Abstract
Aliphatic glucosinolate biosynthesis is highly compartmentalized, requiring import of 2-keto acids or amino acids into chloroplasts for side chain elongation and export of the resulting compounds into the cytosol for conversion into glucosinolate. Aliphatic glucosinolate biosynthesis in Arabidopsis thaliana is regulated by three R2R3-MYB transcription factors, the major player being High Aliphatic Glucosinolate 1 (HAG1/MYB28). Here, we show that BAT5, which belongs to the putative bile acid transporter family, is the only member of this family that is transactivated by HAG1/MYB28, HAG2/MYB76, and HAG3/MYB29. Furthermore, two isopropylmalate isomerases genes, IPMI1 and IPMI2, and the isopropylmalate dehydrogenase gene, IPMDH1, were identified as targets of HAG1/MYB28 and the corresponding proteins localized to plastids, suggesting a role in plastidic chain elongation reactions. The BAT proteins also localized to plastids; however, only mutants defective in BAT5 function contained strongly reduced levels of aliphatic glucosinolates. The bat5 mutant chemotype was rescued by induced overexpression of BAT5. Feeding experiments using 2-keto acids and amino acids of different chain length suggest that BAT5 is a plastidic transporter of (chain-elongated) 2-keto acids. Mechanical stimuli and methyl jasmonate transiently induced BAT5 expression in inflorescences and leaves. Thus, BAT5 was identified as the first transporter component of the aliphatic glucosinolate biosynthetic pathway.
INTRODUCTION
Aliphatic glucosinolates mainly present in the Brassicaceae family, including the model plant Arabidopsis thaliana, comprise primarily Met-derived secondary metabolites (Kliebenstein et al., 2001; Mithen, 2001). These compounds and their breakdown products, for example, isothiocyanates, protect plants against herbivores and pathogens (Giamoustaris and Mithen, 1995; Mari et al., 1996; Manici et al., 2000; Reymond et al., 2004; Chung et al., 2005; Mewis et al., 2005; Brader et al., 2006) and are known to serve as human anticancer agents (Talalay and Fahey, 2001; Traw et al., 2003; Hayes et al., 2008). After Met-derived glucosinoates, indolic and benzyl glucosinolates are the most abundant glucosinolates in Arabidopsis. Although the metabolism of aliphatic glucosinolate has been extensively studied in Arabidopsis, not all genes in the biosynthetic pathway have been characterized yet.
The biosynthesis of Met-derived glucosinolates starts in the cytosol, where Met is transaminated by a recently characterized aminotransferase, BCAT4 (Schuster et al., 2006). The resulting 2-keto acid, 4-methylthio-2-oxobutanoate (MTOB), is thought to be imported into the chloroplast by a yet unidentified transporter, where it reacts with acetyl-CoA in a condensation reaction catalyzed by the methylthiomalate synthases MAM1 to MAM3 to form 2-(2′-methylthio) ethylmalate, a 2-alkylmalic acid (Kroymann et al., 2001; Textor et al., 2004; Textor et al., 2007). The 2-alkylmalic acid is converted to 3-(2′-methylthio) ethylmalate, a 3-alkylmalic acid by a still unidentified isomerase, most probably an isopropylmalate isomerase (IPMI; Schuster et al., 2006). The 3-alkylmalic acid undergoes oxidative decarboxylation to yield 5-methylthio-2-oxopentanoate (MTOP), a 2-keto acid with one additional carbon in the side chain. This reaction is assumed to be catalyzed by a yet unidentified isopropylmalate dehydrogenase (IPMDH). The resulting 2-keto acid can then enter a new condensation cycle, thus creating homoketo acids of increasing side chain length (Grubb and Abel, 2006; Halkier and Gershenzon, 2006). The homoketo acids could be subsequently transaminated to corresponding chain-elongated amino acids, in the case of Met, resulting in homomethionine. These reactions are assumed to be catalyzed by the chloroplastic BCAT3 or the cytosolic BCAT4, which have been recently shown to transaminate short-chained homoketo acids generated in the glucosinolate biosynthetic pathway (Knill et al., 2008). It is therefore expected that either the Met-derived chain-elongated 2-keto acid (if the second transamination step is performed in the cytosol by BCAT4 or a yet unknown BCAT) or the chain-elongated amino acid (if the second transamination is performed by the plastidic BCAT3) must be exported from the chloroplast into the cytosol by so far unknown transporter(s) to be further converted into glucosinolates.
The following cytosolic steps involve oxidation of the chain-elongated amino acid to an aldoximine catalyzed by the cytochrome P450 monooxygenases CYP79F1 and CYP79F2 (Hansen et al., 2001; Reintanz et al., 2001; Chen et al., 2003; Tantikanjana et al., 2004). A second oxidation step leads to an aci-nitro compound, catalyzed by CYP83A1 (Hemm et al., 2003; Naur et al., 2003), which is then conjugated with a sulfur donor into S-alkyl thiohydroximate. The final biosynthetic steps comprise consecutive actions catalyzed by C-S lyases (Mikkelsen et al., 2004), UDP-glucosyltransferases (Grubb et al., 2004), and sulfotransferases (ST5b and ST5c; Piotrowski et al., 2004). Recently, the R2R3-MYB transcription factors HAG1/MYB28, HAG2/MYB76, and HAG3/MYB29 were identified as regulators of Met-derived glucosinolate biosynthesis, with High Aliphatic Glucosinolate 1 (HAG1/MYB28) being the main player (Gigolashvili et al., 2007, 2008; Hirai et al., 2007; Sønderby et al., 2007). Here, we describe the identification of novel target genes of the HAG1/MYB28 transcription factor and provide evidence that BAT5 annotated as a bile acid transporter is involved in the transport of 2-keto acids between chloroplasts and the cytosol.
RESULTS
Identification of Novel HAG1/MYB28 Target Genes Involved in the Met Chain Elongation Pathway
As demonstrated recently, the R2R3-MYB transcription factor HAG1/MYB28 is the key player in the regulation of aliphatic glucosinolate biosynthesis (Gigolashvili et al., 2007; Hirai et al., 2007). To identify novel target genes of HAG1/MYB28, we analyzed publicly available transcriptome gene coexpression profiles for putative interacting factors (Table 1). Coexpressed genes include known genes of the aliphatic glucosinolate biosynthetic pathway, such as BCAT4, MAM1, CYP83A1, CYP79F1, BCAT3, the sulfotransferases ST5b/c, and the C-S lyase, but also putatively involved genes, such as the IPMIs IPMI1 and IPMI2, the isopropropylmalate dehydrogenase IPMDH1, and a gene encoding a putative transporter, BAT5, which has been annotated as a bile acid transporter and belongs to a small gene family comprising five members (BAT1 to BAT5). Several genes identified by this approach were subjected to a real-time PCR analysis using mRNA of wild-type and HAG1/MYB28 overexpression plants to verify putative target genes.
