Abstract
In most algae, the chloroplast division rate is held constant to maintain the proper number of chloroplasts per cell. By contrast, land plants evolved cell and chloroplast differentiation systems in which the size and number of chloroplasts change along with their respective cellular function by regulation of the division rate. Here, we show that PLASTID DIVISION (PDV) proteins, land plant–specific components of the division apparatus, determine the rate of chloroplast division. Overexpression of PDV proteins in the angiosperm Arabidopsis thaliana and the moss Physcomitrella patens increased the number but decreased the size of chloroplasts; reduction of PDV levels resulted in the opposite effect. The level of PDV proteins, but not other division components, decreased during leaf development, during which the chloroplast division rate also decreased. Exogenous cytokinins or overexpression of the cytokinin-responsive transcription factor CYTOKININ RESPONSE FACTOR2 increased the chloroplast division rate, where PDV proteins, but not other components of the division apparatus, were upregulated. These results suggest that the integration of PDV proteins into the division machinery enabled land plant cells to change chloroplast size and number in accord with the fate of cell differentiation.
INTRODUCTION
Chloroplasts originally derived from a bacterium related to extant cyanobacteria, which was engulfed by a primary nonphotosynthetic eukaryotic host cell more than a billion years ago. Over time, the engulfed bacterial endosymbionts have been reduced to chloroplasts and vertically transmitted to subsequent generations (Reyes-Prieto et al., 2007; Gould et al., 2008). Reminiscent of their free-living ancestor, chloroplasts multiply by division (Possingham and Lawrence, 1983; Boffey and Lloyd, 1988; Kuroiwa et al., 1998). However, most of the genes once present in the engulfed bacterial endosymbiont have been lost or transferred to the host nuclear genome; those still used by the chloroplasts are translated by the host and targeted back into the chloroplasts to express their functions. Therefore, chloroplasts cannot divide by themselves, and the division is performed by nucleus-encoded proteins.
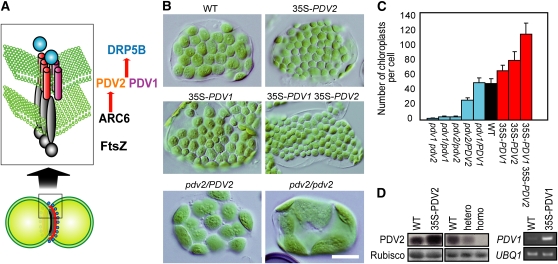
Chloroplast division is performed by ring structures at the division site, spanning both the inside and the outside of the two chloroplast envelope membranes. (Yoshida et al., 2006; Maple and Moller, 2007; Yang et al., 2008). The ring structures were identified by earlier electron microscopic studies (Kuroiwa et al., 1998; Miyagishima et al., 2001; Yoshida et al., 2006), and recent studies have identified several proteins that form a complex at the division site. Consistent with the endosymbiotic origin of chloroplasts, the division complex includes FtsZ, a self-assembling tubulin-like GTPase (Osteryoung and Vierling, 1995; Osteryoung et al., 1998; Mori et al., 2001; Vitha et al., 2001; Kuroiwa et al., 2002), and ACCUMULATION AND REPLICATION OF CHLOROPLASTS6 (ARC6), a J-domain containing protein, both of which are descended from the cell division machinery of the engulfed cyanobacterium (Vitha et al., 2003). In addition, the division complex includes DYNAMIN-RELATED PROTEIN 5B (DRP5B) (also known as ARC5), a member of the dynamin family of self-assembling GTPase proteins (Gao et al., 2003; Miyagishima et al., 2003), and PLASTID DIVISION1 (PDV1) and PDV2 proteins, which contain coiled-coil domains on the cytosolic side (Miyagishima et al., 2006; Glynn et al., 2008). DRP5B is specific to plants and algae and is suggested to have evolved from a dynamin-related protein that is involved in eukaryotic cytokinesis (Miyagishima et al., 2008). PDV1 and PDV2 are specific to land plants (Miyagishima et al., 2006; Glynn et al., 2008).
Chloroplast division in land plants is initiated by stromal FtsZ ring formation at the division site (containing FtsZ1 and FtsZ2, which arose by genetic duplication after the cyanobacterial endosymbiosis) (Vitha et al., 2001; Kuroiwa et al., 2002), which is stabilized by the inner envelope spanning protein ARC6 (Vitha et al., 2003). Then, the outer envelope–spanning proteins PDV1 and PDV2, which are paralogs of each other (Miyagishima et al., 2006), are recruited to the division site through direct interaction between PDV2 and ARC6 (Glynn et al., 2008). In addition, a recent study showed that the recruitment of PDV1 is mediated by PARC6 (paralog of ARC6 unique to vascular plants; Glynn et al., 2009). Finally, the dynamin-related protein DRP5B is recruited by PDV1 and PDV2 (Miyagishima et al., 2006; Glynn et al., 2008), and the entire division complex is involved in the fission of the chloroplast at the division site (Figure 1A).
Figure 1.
Overexpression of PDV1 and PDV2 Increases the Number and Decreases the Size of Chloroplasts.
(A) Diagram showing the pathway of chloroplast division complex assembly (Yang et al., 2008). Only the known division site–localized components are shown. FtsZ, homolog of the tubulin-like bacterial division GTPase, self-assembles into a ring structure at the stromal side of the division site (Vitha et al., 2001; Kuroiwa et al., 2002). The positioning of the FtsZ ring mid-chloroplast is regulated by MinD, MinE, ARC3, and MCD1 (Colletti et al., 2000; Itoh et al., 2001; Shimada et al., 2004; Maple et al., 2007; Nakanishi et al., 2009). FtsZ filaments are stabilized by cyanobacteria-descended inner envelope spanning protein ARC6 (Vitha et al., 2003) through interaction with FtsZ (Glynn et al., 2008). ARC6 recruits the outer envelope spanning proteins PDV1 and PDV2 through direct interaction with PDV2 (Glynn et al., 2008). PDV1 and PDV2 are required for recruitment of the cytosolic dynamin-related GTPase DRP5B at the division site (Miyagishima et al., 2006). FtsZ and ARC6 are descendants of cyanobacterial cell division machinery, while DRP5B is derived from eukaryotic membrane fission machinery, and PDV proteins are specific to land plants.
(B) Chloroplasts of the wild type, transgenic plants overexpressing PDV1, PDV2, and both PDV1 and PDV2 (35S-PDV1, 35S-PDV2, and 35S-PDV1 35S-PDV2) , and pdv2/PDV2 and pdv2/pdv2 T-DNA insertional mutants. Tips of the first true leaves were cut from ∼3-week-old plants grown on agar plates. Single leaf mesophyll cells observed by Nomarski optics are shown. There are no visible differences in growth among these lines. Bar = 20 μm.
(C) Statistical comparison of the number of chloroplasts per mesophyll cell. Error bars represent sd (n = 50 cells).
(D) Immunoblot and RT-PCR analyses showing the levels of PDV2 protein and the PDV1 transcript. The same amount of total protein extracted from rosettes was analyzed by anti-PDV2 antibodies. The ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) large subunit stained by Ponceau S is shown as the loading control. Three biological replicates showed the same result for the immunoblot analyses. Total RNA extracted from rosettes was used for RT-PCR to examine the PDV1 transcript level. UBQ1 was used as the internal control. The number of PCR cycles was 28 for PDV1 and 26 for UBQ1. The signals were estimated by ethidium bromide staining. Two biological replicates showed the same result.
[See online article for color version of this figure.]
In algae and meristematic cells in land plants, chloroplast (or plastid) division keeps pace with cell division to ensure their passage to daughter cells during cell division. By contrast, cells of land plants regulate the chloroplast division rate in accord with cell differentiation, thereby controlling the distinct size and number of chloroplasts (Possingham and Lawrence, 1983; Boffey and Lloyd, 1988). For example, small developing chloroplasts in young emerged leaves actively divide while the division rate slows down as leaves and chloroplasts mature (Boffey and Lloyd, 1988). Although several components that drive chloroplast division have been characterized, little is known about how the division machinery is controlled so as to modulate the rate of division in land plants.
Here, we report that the level of PDV proteins, land plant–specific components of the division apparatus, determines the rate of chloroplast division in the angiosperm Arabidopsis thaliana and the moss Physcomitrella patens. The analyses also show that the PDV level is upregulated by the plant hormone cytokinin. Our results suggest that acquisition of PDV proteins by the common ancestor of land plants has linked the cell differentiation program and chloroplast division and has enabled land plant cells to change the size and number of chloroplasts based on cell differentiation.
