Abstract
Transcription of Arabidopsis thaliana seed maturation (MAT) genes is controlled by members of several transcription factor families, such as basic leucine zippers (bZIPs), B3s, MYBs, and DOFs. In this work, we identify Arabidopsis bZIP53 as a novel transcriptional regulator of MAT genes. bZIP53 expression in developing seeds precedes and overlaps that of its target genes. Gain- and loss-of-function approaches indicate a correlation between the amount of bZIP53 protein and MAT gene expression. Specific in vivo and in vitro binding of bZIP53 protein to a G-box element in the albumin 2S2 promoter is demonstrated. Importantly, heterodimerization with bZIP10 or bZIP25, previously described bZIP regulators of MAT gene expression, significantly enhances DNA binding activity and produces a synergistic increase in target gene activation. Full-level target gene activation is strongly correlated with the ratio of the correspondent bZIP heterodimerization partners. Whereas bZIP53 does not interact with ABI3, a crucial transcriptional regulator in Arabidopsis seeds, ternary complex formation between the bZIP heterodimers and ABI3 increases the expression of MAT genes in planta. We therefore propose that heterodimers containing bZIP53 participate in enhanceosome formation to produce a dramatic increase in MAT gene transcription.
INTRODUCTION
As an outstanding adaptation of terrestrial plants, seed formation favors dispersal of species and allows the interruption of the life cycle and its resumption once optimal growth conditions are newly established (for review, see Vicente-Carbajosa and Carbonero, 2005; Weber et al., 2005; Santos-Mendoza et al., 2008). Seeds are formed after a double fertilization event, triggering the development of a complex organ, which comprises the embryo, the endosperm, and the seed coat derived from the integuments and other surrounding layers of maternal origin. Seed development can be divided into three phases: first, embryogenesis is characterized by cell division and differentiation until embryo morphology is established. Second, the maturation phase is dominated by storage compound accumulation, growth arrest, and acquisition of desiccation tolerance. Third, the embryo can enter into a dormancy state that is broken upon germination. With respect to seed morphology, physiology, and gene regulation, considerable variations occur among species. Arabidopsis thaliana has been developed as a well-established model system for dicot seed development, and several similarities and differences with monocot model systems have been described (Vicente-Carbajosa and Carbonero, 2005; Santos-Mendoza et al., 2008).
Important programs of gene expression related to the metabolic changes that occur during seed maturation are highly coordinated and tightly regulated (Gutierrez et al., 2007). An understanding of gene expression control in the seed was tackled from early studies in plant molecular biology, with maize (Zea mays) Opaque2 (O2) representing a hallmark as one of the first plant transcription factor (TF) genes cloned and characterized (Hartings et al., 1989; Schmidt et al., 1990). Similarly, orthologous genes from wheat (Triticum aestivum) (SPA) and barley (Hordeum vulgare) (BLZ2) play the same roles as O2 in their corresponding species (Albani et al., 1997; Oñate et al., 1999). In dicot species, key TFs have been characterized that control gene expression programs during seed maturation.
The class of maturation genes (MAT) expressed during seed maturation typically includes seed storage protein (SSP) genes, such as albumin and cruciferin genes, which are induced in early or mid-maturation phase. The late embryogenesis abundant (LEA) genes are induced at later stages of maturation and include genes proposed to be involved in acquisition of desiccation tolerance (for review, see Tunnacliffe and Wise, 2007). MAT promoter analyses have revealed several conserved cis-regulatory elements with functional relevance in the control of gene expression during seed maturation. Among them, G-box-related ACGT elements, RY (CATGCA), AACA, and CTTT motifs are the best described examples (for review, see Vicente-Carbajosa and Carbonero, 2005). The corresponding associated TFs belong to the basic leucine zipper (bZIP), B3, MYB, and DOF TF families, respectively. Cooperation of these regulatory units in the control of gene expression appears to be an evolutionarily conserved pattern that can be traced back to the origins of the Spermaphyta (Vicente-Carbajosa and Carbonero, 2005; Schallau et al., 2008).
TFs of the bZIP class, related to cereal O2-type TFs, have been identified in Arabidopsis (Lara et al., 2003), namely, bZIP10 and bZIP25, which have been classified into group C of the Arabidopsis bZIP TF family (Jakoby et al., 2002). Expression during seed development, specific binding to G-box-like ACGT elements of the albumin and cruciferin promoters, and in vivo regulation of these target genes have been demonstrated (Lara et al., 2003). A peculiarity exists in that none of the four genes in the bZIP group C in Arabidopsis shows seed-specific expression, in contrast with characterized members of the cereal O2-type TFs. This suggests that in Arabidopsis, combinatorial interactions and expression levels of different TFs may be of major relevance in the induction of seed-specific gene expression patterns.
ABSCISIC ACID INSENSITIVE3 (ABI3), FUSCA3 (FUS3), and LEAFY COTYLEDON2 (LEC2) have been implicated in seed development and belong to the B3 family of TFs (Parcy et al., 1994; Kroj et al., 2003; Braybrook et al., 2006; Santos-Mendoza et al., 2008). Mutations in any of these genes result in pleiotropic phenotypes affecting SSP accumulation and acquisition of desiccation tolerance. Recent genetic analysis demonstrated that these B3-type TFs are controlled by a complex self-regulating network including LEC1, LEC2, FUS3, and ABI3 (To et al., 2006). The corresponding genes clearly differ in expression and function despite being partially redundant. Although binding of ABI3, FUS3, and LEC2 to the RY element, mediated by the B3 DNA binding domain, has been shown in vitro and in yeast systems (Kroj et al., 2003; Mönke et al., 2004; Braybrook et al., 2006), RY motifs are not sufficient to confer seed-specific expression patterns to a target promoter (Ezcurra et al., 2000; Nakashima et al., 2006). Cooperation with neighboring cis-elements and the cognate TFs has been demonstrated to be important to establish seed-specific transcriptional patterns. bZIP10 and bZIP25 physically interact with ABI3, which in turn enhances in vitro DNA binding of the bZIP proteins to SSP promoters as well as their in vivo activation capacity (Lara et al., 2003). Therefore, cooperation of transcriptional regulators by protein–protein interactions provides an efficient mechanism to control gene expression in seeds and explains some of the molecular mechanisms underlying SSP gene expression. However, our knowledge on the protein partners and their interplay remains limited.
In general, bZIP TFs bind DNA as homo- or heterodimers (Ellenberger et al., 1992). Recently, we demonstrated that group C bZIPs, such as bZIP10 and bZIP25, preferentially interact with group S1 bZIPs, resulting in a set of specific heterodimers designated the C/S1 network of bZIP TFs (Ehlert et al., 2006). In particular, members of the C/S1 network have been implicated in energy homeostasis (Baena-Gonzalez et al., 2007; Usadel et al., 2008), amino acid metabolism (Weltmeier et al., 2006; Hanson et al., 2008), stress response (Kaminaka et al., 2006), and sink-specific gene expression (Rook et al., 1998; Weltmeier et al., 2009). Importantly, these heterodimers were shown to convey synergistic activation properties to target genes, suggesting that heterodimerization serves as an efficient mechanism of signal integration (Weltmeier et al., 2006). With respect to the regulation of the seed gene Em, the importance of bZIP heterodimers and the ABI3 othologous regulator VP1 has already been proposed in rice (Oryza sativa) (Nantel and Quatrano, 1996).
In this work, we aimed to identify S1 bZIP members that could be involved in seed gene regulation as important potential partners of previously described group C bZIPs. We define the S1 TF bZIP53 as a key regulator of MAT gene transcription. bZIP53 enhances MAT gene expression by specific heterodimerization with bZIP10 or bZIP25. Furthermore, these bZIP heterodimers interact with ABI3, which further increases MAT gene activation. Therefore, we propose that bZIP53 plays a pivotal and crucial role in quantitative control of MAT gene transcription levels by cooperation with several TFs forming enhanceosome-like protein complexes.
RESULTS
bZIP53 Is Expressed in Seed Tissues during Maturation
We have previously shown that Arabidopsis S1 bZIP proteins support strong heterodimerization with members of the group C bZIPs using two-hybrid systems in yeast and protoplasts and bimolecular fluorescence complementation (BiFC) techniques (Ehlert et al., 2006; Weltmeier et al., 2006). Since two group C members, bZIP10 and bZIP25, are known activators of SSP genes (Lara et al., 2003), we surveyed the expression patterns of S1 members during seed development to identify possible partners for C-S1 interactions in this process. According to the mRNA profiles of S1 bZIPs during silique and seed development derived from public AtGenExpress microarray data (http://www.genomforschung.uni-bielefeld.de/GF-research/AtGenExpress-SeedsSiliques.html), bZIP53 is the S1 member with the highest expression in mid and late maturation phases. Induction of bZIP53 precedes and overlaps the activation of typical SSP and LEA genes, supporting the hypothesis that bZIP53 might be involved in MAT gene regulation.
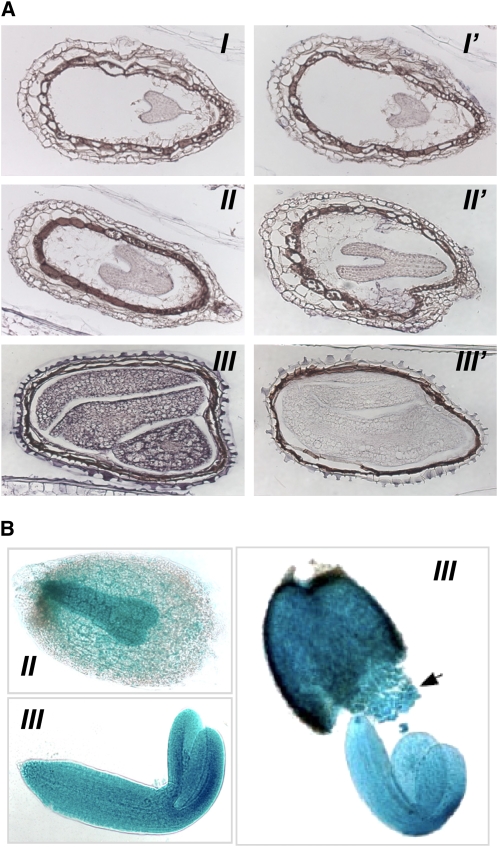
Confirmation of the bZIP53 temporal pattern of expression and its localization within the seed were obtained by in situ hybridization studies (Figure 1A). bZIP53 mRNAs produced a strong signal in the embryo cotyledons during late maturation. In addition, we performed histochemical localization of β-glucuronidase (GUS) activity in seeds of Arabidopsis transgenic plants expressing the GUS reporter gene under control of the bZIP53 gene promoter (ProbZIP53:GUS). The results presented here were confirmed in three independent transgenic lines, all of which showed the same profiles of GUS expression. As seen in Figure 1B, GUS staining in the developing embryo was observed from the torpedo to the green cotyledon stages during seed development (Bowman, 1994). Altogether, these results demonstrate that bZIP53 expression increases during seed development and localizes to the embryo and endosperm during the maturation phase.