Table 1.
Putative Target Genes of HAG1/MYB28 with Defined or Suggested Functions in GS Metabolism Identified by Gene Coexpression Analysis Using a Publicly Available Microarray Data Set (http://atted.jp/)
| AGI | NAME | References | CORa |
|---|---|---|---|
| At5g61420 | Transcription factor HAG1/MYB28 | Hirai et al. (2007) | |
| Gigolashvili et al. (2007) | |||
| Sønderby et al. (2007) | |||
| At4g13770 | CYP83A1 | Hemm et al. (2003) | 0.80 |
| Naur et al. (2003) | |||
| At3g58990 | IPMI1 | Actual work | 0.78 |
| At4g12030 | Putative bile acid transporter (BAT5) | Actual work | 0.77 |
| At1g74090 | St5b | Piotrowski et al. (2004) | 0.76 |
| At5g23010 | MAM1 | Kroymann et al. (2001) | 0.70 |
| At2g43100 | IPMI2 | Actual work | 0.74 |
| At2g31790 | UDP glucosyltransferare | Grub and Abel. (2006) | 0.72 |
| At3g19710 | BCAT4 | Schuster et al. (2006) | 0.68 |
| At1g16410 | CYP79F1 | Reintanz ezt al. (2001) | 0.67 |
| At1g62650 | Flavin-containing monooxygenase | Hansen et al. (2001) | 0.67 |
| Chen et al. (2003) | |||
| Li et al. (2008) | |||
| At1g31180 | IPMDH | Actual work | 0.66 |
| At1g18590 | St5c | Piotrowski et al. (2004) | 0.61 |
| At1g65860 | Flavin-containing monooxygenase FMO3 | Li et al. (2008) | 0.54 |
| At2g20610 | C-S lyase | Mikkelsen et al. (2004) | 0.53 |
| At3g49680 | BCAT3 | Knill et al. (2008) | 0.53 |
| At4g39940 | Adenosine-5-phosphosulfate-kinase | Mugford et al. (2009) | 0.49 |
Pearson's correlation coefficient.
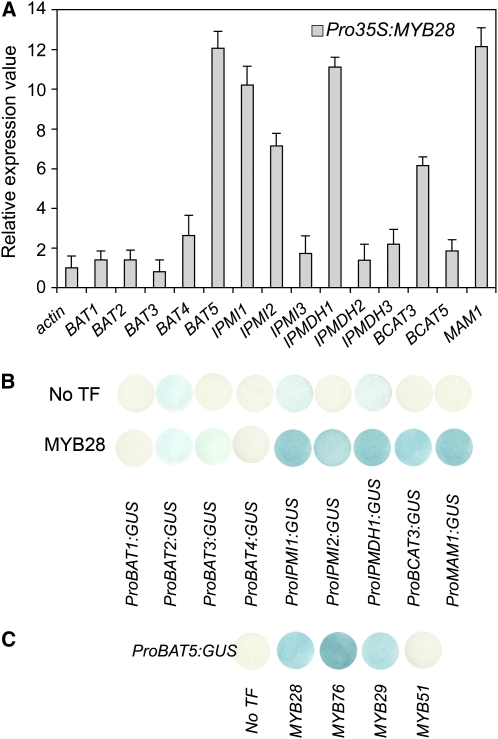
As shown in Figure 1A, the steady state transcript levels of several novel putative glucosinolate biosynthetic genes were significantly increased in the HAG1/MYB28 overexpression lines compared with the wild type. This holds true for the two IPMIs, IPMI1 and IPMI2, the isopropylmalate dehydrogenase IPMDH1, the recently characterized branched-chain aminotransferase BCAT3, and BAT5. The expression of other members of the bile acid transporter family (i.e., BAT1, -2, -3, and -4) and of BCAT5 as well as IPMI3, IPMDH2, and IPMDH3 remained almost unaffected.
Figure 1.
Identification of Novel Genes Involved in Met-Derived Glucosinolate Biosynthesis Using Real-Time PCR Analysis and Cotransformation Assays in Cultured Arabidopsis Cells.
(A) Transcript levels of predicted glucosinolate pathway genes in rosette leaves of 5-week-old HAG1/MYB28 overexpression plants. Relative gene expression values are given compared with the wild type (=1). Means ± sd (n = 3).
(B) Cotransformation assays for the determination of target gene specificity of HAG1/MYB28 (effector) toward target promoters of predicted aliphatic biosynthetic pathway genes are shown. The promoters of BAT1, BAT2, BAT3, BAT4, IPMI1, IPMI2, IPMDH1, and BCAT3 genes were fused to the uidA (GUS) reporter gene (TargetPromoter:GUS vectors). The promoter of the MAM1 gene was used as a positive control. Cultured Arabidopsis cells were inoculated with the supervirulent Agrobacterium strain LBA4404.pBBR1MCS.virGN54D containing either only the reporter construct (TargetPromoter:GUS:pGWB3i) or the reporter construct and the HAG1/MYB28 effector construct (Pro35S:HAG1:pGWB2). GUS staining indicates transactivation of a given promoter by an effector.
(C) Transactivation of ProBAT5:GUS by HAG1/MYB28, HAG2/MYB76, HAG3/MYB29, and HIG1/MYB51.
Furthermore, the ability of HAG1/MYB28 to activate promoters of the identified candidate genes was analyzed in cotransformation assays (Berger et al., 2007). Cultured Arabidopsis Columbia-0 (Col-0) cells were infiltrated with a supervirulent Agrobacterium tumefaciens strain carrying the HAG1/MYB28 construct for effector expression and different reporter constructs containing the uidA (β-glucuronidase [GUS]) gene driven by the promoters of the candidate genes. As shown in Figure 1B, Arabidopsis cells transiently expressing both reporter and effector constructs showed significantly increased GUS activities for IPMI1, IPMI2, IPMDH1, BCAT3, and BAT5, demonstrating the potential of HAG1/MYB28 to transactivate respective promoters of biosynthetic genes as well as that of the putative bile acid transporter BAT5. Finally, it is also evident from Figure 1C that the transactivation capacity of the other two regulators of aliphatic glucosinolate biosynthesis, HAG2/MYB76 and HAG3/MYB29, toward the BAT5 promoter is comparable with that of the main regulator HAG1/MYB28.