RESULTS
Artificial Increase or Decrease of the PDV1 and PDV2 Levels Gives Rise to an Increase or Decrease in the Chloroplast Division Rate in Arabidopsis
To identify the factors that modulate the rate of chloroplast division, we searched for genes that accelerate chloroplast division when the genes are overexpressed. To this end, we screened ∼10,000 independent lines of full-length cDNA-overexpressing (FOX) system of Arabidopsis (Ichikawa et al., 2006). In each FOX line, a full-length cDNA is expressed under the cauliflower mosaic virus 35S promoter (Ichikawa et al., 2006). As a result, we isolated six independent lines that grow normally and contain a larger number of smaller chloroplasts in the expanded leaves than the wild type. Of these, five lines contained a 35S promoter-PDV2 transgene, and the immunoblot analysis using anti-PDV2 antibodies showed that PDV2 protein was overexpressed in the line (see Supplemental Figure 1 online; the absence of the band in the pdv2/pdv2 null mutant in Figure 1D indicate that the antibodies are specific to PDV2). To further confirm whether the phenotype is linked to the overexpression of PDV2, we prepared a PDV2 overexpresser by a newly constructed 35S-PDV2 transgene. An immunoblot analysis showed that PDV2 protein is overexpressed in the line compared with the wild type (Figure 1D) and the plant displayed the same phenotype as the isolated FOX lines (Figures 1B to 1D). These results indicate that PDV2 overexpression accelerates chloroplast division.
Because previous studies showed that PDV2 mediates the recruitment of DRP5B to the chloroplast division site together with PDV1, a protein paralogous to PDV2 (Miyagishima et al., 2006), we also examined the effect of PDV1 overexpression by the 35S promoter on chloroplast division. Leaf cells of the PDV1 overexpresser contained a larger number of smaller chloroplasts than the wild type (Figures 1B to 1D). Although the effect of PDV1 overexpression was less evident than that of PDV2, the difference between the wild type and the PDV1 overexpresser was still significant (t test, one-tailed, P < 0.01; Figure 1C). When both PDV1 and PDV2 were simultaneously overexpressed, the number of chloroplasts further increased and the size was reduced compared with the single gene overexpressers (Figures 1B to 1D). These results indicate that PDV1 and PDV2 independently accelerate chloroplast division when the genes are overexpressed.
It was previously reported that the pdv2/PDV2 heterozygous T-DNA insertional mutant contains a smaller number of larger chloroplasts than the wild type (Miyagishima et al., 2006). The phenotype of pdv2/PDV2 is intermediate between those of the wild-type and pdv2/pdv2 plants (Miyagishima et al., 2006) (Figures 1B to 1D). By immunoblot analysis, we confirmed that PDV2 is absent in pdv2/pdv2 and found that the PDV2 level is reduced in pdv2/PDV2 compared with the wild type (Figure 1D). Taken together, the above results indicate that the levels of PDV1 and PDV2 positively correlate with the rate of chloroplast division. By contrast, overexpression of DRP5B had no effect on chloroplast division (see Supplemental Figure 1 online). Previous studies showed that overexpression of FtsZ1, FtsZ2 (Stokes et al., 2000), or ARC6 (Vitha et al., 2003) in addition to some other proteins related to the division machinery impairs chloroplast division (Colletti et al., 2000; Itoh et al., 2001; Raynaud et al., 2004), resulting in giant chloroplasts. The acceleration of chloroplast division has not been observed in the overexpression or disruption of genes that are reported to be involved in the division process. Taken together, it is suggested that levels of PDV1 and PDV2 determine the rate of chloroplast division.
PDV Levels, but Not Those of Other Division Components, Decrease during Leaf Development along with a Decrease in the Chloroplast Division Rate
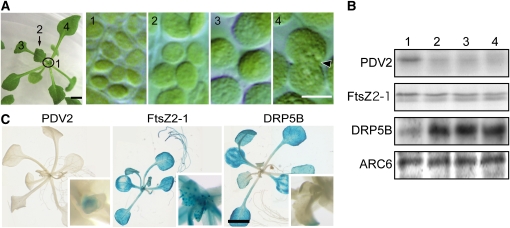
Since these changes in the PDV levels were caused by the expression of transgenes or by mutations, next we examined whether the levels of PDV1 and PDV2 translated from endogenous genes actually change so as to modulate the chloroplast division rate in the wild-type plant. To this end, we examined the levels of PDV and other chloroplast division proteins during leaf development. During leaf development, the frequency of chloroplasts with a division site constriction decreases, while the size of chloroplasts increases as the leaf gets older (Possingham and Lawrence, 1983; Boffey and Lloyd, 1988; Pyke, 1999) (Figure 2A).
Figure 2.
The PDV2 Level, but Not the Other Chloroplast Division Protein Levels, Decreases in Accord with the Increase of Chloroplast Size during Leaf Development.
(A) Change of chloroplast size during leaf development. Chloroplasts in a young emerging leaf (1) and expanding leaves (2 to 4) of the wild-type plant were observed by Nomarski optics. Chloroplasts are observably still dividing in the first true leaf (4, indicated by arrow). Bars = 2 mm (left) and 5 μm (right, panels 1 to 4).
(B) Immunoblot analyses showing the levels of chloroplast division proteins during leaf development. PDV2 and FtsZ2-1 are detected by anti-PDV2 and anti-FtsZ2-1 antibodies, respectively. DRP5B and ARC6 levels were analyzed by anti-GFP antibodies using wild-type plants expressing GFP-DRP5B and ARC6-GFP by their respective promoters. The same amount of protein extracted from leaves corresponding to stages 1 to 4 (indicated in [A], a sample of stage 1 including both the young emerging leaves and shoot apexes) was loaded in each lane. Three biological replicates showed the same result.
(C) Histochemical GUS staining of PDV2 promoter-GUS, FtsZ2-1 promoter-GUS, and DRP5B promoter-GUS transgenic plants. Approximately three-week-old plants grown on agar plates were stained. Magnified images of the centers of the rosettes are also shown (insets). ARC6 promoter-GUS transgenic plants were also prepared, but we could not obtain a staining signal. Three independent transgenic lines for each promoter showed the same results. Bar = 3 mm.
Immunoblot analyses showed that the PDV2 level is highest in the apical meristem and young emerging leaves and decreases during leaf development (Figure 2B). By contrast, the DRP5B level increased, but the FtsZ2-1 (the antibodies are specific to FtsZ2-1 of three FtsZ proteins of Arabidopsis; Suzuki et al., 2009), and ARC6 levels remained constant during leaf development (Figure 2B). Promoter-β-glucuronidase (GUS) fusion assays also showed that the activity of the PDV2 promoter is highest around the shoot apical meristem, in contrast with the FtsZ and DRP5B promoters (Figure 2C). These results indicate that the level of PDV2, but not that of FtsZ, ARC6, and DRP5B, decreases during leaf development.
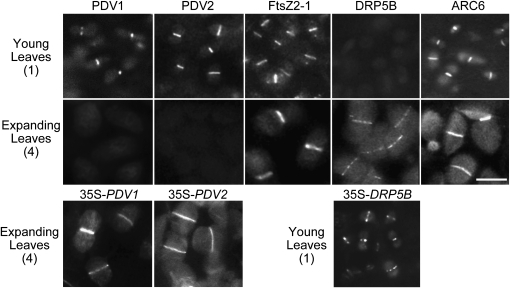
To compare the levels of the chloroplast division proteins at the chloroplast division site, green fluorescent protein (GFP) fusion proteins were expressed by their respective promoters and observed by fluorescence microscopy. Consistent with the results of the immunoblot and GUS analyses, strong fluorescence signals of GFP-PDV1 and GFP-PDV2 at the chloroplast division site were clearly observed in young, emerging leaves, but the signals were hardly detected in older, expanding leaves (Figure 3). By contrast, the FtsZ-GFP and ARC6-GFP signals at the division site were observed in both young and older leaves (Figure 3). Strong GFP-DRP5B signals at the division site were observed in older leaves but hardly at all in young emerging leaves (Figure 3). Despite the absence of signals strong enough to allow the observation of GFP-PDV1 and GFP-PDV2 in expanding leaves, or GFP-DRP5B in young emerging leaves (Figure 3), we detected the signals of these GFP fusion proteins at the chloroplast division site throughout leaf development when the proteins were overexpressed (Figure 3, bottom panels). Therefore, the barely detectable GFP signals by fluorescence microscopy (Figure 3) and faint but still detectable signals by immunoblot analyses (Figure 2B) are probably indicative of low levels of the proteins in the division apparatus, instead of the absence of these proteins.
