The distinctive flavors and anticarcinogenic properties of cruciferous vegetables are largely due to the presence of glucosinolates. These sulfur-rich metabolites are derived from one of several amino acids via three stages: (1) elongation of the amino acid side chain; (2) formation of the core glucosinolate skeleton; and (3) secondary modification of the side chain (reviewed in Grubb and Abel, 2006). Met-derived aliphatic glucosinolates constitute the majority of glucosinolates in many Brassicaceae species, including Arabidopsis (Kliebenstein et al., 2001). Biosynthesis of these glucosinolates is highly compartmentalized, with preliminary reactions and final stages occurring in the cytosol and side chain elongation in the chloroplast.
Prompted by the discovery that the R2R3-MYB transcription factor High Aliphatic Glucosinolate 1 (HAG1/MYB28) is a key regulator of Met-derived glucosinolate biosynthesis in Arabidopsis (Gigolashvili et al., 2007; Hirai et al., 2007), Gigolashvili et al. (pages 1813–1829) set out to identify novel targets of HAG1. Using transcriptome coexpression analysis, they found that BAT5, which encodes a putative bile acid transporter with a hitherto uncharacterized role in plants, is coexpressed with HAG1. Cotransformation assays using a HAG1 effector and a ProBAT5-GUS reporter showed that HAG1 transactivates the BAT5 promoter, confirming that BAT5 is indeed a target of HAG1. Furthermore, the authors established that full-length BAT5-GFP localizes to chloroplasts in transiently transformed Arabidopsis leaves (see figure).
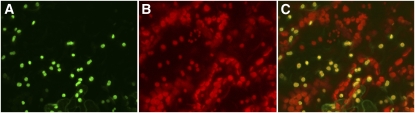
Figure 1.
BAT5-GFP localizes to chloroplasts. BAT5-GFP fluorescence (A) and chloroplast autofluorescence (B) in transgenic Arabidopsis leaves and the overlay (C) of (A) and (B) showing colocalization (yellow) of BAT5-GFP and chloroplasts.
The authors tested whether BAT5 has a role in aliphatic glucosinolate biosynthesis by examining the effect of downregulation of BAT5 on glucosinolate content. Although a homozygous bat5 mutant with vastly reduced steady state levels of BAT5 mRNA appeared to be normal under standard conditions, metabolic analysis revealed that it had significantly reduced levels of aliphatic glucosinolates. Complementation of the bat5 mutant with an estradiol-inducible BAT5 vector rescued the glucosinolate composition to almost wild-type levels, and the degree of induced gene expression correlated with the level of aliphatic glucosinolates produced. The effect on aliphatic glucosinolate content was even more pronounced in BAT5 microRNA overexpressing lines, which have undetectable levels of BAT5 mRNA. The observation that wounding and methyl jasmonate, which are known to induce glucosinolate biosynthesis, upregulate the expression of BAT5, provides further evidence that BAT5 is involved in glucosinolate biosynthesis.
Finally, the authors sought to pinpoint the function of BAT5 in aliphatic glucosinolate biosynthesis by feeding bat5 and wild-type Arabidopsis plants with various intermediates of the Met-derived glucosinolate pathway and then analyzing the glucosinolate content. The results suggest that BAT5 transports Met derivatives into the chloroplast for side chain elongation and exports elongated products back into the cytosol for conversion into glucosinolates. Thus, BAT5 plays an integral role in Met-derived glucosinolate biosynthesis in Arabidopsis.
References
- Gigolashvili, T., Yatusevich, R., Berger, B., Müller, C., and Flügge, U.I. (2007). The R2R3-MYB transcription factor HAG1/MYB28 is a regulator of methionine-derived glucosinolate biosynthesis in Arabidopsis thaliana. Plant J. 51 247–261. [DOI] [PubMed] [Google Scholar]
- Gigolashvili, T., Yatusevich, R., Humphry, M., Gershenzon, J., and Flügge, U.I. (2009). The plastidic bile acid transporter 5 is required for the biosynthesis of methionine-derived glucosinolates in Arabidopsis thaliana. Plant Cell 21 1813–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubb, C.D., and Abel, S. (2006). Glucosinolate metabolism and its control. Trends Plant Sci. 11 89–100. [DOI] [PubMed] [Google Scholar]
- Hirai, M.Y., et al. (2007). Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc. Natl. Acad. Sci. USA 104 6478–6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein, D.J., Kroymann, J., Brown, P., Figuth, A., Pedersen, D., Gershenzon, J., and Mitchell-Olds, T. (2001). Genetic control of natural variation in Arabidopsis glucosinolate accumulation. Plant Physiol. 126 811–825. [DOI] [PMC free article] [PubMed] [Google Scholar]