BAT5 Is Ubiquitously Expressed and Is Localized to Plastids, as Are IPMI1, IPMI2, and IPMDH1
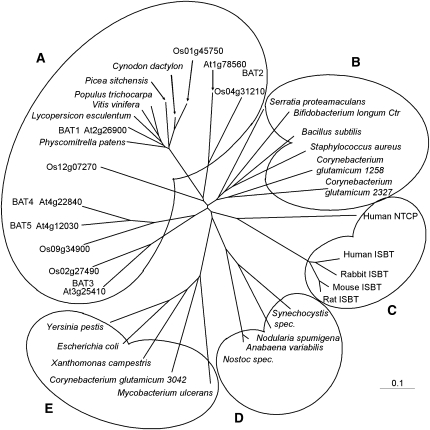
Due to significant sequence similarities to mammalian sodium-coupled bile acid transporters (Hagenbuch et al., 1991; Wong et al., 1994; Trauner and Boyer, 2003), the five BAT proteins were assigned as Arabidopsis bile acid transporters, although their actual function is unknown. All five members of the BAT family in Arabidopsis have eight to nine predicted transmembrane spans (http://aramemnon.botanik.uni-koeln.de/index.ep; Schwacke et al., 2003), indicating that all BAT members presumably function as membrane-integrated transporter proteins. BLAST searches using the Arabidopsis BAT protein sequences revealed that Oryza sativa also possesses five different BAT proteins (Rzewuski and Sauter, 2002) and that a large number of ESTs encoding plant BAT proteins can be found throughout the plant kingdom and in a variety of bacteria (Figure 2; see Supplemental Data Set 1 online). Whereas the only bacterial putative BAT ortholog was recently identified as a cholate transporter named Ctr in Bifidobacterium longum (Price et al., 2006), the function(s) of the plant proteins remains to be elucidated.
Figure 2.
Phylogenetic Relationship Analysis of Bile Acid Transporter(-Like) Proteins.
The corresponding amino acid sequences were aligned with the ClustalX program, and an unrooted tree was calculated using TreeView software. Species and gene names are indicated. Cluster A, plants; B, heterotrophic bacteria group I; C, mammalia; D, cyanobacteria; E, heterotrophic bacteria group II. Ctr, cholate transporter; NTCP, Na+/taurocholate cotransporting peptide; ISBT, ileal sodium-dependent bile acid transporter.
For a detailed analysis of the organ- and tissue-specific expression profile of the BAT5 gene, a translational fusion of the BAT5 promoter containing the first exon with the uidA (GUS) gene was analyzed for GUS activity in several transgenic Arabidopsis lines. As shown in Figure 3, BAT5 is strongly expressed in all organs in young seedlings and in leaves and roots of mature plants. High GUS activity could also be detected in sepals, stamens, and in pollen grains. The GUS expression profiles of the other Arabidopsis BAT genes are given as supplementary material, showing overlapping but distinct expression patterns (see Supplemental Figure 1 online).
Figure 3.
Histochemical GUS Staining in Tissues of ProBAT5:GUS Plants.
(A) A 14-day-old seedling.
(B) Adult leaf with cut site at the petiole.
(C) Flower.
(D) Silique.
(E) Roots of adult plants.
(F) GUS induction at cut site of inflorescence.
Bars = 500 μm in (A) and (F), 1000 μm in (B), and 150 μm in (C) to (E).
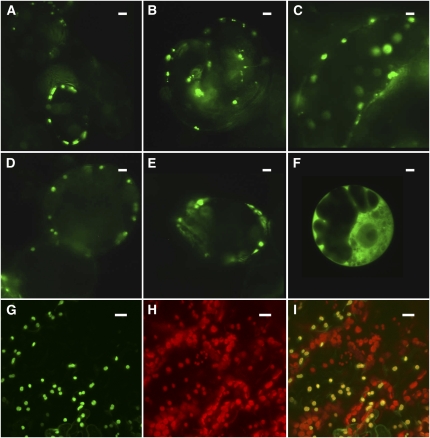
In contrast with the mammalian proteins, the Arabidopsis BATs possess N-terminal extensions of ∼60– to 80–amino acid residues in length that might function to target the proteins to specific cellular compartments. All Arabidopsis BAT proteins were indeed predicted to be localized to plastids and, in some cases, also to mitochondria (http://aramemnon.botanik.uni-koeln.de/index.ep; Schwacke et al., 2003). To assess the subcellular localization of BAT5 along with that of IPMI1, IPMI2, and IPMDH1, which have also been identified as novel target genes of HAG1/MYB28 (Figure 1), the full-length coding sequence of the BAT5 gene (1221 bp) and the N-terminal sequences, including the putative plastidic targeting peptides (as predicted by TargetP) of BAT5 (252 bp), IPM1 (321 bp), IPMI2 (303 bp), and IPMDH1 (288 bp), were translationally fused to green fluorescent protein (GFP). These constructs were introduced into cultured Arabidopsis cells and/or adult leaves of Arabidopsis wild-type plants by infiltration with A. tumefaciens. As demonstrated in Figure 4, GFP fused to N-terminal fragments of BAT5, IPMI1, IPMI2, and IPMDH1, as well as the plastidic triose phosphate/phosphate translocator, which was used as a positive control, all localized to plastids of cultured Arabidopsis cells (Figures 4A to 4F). Likewise, the full-length BAT5-GFP construct localized to chloroplasts when transiently expressed in Arabidopsis leaves (Figures 4G to 4I). Both the GFP fluorescence signal of BAT5-GFP (Figure 4G) and the chlorophyll autofluorescence signal (Figure 4H) colocalized to the same compartments representing chloroplasts (Figure 4I). Notably, the other members of the Arabidopsis BAT family were also shown to be plastid-localized (see Supplemental Figure 2 online).
Figure 4.
BAT5, IPMI1, IPMI2, and IPMDH1 Localize to Plastids.
Plastidic localization of GFP fused to N-terminal fragments of BAT5, IPMI1, IPMI2, and IPMDH1 in cultured Arabidopsis cells ([A] to [F]) and localization of the BAT5 full-length-GFP construct to chloroplasts of Arabidopsis leaves ([G] to [I]). The patterns of fluorescence were analyzed by fluorescence ([A] to [F]) or confocal ([G] to [I]) microscopy.
(A) IPMI1.
(B) IPMI2.
(C) IPMDH1.
(D) BAT5.
(E) Triose phosphate/phosphate translocator (positive control for plastidic localization).
(F) Empty vector pGWB2-GFP (cytosolic localization of GFP).
(G) BAT5-GFP localization to chloroplasts of Arabidopsis leaves.
(H) Autofluorescence of chloroplasts (red).
(I) Overlay for BAT5-GFP and chloroplast autofluorescence (yellow). Bar = 10 μm. For details, see text and Methods.