Figure 3.
Division Site Localization of PDV1, PDV2, FtsZ, DRP5B, and ARC6 during Leaf Development.
Fluorescence microscopy showing the localization of chloroplast division proteins during leaf development. GFP-PDV1, GFP-PDV2, FtsZ-GFP, GFP-DRP5B, and ARC6-GFP expressed by their respective promoters were observed in emerging (top panels) and expanding (middle panels) leaves corresponding to stages 1 and 4 in Figure 2. GFP-PDV1, GFP-PDV2, and GFP-DRP5B were overexpressed in wild-type plants, and the localization was observed by fluorescence microscopy (bottom panels). In emerging (35S-DRP5B) and expanding (35S-PDV1 and 35S-PDV2) leaves corresponding to stages 1 and 4 in Figure 2, these proteins localize at the chloroplast division site. Three independent transgenic lines for each GFP fusion showed the same results. Bars = 5 μm.
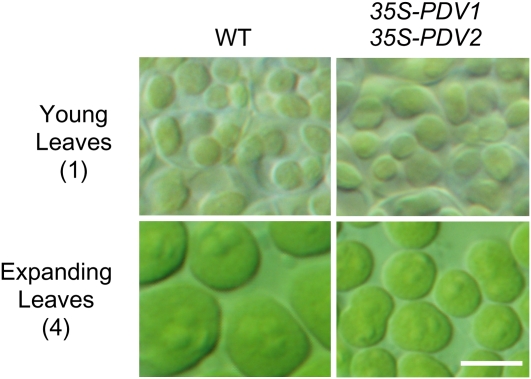
The above results indicate that the PDV levels in the chloroplast division apparatus, but not FtsZ, ARC6, or DRP5B, decrease during leaf development, suggesting that the stoichiometric relationship between the components of the division apparatus changes during chloroplast development. Given the results of overexpression and the reduction of PDV levels (Figure 1), it is suggested that the decrease of PDV levels during leaf development in turn decreases the rate of chloroplast division, thereby increasing the size of chloroplasts. Supporting this conclusion, the size and number of chloroplasts in young emerging leaves are similar in the wild type and the PDV1 and PDV2 overexpressers (Figure 4), but constant overexpression depresses the chloroplast increase in size in the older, expanding leaves (Figure 4).
Figure 4.
Effect of PDV1 and PDV2 Overexpression on the Young Emerging Leaves and Expanding Leaves.
Chloroplasts of the wild type and PDV1 and PDV2 overexpressers in emerging and expanding leaves corresponding to stages 1 and 4 in Figure 2 A. Bar = 5 μm.
[See online article for color version of this figure.]
Overexpression of Cytokinin-Responsive Transcription Factor CRF2 or Exogenous Cytokinin Treatment Increases PDV2 Level, but Not Those of Other Division Proteins, in Parallel with an Increase in the Chloroplast Division Rate
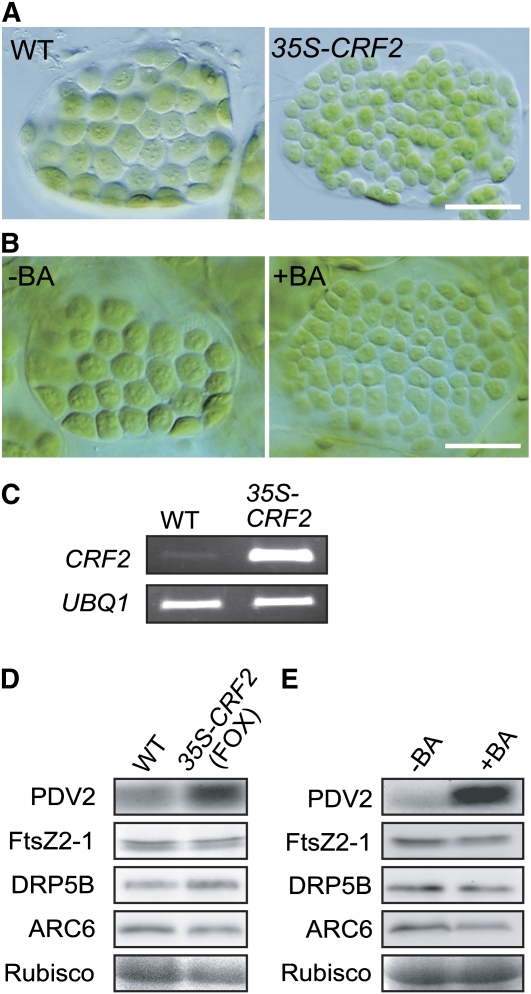
Further supporting the relationship between cell differentiation and change in the chloroplast division rate, another FOX line, in which chloroplast division is accelerated, turned out to be an overexpresser of CRF2 (Rashotte et al., 2006) (see Supplemental Figure 2 online). Transgenic plants overexpressing CRF2 as the result of a newly constructed 35S-CRF2 transgene displayed a similar phenotype (Figures 5A and 5C), indicating that the phenotype is indeed caused by CRF2 overexpression. Immunoblot analyses showed that the PDV2 level, but not the level of FtsZ, DRP5B, or ARC6, was increased in the CRF2 overexpresser (Figure 5D), suggesting that the increase of the PDV2 level is the cause of the acceleration of chloroplast division in the CRF2 overexpresser.
Figure 5.
CRF2 Overexpression or Cytokinin Treatment Increases the PDV2 Level and Increases the Number and Decreases the Size of Chloroplasts.
(A) Chloroplasts of wild-type and transgenic plants overexpressing CRF2 (35S-CRF2) in single leaf mesophyll cells. Tips of the first true leaves were cut from ∼3-week-old plants grown on agar plates. Bar = 20 μm.
(B) Effect of cytokinin treatment on the size and number of chloroplasts. Chloroplasts in single mesophyll cells of the cotyledon are shown. Wild-type seeds were germinated and grown for 10 d on agar plates with (+BA) or without (−BA) 5 μM BA. Bar = 20 μm.
(C) RT-PCR analyses showing the CRF2 transcript was increased in 35S-CRF2 transgenic plants. Total RNA extracted from rosettes was used for RT-PCR. UBQ1 was used as the internal control. The number of PCR cycles was 28 for CRF2 and 26 for UBQ1. The signals were estimated by ethidium bromide staining. Two biological replicates showed the same result.
(D) Immunoblot analyses comparing the levels of the chloroplast division proteins between the wild type and CRF2 overexpresser. The same amount of total protein extracted from ∼3-week-old russets was loaded in each lane. Rubisco large subunit stained by Ponceau S is shown as the loading control. Three biological replicates showed the same result.
(E) Immunoblot analyses comparing the levels of the chloroplast division proteins between wild-type seedlings germinated on medium with (+BA) or without (−BA) BA. The same amount of total protein extracted from 10-d-old seedlings was loaded in each lane. PDV2, FtsZ2-1, GFP-DRP5B, and ARC6-GFP were detected as in Figure 2B. Three biological replicates showed the same result.
[See online article for color version of this figure.]
CRF2 is a putative transcription factor that, together with other paralogous CRF proteins, mediates transcriptional responses to the plant hormone cytokinin (Rashotte et al., 2006). In land plants, cytokinin regulates numerous growth and developmental processes, including cell division, shoot initiation, and apical meristem function (Kieber, 2002; Kakimoto, 2003). Therefore, the above results suggest that the PDV2 level is regulated by a developmental program responsive to cytokinin. To examine the relationship between PDV levels and cytokinin, wild-type seeds were germinated on medium containing cytokinin (6-benzyladenine [BA]). We then compared the size and number of chloroplasts and the PDV2 level between plants treated or untreated with cytokinin. Chloroplasts in the cotyledon cells of cytokinin-treated plants were smaller and more numerous than those grown without cytokinin (Figure 5B). The same pattern of difference was observed when detached true leaves were put on the medium with or without cytokinin. Immunoblot analyses of total plantlets showed that the PDV2 level, but not the level of FtsZ, DRP5B, or ARC6, was increased in plants germinated on the cytokinin-containing medium (Figure 5E). These results suggest that cytokinin, or a developmental program induced by cytokinin, upregulates PDV2, at least in part through an upregulation of CRF2, which leads to an increase in the chloroplast division rate.