Figure 1.
Expression of bZIP53 in Seeds.
(A) In situ mRNA hybridization for bZIP53 at different stages of seed development. Longitudinal sections of siliques with seeds at heart (I and I′), early torpedo (II and II′), and green cotyledon (III and III′) stages of development are shown. Sections were hybridized with a bZIP53 antisense (I, II, and III) or a sense probe (I′, II′, and III′).
(B) Histochemical analysis of Columbia (Col-0) seeds harboring a bZIP53 promoter fused to GUS (ProbZIP53:GUS). GUS staining of late torpedo stage (II) and green cotyledon stage (III) embryo development are shown. The arrow indicates the developing endosperm; to facilitate viewing, the seed has been pressed to push out the embryo.
Ectopic Expression of bZIP53 Results in Abnormal Plant Growth and Expression of Seed-Specific Genes in Leaf Tissue
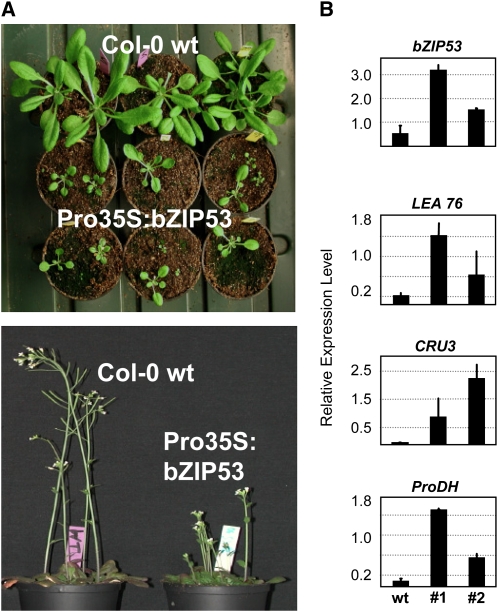
Because of its particularly high levels of expression during seed development, bZIP53 could be expected to participate in seed gene regulation. To address this question, we followed a gain-of-function approach and obtained transgenic Arabidopsis plants with ectopic expression of bZIP53 driven by the cauliflower mosaic virus 35S promoter (Pro35S:bZIP53) (Weltmeier et al., 2006). As seen in Figure 2A, strong overexpression of bZIP53 resulted in phenotypic alterations, including dwarfism and delayed bolting compared with the wild-type plants. To test whether these modifications were associated to altered patterns of seed gene expression, mRNA isolated from wild-type and Pro35S:bZIP53 plants was subjected to quantitative RT-PCR (qRT-PCR) to check for bZIP53, MAT, and LEA gene expression. Figure 2B shows data of wild-type and two independent Pro35S:bZIP53 transgenic lines with increased levels of bZIP53 mRNA as well as induced expression of seed MAT and LEA genes, indicating that bZIP53 is able to activate seed-specific expression in Arabidopsis leaves. Accumulation of SSP transcripts in leaves, as exemplified by the cruciferin (CRU3) and albumin (2S2) genes, was also obtained by overexpression of HA-tagged bZIP53 (see Supplemental Figures 1A and 1B online). The presence of the transgene-encoded protein was confirmed by immunoblot analysis. Further examples of seed genes misexpressed in leaves are legumin-like CRA1 (Wang et al., 2007) and the 11-β-hydroxysteroid dehydrogenase gene (HSD1) (Li et al., 2007; see Supplemental Figure 1C online). In addition, these plants also show enhanced expression of the proline dehydrogenase (ProDH) gene, as previously reported (Weltmeier et al., 2006), or the Asparagine Synthetase1 (ASN1) gene, both typical genes involved in amino acid metabolism (Lam et al., 2003) (Figure 2B; see Supplemental Figure 1C online). Altogether, these data support the assumption that overexpression of bZIP53 in leaves triggers the misexpression of a substantial fraction of seed-specific genes.
Figure 2.
Plants with Constitutive Expression of bZIP53 Have Growth Defects and Increased Levels of Seed Maturation Transcripts.
(A) Plants overexpressing bZIP53 (Pro35S:bZIP53) have a stunted and late flowering phenotype compared with Col wild-type (Col-0 wt) plants.
(B) mRNA samples from 2-week-old wild-type and two Pro35S:bZIP53 lines (#1 and #2) were analyzed by qRT-PCR to quantify the transcript levels of bZIP53, LEA76, CRU3, and ProDH. Expression levels are given relative to a UBIQUITIN gene for normalization. Given are mean values and standard deviation of two to three replicates.
The Albumin 2S2 Gene Is a Direct Target of bZIP53
To elucidate whether the SSP promoters were directly targeted by bZIP53, we performed chromatin immunoprecipitation (ChIP) from leaves of plants expressing ectopical bZIP53 tagged at the N terminus with a 3xHA epitope. These plants have been previously described and display the same phenotype as the untagged Pro35S:bZIP53 plants described in Figure 2A (Weltmeier et al., 2006).
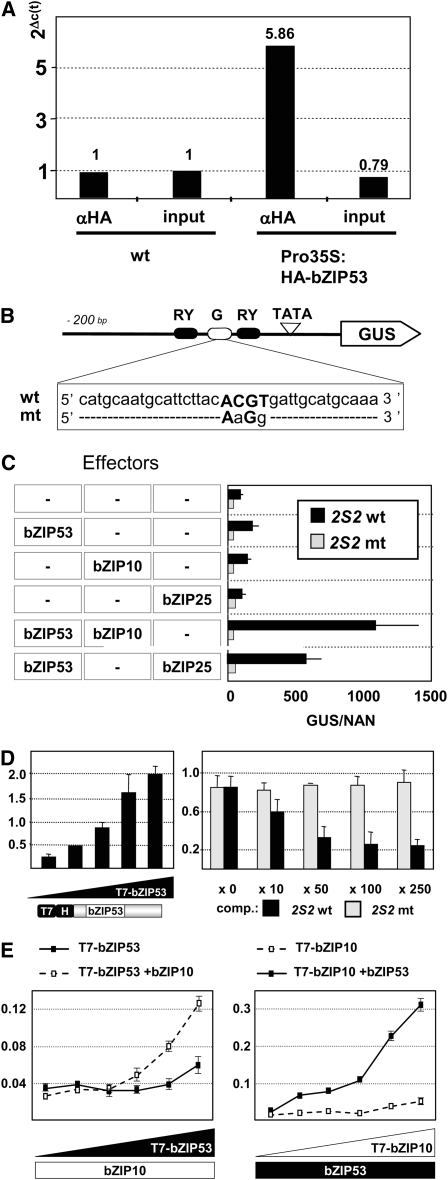
Figure 3A shows the results of qPCR analyses for the 2S2 promoter on chromatin samples isolated from wild-type and Pro35S:HA-bZIP53 plants. The relative enrichment of the 2S2 gene promoter in Pro35S:HA-bZIP53 immunoprecipitated samples supports direct binding of a protein complex that includes bZIP53.
Figure 3.
In Vivo and in Vitro Binding of the bZIP53 Protein to the Albumin 2S2 Promoter and Interaction with bZIP10 and bZIP25.
(A) Chromatin extracts from wild-type plants and plants overexpressing a HA-tagged bZIP53 protein (Pro35S:HA-bZIP53) were subjected to qRT-PCR analysis with 2S2 promoter-specific primers before (input) and after immunoprecipitation with an anti-HA antibody (α-HA). Ct values for Pro35S:HA-bZIP53 samples were subtracted from the Ct values of the equivalent wild type, and the differentials are shown on top of the right bars in the graph. A value of 1 was assigned to the Col-0 samples. For normalization, an actin (ACT7) gene was used (see Methods).
(B) Schematic view of the 2S2 promoter fused to GUS used as reporter in transient expression analysis. Depicted are RY (black) and G-box (white) elements. The sequence of the wild type and mutated G-box (mt) is shown.
(C) Arabidopsis leaves were transformed with the reporter constructs containing sequences described in (B) and effector constructs containing specific bZIP genes under control of the Pro35S in cobombardment experiments as described by Lara et al. (2003). Three microliters of control plasmid Pro35S:NAN was included in all the experiments to normalize GUS expression values for differences in bombardment efficiencies (Kirby and Kavanagh, 2002). The x axis values are expressed as GUS activity relative to NAN. Given are mean values and standard deviation of three independent experiments.
(D) In vitro binding of bZIP53 to the G-box sequence from the 2S2 promoter. A biotinylated oligonucleotide containing the G-box sequence was bound to streptavidin-coated wells and incubated with increasing amounts of a T7-tagged bZIP53 protein (1 to 1:90). Nonbound proteins were removed from the reaction wells, and the amount of T7-bZIP53 protein was quantified by immunodetection with an anti-T7 antibody (α-T7; left panel). The binding specificity of bZIP53 to the 2S2 G-box was analyzed in competition experiments where increasing amounts of unlabeled oligonucleotides (as indicated) containing a wild-type (2S2 wt; black bars) or a mutated 2S2 mt version (gray bars) were incubated with a fixed amount of the T7-bZIP53 protein and the biotinylated oligonucleotide containing the wild-type G-box sequence (right panel).
(E) Effect of heterodimerization on bZIP53 and bZIP10 binding affinity. A biotinylated oligonucleotide containing the G-box sequence was bound to streptavidin-coated wells and incubated with increasing amounts (1 to 1:30) of a T7-bZIP53 protein in the absence (filled squares) or presence of a fixed amount of bZIP10 protein (open squares). Nonbound proteins were removed from the reaction wells, and the amount of T7-bZIP53 protein was quantified by immunodetection with an α-T7 antibody (left panel). The reciprocal experiment was performed with a T7-bZIP10 protein and a nontagged bZIP53 protein (right panel). Given are mean values and standard deviation of three repetitions.