The bat5 Knockdown Mutant, Cultured Pro35S:amiBAT5 Arabidopsis Cells, and Stably Transformed Pro35S:amiBAT5 Plants Contain Reduced Levels of Aliphatic Glucosinolates
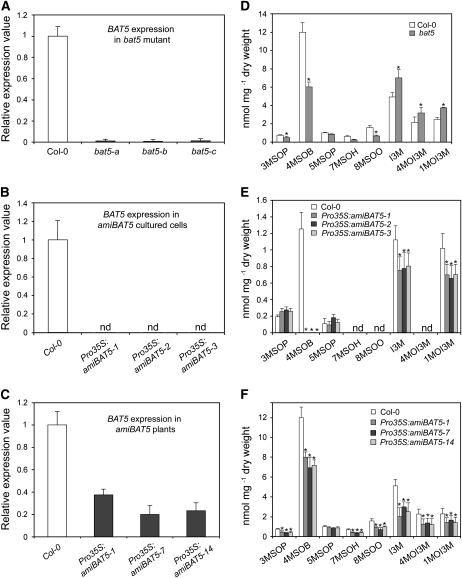
To assess the function of BAT5 in the biosynthesis of aliphatic glucosinolates, two publicly available knockout lines for BAT5, SALK_126525 and SALK_126515, were analyzed at the genomic level. The SALK 126515 line turned out not to contain any T-DNA insertion. On the other hand, a T-DNA insertion in the BAT5 gene within the SALK_126525 line could be confirmed, and a homozygous loss-of-function allele of the BAT5 gene (bat5) could be isolated. Analysis of the bat5 mutant genomic DNA by PCR with gene-specific and T-DNA border-specific primers indicated that the T-DNA insertion was not in the first intron as annotated by SALK, but 180 bp upstream of the start codon of the BAT5 gene (At4g12030.2). However, even this insertion led to drastically reduced steady state levels of BAT5 mRNA in different mutant plants of the same line (bat5-a, -b, and -c), as revealed by real-time PCR analysis (Figure 5A). The bat5 mutant lines showed no visible effects on plant growth and morphology under the given growth conditions.
Figure 5.
Analyses of the bat5 Mutant, Cultured Pro35S:amiBAT5 Arabidopsis Cells, and Stably Transformed Pro35S:amiBAT5 Plants.
(A) to (C) Steady state level of BAT5 mRNA in three different plants of the bat5 mutant (A), cultured Pro35S:amiBAT5 Arabidopsis cells (B), and stably transformed Pro35S:amiBAT5 plants (C). Relative gene expression values are given in comparison with the wild-type (Col-0) plants or wild-type cells (wild type = 1). Means ± sd (n = 3). nd, not detectable.
(D) Contents of GSs (nmol/mg dry weight) in bat5 mutant plants in comparison with the wild type (Col-0). Means ± sd (n = 5).
(E) Contents of GSs (nmol/mg dry weight) in Pro35S:amiBAT5 cultured Arabidopsis cell lines in comparison with wild-type cells. The levels of long-chained aliphatic GSs were below the detection limit in both wild-type cells extracts and in Pro35S:amiBAT5 cells. Means ± sd (n = 3).
(F) Contents of GSs (nmol/mg dry weight) in Pro35S:amiBAT5 stably transformed Arabidopsis plants in comparison with cultured wild-type cells. Means ± sd (n = 5). Asterisk indicates significant difference in comparison to the wild-type Col-0 (Student's t test, P < 0.05).
For metabolite analysis, glucosinolates (GSs) were extracted from freeze-dried rosette leaves of 5-week-old bat5 plants, and the levels of 3-methylsulfinylpropyl-GS (3MSOP), 4-methylsulfinylbutyl-GS (4MSOB), 5-methylsulfinylpentyl-GS (5MSOP), 7-methylsulfinylheptyl-GS (7MSOH), 8-methylsulfinyloctyl-GS (8MSOO), indol-3-ylmethyl-GS (I3M), 4-methoxyindol-3-ylmethyl-GS (4MOI3M), and 1-methoxyindol-3-ylmethyl-GS (1MOI3M) were determined. As shown in Figure 5D, bat5 contained significantly reduced levels of the aliphatic GSs 3MSOB, 4MSOB, 7MSOH, and 8MSOO compared with the wild type. The content of total Met-derived GSs was reduced from 16 nmol/mg dry weight in the wild type to 8 nmol/mg dry weight in the bat5 mutant, corresponding to a 50% reduction, mainly due to the reduced content of the most abundant Met-derived GS 4MSOB. 4MSOB was reduced from 12 nmol/mg dry weight in the wild type to 6 nmol/mg dry weight in bat5 (Figure 5D). By contrast, the content of indolic GSs, I3M, 4MOI3M, and 1MOI3M, was slightly increased in the bat5 mutant, most likely as a result of the negative feedback regulation of Trp- and Met-derived GS biosynthetic pathways (Grubb and Abel, 2006; Gigolashvili et al., 2008).
As no other BAT5 loss-of-function mutant was available for analysis, artificial BAT5 microRNA-overexpressing lines were established (Schwab et al., 2006). Synthetic microRNA for the BAT5 gene (amiBAT5 5′-GCGGUCCUCGUUACACCUAUG-3′) was designed at http://wmd.weigelworld.org/cgi-bin/mirnatools.pl using the WMD version 1 software, and a transgenic Arabidopsis cell culture and stably transformed Arabidopsis plants were generated. In each case, the residual BAT5 transcript levels (Figures 5B and 5C) and the GS content (Figures 5E and 5F) were measured in three representative lines. As shown in Figure 5B, the BAT5 transcripts were below the detection limit in cultured cells (lines Pro35S:amiBAT5-1, -2, and -3) 5 to 7 d after transfection, and the content of the main aliphatic GS, 4MSOB, could not be detected at all in these lines. The stably transformed transgenic lines carrying the Pro35S:amiBAT5 construct (Pro35S:amiBAT5-1, -7, and -14) showed a 60 to 80% decreased steady state level of the BAT5 mRNA (Figure 5C), and the short-chain and long-chain aliphatic GS contents were 35 to 40% those of wild-type levels (Figure 5F). Remarkably, both the Pro35S:amiBAT5 cell lines and the stably transformed artificial micro RNA (ami) plants contained slightly decreased levels of indolic GSs, which was opposite to the bat5 mutant that showed a slight upregulation of indolic GSs (Figure 5D).
The downregulation of indolic GSs in Pro35S:amiBAT5 lines does not seem to be related to the loss of the BAT5 gene, but to side effects of the amiRNA construct that inhibit the biosynthesis of indolic GSs. This suggestion is supported by the observation that stably transformed Pro35S:amiBAT5 lines but not the bat5 mutant contained decreased steady state transcript levels of MYB34, one of the regulators of indolic GS biosynthesis (see Supplemental Figure 3 online). Changes in the transcript levels of other regulators of indolic (i.e., High Indolic Glucosinolate 1 [HIG1/MYB51] and HIG2/MYB122) and aliphatic GS biosynthesis (i.e., HAG1/MYB28, HAG3/MYB29, and HAG2/MYB76) were comparable in both Pro35S:amiBAT5 lines and the bat5 mutant (see Supplemental Figure 3 online). The obtained data are in accordance with the recent finding that MYB34 plays a special role in balancing metabolic networks between aliphatic and indolic GSs (Malitsky et al., 2008). Taken together, both the analysis of the bat5 knockdown mutant and of Pro35S:amiBAT5 lines showed reduced levels of Met-derived GSs compared with the wild type, indicating a role for BAT5 in the transport of chain-elongated Met derivatives between the plastids and the cytosol.