The PDV Protein Level Determines the Rate of Chloroplast Division in the Moss P. patens
The results obtained in Arabidopsis suggest that PDV1 and PDV2 determine the rate of chloroplast division, at least in the angiosperms. Homologs of PDV1 and PDV2 are encoded in the genomes of other lineages, but are only evident in land plants, suggesting that the genes for these proteins were acquired by the ancestral plant during the transition to terrestrial habitats (Miyagishima et al., 2006; Glynn et al., 2008) (Figure 6A; see Supplemental Figure 3 online). To obtain evolutionary insights into the function of PDV proteins, we examined whether PDV proteins also rate-determine chloroplast division in mosses, mosses having branched earliest in land plant evolution (Kenrick and Crane, 1997). To this end, we used the moss P. patens since the draft genome sequence is available and earlier studies showed involvement of FtsZ in chloroplast division by gene disruption in this organism (Strepp et al., 1998).
Figure 6.
PDV2 also Rate-Determines Chloroplast Division in the Moss P. patens.
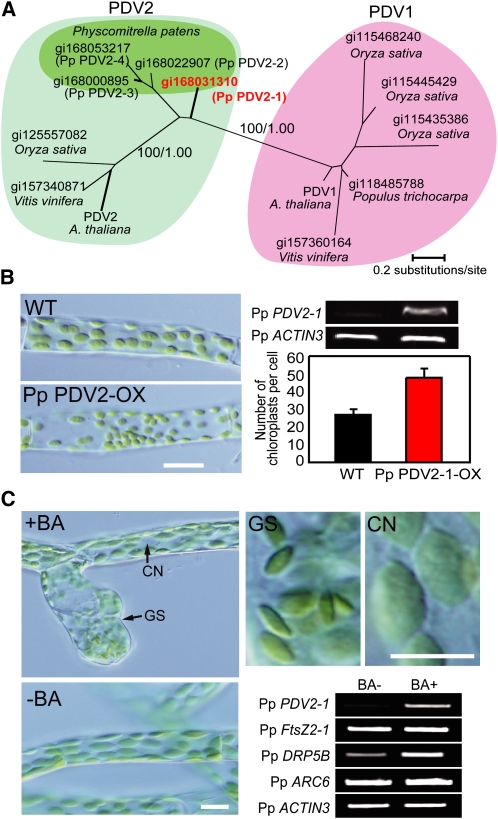
(A) Phylogenetic relationships in the PDV family of proteins. The amino acid sequences were collected from the National Center for Biotechnology Information database of nonredundant protein sequences. The GI numbers of the respective amino acid sequences are indicated. The tree shown is the maximum likelihood tree constructed by the RaxML program (Stamatakis, 2006) based on the alignment of 144 amino acid residues of 13 proteins. The numbers at the selected nodes are local bootstrap values (left) and Bayesian posterior probabilities (right) calculated by the maximum-likelihood method and Bayesian inference analyses, respectively.
(B) Number of chloroplasts per chloronema cells of the wild type and Pp PDV2-1 overexpresser (Pp PDV2-OX) (n = 30 cells). In P. patens, protonema cells are classified into chloronema and caulonema. Chloronema, which contains round chloroplasts, develops to caulonema, which contains spindle-shaped chloroplasts. An increase in number of chloroplasts was also observed in caulonemal cells of the Pp PDV2-1 overexpresser. RT-PCR analyses showing the Pp PDV2-1 transcript is increased in the transgenic line. Total RNA extracted from protonemal colonies was used for RT-PCR. Pp ACTIN3 was used as the internal control. The number of PCR cycles was 28 for Pp PDV2-1 and 28 for Pp ACTIN3. The signals were estimated by ethidium bromide staining. The same results were obtained in four independent transformants. Bar = 10 μm.
(C) Effect of cytokinin treatment on chloroplasts and expression of chloroplast division genes. RT-PCR analyses comparing transcript levels of chloroplast division genes between cells grown on medium with (+BA) or without (−BA) BA. Four-day-old protonemal cells were transferred onto medium with or without 5 μM BA and grown for 4 d. Pp ACTIN3 was used as the internal control. The number of PCR cycles was 28 for each gene. The signals were estimated by ethidium bromide staining. Two biological replicates showed the same result. CN, caulonema; GS, gametophore shoot apical cell of a bud induced by cytokinin. Bars = 10 μm.
[See online article for color version of this figure.]
By BLAST and PSI-BLAST (Altschul et al., 1997) searches, we identified four P. patens genes encoding proteins homologous to angiosperm PDV1 and PDV2 (Figure 6A; see Supplemental Figure 3 online). Phylogenic analyses revealed the conservation of PDV1 and PDV2 in angiosperms (Figure 6A). By contrast, the amino acid sequence alignment showed that all the moss PDV proteins bear regions found in angiosperm PDV2 but not in angiosperm PDV1; these regions flank both sides of the predicted-membrane spanning domain (i.e., both cytosolic and the intermembrane space; see Supplemental Figure 3 online). The alignment along with the pattern of the tree topology suggest that all the moss proteins are orthologous to angiosperm PDV2. Given the recent report that recruitment of PDV1 but not PDV2 is mediated by PARC6 and that PARC6 is unique to vascular plants (Glynn et al., 2009), it is suggested that PDV1, PARC6, and their relationship arose by gene duplications and diversification in ancestral vascular plants after mosses branched out. When the Pp PDV2-1 gene was overexpressed by an E7113 promoter (consisting of seven 35S promoters and additional enhancers; Mitsuhara et al., 1996) PDV2-1 cDNA transgene, the protonemal cells contained a greater number of smaller sized chloroplasts than did the wild type (Figure 6B), as did the Arabidopsis PDV2 overexpresser (Figure 1B).
In contrast with Arabidopsis (Figure 5B), exogenous cytokinin treatment had no effect on the size or number of protonemal chloroplasts. However, the treatment induced the formation of buds from which gametophytes arise, as reported previously (Reski and Abel, 1985) (Figure 6C). The gametophore shoot apical meristem contained chloroplasts more numerous (per volume) and smaller than those in caulonemal cells (Figure 6C). These results suggest that rate of chloroplast division is increased during the course of bud induction by cytokinin. RT-PCR analyses of the chloroplast division genes showed that the Pp PDV2-1 level was increased by cytokinin treatment (Figure 6C). By contrast, the DRP5B level was slightly upregulated, and the levels of FtsZ2-1 and ARC6 were not changed by the treatment (Figure 6C). We also confirmed that the cytokinin treatment upregulated Pp PDV2-2, Pp PDV2-3, and Pp PDV2-4 but did not change levels of all of the five FtsZ genes (Martin et al., 2009) encoded in the P. patens genome (see Supplemental Figure 4 online). These results suggest that the PDV2 level changes to modulate the rate of chloroplast division during bud formation in the moss P. patens, and PDV protein function serving as a rate-limiting component of chloroplast division machinery is conserved in land plants.
DISCUSSION
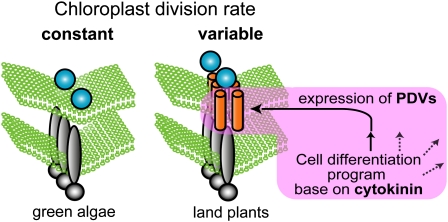
Recent studies have reported several components of the plastid division apparatus. Of these components, FtsZ and ARC6, which function inside chloroplasts, are descended from a cyanobacterial ancestor, and cytosolic DRP5B evolved from a eukaryotic membrane fission system (Figure 1A). These three components are well conserved in green algae and land plants (Miyagishima, 2005; Maple and Moller, 2007; Yang et al., 2008) (Figure 7). DRP5B is also conserved in red algae and stramenopiles, the chloroplast in which was established by secondary endosymbiosis of a red algae (Yoon et al., 2002; Reyes-Prieto et al., 2007; Gould et al., 2008), suggesting that DRP5B became integrated in the division apparatus prior to the branching of the green and red lineages of photosynthetic eukaryotes ∼1 billion years ago (Miyagishima, 2005). Given the conservation of FtsZ, ARC6, and DRP5B, it is suggested that the division apparatus consisting of these three components was established at a relatively early point in chloroplast evolution. By contrast, PDV proteins, connecting the bacterial and eukaryotic division components, are specific to land plants, suggesting that they became integrated into the division apparatus when land plants evolved from green algae (Miyagishima et al., 2006; Glynn et al., 2008) (Figures 6A and 7), which is estimated to be ∼0.4 to 0.5 billion years ago (Kenrick and Crane, 1997).
Figure 7.
Schematic Representation of PDV-Mediated Control of the Chloroplast Division Rate That Evolved in Land Plants.