Heterodimerization of bZIP53 with Group C bZIP10 or bZIP25 Promotes Strong Activation of Seed-Specific Genes
Activation of seed-specific genes in leaves of Pro35S:bZIP53 plants as well as ChIP analyses suggest that bZIP53 is directly involved in this regulation. We studied the activation properties of bZIP53 in Arabidopsis leaves transformed by particle bombardment (Lara et al., 2003). Figures 3B and 3C show an example of 2S2 promoter constructs driving the expression of a GUS reporter that is cotransformed with effector plasmids for bZIP53, bZIP10, and bZIP25. Individually, none of these bZIPs is able to produce a significant increase in the basal activity of the reporter. Previous studies showed no significant heterodimerization for bZIP10 and bZIP25, whereas strong heterodimerization of group C bZIPs and bZIP53 was reported (Ehlert et al., 2006). Accordingly, when bZIP53 was cotransformed with bZIP10 or bZIP25, a dramatic increase in the reporter activity was observed. These results were confirmed in transiently transformed leaf mesophyll protoplasts using two different seed-specific reporter constructs, driven by the 2S2 or the CRU3 promoter (see Supplemental Figure 2 online). Similarly, immunoblot analysis confirmed expression of the bZIP genes (see Supplemental Figure 3A online) in transient assays in protoplasts. Unexpectedly, we observed that coexpression of bZIP10 and bZIP53 leads to enhanced protein levels, suggesting that heterodimer formation might stabilize the bZIP proteins from degradation. This data emphasize the importance of heterodimer formation between bZIP53 and group C bZIPs in the observed enhancement of gene activation.
Activation of the 2S2 Seed Storage Protein Gene by bZIP53 and Its Heterodimers Relies on the G-Box Promoter Element
The in vivo regulation of bZIP53 and its heterodimers was further investigated on the 2S2 promoter. A modified version of the native promoter was used, in which the G-box element was mutated to a sequence that prevents bZIP DNA binding (Figure 3B; Lara et al., 2003). Different from the behavior of the native promoter, the activation mediated by the bZIP TFs is completely abolished in this mutant (Figure 3C), indicating that the regulation of the 2S2 promoter by these bZIP proteins requires an intact G-box motif.
bZIP53 Binds in Vitro to the G-Box Present in the 2S2 Promoter, and Heterodimerization with bZIP10 or bZIP25 Increases Its Binding Activity
To establish if bZIP53 was able to bind to seed-specific gene promoters, we performed in vitro DNA binding experiments with an epitope-tagged bZIP53 (T7-bZIP53) protein. A native-sequence oligonucleotide (wild type) containing a functional bZIP (G-box) binding site (Ezcurra et al., 2000) derived from the 2S2 promoter and a mutated version were used (Figure 3B). Double-stranded oligonucleotides were covalently attached to 96-well ELISA plates before doing the binding assays as described in Methods. Figure 3D shows a positive correlation between the amount of T7-bZIP53 detected with the epitope-specific antibody and the amount of T7-bZIP53 protein extract added to the wells, indicating T7-bZIP53 binding to the wild-type oligonucleotide in a concentration-dependent manner. To analyze the specificity of this binding, competition experiments were performed with increasing amounts of the wild-type or mutated oligonucleotides added to the binding reactions. As shown in Figure 3D, the wild-type, but not the mutated oligonucleotide, was able to reduce the amount of T7-bZIP53 bound to the plate, thus confirming the specificity of the binding.
We have shown in previous work and in this study that bZIP53 heterodimerization with bZIP10 or bZIP25 increases its transactivation properties (Ehlert et al., 2006; Weltmeier et al., 2006). To determine if the heterodimerization was also affecting the binding capabilities of these proteins, we incubated increasing amounts of T7-bZIP53 in the wild-type oligo-coated wells in the presence or absence of fixed amounts of bZIP10. Figure 3E illustrates the effect of heterodimerization between bZIP53 and bZIP10 and shows that the binding activity for the concentration series of bZIP53 is higher in the presence (than in the absence) of bZIP10. Likewise, the same behavior is observed in the reciprocal experiment for a bZIP10 series, in support of an increased binding activity of the heterodimers.
ABI3 Does Not Interact with bZIP53 Alone, but Heterodimers with bZIP10 or bZIP25 Form a Ternary Complex in Yeast and in Planta
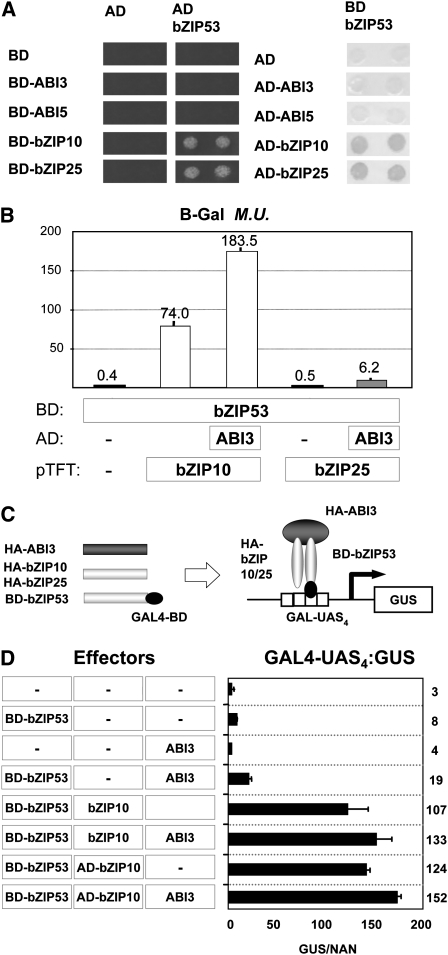
ABI3 is an important regulator of gene expression in seeds of Arabidopsis (Giraudat et al., 1992; Parcy et al., 1994) and is able to interact with bZIP10 or bZIP25 to increase their activation capacity on the 2S2 promoter (Lara et al., 2003). We used a yeast two-hybrid system to determine if bZIP53 was also able to interact with ABI3. We expressed the ABI3 and bZIP53 proteins as fusions to the GAL4 binding (BD) or activation (AD) domains, respectively, and introduced them into yeast strains containing LacZ or HIS reporter genes under the control of GAL4 binding sites. Different combinations of ABI3 and bZIP53 were tested for possible interactions in this system, alongside ABI5, another bZIP TF that has been implicated in seed gene expression serves here as a negative control (Finkelstein and Lynch, 2000). As seen in Figure 4A, no interaction could be detected between bZIP53 and ABI3 or between bZIP53 and ABI5 in any of the experimental systems used. As previously reported, positive interactions between bZIP53 and bZIP10 or bZIP25 confirmed their capacity to heterodimerize. To further analyze the possibility that these heterodimers could still interact with ABI3, three-hybrid system experiments were devised. Yeast cells expressing BD-bZIP53 were cotransformed with either AD-bZIP10 or AD-bZIP25 in the presence or absence of ABI3, and β-galactosidase reporter activities were quantified (see Supplemental Figure 4 online). As previously observed, the interaction between BD-bZIP53 and either AD-bZIP10 or AD-bZIP25 was confirmed by an increase of the reporter activity. However, when a construct designed to express ABI3 was cotransformed into the strains, a further increase of the β-galactosidase activity was observed, in support of a trimeric interaction. As expected for a negative control, the bZIP factor ABI5, which interacts with ABI3 (Nakamura et al., 2001) but does not heterodimerize with AtbZIP53, did not show activation indicative of ternary complex formation.
Figure 4.
Interaction of bZIP53, bZIP10, and bZIP25 Homo- or Heterodimers with ABI3.
(A) Protein interactions in yeast two- and three-hybrid systems. ABI3, ABI5, bZIP10, and bZIP25 proteins were fused to the GAL4 DNA BD or AD and introduced separately into a yeast strain containing the AD-bZIP53 (HF7c) or BD-bZIP53 (SFY526) protein, respectively. Activation of the reporter genes HIS3 (growth in a His-depleted medium; left panel) and LacZ (blue colored colonies; right panel) indicates positive protein–protein interactions.
(B) Yeast strains (SFY526) expressing different combinations of BD-bZIP53, AD-ABI3, and bZIP10 or bZIP25 were assayed for β-galactosidase activity. The latter were provided in the three-hybrid vector pTFT (Egea-Cortines et al., 1999). Average values (Miller units) and standard errors from six replicates and two independent experiments are shown.
(C) Schematic overview of a three-hybrid assay in Arabidopsis protoplasts. Constructs are shown on the left, and a model of reporter gene activation is shown on the right.
(D) Activation of the GAL4-UAS4:GUS reporter after cotransfection with the constructs indicated in (C). The x axis values are expressed as GUS activity relative to NAN (Ehlert et al., 2006). Average values and standard errors from four transfections are shown. Numbers along the y axis represent fold induction values relative to nontransfected control cells. The experiments were repeated three times with similar results.
Reciprocal three-hybrid analysis where the noninteracting proteins bZIP53 and ABI3 were fused to the Gal4BD and Gal4AD, respectively, showed also an increase of reporter activity when assayed in the presence of bZIP10 or bZIP25 (Figure 4), in support of the proposed trimeric interaction. Altogether, these results indicate that ABI3 can be brought into the heterodimer complex, probably by its interaction with bZIP10 and/or bZIP25. Importantly, bZIP53 does not interfere with the interaction between ABI3 and bZIP10 or bZIP25.
Ternary complex formation in plant cells was supported by a similar three-hybrid approach developed in Arabidopsis protoplasts. Figure 4C shows a schematic overview of the experimental setup and the structure of the expressed proteins used to transactivate a reporter construct containing the GUS gene under the control of a minimal promoter bearing a tetramer of the GAL4 upstream activating sequence (GAL4-UAS4:GUS) (Ehlert et al., 2006). As shown in Figure 4D, comparable results to the yeast system were achieved. Transactivation of the reporter with BD-bZIP53 alone results in low levels of GUS activity, whereas cotransformation with either HA-bZIP10 or AD-bZIP10 displayed enhanced activity indicative of heterodimer formation as previously observed in the yeast two-hybrid system. Expression of the corresponding proteins was assayed by immunoblot analysis with HA-tag and BD-specific antibodies (see Supplemental Figure 3B online). Equally, in both systems, the supplementary inclusion of HA-ABI3 resulted in an additional increase of reporter activity, in support of its capacity to interact with the heterodimers in plant cells.