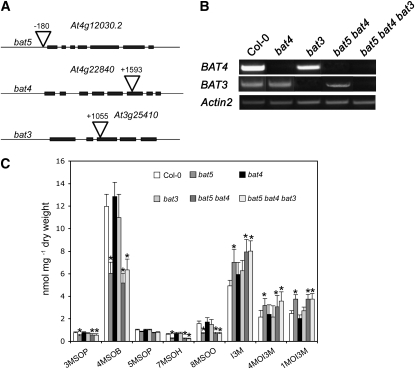
Double bat5 bat4 and Triple bat5 bat4 bat3 Mutants Contain GS Levels Comparable to Those of the bat5 Mutant
Although the overexpression of HAG1/MYB28 lead neither to an increase in the steady state mRNA levels of other BAT genes (Figure 1A) nor to the transactivation of these genes (Figure 1B), some functional redundancy within the BAT family cannot be excluded. To analyze whether or not the closest homologs of BAT5 (i.e., BAT4 and BAT3; Figure 2), are involved in the biosynthesis of GSs, homozygous knockout lines for BAT4 (SALK_044369) and BAT3 (Gabi-Kat479D02) were isolated, and double bat5 bat4 and triple bat5 bat4 bat3 homozygous knockout mutants were generated (Figures 6A and 6B). Homozygous double and triple knockout lines were confirmed by PCR using genomic DNA of mutants, and the loss of transcripts was confirmed by the analysis of mRNA levels of the corresponding genes by RT-PCR (Figure 6B). Single bat4 and bat3 T-DNA insertion lines as well as double bat5 bat4 and triple bat5 bat4 bat3 mutants did not show any phenotypic differences compared with Col-0. As shown in Figure 6C, bat4 and bat3 single mutants contained wild-type levels of GSs, whereas bat5 bat4 and bat5 bat4 bat3 showed GS profiles resembling those of the bat5 single mutant. Together, these results demonstrate that BAT5 is the only member of the BAT family that is activated by MYB transcription factors and is involved in the biosynthesis of aliphatic GSs.
Figure 6.
BAT4 and BAT3 Are Not Involved in Aliphatic GS Biosynthesis.
(A) Positions of T-DNA insertions in BAT5, BAT4, and BAT3 genes.
(B) Conformation of the knockout status of plants homozygous for the T-DNA insertion by RT-PCR. The cDNAs from mutant plants were amplified using primers specific to the coding regions of the corresponding BAT genes. The ACTIN2 gene was used as an endogenous control.
(C) Contents of GSs (nmol/mg dry weight) in bat4, bat3, bat5 bat4, and bat5 bat4 bat3 mutant plants in comparison with bat5 and wild-type plants (Col-0). Means ± sd (n = 5). Asterisk indicates significant difference in comparison to the wild-type Col-0 (Student's t test, P < 0.05).
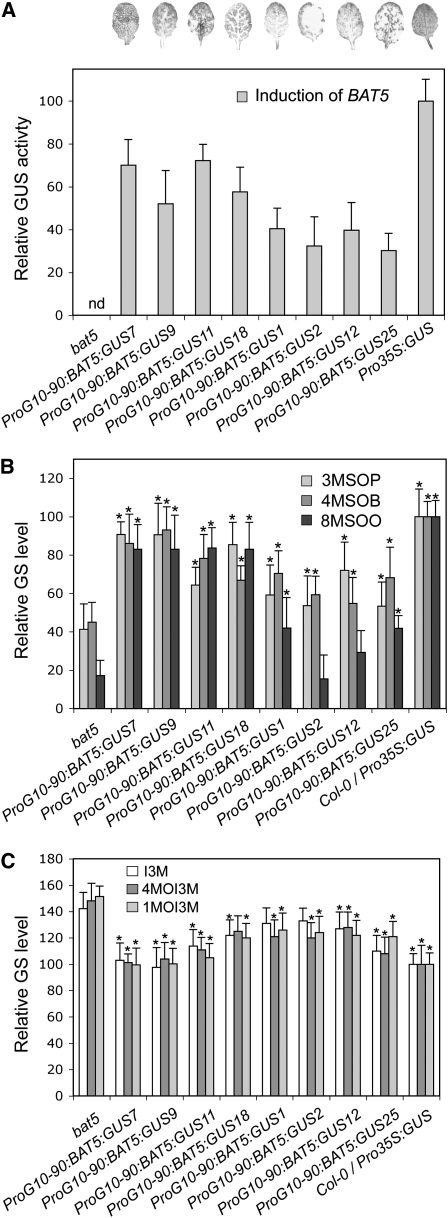
Induced Expression of BAT5 Leads to an Increased Production of Aliphatic GSs and Complementation of the bat5 Mutant Chemotype
To restore BAT5 activity in the bat5 mutant, a gene complementation experiment was performed. Because constitutive overexpression of genes often leads to pleiotropic effects, BAT5 expression in the bat5 background was controlled by the estrogen receptor-based inducible system. The binary gateway vector used for plant transformation harbored the BAT5-uidA gene within recombination attR sites driven by the XVE-inducible expression system and the constitutive G10-90 promoter. This estradiol-inducible vector is identical to pER8 (Zuo et al., 2000), except that the multiple cloning site also contains recombination sites for Gateway cloning. The chemical induction followed by GUS staining of transformed plants thus allowed us to estimate the efficiency of BAT5 induction. After treating plants with β-estradiol, leaves of 35 independent transgenic lines were tested for GUS activity, and 15 lines showing moderate to high uidA expression levels were selected for chemotype analysis. The results for several representative lines (complemented bat5/BAT5 lines) are shown in Figure 7.
Figure 7.
Gene Complementation Analysis of bat5 Mutant Plants Using an Inducible Promoter BAT5 Construct Fused to the GUS Gene.
Relative GUS activity values (A) and levels of aliphatic (B) and indolic (C) GSs after induction with β-estradiol are shown. Wild-type levels were set to 100%. Asterisk indicates significant difference (Student's t test, P < 0.05) in complemented lines compared with the bat5 mutant.
Generally, the level of GUS expression (Figure 7A) correlated well with the levels of aliphatic GSs (Figure 7B) in complemented bat5 lines. The content of total GSs increased from 50% in the bat5 mutant to almost wild-type levels in complemented bat5/BAT5 lines with moderate or high GUS expression levels. As evident from Figure 7B, the majority of the complemented lines contained increased levels of both short- and long-chain Met-derived GSs, including of the most abundant 4MSOB. Remarkably, the slightly increased levels of indolic GSs in the bat5 mutant were also restored to wild-type levels in the complemented bat5/BAT5 lines (Figure 7C).