In a common ancestor of land plants, PDV proteins became inserted into the chloroplast division apparatus descended from green algae. PDV mediate recruitment of the eukaryotic DRP5B to the division site in which cyanobacteria-descended FtsZ and ARC6 have been assembled. Levels of PDV expression are regulated by a cytokinin-dependent cell differentiation program, and the PDV level determines the rate of division site constriction.
[See online article for color version of this figure.]
Our results show that increases or decreases of the PDV levels increase or decrease the rate of chloroplast division (Figures 1B to 1D), although changes in the levels of the other division components either impair division or have no effect. The levels of PDV translated from the nuclear genome, but not those of other division components, actually decreased during leaf development, in parallel with the decrease of the division rate (Figure 2B). Cytokinin treatment or CRF2 overexpression increases the PDV levels, but not those of the other division components, in parallel with the increase in the division rate (Figure 5). These results suggest that PDV proteins determine the rate of chloroplast division based on the cell differentiation system that evolved in the ancestor of land plants. Given the evolutionary distribution of the chloroplast division proteins and the conservation of PDV function as the rate-determining factor of chloroplast division in the moss P. patens (Figure 6), it is suggested that the integration of PDV proteins into the division machinery enabled land plant cells to change chloroplast size and number in accord with the fate of cell differentiation (Figure 7).
The PDV2 level was increased by cytokinin treatment in both Arabidopsis (Figure 5E) and P. patens (Figure 6C). CRF2 overexpression increased the PDV2 level; this suggests that induction of PDV2 is in a pathway downstream of CRF. Cytokinin accelerated leaf chloroplast division in Arabidopsis (Figure 5), whereas the size and number of chloroplasts in the moss protonema were unchanged by cytokinin treatment (Figure 6). Instead, cytokinin induced the formation of buds containing numerous smaller chloroplasts (Figure 6). Although the reason for the difference between Arabidopsis and P. patens is unclear at present, our results indicate that the PDV levels are regulated by a cell differentiation system responsive to cytokinin. The highest expression of PDV proteins around the shoot apical meristem and young emerging leaves in Arabidopsis (Figure 2) is consistent with a previous observation that content of cytokinin is highest in meristems (Jacqmard et al., 2002). The induction of PDV expression by cytokinin might be related to the observation that cytokinin addition or expression of the ipt gene, whose product catalyzes the rate-limiting step in cytokinin synthesis, complements a P. patens mutant defective in chloroplast division (Reski et al., 1991; Reutter et al., 1998).
In P. patens, a previous study showed that slightly enhanced levels of FtsZ by expression of transgenes seem to accelerate chloroplast division, although strong overexpression of FtsZ impaired the division as observed in Arabidopsis (Kiessling et al., 2000). However, it is still not known whether FtsZ proteins translated from endogenous genes are actually upregulated to increase the rate of chloroplast division in some points of the P. patens life cycle. Since all the four Pp PDV2 genes, but not the all the five Pp FtsZ genes, are upregulated by cytokinin treatment (Figure 6; see Supplemental Figure 4 online), increase of FtsZ is not likely to be involved in the acceleration of chloroplast division during bud formation.
In vascular plants, all plastids, including chloroplasts, are derived from small, non-green proplastids in meristematic cells (Pyke, 1999; Lopez-Juez and Pyke, 2005). The decrease of PDV levels during leaf development (Figure 2) and the highest promoter activity of PDV2 around the shoot apical meristem (Figure 2C) appear to correlate with the development of leaf chloroplasts from meristematic proplastids in Arabidopsis. However, the overexpression of PDV proteins increased the division rate of photosynthetic chloroplasts (Figure 1). In addition, our results suggest that PDV proteins also rate-determine chloroplast division in the mosses (Figure 6), which contain chloroplasts throughout the life cycle, as do algae (Boffey and Lloyd, 1988). Therefore, PDV proteins were probably acquired to modulate photosynthetic chloroplast division prior to the evolutionary emergence of the differentiation system based on the proplastids. It is also suggested that the modulation of chloroplast division rate by PDV proteins might be a critical step for the evolution of the plastid differentiation system in vascular plants.
The change in the cellular levels of PDV proteins gives rise to two possibilities. One is that the ratio of chloroplasts in which the PDV proteins are integrated changes. The other is that the amount of PDV proteins in the division complex, that is, the stoichiometrical relationship of PDV proteins to other division components changes. Almost every chloroplast in expanding leaves exhibited FtsZ and ARC6 localization at the division site, and the fluorescent signals of these proteins at the division site appeared to be constant during leaf development (Figure 3). By contrast, the fluorescent signals of GFP-PDV1 and GFP-PDV2 were hardly detected in any chloroplast in expanding leaves in which the chloroplasts were still dividing (Figure 3). These results suggest that the state of the chloroplast division complex changes as a result of the change in the PDV levels in each division complex. Chloroplasts in the pdv1 and pdv2 mutants reportedly frequently display constriction at the division site (Miyagishima et al., 2006; Glynn et al., 2008) (Figure 1; see Supplemental Figure 1 online), suggesting that constriction is delayed in the mutants and that the levels of PDV proteins likely determine the rate of division site constriction. To understand how the division machinery is affected by the change in the PDV levels, further structural and biochemical studies are required. In this regard, the phenotypes of the pdv1 and pdv2 mutants indicated that PDV1 and PDV2 are required for cytosolic DRP5B localization after stromal FtsZ and inner envelope ARC6 assembly at the division site (Miyagishima et al., 2006; Glynn et al., 2008). However, the exact biochemical relationship between PDV proteins and DRP5B is not yet clear (i.e., whether there is direct interaction between PDV proteins and DRP5B or a requirement of other intermediate components). Identification and characterization of cytosolic proteins that directly interact with PDV proteins will provide further insights into that regulation of the chloroplast division rate by PDV proteins.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana (Columbia-0) was used as the wild type. Arabidopsis seeds were surface-sterilized, sown on Murashige and Skoog (MS) plates, and stratified at 4°C for 48 h in the dark before germination. Plants were grown in controlled-environment chambers with 16 h of light (100 μmol/m2s) and 8 h of dark at 20°C. For cytokinin treatment, sterilized seeds were sown on MS plates supplemented with 5 μM BA and grown for 10 d after germination. The pdv1 and pdv2 mutants used in this study are pdv1-1 (has a premature stop codon close to the start codon) and pdv2-1 (T-DNA is inserted into the first intron), respectively (Miyagishima et al., 2006). Physcomitrella patens Bruch and Schimp subsp patens Tan was grown in controlled-environment chambers with 16 h of light (100 μmol/m2s) and 8 h of dark at 20°C on the minimal medium (BCD medium) supplemented with 0.5% glucose, 1 mM CaCl2, and 5 mM diammonium (+)-tartrate agar plate as described (Nishiyama et al., 2000). For cytokinin treatment, protonema grown on BCD medium were transferred and then grown on BCD medium containing 5 μM BA for 4 d.
Analyses of Arabidopsis FOX Lines
Seeds of ∼10,000 independent FOX lines (Ichikawa et al., 2006) (Columbia-0 background) were separated to 200 pools, in which ∼50 independent lines were mixed in each pool. One hundred seeds from each pool were germinated and grown for 3 weeks on MS plates. Tips from expanding leaves were put on a glass slide without fixation, covered with a cover slip, and were observed with Nomarski optics. As a result, leaf cells of 13 plants contained a larger number of smaller chloroplasts than the wild type. Among them, seven lines also showed dwarf phenotypes, while growth and shape of plantlets were normal in six lines. The inserted cDNA of the six lines was amplified by T-DNA–specific primers FOX.F and FOX.R and sequenced by primer FOX-seq.
Constructing Overexpression and Fusion Constructs and Generating Transgenic Arabidopsis
For overexpression of PDV1, PDV2, and CRF2, fragments containing respective open reading frames (orf) franked by ∼0.1 kb 5′ upstream sequences were amplified by primers: PDV1-ox.F and PDV1-ox.R for PDV1, PDV2-ox.F and PDV2-ox.R for PDV2, and CRF2-ox.F and CRF2-ox.R for CRF2. The amplified PDV2 and CRF2 fragments were digested with XbaI and SmaI, and the PDV1 fragment was digested with NheI and SmaI (the recognition sequences of these enzymes are underlined in the sequences of respective primers). The digested products were inserted between XbaI and blunting SacI sites (downstream of cauliflower mosaic virus 35S promoter) of pBI121 vector (conferring resistance to kanamycin; Clontech).
For overexpression of GFP-DRP5B, DRP5B promoter of DRP5B promoter-GFP-DRP5B construct (Miyagishima et al., 2006) was replaced by 35S promoter amplified from pBI121 vector. The generated constructs were transformed into the wild type.
