruch P. Lecture Notes in Computer Science, Vol. 3491: Cross Language Evaluation Forum (CLEF 2004) Berlin: Springer Verlag; 2005. ImageCLEF 2004: Combining image and multilingual search for medical image retrieval; pp. 718–727. [Google Scholar]
- Müller H, Michoux N, Bandon D, Geissbuhler A. A review of content-based image retrieval systems in medical applications Clinical benefits and future directions. International Journal of Medical Informatics. 2004;73:1–23. doi: 10.1016/j.ijmedinf.2003.11.024. [DOI] [PubMed] [Google Scholar]
- Névéol A, Douyère M, Rogozan A, Darmoni SJ. Construction de ressources terminologiques en santé [Development of health terminology resources] In: Koeva S, Maurel D, Silberztein M, editors. Formaliser les langues avec l’ordinateur [Formalizing languages with the computer] Besançon, France: Presses Universitaires de Framche-Comté; 2007. pp. 171–187. [Google Scholar]
- Niblack W, Barber R, Equitz W, Flickner MD, Glasman EH, Petkovic D, et al. The QBIC project: Querying images by content using color, texture, and shape; Proceedings of the International Society for Optical Engineering (SPIE); 1993. pp. 173–187. [Google Scholar]
- Pastra K, Wilks Y. Image-language multimodal corpora: Needs, lacunae, and an AI synergy for annotation; Proceedings of the Language Resources and Evaluation Conference; 2004. pp. 767–770. [Google Scholar]
- Rafkind B, Lee M, Chang SF, Yu H. Exploring text and image features to classify images in bioscience literature; Proceedings of the BioNLP Workshop on Linking Natural Language Processing and Biology at HLT-NAACL; 2006. pp. 73–80. [Google Scholar]
- Rius S. Vers une représentation riche des images [Towards a rich representation of images] Rennes: Institut de recherche en informatique et systèmes aléatoires (IRISA); 2006. Unpublished master's thesis. [Google Scholar]
- Robertson SE, Walker S, Beaulieu M. OKAPI at TREC-7: Automatic Ad Hoc, Filtering, VLC, and Interactive; Proceedings of the Seventh Text REtrieval Conference (TREC 7) National Institute of Standards and Technology Special Publication 500-242; Gaithersburg, MD: NIST; 1998. pp. 253–264. [Google Scholar]
- Rowe NC, Guglielmo EJ. Exploiting captions in retrieval of multimedia data. Information Processing & Management. 1993;29:453–461. [Google Scholar]
- Ruiz ME, Srikanth M. UB at CLEF 2004: Part 2 cross language medical image retrieval; Proceedings of the Cross-Language Evaluation Forum 2004 Workshop; 2004. pp. 773–780. [Google Scholar]
- Shatkay H, Chen N, Blostein D. Integrating image data into biomedical text categorization. Bioinformatics. 2006;22(14):e446–e453. doi: 10.1093/bioinformatics/btl235. [DOI] [PubMed] [Google Scholar]
- Smeulders AW, Worring M, Santini S, Gupta A, Jain R. Content-based image retrieval at the end of the early years. IEEE Transactions on Pattern Analysis & Machine Intelligence. 2000;22:1349–1380. [Google Scholar]
- Srihari RK, Zhang Z, Rao A. Intelligent indexing and semantic retrieval of multimodal documents. Information Retrieval. 2000;2:245–275. [Google Scholar]
- Tamura H, Mori S, Yamawaki T. Textural features corresponding to visual perception. IEEE Transactions on Systems, Man, and Cybernetics. 1978;8:460–473. [Google Scholar]
- Zhao R, Grosky WI. Narrowing the semantic gap: Improved text-based Web document retrieval using visual features. IEEE Transactions on Multimedia. 2002;4:189–200. [Google Scholar]