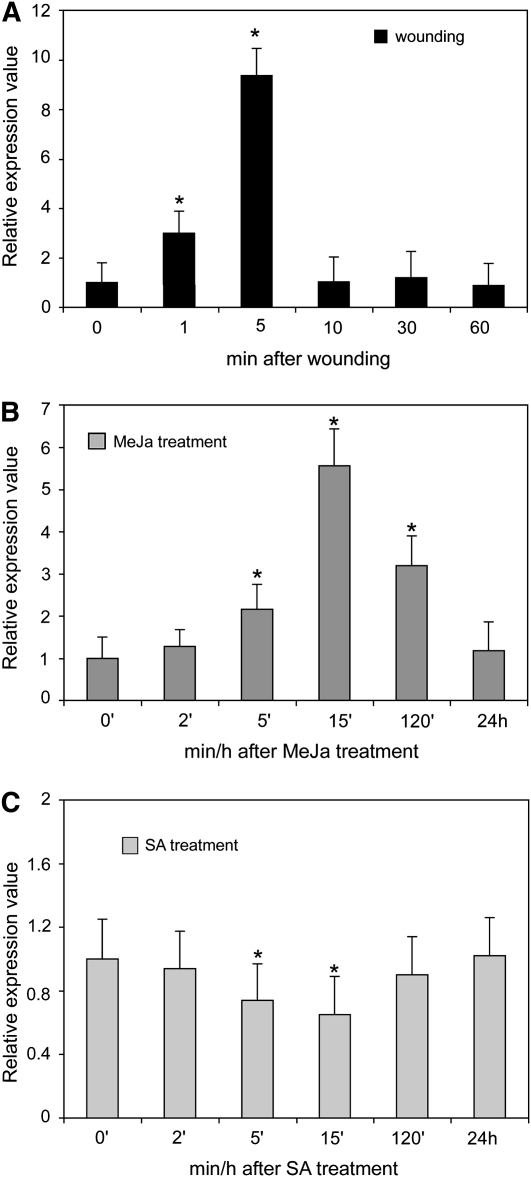
Transcription of BAT5 Is Triggered by Wounding and Induced by Methyl Jasmonate
Transcription of GS biosynthetic genes and of the regulators of GS biosynthesis has been shown to be induced by wounding (Schuster et al., 2006; Gigolashvili et al., 2007a, 2007b, 2008). To analyze whether BAT5 promoter activity is indeed induced by wounding, leaves and inflorescences of ProBAT5:GUS plants were examined at cut sites for GUS activity. As shown in Figures 3B and 3F, GUS staining was increased at the cut surface of the petiole and inflorescence close to the wounding sites.
To get more information about the time dependence of the BAT5 response, a real-time RT-PCR analysis of wounded wild-type inflorescences was performed. Figure 8A shows that the BAT5 expression level increased by ∼10-fold within 5 min of wounding and started to drop to the initial level after 10 min.
Figure 8.
Induction of BAT5 Expression by Wounding, MeJA, and SA.
(A) Wounding experiments. The plant material (inflorescences) were punctured and, after times indicated, used for real-time RT-PCR experiments (means ± sd, n = 3). Relative expression values are given compared with nonwounded tissue (=1). For details, see Methods.
(B) and (C) Hormone experiments. Three-week-old seedlings were exposed in aqueous solutions to 10 μM MeJA (B) or SA (C). Samples were taken after 0, 2, 5, 10, 15, and 120 min and 24 h (means ± sd, n = 3). Relative gene expression values are shown compared with noninduced plants (noninduced = 1). Asterisk indicates significant difference (Student's t test; P < 0.05) in comparison to the noninduced tissue.
Since GS biosynthesis is known to be induced by methyl jasmonate (MeJa) and salicylic acid (SA) treatment (Brader et al., 2001; Mikkelsen and Halkier, 2003; Cipollini et al., 2004; Devoto and Turner, 2005; Mewis et al., 2005; Sasaki-Sekimoto et al., 2005), and as HAG3/MYB29, an important regulator of Met-derived GS biosynthesis, is induced by MeJa (Gigolashvili et al., 2008), we asked whether or not BAT5 is part of these signaling pathways. Therefore, wild-type plants were treated with MeJa and SA, and the response of BAT5 transcription was monitored using real-time RT-PCR. As shown in Figure 8B, an induction of the BAT5 transcript could be observed within 5 to 120 min of application of MeJA, and levels returned to basic levels within 24 h. Conversely, treatment with SA caused an opposite effect and led to decreased steady state BAT5 mRNA levels within 5 and 15 min (Figure 8C).
MTOB Transport Experiments
We have intensely tried to directly assess the transport characteristics of BAT5 by measuring the transport of [35S]-labeled MTOB (prepared as described in Ogier et al., 1993) into chloroplasts from wild-type and bat5 mutant plants. Arabidopsis chloroplasts were isolated according to Kunst et al. (1988) and used for transport studies in silicone oil filtering centrifugation experiments (Fliege et al., 1978). BAT5 was also expressed in various heterologous and homologous systems, namely, in Escherichia coli, yeast cells, and plant leaves. Membranes isolated from these systems and/or the recombinant transporter purified using the His-Tag system were reconstituted into artificial membranes prepared from acetone-washed soybean (Glycine max) phospholipids to assess transport characteristics (Loddenkötter et al., 1993). Unfortunately, all these experiments failed, most probably due to the hydrophobicity of the 2-keto acids, which leads to unspecific membrane binding and permeation, thus covering specific transport events. It therefore appears generally not feasible to directly measure transport of these substrates.