For expression of GFP-PDV2 by the PDV2 promoter, two unique restriction sites were added between the PDV2 promoter and the start codon by overlap-extension PCR. We amplified a 1.2-kb 5′ upstream sequence of PDV2, including the start codon, by primers GFP-PDV2.F1 and GFP-PDV2.R1. A PDV2 orf flanked by a 60-bp 3′ downstream sequence was amplified by primers GFP-PDV2.F2 and GFP-PDV2.R2. These two amplified fragments were mixed and fused by PCR using primers GFP-PDV2.F1 and GFP-PDV2.R2. The fused fragment was cloned into pGEM-T Easy (Promega). An orf of GFP was amplified by primers 5′-GFP-BamHI.F and GFP-KpnI.R, digested with BamHI and KpnI, and cloned between a 5′ flanking region and an orf of PDV2. The resulting PDV2 promoter-GFP-PDV2 fusion was excised with NotI and then transferred into pMLBART (conferring resistance to glufosinate ammonium; Vitha et al., 2001). The construct was transformed into the wild type.
For expression of GFP-PDV1 by the 35S promoter, GFP-PDV1 fusion (Miyagishima et al., 2006) was cut out with BamHI and EcoRI and inserted into BamHI and EcoRI sites of pBI121. For expression of GFP-PDV2 by the 35S promoter, GFP-PDV2 was amplified using the PDV2 promoter-GFP-PDV2 fusion described above as template by primers PDV2-ox.F and PDV2-ox.R. The amplified fragment was digested with XbaI and SmaI and inserted into XbaI and blunted SacI sites of pBI121. The generated construct was transformed into the wild type.
For expression of ARC6-GFP by the ARC6 promoter, ARC6 orf franked by ∼1.0-kb 5′ upstream sequence was amplified by primers ARC6-GFP.F and ARC6-GFP.R and was cloned into pGEM-T Easy. An orf of GFP (S65T) was amplified by primers GFP-KpnI.F and GFP-SacII.R, digested with KpnI and SacII, and cloned downstream of the ARC6 orf. The resulting ARC6 promoter-ARC6-GFP fusion was excised with NotI and then transferred into pMLBART. The generated construct was transformed into the wild type.
FtsZ promoter-GFP-FtsZ (Nakanishi et al., 2009) and DRP5B promoter-GFP-DRP5B (Miyagishima et al., 2006) transformants were previously generated as described.
To create promoter-GUS fusion, 0.6- to 1.5-kb 5′ regions of PDV2, FtsZ2-1, and DRP5B were amplified by primers: PDV2-GUS.F and PDV2-GUS.R for PDV2, FtsZ2-1-GUS.F and FtsZ2-1-GUS.R for FtsZ2-1, and DRP5B-GUS.F and DRP5B-GUS.R for DRP5B. PDV2 fragment was digested with BamHI and FtsZ2-1, and DRP5B fragments were digested with XbaI (the recognition sequences of these enzymes are underlined in the sequences of respective primers). These fragments were cloned into the BamHI site or the XbaI site of pBI101 (conferring resistance to kanamycin; Clontech). The constructs were transformed into the wild type.
All constructs were transferred to Agrobacterium tumefaciens GV3101 and introduced into Arabidopsis as described (Clough and Bent, 1998). T1 plants were selected by resistance to glufosinate or kanamycin as described (Miyagishima et al., 2006). To overexpress both PDV1 and PDV2, a PDV1 overexpresser line was crossed with a PDV2 overexpresser line and the next generation was examined. To express GFP-DRP5B and ARC6-GFP in CRF2 overexpresser, GFP-DRP5B and ARC6-GFP expressing the wild type, respectively, were crossed with 35S-CRF2 plants, and the next generation was used for further analyses. Before using each transgenic plant for further analyses, the existence of respective transgene(s) was confirmed by PCR analyses.
Construction and Generating Transgenic P. patens
For overexpression of P. patens PDV2-1 (GI 168031309), a fragment containing the orf was amplified by primers Pp_PDV2-1-ox.F and Pp_PDV2-1-ox.R from genomic DNA (the SmaI sites are underlined). The amplified product was digested with SmaI and was inserted into SmaI site of the expression vector pPpMADS2-7113 with E7113 promoter (Mitsuhara et al., 1996). The construct was cut out by NotI and was introduced into the protoplasts of P. patens by polyethylene glycol–mediated protocol (Nishiyama et al., 2000). The protoplasts were grown in the regeneration medium for 3 d and then transferred onto BCDAT medium (Nishiyama et al., 2000) containing 50 μg mL−1 G418 to select transformants. The selected plants were transferred onto a medium without G418 and allowed to grow for 1 week. Then, they were transferred again onto the selection medium.
Microscopy
For observation of chloroplasts in Arabidopsis leaf cells, tips from expanding leaves from ∼3-week-old plants (or other stages where indicated) grown on MS plates were cut and fixed with 3.5% glutaraldehyde and then incubated in 0.1 M Na2-EDTA, pH 9.0, for 15 min at 50°C. Chloroplasts of P. patens in protonemal cells were observed without fixation. Samples were observed with Nomarski optics. GFP fluorescence was visualized without fixation in young and expanding leaves from ∼3-week-old plants.
Antibodies and Immunoblotting
The polyclonal rabbit antibodies against Arabidopsis PDV2 were raised against recombinant PDV2. The full-length PDV2 coding region was amplified from Arabidopsis cDNA by primers His-PDV2.F and His-PDV2.R and was cloned into pET100 expression vector (Invitrogen). Six-His fusion proteins were expressed in Escherichia coli (Rosetta 2 DE3; Novagen), purified, and injected to rabbits to stimulate antibody production. The antisera were purified by N-hydroxysuccinimide–activated column (HiTrap NHS-activated HP; GE Healthcare) conjugated with the recombinant PDV2 according to the manufacturer's protocol.
Approximately three-week-old plants grown on MS plates were frozen in liquid nitrogen, ground with pestles, and homogenized in extraction buffer (50 mM Tris, pH 7.5, 2 mM MgCl2, 5 mM EDTA, and a protease inhibitor mixture [P2714; Sigma]). The homogenate was filtered through Miracloth (Calbiochem). Protein concentration of the homogenate was determined and then subjected to immunoblotting. To detect proteins during leaf development, leaves were separately collected in order of size and extracted. Samples containing 50 μg of proteins were subjected to immunoblot analyses.
Immunoblotting assays were performed as previously described (Nakanishi et al., 2009). Anti-PDV2 antibodies, anti-FtsZ2-1 antibodies (Nakanishi et al., 2009), and anti-GFP mouse monoclonal antibody (JL-8; Invitrogen) were diluted 1:20,000, 1:10,000, and 1:1000, respectively. The primary antibody was detected by horseradish peroxidase–conjugated goat anti-mouse or anti-rabbit antibody diluted at 1:20,000. The signal was detected by SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific) and the VersaDoc 5000 imaging system (Bio-Rad). We confirmed that the signal was not saturated by comparison of the images from longer and shorter exposure times.
Analyses of Gene Expression by RT-PCR and GUS Staining
Total RNA of Arabidopsis was extracted from ∼3-week-old plants using an RNeasy mini kit (Qiagen). Total RNA of P. patens was extracted from 8-d-old colonies using the RNeasy mini kit. DNase-treated RNA (1 μg) was reverse-transcribed with oligo(dT) (15 nucleotides) primer, and resulting cDNA was used as template for PCR. Before comparison of expression levels, we confirmed that the amplification was in the linear range by comparing different cycles of amplification. PCR was performed using primer sets as follows: PDV1-rt.F and PDV1-rt.R for Arabidopsis PDV1, CRF2-rt.F and CRF2-rt.R for Arabidopsis CRF2, Pp_PDV2-1-rt.F and Pp_PDV2-1-rt.R for P. patens PDV2-1, Pp_PDV2-2-rt.F and Pp_ PDV2-2-rt.R for P. patens PDV2-2, Pp_PDV2-3-rt.F and Pp_PDV2-3-rt.R for P. patens PDV2-3, Pp_PDV2-4-rt.F and Pp_PDV2-4-rt.R for P. patens PDV2-4, Pp_FtsZ1-1-rt.F and Pp_FtsZ1-1-rt.F for P. patens FtsZ1-1 (GI 168056460), Pp_FtsZ1-2-rt.F and Pp_FtsZ1-2-rt.R for P. patens FtsZ1-2 (GI 168033106), Pp_FtsZ2-1-rt.F and Pp_FtsZ2-1-rt.R for P. patens FtsZ2-1 (GI 168026867), Pp_FtsZ2-2-rt.F and Pp_FtsZ2-2-rt.R for P. patens FtsZ2-2 (GI 168028518), Pp_FtsZ3-rt.F and Pp_FtsZ3-rt.R for P. patens FtsZ3 (GI 168025379), Pp_DRP5B-rt.F and Pp_DRP5B-rt.R for P. patens DRP5B (GI 76880153), and Pp_ARC6-rt.F and Pp_ARC6-rt.R for P. patens ARC6 (GI 168052683). As a control for Arabidopsis, an UBQ1 cDNA was amplified by primers UBQ1-rt.F and UBQ1-rt.R. As a control for P. patens, ACTIN3 cDNA was amplified by primers Pp_ACTIN3-rt.F and Pp_ACTIN3-rt.R.