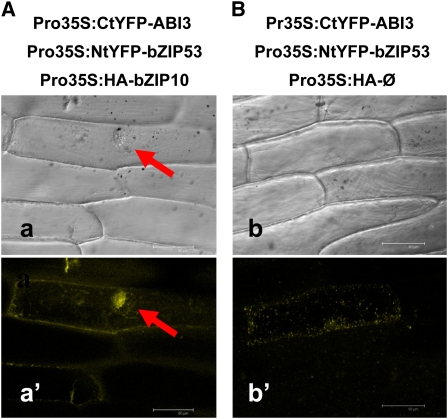
BiFC (Walter et al., 2004) was used as an additional experimental system to verify the detected ternary interactions in plant cells. bZIP53 and ABI3 proteins fused to different domains of yellow fluorescent protein (YFP) were coexpressed in onion cells in distinct combinations, rendering no reconstruction of YFP activity. However, when a third construct expressing bZIP10 protein was cotransfected, YFP fluorescence was observed in support of a tethering function of this protein to promote a ternary complex formation (Figure 5).
Figure 5.
bZIP53, bZIP10, and ABI3 Protein–Protein Interaction Studied by BiFC (Walter et al., 2004).
Onion epidermis cells have been transiently transformed by particle bombardment. Fusion proteins of the C-terminal part of YFP (CtYFP) and ABI3 and the N-terminal part of YFP (NtYFP) and bZIP53 have been coexpressed with HA-tagged bZIP10 (HA-bZIP10) (a and a′) or an empty vector control (b and b′). Bright-field images (top panels) and epifluorescence images taken by confocal microscopy (bottom panels) are shown.
Heterodimer Formation and Interaction with ABI3 Regulate the Activation Properties of bZIP53 on SSP Gene Expression in Plant Cells
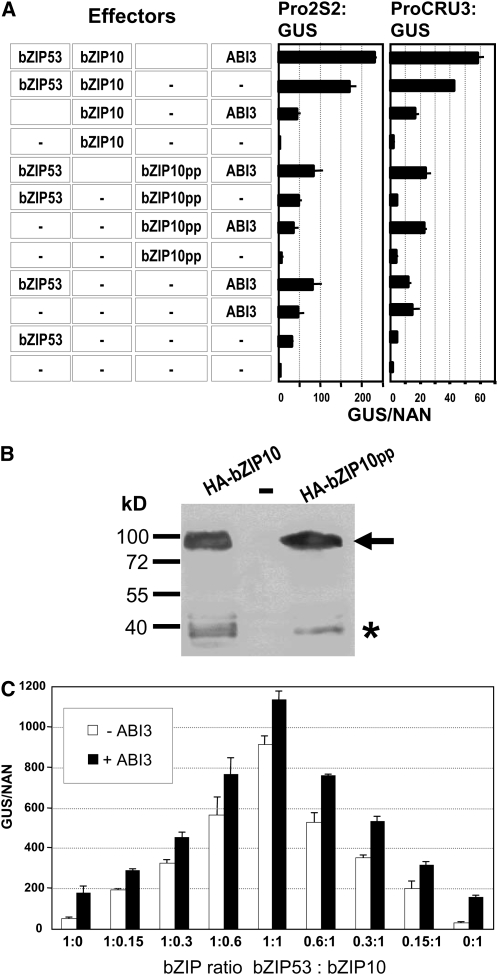
To study the functional implications of the interaction between ABI3 and bZIP heterodimers in plant cells, we performed transient expression analysis in Arabidopsis protoplasts using two different SSP promoters, corresponding to the 2S2 and CRU3 genes. Figure 6A shows that the use of bZIP effectors individually or in combination with ABI3 resulted in limited activation capacity on both promoters. Combination of bZIP53 and bZIP10 led to an important increase in the activity, in accordance with previous results. Moreover, the inclusion of ABI3 in this combination produced an additional increase in the activation of both promoters.
Figure 6.
Effect of ABI3 on the Transcriptional Activation Mediated by the bZIP53/bZIP10 Heterodimers.
(A) GUS reporter activity under the control of the 2S2 or CRU3 promoter was measured in transiently transformed Arabidopsis protoplasts after cotransfection of the effector constructs indicated. bZIP10, bZIP53, and ABI3 are expressed under the control of Pro35S. For immunodetection, 3xHA-epitope-tagged derivatives were used (see Supplemental Figure 3 online). The importance of bZIP heterodimerization was demonstrated by including bZIP10pp, which is impaired in zipper-mediated dimerization (Weltmeier et al., 2006). Given are mean values and standard deviation of four independent transfections.
(B) Immunoblot analysis of transiently transformed protoplasts confirms that expression of HA-bZIP10 is comparable to HA-bZIP10pp-untransformed control protoplasts. HA epitope-tagged proteins were detected using an α-HA antiserum. The arrow indicates HA-bZIP10 and bZIP10pp proteins, and the asterisk shows degradation products.
(C) Transient expression using a GUS reporter gene under the control of the 2S2 promoter. Combinations of bZIP53 and bZIP10 at different ratios (indicated are ratios of input DNA) in the presence (black bars) or absence (white bars) of ABI3 were used as effectors. The x axis values are expressed as GUS activity relative to NAN as described by Ehlert et al. (2006). Average values and standard errors from four transfections are shown.
This reporter gene activation cannot be observed when using an effector construct expressing bZIP10 with two Pro exchange mutations (bZIP10pp); these mutations disrupt the zipper dimerization surface and therefore abolish the interaction with bZIP53 (Weltmeier et al., 2006). Expression of bZIP10pp was found to be comparable to the wild-type protein as confirmed by immunoblot analysis (Figure 6B). Accordingly, disruption of the bZIP heterodimer using bZIP10pp abolishes this induced gene activation both in the presence or absence of ABI3. These results indicate a functional interaction between bZIP53/bZIP10 heterodimer and ABI3 and confirm a positive effect of ABI3 on heterodimer-mediated transcription.
To further investigate this effect, a comprehensive analysis of ABI3 action on the heterodimer was performed. In Figure 6C, different ratios of bZIP53 and bZIP10 were used to transactivate the 2S2 promoter in Arabidopsis protoplasts, either in the presence or in the absence of ABI3. In all cases, the activation effect of the heterodimer was enhanced in the presence of ABI3 by a similar increment. However, maximum promoter activity was greatly determined by the ratio of the heterodimerization partners. A 1:1 ratio, which enables optimal heterodimerization, results in maximum promoter activation.
bZIP53 Loss-of-Function Plants Display Reduced Expression of SSP Genes
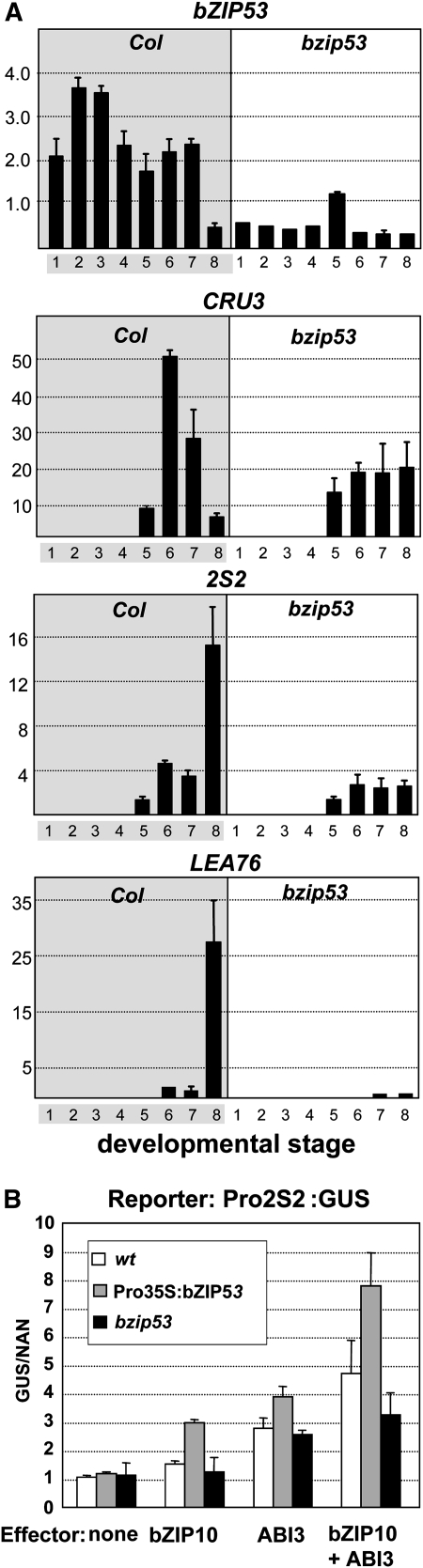
Results from the analysis of ectopic expression of bZIP53 suggested a regulatory role in SSP gene expression. We used a complementary loss-of-function strategy making use of a bzip53 T-DNA insertion mutant (Weltmeier et al., 2006) to corroborate the functional relevance of bZIP53 for SSP expression. We quantified SSP gene expression in bzip53 plants during seed development. Siliques of wild-type and bzip53 plants were collected at different stages of seed development, and total RNA was isolated and used in qRT-PCR experiments. Figure 7A shows that levels of bZIP53 mRNA were greatly reduced throughout all the stages of silique development in the bzip53 plants. However, residual levels of bZIP53 were detected, probably due to the leaky nature of the bzip53 mutation since the T-DNA is inserted in the promoter region (Weltmeier et al., 2006). Observed differences in the timing of RNA accumulation between CRU3, 2S2, and LEA76 genes were in accordance with those previously described for MAT and LEA genes (Parcy et al., 1994). Comparisons between wild-type and bzip53 plants show significantly reduced levels of SSP transcript in the latter. Although the timing of mRNA accumulation for these genes in the bzip53 samples was not significantly altered, the mRNA levels were reduced throughout the maturation stages and no peaks of RNA accumulation were detected, in contrast with the wild-type samples (Figure 7A).
Figure 7.
Effect of a bZIP53 T-DNA Insertion Mutant on Seed-Specific Gene Regulation.