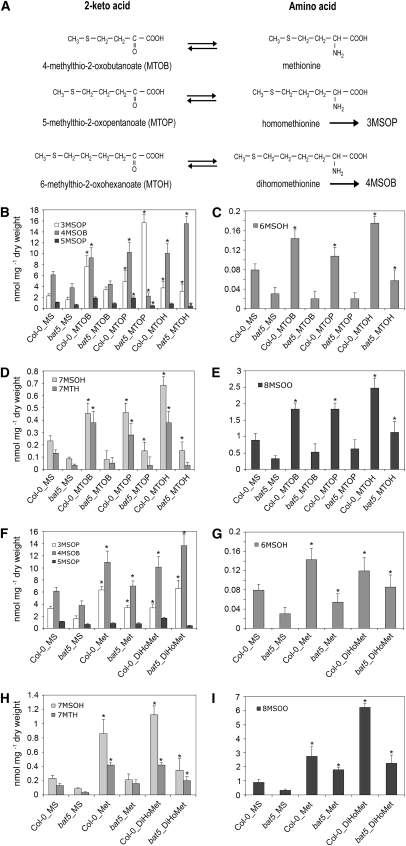
Feeding of bat5 Mutant Plants with Various Met-Derived Intermediates Indicates That BAT5 Is a Chloroplastidic Transporter of Short- and Long-Chain 2-Keto Acids
To gain information on the transport characteristics of BAT5, feeding experiments were performed with bat5 and Col-0 wild-type plants using Met-derived 2-keto acids and amino acids of different chain length. The starting molecule for the biosynthesis of Met-derived GSs in the cytosol is the transamination product of Met, MTOB, which has to be imported into chloroplasts. BAT5 could thus function as an importer of MTOB, and we therefore first performed feeding experiments with both MTOB and Met. On the other hand, 2-keto acids, which are chain-elongated in chloroplasts by MAM1 or MAM3, could be further transaminated either in the chloroplasts by the plastidic BCAT3 (Knill et al., 2008) or, following export of the corresponding 2-keto acid, in the the cytosol by BCAT4 or yet unknown BCATs (Schuster et al., 2006; Knill et al., 2008) before being converted to aliphatic GSs. However, chain-elongated 2-keto acids or amino acids could also reenter the chloroplasts for additional side chain elongation reactions. We therefore also fed bat5 and wild-type plants with the chain-elongated 2-keto acid MTOP, which is converted to the C3 GS 3MSOP) and, in addition, with 6-methylthio-2-oxohexanoate (MTOH) and the chain-elongated amino acid dihomomethionine, which both give rise to the C4 GS, 4MSOB (Figure 9A).
Figure 9.
Metabolite Complementation Analysis of the bat5 Mutant.
(A) Met-derived amino acids and the corresponding 2-keto acids. Chain-elongation of 2-keto acids proceeds in plastids, and the transamination reactions are catalyzed by BCATs. Homomethionine/MTOP and dihomomethionine/MTOH are converted to the Met-derived GSs 3MSOP and 4MSOB, respectively.
(B) to (I) Two-week-old seedlings were fed with 2-keto acids or amino acids on MS plates. Feeding was performed with MTOB, MTOP, and MTOH ([B] to [E]) and Met and dihomomethionine (DiHoMet) ([F] to [I]). Means ± sd (n = 5). Contents of short- and long-chained aliphatic GSs (3MSOP, 4MSOB, 5MSOP, 6MSOH, 7MSOH, 7MTH, and 8MSOO) are given in nmol/mg dry weight in comparison with Col-0. Asterisk indicates significant difference in comparison to the wild type or bat5, respectively (Student's t test, P < 0.05). Plants were germinated on half-strength Murashige and Skoog (MS) plates with agar, and 2-week-old wild-type and bat5 seedlings were transferred to media supplemented with 0.2 mM MTOB, MTOP, MTOH, Met, or dihomomethionine, respectively, for 14 additional days, followed by analyses of aliphatic GSs.
As shown in Figures 9B to 9E, feeding with all three 2-keto acids (MTOB, MTOP, and MTOH) caused a drastic increase in the content of both short- and long-chain aliphatic GSs in wild-type plants. These increases can be attributed to the successful incorporation of the fed substances into the GS structure, which would imply entry of the 2-keto acids into plastids, several (maximum of six) side chain elongation reactions, and exit from the chloroplasts as chain-elongated 2-keto acids or amino acids. Remarkably, and in contrast with wild-type plants, the content of both short- and long-chain aliphatic GSs remained almost unaltered in the bat5 mutant after feeding with MTOB. This observation indicates that bat5 is impaired in the transport of MTOB into chloroplasts.
As also shown in Figures 9B to 9E, feeding of wild-type plants with MTOP and MTOH resulted in elevated levels of all measured aliphatic GSs. By contrast, the bat5 mutant showed generally only a significant increase in the GS that is a direct derivative of the corresponding 2-keto acid. For example, feeding of the bat5 mutant with MTOP resulted in an approximately eightfold increased level of 3MSOP and feeding with MTOH in five times higher levels of 4MSOB compared with bat5 (Figures 9B to 9E). The increased synthesis of 3MSOP and 4MSOB in the bat5 mutant is due to the fact that these aliphatic GSs can be directly synthesized in the cytosol from the corresponding 2-keto acids MTOP and MTOH (i.e., they can be transaminated by cytosolic BCATs and thus bypass transport into plastids and subsequent side chain elongation reactions). The slight increase in the levels of some long-chain aliphatic GSs in response to MTOP and MTOH feeding could be explained by the remaining BAT5 transcript present in bat5 plants (Figure 5A) that could support a residual uptake of 2-keto acids into plastids. Alternatively, because the uptake of 2-keto acids is impaired due to a defect in BAT5, they could be transaminated in the cytosol by BCATs into the corresponding amino acids, which are then transported into the plastids via a yet unknown amino acid transporter and deaminated by BCAT3 before entering the side chain elongation cycle.
Finally, feeding with Met also led to elevated levels of short- and long-chain aliphatic GSs in wild-type plants, but not in the bat5 mutant, thus nicely corroborating observations obtained by feeding with 2-keto acids. Additionally, feeding with the chain-elongated amino acid dihomomethionine, the transamination product of MTOH, which could directly and without additional transamination be directed into the cytosolic aliphatic GS biosynthetic pathway, also led to threefold elevated levels of 4MSOB in the bat5 mutant (Figures 9F to 9I). The level of 4MSOB in bat5 was even 25% higher than in the wild type, as all dihomomethionine taken up by bat5 could only be converted into 4MSOB but to a lesser extent into longer-chained GSs. Surprisingly, not only 4MSOB, but also the 3MSOP level was moderately increased in the bat5 mutant, which could probably be explained by a metabolic jam in the side chain elongation reaction of endogenously produced MTOP and homomethionine. If the long-chain amino acid dihomomethionine or the 2-keto acid MTOH is supplied externally, endogenously generated MTOP could not be further metabolized into long-chain 2-keto acids.
It should be noted that the feeding experiments had no significant effect on the production of indolic GSs (see Supplemental Figure 4 online). However, in all feeding experiments, the production of aliphatic GSs was significantly higher in wild-type plants than in bat5 (Figure 9). In summary, we conclude that the bat5 mutant is defective in the transport of 2-keto acids across the chloroplast envelope membrane.