GUS expression analyses were performed as described (Jefferson et al., 1987) with some modifications. Arabidopsis grown on MS plates were soaked in the GUS assay solution [0.5 mg/mL 5-bromo-4-chloro-3-indolylglucronide, 0.5 mM K3Fe(CN)6, 0.5 mM K4Fe(CN)6, 0.1% (v/v) Triton X-100, and 100 mM Pi-buffered saline] and incubated at 37°C overnight for PDV2-GUS and for 3 h for the others. Then the samples were washed by 70% ethanol and soaked in ethanol and acetic acid mixture (6:1, by volumes) to stop the reaction and remove chlorophylls.
Phylogenetic Analyses
Deduced amino acid sequences of PDV1 and PDV2 homologs encoded by the 13 genes (GI numbers are indicated in Figure 6A) were collected by BLAST searches. The sequences were aligned by Clustal X 2.0 (Larkin et al., 2007) and manually refined, and 144 amino acid residues were used for the phylogenetic analyses. Maximum likelihood trees were constructed using RaxML 7.0.4 (Stamatakis, 2006) with 100 replicates using the WAG matrix of amino acid replacements assuming a proportion of invariant positions and four γ-distributed rates (WAG+I+gamma model). Bayesian inference was performed with the program MrBayes version 3.1.2 (Ronquist and Huelsenbeck, 2003) using the WAG+I+gamma model. For the MrBayes consensus trees, 1,000,000 generations were completed with trees collected every 100 generations.
Accession Numbers
Sequence data from this work can be found in the Arabidopsis Genome Initiative or GenBank databases under the following accession numbers and GI numbers: Arabidopsis PDV1 (At5g53280), Vitis vinifera PDV1 (CA069353/gi:157360164), Populus trichocarpa PDV1 (ABK04742/gi:118485788), Oryza sativa PDV1 (NP_001057719/gi:115468240, NP_001046494/gi:115445429, and NP_001042451/gi:115435386), Arabidopsis PDV2 (At2g16070), V. vinifera PDV2 (CA047676/gi:157340871), O. sativa PDV2 (EAZ02618/gi:125557082), P. patens PDV2 (Pp PDV2-1, XP_001768164/gi:168031310; Pp PDV2-2, XP_001763980/gi:168022907; Pp PDV2-3, XP_001753151/gi:168000895; and Pp PDV2-4, XP_001768164/gi:168053217), Arabidopsis ARC6 (At5g42480), Arabidopsis DRP5B (At3g19720), Arabidopsis FtsZ2-1 (At2g36250), Arabidopsis CRF2 (At4g23750), P. patens ARC6 (XP_001778770), P. patens DRP5B (AB426132), P. patens FtsZ1-1 (XM_001780186), P. patens FtsZ1-2 (XM_001769006), P. patens FtsZ2-1 (XP_001765953), P. patens FtsZ2-2 (XM_001766723), P. patens FtsZ3 (XM_001765160), and P. patens ACTIN3 (AW698983).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Phenotypes of the PDV2 Overexpresser Isolated from Full-Length cDNA Overexpressing (FOX) Lines, the DRP5B Overexpresser, pdv1/PDV1, pdv1/pdv1, and pdv1/pdv1 pdv2/pdv2.
Supplemental Figure 2. Phenotypes of the CRF2 Overexpresser Isolated from FOX Lines.
Supplemental Figure 3. Amino Acid Sequence Alignment of the PDV Family of Proteins.
Supplemental Figure 4. Effect of Cytokinin Treatment on Expression of Pp FtsZ and Pp PDV2 Genes.
Supplemental Table 1. Primers Used in This Study.
Supplementary Material
Acknowledgments
We thank R. Kofuji and M. Hasebe for providing us with pPpMADS2-7113 plasmid. We also thank A. Minoda, A. Nakabachi, and C. Saito for useful discussions, and we thank Y. Ono for technical support. This work was supported by a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (to S.M. and H.N.) and the Sumitomo Foundation (to S.M.).
Author Contributions
K.O. and Y.K. contributed equally to experiments of PDV and cytokinin and wrote the draft of the manuscript. H.N., T.I., M.M., and S.M. designed the screening of Arabidopsis FOX lines, and H.N. and K.S. screened the mutants and identified inserted cDNA by supervision of T.I. and M.M. S.M. supervised the whole project with help from T.M., M.M., and T.I. S.M. and T.M. prepared the final version of the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Shin-ya Miyagishima (smiyagi@riken.jp.).
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boffey, S.A., and Lloyd, D. (1988). Division and Segregation of Organelles. (Cambridge, UK: Cambridge University Press).
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Colletti, K.S., Tattersall, E.A., Pyke, K.A., Froelich, J.E., Stokes, K.D., and Osteryoung, K.W. (2000). A homologue of the bacterial cell division site-determining factor MinD mediates placement of the chloroplast division apparatus. Curr. Biol. 10 507–516. [DOI] [PubMed] [Google Scholar]
- Gao, H., Kadirjan-Kalbach, D., Froehlich, J.E., and Osteryoung, K.W. (2003). ARC5, a cytosolic dynamin-like protein from plants, is part of the chloroplast division machinery. Proc. Natl. Acad. Sci. USA 100 4328–4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn, J.M., Froehlich, J.E., and Osteryoung, K.W. (2008). Arabidopsis ARC6 coordinates the division machineries of the inner and outer chloroplast membranes through interaction with PDV2 in the intermembrane space. Plant Cell 20 2460–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn, J.M., Yang, Y., Vitha, S., Schmitz, A.J., Hemmes, M., Miyagishima, S., and Osteryoung, K.W. (May 29, 2009). PARC6, a novel chloroplast division factor, influences FtsZ assembly and is required for recruitment of PDV1 during chloroplast division in Arabidopsis. Plant J. http://dx.doi.org/10.1111/j.1365-313X.2009.03905.x. [DOI] [PubMed]
- Gould, S.B., Waller, R.F., and McFadden, G.I. (2008). Plastid evolution. Annu. Rev. Plant Biol. 59 491–517. [DOI] [PubMed] [Google Scholar]
- Ichikawa, T., et al. (2006). The FOX hunting system: An alternative gain-of-function gene hunting technique. Plant J. 48 974–985. [DOI] [PubMed] [Google Scholar]
- Itoh, R., Fujiwara, M., Nagata, N., and Yoshida, S. (2001). A chloroplast protein homologous to the eubacterial topological specificity factor minE plays a role in chloroplast division. Plant Physiol. 127 1644–1655. [PMC free article] [PubMed] [Google Scholar]
- Jacqmard, A., Detry, N., Dewitte, W., Van Onckelen, H., and Bernier, G. (2002). In situ localisation of cytokinins in the shoot apical meristem of Sinapis alba at floral transition. Planta 214 970–973. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakimoto, T. (2003). Perception and signal transduction of cytokinins. Annu. Rev. Plant Biol. 54 605–627. [DOI] [PubMed] [Google Scholar]
- Kenrick, P., and Crane, P.R. (1997). The origin and early evolution of plants on land. Nature 389 33–39. [Google Scholar]
- Kieber, J.J. (2002). Cytokinins. In The Arabidopsis Book, C.R. Somerville and E.M. Meyerowitz, eds (Rockville, MD: American Society of Plant Biologists), doi/, http://www.aspb.org/publications/arabidopsis/.