(A) mRNA levels detected of the seed maturation genes indicated by qRT-PCR in Col wild-type and bzip53 siliques at different stages of development. Lane 1, 0 d after flowering (DAF); lane 2, 2 DAF; lane 3, 4 DAF; lane 4, 6 DAF; lane 5, 9 DAF; lane 6, 12 DAF; lane 7, 15 DAF; lane 8, ≥18 DAF. Data are normalized using UBIQUITIN expression values. Average values and standard errors from at least two technical replicates are shown.
(B) Transient expression by microparticle bombardment of Arabidopsis leaves from the wild type (white bars) and plants with increased (Pro35S:bZIP53; gray bars) or decreased (bzip53; black bars) expression of bZIP53. Effector constructs containing bZIP10 or ABI3 under the control of a 35S promoter (Pro35S) and a GUS reporter gene under the control of the 2S2 promoter were used. The x axis values are expressed as GUS activity relative to NAN (Kirby and Kavanagh, 2002). Average values and standard errors from four replicates and two different experiments are shown.
As seen above, impaired bZIP53 activity leads to decreased SSP gene expression in the seed, probably by limiting the activation capacity of the regulatory complex involving bZIP10/bZIP25 and ABI3. To test this hypothesis, we performed transactivation experiments using a 2S2 promoter driving a GUS reporter in different plant backgrounds. Figure 7B shows results of particle bombardment experiments on Arabidopsis leaves from wild-type, Pro35S:bZIP53, and bzip53 plants. As previously reported (Lara et al., 2003), the use of bZIP10 and ABI3 as effectors in this system resulted in the activation of the 2S2 promoter. This activation was significantly enhanced in Pro35S:bZIP53 plants and reduced in the bzip53 knockdown mutant line, consistent with the participation of bZIP53 in this regulatory complex. The same results were obtained for bZIP25 in combination with ABI3 (see Supplemental Figure 5 online).
DISCUSSION
Seed gene expression relies on specific TFs acting in a combinatorial fashion. Both in monocot and dicot species, group C bZIP factors related to maize O2 have been shown to be important players in this regulation. However, expression of the corresponding Arabidopsis bZIP genes is not seed specific. The recently described functional interaction between group C and S1 bZIPs (Ehlert et al., 2006) prompted us to investigate the possible participation of S1 members in seed gene regulation. In this study, we identified bZIP53 as the major S1 member expressed during seed maturation. Evidence of its participation in seed gene regulation was obtained by different approaches, including the analysis of plants with bZIP53 gain and loss of function. Moreover, functional studies show that bZIP53 plays a key role in the strong activation of seed-specific genes, directly involved in a seed regulatory protein complex encompassing the group C bZIP10 and bZIP25 and the B3 seed regulator ABI3.
bZIP53 Constitutes a Transcriptional Regulator of Seed Maturation Genes
Analyses of transcriptomic data of S1 bZIPs from repository databases show an increase in expression of bZIP53 (and to a lesser extent for bZIP1) during intermediate and late stages of seed development. This suggests a prominent role of bZIP53 in regulating seed gene expression in conjunction with the group C members. We further confirmed the precise pattern of expression of bZIP53 in seed tissues by mRNA in situ hybridization and promoter reporter fusions in transgenic Arabidopsis plants.
Gain-of-function approaches in transgenic plants and Arabidopsis leaf protoplasts ectopically expressing bZIP53 demonstrate that bZIP53 is able to activate several seed MAT and LEA genes, such as 2S2, CRU3, CRA1, HSD1, and LEA76. However, as observed before, the presence of MAT RNA does not necessarily result in the detection of the corresponding proteins. Additional cellular requirements and tissue-specific protein degradation determine high-level accumulation of MAT proteins (Gruis et al., 2004)
Complementing the gain-of-function results, reduced expression of bZIP53 in a knockdown line that harbors a T-DNA insertion in the promoter leads to strong reduction in the expression of the corresponding target genes. However, these plants still set viable seeds, which might be due to a residual amount of bZIP53 protein. The lack of complete knockout lines in mutant collections might be explained by an essential function of this gene. Alternatively, other genes among the 75 bZIP TFs identified in Arabidopsis (Jakoby et al., 2002) may partly substitute for bZIP53 function. Within the group S1, which harbors the closest homologs of bZIP53, only bZIP1 is intermediately induced during seed maturation with timing comparable to bZIP53. However, ectopic overexpression of At bZIP1 does not lead to activation of MAT genes, indicating a lack of functional redundancy of these genes in MAT gene regulation (Weltmeier et al., 2009).
High-level ectopic expression of bZIP53 results also in a dwarf growth phenotype in vegetative phases. Based on these observations, it can be assumed that regulation of SSP genes reflects only a subset of bZIP53 targets. In previous work, we and others demonstrated that bZIP53 is also involved in regulating hypo-osmolarity responses (Satoh et al., 2004; Weltmeier et al., 2006). In particular, the promoter of the ProDH gene, which is involved in regulating amino acid metabolism, has been defined as an in vivo target of bZIP53. Furthermore, a second amino acid metabolism gene, ASN1, was shown to be regulated by bZIP53 (this study) and other members of the C/S1 network (Baena-Gonzalez et al., 2007; Hanson et al., 2008). Amino acid synthesis, transport, and SSP synthesis are linked processes that have to be coordinated in vegetative tissues as well as in the seed. In accordance, ectopic overexpression of ASN1 enhances the SSP content and nitrogen status of the seed (Lam et al., 2003). Further studies are necessary to test whether bZIP53 is coordinating other pathways in addition to SSP synthesis, such as amino acid metabolism during seed and vegetative development.
bZIP53 expression is not restricted to the seed and can be observed in vegetative tissues, particularly under certain stress conditions. Interestingly, salt treatment dramatically induces bZIP53 transcription in roots (Weltmeier et al., 2009). In the context of the seed, it is worth mentioning that besides storage compound accumulation, dehydration occurs during late stages of the maturation phase. Accordingly, stress-related programs of gene expression are also important in the establishment of desiccation tolerance. We have determined that bZIP53 can also control the expression of the seed-specific gene LEA76. Although our knowledge of LEA76 function is limited, LEA proteins are known to accumulate in seed and vegetative plant tissues following environmental stress (Xu et al., 1996) and also in desiccation-tolerant bacteria and invertebrates (Bies-Etheve et al., 2008; Hundertmark and Hincha, 2008). The hypothesis whether bZIP53 is involved in establishing stress tolerance in vegetative tissues requires further testing.
Specific Heterodimerization of bZIP53 and Group C bZIPs Executes an Efficient Mechanism to Enhance Transcription of Seed Maturation Genes
MAT gene regulation is one of the early investigated areas in plant transcriptional control. The importance of G-box cis-elements in MAT gene promoters has been determined both in monocot and dicot species (Vicente-Carbajosa and Carbonero, 2005; Santos-Mendoza et al., 2008), and its regulation by seed-specific O2-type bZIPs has been firmly established in cereals. By contrast, genome scale transcriptomic data in Arabidopsis do not reveal a seed-specific candidate among all bZIPs, indicating that in this species (and probably in nonendospermic seeds from dicot plants), combinatorial effects are likely of major importance in the regulation of seed gene expression. In this framework, the group C bZIPs related to O2, bZIP10, and bZIP25 are able to interact with ABI3 and have been proposed as Arabidopsis counterparts of the monocot bZIP seed regulator (Schmidt et al., 1990; Lara et al., 2003). The corresponding genes, which are not seed specific, are constitutively expressed at low levels in other tissues, and bZIP10 has been implicated in plant stress responses to pathogen attack in vegetative tissues (Kaminaka et al., 2006). Previous work has defined a specific heterodimerization network of group C members with group S1 bZIPs, such as bZIP53 (Ehlert et al., 2006). The expression of this gene increases during seed maturation, and it strongly heterodimerizes with bZIP10 and bZIP25 as demonstrated in yeast and plant protoplasts (Ehlert et al., 2006), as well as by BiFC. Additionally, in vitro binding studies show that they can bind DNA as heterodimers (Weltmeier et al., 2006).
Efficient methods for in vivo DNA binding studies in Arabidopsis seeds have not been established. However, here, we demonstrated binding of bZIP53 to the 2S2 promoter using leaf-derived chromatin from Pro35S:HA-bZIP53 plants, which in turn show vegetative transcription of the 2S2 gene. Although this approach does not unequivocally confirm participation of bZIP53 in regulation of MAT genes, it provides circumstantial evidence in line with other results presented in this study. Mutational approaches confirm the 2S2 G-box as the in vitro binding site and also as the relevant in vivo cis-regulatory element. At high protein concentration, bZIP53 displays interaction with G-box elements in vitro, but titration experiments demonstrate that binding of bZIP53 in the presence of bZIP10 or bZIP25, presumably by heterodimer formation, is significantly enhanced. The in vivo relevance of heterodimer formation is further supported by expression of bZIP mutants, which are impaired in zipper-mediated dimerization, and consequently target gene activation. Detailed titration experiments in protoplasts also demonstrate that the level of target gene expression is determined by the ratio of the bZIP heterodimerization partners. The 1:1 ratio of bZIP53 and bZIP10 leads to the highest target gene activity. Altogether, these data provide conclusive evidence that bZIP heterodimers show enhanced binding to and gene activation through G-box-like cis-elements. Importantly, heterodimerization provides an efficient mechanism by which bZIP53 drives MAT gene expression.
Possible mechanisms of heterodimer-enhanced transactivation have been described elsewhere (Weltmeier et al., 2006), including (1) increased affinity for binding target sites, (2) stabilization of dimer structures (as effective DNA binding forms) via equilibrium displacement in dimer-monomer concentrations, and (3) heterodimer interactions with other protein factors that increase activation. Here, we showed that the second and third possibilities must be operating in vivo in the regulation of MAT genes. In support of this is the increased activation observed in protoplast systems for the heterodimers, which is independent of their DNA binding when used as GAL4-BD protein fusions (Figure 4E), and their interaction and enhanced gene activation when assayed with ABI3 (Figure 6). In addition, heterodimer formation seems to protect bZIP proteins from degradation (see Supplemental Figure 3 online) and therefore provides an additional mechanism that contributes to the observed synergistic effect on transcription. Although not investigated in this study, the first possibility might also explain how diversification in target binding sites can lead to variations in the expression of different SSP genes (Conceicao Ada and Krebbers, 1994), depending on the binding properties of the heterodimers prevailing temporally and spatially in the seed. In conclusion, the concurrence of several regulatory mechanisms must be operating on the underlying bZIP network.
bZIP53, Group C bZIPs, and ABI3 Physically and Functionally Interact in the Control of Seed Maturation Genes
Current models of regulation of MAT gene expression in the seed entail the participation of bZIP and B3 interacting proteins, particularly bZIP10 and 25 and ABI3. We have demonstrated that bZIP53 is a positive factor in this regulation and that its interaction with group C bZIPs results in enhanced activation of the heterodimers. Although yeast and protoplast two-hybrid data indicate that bZIP53 on its own is not able to interact with ABI3, heterodimerizations should not preclude the group C bZIP-ABI3 interactions. Three-hybrid and BiFC analyses confirmed that bZIP heterodimerization of bZIP10 or bZIP25 with bZIP53 does not interfere with ABI3 interaction and supports the existence of a ternary complex. ABI3 has been shown to bind to the RY element (Mönke et al., 2004), and a functional interplay between G-box and RY-element in MAT gene regulation has been previously described (Ezcurra et al., 2000; Lara et al., 2003). In this report, we clearly demonstrate functional interplay between bZIP heterodimers and ABI3, presumably by ternary complex formation.
By altering the bZIP expression levels in leaves, we can still see functionality of the bZIP heterodimers despite the absence of the ABI3 protein (Parcy et al., 1994), though their mediated activation occurs at a significantly lower level. Further experiments in bZIP53 overexpressing and knockdown plants show that although bZIP10 and/or ABI3 can enhance MAT gene activation to a certain extent, full-level expression clearly depends on the presence of bZIP53 (see Figure 7B). In this respect, nonseed model systems, such as protoplasts and bombarded leaves, might not fully reflect the native situation in seeds. Nevertheless, all results obtained in these systems are in agreement with the intrinsic reduction of MAT gene expression observed in seeds of mutant bzip53 plants. On the other hand, similar experiments performed in cereal systems that allowed the postulation of regulatory models have been confirmed in null mutants and transgenic plants (Norre et al., 2002). In summary, the output in MAT gene expression depends on all partners, ABI3 and bZIPs, and is quantitatively triggered by the amount of bZIP heterodimers.
Complex formation between the bZIP TF ABI5 and ABI3 have been recognized in LEA but not in MAT gene regulation (Bensmihen et al., 2002). Our findings suggest that the same principle based on ABI3 protein interactions operates on MAT expression, but inclusion of different bZIP partners defines distinct subsets of target genes. Therefore, this study provides further insight into the synergistic action of TFs and formation of enhanceosome-like protein-DNA complexes, which control seed gene transcription.
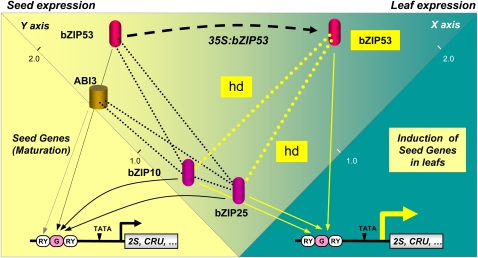
A Model of Seed Maturation Gene Expression Triggered by bZIP53 Heterodimerization and Protein Complex Formation
Based on previous findings and data from this study, the working model in Figure 8 is proposed. The function of the previously described group C bZIP factors bZIP10 and bZIP25 in seed maturation strongly depends on bZIP53 as a heterodimerization partner. Spatial and developmentally controlled expression of bZIP53 is precisely timed, leading to increased amounts in the seed from mid maturation. Subsequent accumulation of bZIP53 would trigger heterodimer formation with bZIP10 or bZIP25 as prerequisite for efficient promoter binding and target SSP gene activation. ABI3 protein, which also accumulates during seed maturation, interacts with the bZIP heterodimer via bZIP10 or bZIP25 and provides an additional increment in promoter activation. Hence, SSP gene regulation is driven by the interplay of (at least) two maturation-dependent transcriptional regulators, namely, bZIP53 and ABI3. The crucial role of ABI3 in this process was largely uncovered through the characterization of abi3 loss-of-function mutants (Parcy et al., 1994). This factor has a prominent role in seed maturation and germination gene expression programs. In this respect, its participation in a ternary complex with C/S bZIPs might partly explain many of numerous pleotropic effects observed in abi3 mutants. Interestingly, transcriptomic data suggest that bZIP53 and bZIP10 expression levels are reduced in mutant abi3-4 developing seeds (Carrera et al., 2008).
Figure 8.
Model for bZIP53 Regulation of Seed Maturation Gene Expression.
Yellow and blue backgrounds represent expression in the seed and leaf, respectively. Positions of the indicated TFs were represented in a coordinate system according to relative expression values in seeds (y axis) and leaves (x axis) (data derived from AtGenExpress). Structure of a typical SSP promoter-like albumin (2S2) or cruciferin (CRU3) is depicted, indicating regulatory elements in their promoters (RY, RY-box; G, G-box; TATA, TATA-box). Continuous and dotted lines indicate DNA–protein interactions and protein–protein interactions, respectively. A displacement arrow (open line) indicates overexpression of bZIP53 in Pro35S:bZIP53 plants. bZIP53 increased expression in leaves favors heterodimer formation (hd) with group C bZIP10 and bZIP25 and triggers induction of seed gene expression in this organ.
In vegetative tissues, where bZIP10 and bZIP25 are constitutively present at low levels, the ectopic high-level expression of ABI3 can lead to the activation of SSP genes (Parcy et al., 1994; Parcy and Giraudat, 1997). Likewise, ectopic high-level expression of bZIP53 can substitute for the presence of ABI3, triggering SSP gene expression even more effectively than this factor. In summary, we propose that bZIP53 acts as a key regulator, which drives a subset of MAT genes mainly by the formation of heterodimers with a high activation capacity. Although further research is needed to elucidate distinct interacting partners and target genes both in vegetative and seed tissues, bZIP53 might represent an important target for manipulating seed storage compound accumulation and establishment of desiccation tolerance.
METHODS
Plant Materials and Transformation
For plant and protoplast transformation, particle bombardment, and ChIP experiments, Arabidopsis thaliana ecotype Col-0 was grown on soil or Murashige and Skoog plates under controlled environmental conditions at 16-h-light/8-h-dark cycles. To improve germination uniformity, seeds were pretreated at 4°C for 2 to 4 d. Floral dip transformations were performed using the Agrobacterium tumefaciens strain GV3101 (Weigel and Glazebrook, 2002). Pro35S:bZIP53, Pro35S:HA-bZIP53, ProbZIP53:GUS, and bzip53 (NASC ID: N569883) plants were described by Weltmeier et al. (2006).
In Situ Hybridization and Histochemistry
In situ hybridization was performed according to Ferrandiz et al. (2000). Forward 5′-TTGTCCAATGCAACCCAATCA-3′ and reverse 5′-ACAAGACTAGAGGACTGAGGCT-3′ bZIP53-specific oligonucleotides were used to amplify a 200-bp fragment from the 3′ untranslated region. The fragment was inserted into pGEM-T easy (Promega), and the sense and antisense probes were generated as specified by Lara et al. (2003). The construction of ProbZIP53:GUS plants has been described by Weltmeier et al. (2006). GUS staining was performed according to Stangeland and Salehian (2002) with some modifications. The siliques were incubated in GUS assay buffer overnight at room temperature under vacuum and transferred to an ethanol/acetic acid (1:1) solution for 4 h (young seeds) or 12 h (mature seeds). Clearing was performed by incubating tissues in Hoyer's light medium (100 g chloral hydrate in 60 mL water) for 12 h (young seeds) or 24 h (mature seeds) at room temperature. Samples were analyzed with a Zeiss Axiophot microscope.
qRT-PCR
RNA was isolated from seedlings and siliques as described by Oñate-Sánchez and Vicente-Carbajosa (2008). cDNA synthesis was performed according to Oñate-Sánchez and Singh (2002), and between 8 and 16 ng were used as a template for qRT-PCR together with forward and reverse oligonucleotides (0.5 μM each) in 1× Power SYBR Green PCR Master mix (Applied Biosystems). Cycling conditions (ABI Prism 7300; Applied Biosystems) were as follows: 10 min at 95°C, 50 cycles of 15 s at 95°C, and 60 s at 60°C, linked to a default dissociation stage program to detect nonspecific amplification. Primers are provided in Supplemental Table 1 online. At least two technical and two biological replicates were analyzed in all the experiments
Transient Expression Assays and Constructs
Protoplast isolation, transformation, construction of effector plasmids, and immunoblot analysis were performed according to Ehlert et al. (2006) and Weltmeier et al. (2006). For reporter gene assays, 9 μg reporter and 14 μg of each effector were used if not stated otherwise. Three micrograms of Pro35S:NAN plasmid was added for normalization (Kirby and Kavanagh, 2002). For three-hybrid analysis, 9 μg of each reporter, BD-, AD-, and HA-plasmid were applied. The overall amount of DNA was set to 40 μg by adding pUC19 plasmid DNA. Particle bombardment was performed according to Lara et al. (2003). Data presented for the different assayed constructs are derived from three independent experiments, including six plates with 10 leaves per plate. Pro2S2:GUS reporter vector is a pUC19-derived plasmid containing the GUS reporter gene under the control of the 2S2 promoter and fused to the 3′-terminator of the nopaline synthase gene (3′-nos) (Lara et al., 2003). Effector plasmids for the expression of bZIP53, bZIP10, bZIP25, and ABI3 were generated by cloning the corresponding coding sequence under the control of the Pro35S and a 3′-nos terminator (Lara et al., 2003) in the pJIT60 plasmid (Guerineau, 1995). Three micrograms of Pro35S:NAN plasmid was added for normalization. GUS and NAN enzyme assays were performed according to Kirby and Kavanagh (2002). Immunoblot analysis was performed according to Weltmeier et al. (2006) using α-HA and α-BD antisera (SantaCruz).
ChIP
ChIP was performed as described by Weltmeier et al. (2006) using an HA-specific antibody (SantaCruz). 2S2 primers were used to amplify a 306-bp fragment of the 2S2 promoter (5′-GACCGGTGACCTGCGTGTA-3′ and 5′-GACTTGCATGGAGTTCACGTG-3′). The difference between the resulting C(t) values of wild-type and Pro35S:HA-bZIP53 overexpressor was calculated and normalized with the input controls of these samples that were analyzed with the same primers. For normalization, PCR was performed with unspecific actin (ACT7) promoter primers (5′-CGTTTCGCTTTCCTTAGTGTTAGCT-3′ and 5′-AGCGAACGGATCTAGAGACTCACCTTG-3′).