- Kiessling, J., Kruse, S., Rensing, S.A., Harter, K., Decker, E.L., and Reski, R. (2000). Visualization of a cytoskeleton-like FtsZ network in chloroplasts. J. Cell Biol. 151 945–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroiwa, H., Mori, T., Takahara, M., Miyagishima, S., and Kuroiwa, T. (2002). Chloroplast division machinery as revealed by immunofluorescence and electron microscopy. Planta 215 185–190. [DOI] [PubMed] [Google Scholar]
- Kuroiwa, T., Kuroiwa, H., Sakai, A., Takahashi, H., Toda, K., and Itoh, R. (1998). The division apparatus of plastids and mitochondria. Int. Rev. Cytol. 181 1–41. [DOI] [PubMed] [Google Scholar]
- Larkin, M.A., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23 2947–2948. [DOI] [PubMed] [Google Scholar]
- Lopez-Juez, E., and Pyke, K.A. (2005). Plastids unleashed: Their development and their integration in plant development. Int. J. Dev. Biol. 49 557–577. [DOI] [PubMed] [Google Scholar]
- Maple, J., and Moller, S.G. (2007). Plastid division: Evolution, mechanism and complexity. Ann. Bot. (Lond.) 99 565–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maple, J., Vojta, L., Soll, J., and Moller, S.G. (2007). ARC3 is a stromal Z-ring accessory protein essential for plastid division. EMBO Rep. 8 293–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, A., Lang, D., Heckmann, J., Zimmer, A.D., Vervliet-Scheebaum, M., and Reski, R. (2009). A uniquely high number of ftsZ genes in the moss Physcomitrella patens. Plant Biol. http://dx.doi.org/10.1111/j.1438–8677.2008.00174. [DOI] [PubMed]
- Mitsuhara, I., et al. (1996). Efficient promoter cassettes for enhanced expression of foreign genes in dicotyledonous and monocotyledonous plants. Plant Cell Physiol. 37 49–59. [DOI] [PubMed] [Google Scholar]
- Miyagishima, S. (2005). Origin and evolution of the chloroplast division machinery. J. Plant Res. 118 295–306. [DOI] [PubMed] [Google Scholar]
- Miyagishima, S., Froehlich, J.E., and Osteryoung, K.W. (2006). PDV1 and PDV2 mediate recruitment of the dynamin-related protein ARC5 to the plastid division site. Plant Cell 18 2517–2530.16998069 [Google Scholar]
- Miyagishima, S., Kuwayama, H., Urushihara, H., and Nakanishi, H. (2008). Evolutionary linkage between eukaryotic cytokinesis and chloroplast division by dynamin proteins. Proc. Natl. Acad. Sci. USA 105 15202–15207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagishima, S., Nishida, K., Mori, T., Matsuzaki, M., Higashiyama, T., Kuroiwa, H., and Kuroiwa, T. (2003). A plant-specific dynamin-related protein forms a ring at the chloroplast division site. Plant Cell 15 655–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagishima, S., Takahara, M., and Kuroiwa, T. (2001). Novel filaments 5 nm in diameter constitute the cytosolic ring of the plastid division apparatus. Plant Cell 13 707–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori, T., Kuroiwa, H., Takahara, M., Miyagishima, S., and Kuroiwa, T. (2001). Visualization of an FtsZ ring in chloroplasts of Lilium longiflorum leaves. Plant Cell Physiol. 42 555–559. [DOI] [PubMed] [Google Scholar]
- Nakanishi, H., Suzuki, K., Kabeya, Y., and Miyagishima, S. (2009). Plant-specific protein MCD1 determines the site of chloroplast division in concert with bacteria-derived MinD. Curr. Biol. 19 151–156. [DOI] [PubMed] [Google Scholar]
- Nishiyama, T., Hiwatashi, Y., Sakakibara, I., Kato, M., and Hasebe, M. (2000). Tagged mutagenesis and gene-trap in the moss, Physcomitrella patens by shuttle mutagenesis. DNA Res. 7 9–17. [DOI] [PubMed] [Google Scholar]
- Osteryoung, K.W., Stokes, K.D., Rutherford, S.M., Percival, A.L., and Lee, W.Y. (1998). Chloroplast division in higher plants requires members of two functionally divergent gene families with homology to bacterial ftsZ. Plant Cell 10 1991–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osteryoung, K.W., and Vierling, E. (1995). Conserved cell and organelle division. Nature 376 473–474. [DOI] [PubMed] [Google Scholar]
- Possingham, J.V., and Lawrence, M.E. (1983). Controls to plastid division. Int. Rev. Cytol. 84 1–56. [Google Scholar]
- Pyke, K.A. (1999). Plastid division and development. Plant Cell 11 549–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashotte, A.M., Mason, M.G., Hutchison, C.E., Ferreira, F.J., Schaller, G.E., and Kieber, J.J. (2006). A subset of Arabidopsis AP2 transcription factors mediates cytokinin responses in concert with a two-component pathway. Proc. Natl. Acad. Sci. USA 103 11081–11085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynaud, C., Cassier-Chauvat, C., Perennes, C., and Bergounioux, C. (2004). An Arabidopsis homolog of the bacterial cell division inhibitor SulA is involved in plastid division. Plant Cell 16 1801–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reski, R., and Abel, W.O. (1985). Induction of budding on chloronemata and caulonemata of the moss, Physcomitrella patens, using isopentenyladenine. Planta 165 354–358. [DOI] [PubMed] [Google Scholar]
- Reski, R., Wehe, M., Hadeler, B., Marienfeld, J.R., and Abel, W.O. (1991). Cytokinin and light quality interact at the molecular level in the chloroplast-mutant PC22 of the moss Physcomitrella. J. Plant Physiol. 138 236–243. [Google Scholar]
- Reutter, K., Atzorn, R., Hadeler, B., Schmülling, T., and Reski, R. (1998). Expression of the bacterial ipt gene in Physcomitrella rescues mutations in budding and in plastid division. Planta 206 196–203. [Google Scholar]
- Reyes-Prieto, A., Weber, A.P., and Bhattacharya, D. (2007). The origin and establishment of the plastid in algae and plants. Annu. Rev. Genet. 41 147–168. [DOI] [PubMed] [Google Scholar]
- Ronquist, F., and Huelsenbeck, J.P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19 1572–1574. [DOI] [PubMed] [Google Scholar]
- Shimada, H., Koizumi, M., Kuroki, K., Mochizuki, M., Fujimoto, H., Ohta, H., Masuda, T., and Takamiya, K. (2004). ARC3, a chloroplast division factor, is a chimera of prokaryotic FtsZ and part of eukaryotic phosphatidylinositol-4-phosphate 5-kinase. Plant Cell Physiol. 45 960–967. [DOI] [PubMed] [Google Scholar]
- Stamatakis, A. (2006). RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22 2688–2690. [DOI] [PubMed] [Google Scholar]
- Stokes, K.D., McAndrew, R.S., Figueroa, R., Vitha, S., and Osteryoung, K.W. (2000). Chloroplast division and morphology are differentially affected by overexpression of FtsZ1 and FtsZ2 genes in Arabidopsis. Plant Physiol. 124 1668–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strepp, R., Scholz, S., Kruse, S., Speth, V., and Reski, R. (1998). Plant nuclear gene knockout reveals a role in plastid division for the homolog of the bacterial cell division protein FtsZ, an ancestral tubulin. Proc. Natl. Acad. Sci. USA 95 4368–4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, K., Nakanishi, H., Bower, J., Yoder, D.W., Osteryoung, K.W., and Miyagishima, S. (2009). Plastid chaperonin proteins Cpn60 alpha and Cpn60 beta are required for plastid division in Arabidopsis thaliana. BMC Plant Biol. 9 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitha, S., Froehlich, J.E., Koksharova, O., Pyke, K.A., van Erp, H., and Osteryoung, K.W. (2003). ARC6 is a J-domain plastid division protein and an evolutionary descendant of the cyanobacterial cell division protein Ftn2. Plant Cell 15 1918–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitha, S., McAndrew, R.S., and Osteryoung, K.W. (2001). FtsZ ring formation at the chloroplast division site in plants. J. Cell Biol. 153 111–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y., Glynn, J.M., Olson, B.J., Schmitz, A.J., and Osteryoung, K.W. (2008). Plastid division: Across time and space. Curr. Opin. Plant Biol. 11 577–584. [DOI] [PubMed] [Google Scholar]
- Yoon, H.S., Hackett, J.D., and Bhattacharya, D. (2002). A single origin of the peridinin- and fucoxanthin-containing plastids in dinoflagellates through tertiary endosymbiosis. Proc. Natl. Acad. Sci. USA 99 11724–11729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, Y., Kuroiwa, H., Misumi, O., Nishida, K., Yagisawa, F., Fujiwara, T., Nanamiya, H., Kawamura, F., and Kuroiwa, T. (2006). Isolated chloroplast division machinery can actively constrict after stretching. Science 313 1435–1438. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.