ELISA-Based Protein-DNA Binding Assays
The cDNAs encoding bZIP10, bZIP25, bZIP53, and ABI3 proteins were cloned into the expression vectors pET23a (Novagen) to generate T7-epitope tagged proteins (Lara et al., 2003). Expression in Escherichia coli and preparation of protein extracts were performed as previously described (Lara et al., 2003). Biotinylated complementary oligonucleotides derived from the At2S2 promoter containing a G-box sequence (5′-CATGCAATGCATTCTTACACGTGATTGCATGCAAA-3′ and 5′-TTTGCATGCAATCACGTGTAAGAATGCATTGCATG-3′) or a mutated version (5′-CATGCAATGCATTCTTACAaGgGATTGCATGCAAA-3′ and 5′-TTTGCATGCAATCCCtTgTAAGAATGCATTGCATG-3′) were annealed in TES buffer (10 mM Tris, pH 8, 1 mM EDTA, and 300 mM NaCl). The appropriate double-stranded DNA (2 pmoles; target) in 60 μL of TBS-T buffer (20 mM Tris, pH 7.4, 180 mM NaCl, and 0.1% Tween 20) was added to each well of a streptavidin-coated plate (Nunc Inmobilizer) and incubated 1 h at 37°C, and nonbound targets were removed by washing three times with TBS-T. Wells were blocked by incubating 30 min at room temperature and gentle shaking (150 rpm) with 60 μL of 1% blocking agent (Roche) in TBS-T and washed three times with TBS-T. Twenty microliters of the protein extracts were mixed with 40 μL of binding buffer (4 mM HEPES, pH 7.5, 100 mM KCl, 0.2% BSA, 8% glycerol, and 5 mM DTT) in microfuge tubes and incubated 15 min on ice. The mixes were added to the wells of the plate, the binding reactions incubated for 1 h at room temperature with gentle shaking, and well washed three times with TBS-T. For epitope detection, we added 60 μL of a 1:5000 dilution of the antibody (T7 tag horseradish peroxidase conjugate; Novagen) in TBS-T to each well. Following incubation for 1 h at room temperature at 150 rpm, the wells were four times washed with TBS-T. The antibody peroxidase-conjugated reaction was performed according to manufacturer's instructions, and the activity was measured at 492 nm using an ELISA plate reader (Tecan) with filter setting at 650 for reference.
Yeast Two- and Three-Hybrid Analyses
Yeast two-hybrid analyses were done as described previously (Lara et al., 2003). BD and AD constructs were generated using appropriate restriction enzymes and cloning the indicated open reading frames into the pGBT9 and pGAD424 plasmids (Clontech), respectively, as follows: EcoRI/SalI sites for the BD-bZIP53, AD-bZIP53, BD-ABI5, and AD-ABI5 constructs and BamHI/SalI sites for the BD-ABI3 and AD-ABI3 constructs. BD-bZIP10, AD-bZIP10, BD-bZIP25, and AD-bZIP25 were described by Lara et al. (2003). To analyze ternary complex formation in yeast, we used BD and AD constructs from the two-hybrid assays and expressed a third protein whose coding sequences were inserted into the EcoRI/SalI sites of the pTFT1 plasmid (Egea-Cortines et al., 1999).
BiFC
Experimental procedures were according to Moreno-Risueno et al. (2007). In general, at least two independent assays were performed that included four independent plates bombarded for every individual construct or the different combinations. Comparisons were based on transformation efficiencies estimated to Pro35S:green fluorescent protein as a reference. Vectors used have been described by Weltmeier et al. (2006).
Accession Numbers
Arabidopsis Genome Initiative identifiers for the genes mentioned in this article are as follows: bZIP53 (At3g62420), bZIP10 (At4g02640), bZIP25 (At3g54620), bZIP1 (At5g49450), 2S2 (At4g27150), CRU3 (At4g28520), LEA76 (At3g15670), ProDH (At5g38710), UBI (At5g25760), ACT7 (At5g09810), ABI3 (At3g24650), ABI5 (At2g36270), ASN1 (At3g47340), CRA1 (At5g44120), and At HSD1 (At5g50600).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Analysis of Putative bZIP53 Target Genes in Plants Constitutively Expressing HA-bZIP53 or bZIP53.
Suplemental Figure 2. Transient Expression of bZIP53 in Combination with bZIP10 or bZIP25 Synergistically Enhances Transactivation of CRU3 and 2S2 Promoters.
Supplemental Figure 3. Analysis of Effector Proteins Transiently Expressed in Arabidopsis Protoplasts.
Supplemental Figure 4. Ternary Protein Interaction Studied in a Yeast Three-Hybrid System.
Supplemental Figure 5. Transient Expression by Microparticle Bombardment of Arabidopsis Leaves from Columbia Wild Type and Plants with Increased (Pro35S:bZIP53) or Decreased (bzip53) Expression of bZIP53.
Supplemental Table 1. Primers Used in This Study.
Supplementary Material
Acknowledgments
We thank Mar Gonzalez and Anna Herman for their valuable technical assistance and Caroline Carsjens and Louise Thatcher for proofreading. Critical comments and suggestions from anonymous referees and the Scientific Editor greatly improved previous versions of this study. Financial support from the Ministerio de Educación y Ciencia, Spain (GEN2003-20859 and BIO2007-68073) is acknowledged. R.A. is the recipient of a predoctoral fellowship from the Spanish Agency for International Cooperation. L.O.-S. was supported by the Spanish Ministry for Education and Science with a Ramón and Cajal Contract and funded with a Marie Curie International Reintegration Grant (036524). W.D.-L. is supported by Deutsche Forschungsgemeinschaft and Deurscher Akademische Auslausik Dienst.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Wolfgang Dröge-Laser (wdroege@gwdg.de).
Online version contains Web-only data.
References
- Albani, D., Hammond Kosack, M.C., Smith, C., Conlan, S., Colot, V., Holdsworth, M., and Bevan, M.W. (1997). The wheat transcriptional activator SPA: A seed-specific bZIP protein that recognizes the GCN4-like motif in the bifactorial endosperm box of prolamin genes. Plant Cell 9 171–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-Gonzalez, E., Rolland, F., Thevelein, J.M., and Sheen, J. (2007). A central integrator of transcription networks in plant stress and energy signalling. Nature 448 938–942. [DOI] [PubMed] [Google Scholar]
- Bensmihen, S., Rippa, S., Lambert, G., Jublot, D., Pautot, V., Granier, F., Giraudat, J., and Parcy, F. (2002). The homologous ABI5 and EEL transcription factors function antagonistically to fine-tune gene expression during late embryogenesis. Plant Cell 14 1391–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bies-Etheve, N., Gaubier-Comella, P., Debures, A., Lasserre, E., Jobet, E., Raynal, M., Cooke, R., and Delseny, M. (2008). Inventory, evolution and expression profiling diversity of the LEA (late embryogenesis abundant) protein gene family in Arabidopsis thaliana. Plant Mol. Biol. 67 107–124. [DOI] [PubMed] [Google Scholar]
- Bowman, J. (1994). Arabidopsis: An Atlas of Morphology and Development. (New York: Springer-Verlag).
- Braybrook, S.A., Stone, S.L., Park, S., Bui, A.Q., Le, B.H., Fischer, R.L., Goldberg, R.B., and Harada, J.J. (2006). Genes directly regulated by LEAFY COTYLEDON2 provide insight into the control of embryo maturation and somatic embryogenesis. Proc. Natl. Acad. Sci. USA 103 3468–3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrera, E., Holman, T., Medhurst, A., Dietrich, D., Footitt, S., Theodoulou, F.L., and Holdsworth, M.J. (2008). Seed after-ripening is a discrete developmental pathway associated with specific gene networks in Arabidopsis. Plant J. 53 214–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conceicao Ada, S., and Krebbers, E. (1994). A cotyledon regulatory region is responsible for the different spatial expression patterns of Arabidopsis 2S albumin genes. Plant J. 5 493–505. [DOI] [PubMed] [Google Scholar]
- Egea-Cortines, M., Saedler, H., and Sommer, H. (1999). Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of architecture in Antirrhinum majus. EMBO J. 18 5370–5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlert, A., Weltmeier, F., Wang, X., Mayer, C.S., Smeekens, S., Vicente-Carbajosa, J., and Dröge-Laser, W. (2006). Two-hybrid protein-protein interaction analysis in Arabidopsis protoplasts: Establishment of a heterodimerisation map of group-C and S bZIP transcription factors. Plant J. 46 890–900. [DOI] [PubMed] [Google Scholar]
- Ellenberger, T.E., Brandl, C.J., Struhl, K., and Harrison, S.C. (1992). The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted alpha helices: Crystal structure of the protein-DNA complex. Cell 71 1223–1237. [DOI] [PubMed] [Google Scholar]
- Ezcurra, I., Wycliffe, P., Nehlin, L., Ellerstrom, M., and Rask, L. (2000). Transactivation of the Brassica napus napin promoter by ABI3 requires interaction of the conserved B2 and B3 domains of ABI3 with different cis-elements: B2 mediates activation through an ABRE, whereas B3 interacts with an RY/G-box. Plant J. 24 57–66. [DOI] [PubMed] [Google Scholar]
- Ferrandiz, C., Gu, Q., Martienssen, R., and Yanofsky, M.F. (2000). Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development 127 725–734. [DOI] [PubMed] [Google Scholar]
- Finkelstein, R.R., and Lynch, T.J. (2000). The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudat, J., Hauge, B.M., Valon, C., Smalle, J., Parcy, F., and Goodman, H.M. (1992). Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4 1251–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruis, D., Schulze, J., and Jung, R. (2004). Storage protein accumulation in the absence of the vacuolar processing enzyme family of cysteine proteases. Plant Cell 16 270–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerineau, F. (1995). Tools for expressing foreign genes in plants. Methods Mol. Biol. 49 1–32. [DOI] [PubMed] [Google Scholar]
- Gutierrez, L., Van Wuytswinkel, O., Castelain, M., and Bellini, C. (2007). Combined networks regulating seed maturation. Trends Plant Sci. 12 294–300. [DOI] [PubMed] [Google Scholar]
- Hanson, J., Hanssen, M., Wiese, A., Hendriks, M.M.W.B., and Smeekens, S. (2008). The sucrose regulated transcription factor bZIP11 affects amino acid metabolism by regulating the expression of Asparagin Synthase1 and Proline Dehydrogenase 2. Plant J. 53 935–949. [DOI] [PubMed] [Google Scholar]
- Hartings, H., Maddaloni, M., Lazzaroni, N., Di Fonzo, N., Motto, M., Salamini, F., and Thompson, R. (1989). The O2 gene which regulates zein deposition in maize endosperm encodes a protein with structural homologies to transcriptional activators. EMBO J. 8 2795–2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundertmark, M., and Hincha, D.K. (2008). LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics 9 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoby, M., Weisshaar, B., Dröge-Laser, W., Vicente-Carbajosa, J., Tiedemann, J., Kroj, T., and Parcy, F. (2002). bZIP transcription factors in Arabidopsis. Trends Plant Sci. 7 106–111. [DOI] [PubMed] [Google Scholar]
- Kaminaka, H., Nake, C., Epple, P., Dittgen, J., Schütze, K., Chaban, C., Holt III, B.F., Merkle, T., Schafer, E., Harter, K., and Dangl, J.L. (2006). bZIP10-LSD1 antagonism modulates basal defense and cell death in Arabidopsis following infection. EMBO J. 25 4400–4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby, J., and Kavanagh, T.A. (2002). NAN fusions: A synthetic sialidase reporter gene as a sensitive and versatile partner for GUS. Plant J. 32 391–400. [DOI] [PubMed] [Google Scholar]
- Kroj, T., Savino, G., Valon, C., Giraudat, J., and Parcy, F. (2003). Regulation of storage protein gene expression in Arabidopsis. Development 130 6065–6073. [DOI] [PubMed] [Google Scholar]
- Lam, H.M., Wong, P., Chan, H.K., Yam, K.M., Chen, L., Chow, C.M., and Coruzzi, G.M. (2003). Overexpression of the ASN1 gene enhances nitrogen status in seeds of Arabidopsis. Plant Physiol. 132 926–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara, P., Oñate-Sánchez, L., Abraham, Z., Ferrandiz, C., Diaz, I., Carbonero, P., and Vicente-Carbajosa, J. (2003). Synergistic activation of seed storage protein gene expression in Arabidopsis by ABI3 and two bZIPs related to OPAQUE2. J. Biol. Chem. 278 21003–21011. [DOI] [PubMed] [Google Scholar]
- Li, F., Asami, T., Wu, X., Tsang, E.W., and Cutler, A.J. (2007). A putative hydroxysteroid dehydrogenase involved in regulating plant growth and development. Plant Physiol. 145 87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mönke, G., Altschmied, L., Tewes, A., Reidt, W., Mock, H.P., Bäumlein, H., and Conrad, U. (2004). Seed-specific transcription factors ABI3 and FUS3: Molecular interaction with DNA. Planta 219 158–166. [DOI] [PubMed] [Google Scholar]
- Moreno-Risueno, M.A., Diaz, I., Carrillo, L., Fuentes, R., and Carbonero, P. (2007). The HvDOF19 transcription factor mediates the abscisic acid-dependent repression of hydrolase genes in germinating barley aleurone. Plant J. 51 352–365. [DOI] [PubMed] [Google Scholar]
- Nakamura, S., Lynch, T.J., and Finkelstein, R.R. (2001). Physical interactions between ABA response loci of Arabidopsis. Plant J. 26 627–635. [DOI] [PubMed] [Google Scholar]
- Nakashima, K., Fujita, Y., Katsura, K., Maruyama, K., Narusaka, Y., Seki, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2006). Transcriptional regulation of ABI3- and ABA-responsive genes including RD29B and RD29A in seeds, germinating embryos, and seedlings of Arabidopsis. Plant Mol. Biol. 60 51–68. [DOI] [PubMed] [Google Scholar]
- Nantel, A., and Quatrano, R.S. (1996). Characterization of three rice basic/leucine zipper factors, including two inhibitors of EmBP-1 DNA binding activity. J. Biol. Chem. 271 31296–31305. [DOI] [PubMed] [Google Scholar]
- Norre, F., Peyrot, C., Garcia, C., Rance, I., Drevet, J., Theisen, M., and Gruber, V. (2002). Powerful effect of an atypical bifactorial endosperm box from wheat HMWG-Dx5 promoter in maize endosperm. Plant Mol. Biol. 50 699–712. [DOI] [PubMed] [Google Scholar]
- Oñate, L., Vicente Carbajosa, J., Lara, P., Diaz, I., and Carbonero, P. (1999). Barley BLZ2, a seed-specific bZIP protein that interacts with BLZ1 in vivo and activates transcription from the GCN4-like motif of B-hordein promoters in barley endosperm. J. Biol. Chem. 274 9175–9182. [DOI] [PubMed] [Google Scholar]
- Oñate-Sánchez, L., and Singh, K.B. (2002). Identification of Arabidopsis ethylene-responsive element binding factors with distinct induction kinetics after pathogen infection. Plant Physiol. 128 1313–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oñate-Sánchez, L., and Vicente-Carbajosa, J. (2008). DNA-free RNA isolation protocols for Arabidopsis thaliana, including seeds and siliques. BMC Res Notes. 1 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parcy, F., and Giraudat, J. (1997). Interactions between the ABI1 and the ectopically expressed ABI3 genes in controlling abscisic acid responses in Arabidopsis vegetative tissues. Plant J. 11 693–702. [DOI] [PubMed] [Google Scholar]
- Parcy, F., Valon, C., Raynal, M., Gaubier-Comella, P., Delseny, M., and Giraudat, J. (1994). Regulation of gene expression programs during Arabidopsis seed development: Roles of the ABI3 locus and of endogenous abscisic acid. Plant Cell 6 1567–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rook, F., Gerrits, N., Kortstee, A., van Kampen, M., Borrias, M., Weisbeek, P., and Smeekens, S. (1998). Sucrose-specific signalling represses translation of the Arabidopsis ATB2 bZIP transcription factor gene. Plant J. 15 253–263. [DOI] [PubMed] [Google Scholar]
- Santos-Mendoza, M., Dubreucq, B., Baud, S., Parcy, F., Caboche, M., and Lepiniec, L. (2008). Deciphering gene regulatory networks that control seed development and maturation in Arabidopsis. Plant J. 54 608–620. [DOI] [PubMed] [Google Scholar]
- Satoh, R., Fujita, Y., Nakashima, K., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2004). A novel subgroup of bZIP proteins functions as transcriptional activators in hypoosmolarity-responsive expression of the ProDH gene in Arabidopsis. Plant Cell Physiol. 45 309–317. [DOI] [PubMed] [Google Scholar]
- Schallau, A., Kakhovskaya, I., Tewes, A., Czihal, A., Tiedemann, J., Mohr, M., Grosse, I., Manteuffel, R., and Bäumlein, H. (2008). Phylogenetic footprints in fern spore- and seed-specific gene promoters. Plant J. 53 414–424. [DOI] [PubMed] [Google Scholar]
- Schmidt, R.J., Burr, F.A., Aukerman, M.J., and Burr, B. (1990). Maize regulatory gene opaque-2 encodes a protein with a “leucine-zipper” motif that binds to zein DNA. Proc. Natl. Acad. Sci. USA 87 46–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stangeland, B., and Salehian, Z. (2002). An improved clearing method for GUS assay in Arabidopsis endosperm and seeds. Plant Mol. Biol. Rep. 20 107–114. [Google Scholar]
- To, A., Valon, C., Savino, G., Guilleminot, J., Devic, M., Giraudat, J., and Parcy, F. (2006). A network of local and redundant gene regulation governs Arabidopsis seed maturation. Plant Cell 18 1642–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunnacliffe, A., and Wise, M.J. (2007). The continuing conundrum of the LEA proteins. Naturwissenschaften 94 791–812. [DOI] [PubMed] [Google Scholar]
- Usadel, B., Blasing, O.E., Gibon, Y., Retzlaff, K., Hohne, M., Gunther, M., and Stitt, M. (2008). Global transcript levels respond to small changes of the carbon status during progressive exhaustion of carbohydrates in Arabidopsis rosettes. Plant Physiol. 146 1834–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Carbajosa, J., and Carbonero, P. (2005). Seed maturation: Developing an intrusive phase to accomplish a quiescent state. Int. J. Dev. Biol. 49 645–651. [DOI] [PubMed] [Google Scholar]
- Walter, M., Chaban, C., Schütze, K., Batistic, O., Weckermann, K., Nake, C., Blazevic, D., Grefen, C., Schumacher, K., Oecking, C., Harter, K., and Kudla, J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40 428–438. [DOI] [PubMed] [Google Scholar]
- Wang, H.W., Zhang, B., Hao, Y.J., Huang, J., Tian, A.G., Liao, Y., Zhang, J.S., and Chen, S.Y. (2007). The soybean Dof-type transcription factor genes, GmDof4 and GmDof11, enhance lipid content in the seeds of transgenic Arabidopsis plants. Plant J. 52 716–729. [DOI] [PubMed] [Google Scholar]
- Weber, H., Borisjuk, L., and Wobus, U. (2005). Molecular physiology of legume seed development. Annu. Rev. Plant Biol. 56 253–279. [DOI] [PubMed] [Google Scholar]
- Weigel, R., and Glazebrook, J. (2002). Arabidopsis: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Weltmeier, F., Ehlert, A., Mayer, C.S., Dietrich, K., Wang, X., Schütze, K., Harter, K., Vicente-Carbajosa, J., and Dröge-Laser, W. (2006). Combinatorial control of Arabidopsis proline dehydrogenase transcription by specific heterodimerisation bZIP transcription factors. EMBO J. 25 3133–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weltmeier, F., Rahmani, F., Ehlert, A., Dietrich, K., Schütze, K., Wang, X., Chaban, C., Hanson, J., Teige, M., Harter, K., Vicente-Carbajosa, J., Smeekens, S., and Dröge-Laser, W. (2009). Expression patterns within the Arabidopsis C/S1 bZIP transcription factor network: Availability of heterodimerization partners controls gene expression during stress response and development. Plant Mol. Biol. 69 107–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, D., Duan, X., Wang, B., Hong, B., Ho, T., and Wu, R. (1996). Expression of a late embryogenesis abundant protein gene, HVA1, from barley confers tolerance to water deficit and salt stress in transgenic rice. Plant Physiol. 110 249–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.