DISCUSSION
Chloroplastidic IMPI1, IMPI2, IPMDH1, and BAT5 Are Involved in the Met Side Chain Elongation of GSs
The biosynthetic pathway leading to Met-derived aliphatic GSs is highly compartmentalized and requires the participation of plastids for chain elongation reactions of corresponding 2-keto acids. These plastidic reactions include the condensation of the transaminated amino acid with acetyl-CoA, catalyzed by MAM enzymes, the isomerization of the resulting 2-alkylmalic acid to yield a 3-alkylmalic acid, and the subsequent oxidative decarboxylation to a chain-elongated 2-keto acid (Figure 10). Here, we show that the IPMI1 and IPMI2 isomerases and IPMDH1, which catalyzes the oxidative decarboxylation step, are plastid localized and are targets of HAG1/MYB28, the key regulator of the aliphatic GS biosynthetic pathway (Figures 1 and 4). BAT5, a member of the putative bile acid transporter family (Figures 1 and 4), is also a target of HAG1/MYB28. These findings indicate that IPMI1, IPMI2, IPMDH1, and BAT5 are involved in the biosynthesis of aliphatic GSs and that the final product of MAM-mediated condensation, IPMI1/2-mediated isomerization, and IPMDH1-mediated oxidative decarboxylation are exported from the chloroplasts. The observation that IPMI3, IPMDH2, and IPMDH3 as well as the other members of the BAT family (BAT1 to BAT4) are not regulated by HAG1/MYB28 suggests that these paralogs (http://aramemnon.botanik.uni-koeln.de/) are components of alternative pathways.
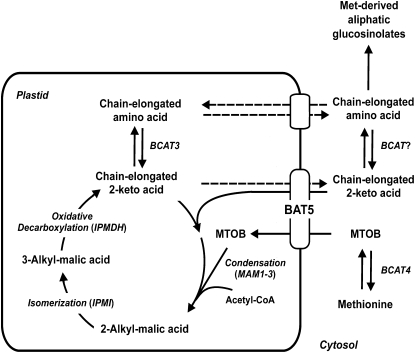
Figure 10.
Schematic Representation of the Role of BAT5 in the Transport of 2-Keto Acids, Side Chain Elongation of 2-Keto Acids, and Biosynthesis of Met-Derived GSs.
BAT5 mediates the transport of MTOB and of chain-elongated 2-keto acids. Dotted lines indicate proposed functions yet lacking experimental evidence. For details, see text.
The chain-elongated 2-keto acids are further transaminated to the corresponding amino acids. It has recently been demonstrated that plastid-located BCAT3 converts MTOP and MTOH into the corresponding amino acids homomethionine and dihomomethionine (Knill et al., 2008). As shown in Figure 1, plastidic BCAT3 and cytosolic BCAT4 are targets of HAG1/MYB28. It is therefore conceivable that the 2-keto acids could be transaminated by BCATs before being imported into the plastids, as well as after being exported into the cytosol for further conversion into aliphatic GSs (Figure 10).
Generation of Met-Derived GSs Is Impaired in the bat5 Mutant and Pro35S:amiBAT5 Lines
In the bat5 mutant, the level of total aliphatic GSs was not completely abolished (Figure 5B) but reduced to 50% of the wild-type level. The remaining BAT5 transcript (Figure 5A) seems to maintain a certain level of side chain elongation and GS biosynthesis. A conceivable functional redundancy by other members of the BAT family (i.e., BAT3 and BAT4) could be excluded (Figure 6). Thus, BAT5 is the main player involved in side chain elongation reactions of Met/MTOB in chloroplasts. In addition, cultured Pro35S:amiBAT5 cells contained 75 to 80% reduced levels of aliphatic GSs and completely abolished levels of the main aliphatic GS 4MSOB (Figure 5C). The complete absence of 4MSOB in Pro35S:amiBAT5 cultured root cells (Figure 5E), which show a lower rate of GS biosynthesis compared with leaves (Brown et al., 2003), further supports the assumption that BAT5 is the only BAT family member involved in the biosynthesis of Met-derived GSs in Arabidopsis. In addition, stably transformed Pro35S:amiBAT5 lines (Pro35S:amiBAT5-1, -7, and -14) also contained reduced levels of aliphatic GSs (Figure 5D), thus corroborating the assumption that BAT5 functions as a transporter of GS biosynthesis intermediates.
Interestingly, the bat5 mutant does not only contain reduced levels of aliphatic GSs, but also slightly increased levels of indolic GSs (Figure 5B). This observation could be explained by the previously reported reciprocal negative feedback regulation of the two branches of aliphatic and indolic GS biosynthesis (Grubb and Abel, 2006; Gigolashvili et al., 2008). Remarkably, the increased level of indolic GSs in the bat5 mutant could be reverted to wild-type levels in complementation experiments using the inducible vector system (Figure 7C). Interestingly, both Pro35S:amiBAT5 transient cultured lines and stably transformed plants contained not only decreased levels of aliphatic, but also of indolic GSs. This observation could be a side effect of the amiRNA construct and suggests that the artificial BAT5 amiRNA does not only downregulate the expression of BAT5 but also initiates the repression of indolic GS biosynthesis, most probably by the negative regulation of MYB34 in BAT5 amiRNA plants but not in bat5 plants (see Supplemental Figure 3 online). MYB34 was shown to be coregulated with HAG1/MYB28 (Malitsky et al., 2008), which is also repressed in Pro35S:amiBAT5 and bat5 lines.
Overall, the data indicate that BAT5 is a key component in the biosynthesis of Met-derived GSs and is essential for the generation of aliphatic GSs.
BAT5 Is Active at the Site of Aliphatic GS Biosynthesis and Is Induced by Wounding and MeJa
Further evidence for BAT5 being involved in aliphatic GS biosynthesis is provided by the tissue-specific expression pattern of BAT5, which overlaps with those of aliphatic GS biosynthetic genes (Figure 3; CYP79F1 and CYP79F2; Reintanz et al., 2001; BCAT4, Schuster et al., 2006) and aliphatic GS regulators (Gigolashvili et al., 2007, 2008). BAT5 expression was present not only in the mesophyll of leaves and the vasculature, in generative plant organs like inflorescences, flowers, and siliques, but also in nonphotosynthetic organs like roots (i.e., at sites where aliphatic GSs are found) (Brown et al., 2003). Environmental stimuli, such as wounding, which are known to have an impact on GS biosynthesis also affected BAT5 expression (Figure 8). Consistent with the wound-induced mRNA accumulation of aliphatic GS regulators (HAG1/MYB28, HAG3/MYB29, and HAG2/MYB76), BAT5 transcript levels increased up to ninefold 5 min after wounding (Figure 8A). Several GS biosynthetic pathway genes like MAM1, BCAT4, BCAT3, and CYP79F1 were also shown to be induced upon mechanical stimuli (Schuster et al., 2006; Gigolashvili et al., 2007; Knill et al., 2008). Furthermore, we demonstrated that BAT5 expression is induced in response to MeJa (Figure 8B) and repressed after SA treatment (Figure 8C), consistent with the increased accumulation of aliphatic GSs (5MSOP and 8MSOO) and the observation that the aliphatic GS regulator, HAG3/MYB29, is upregulated in response to MeJa (Hirai et al., 2007; Gigolashvili et al., 2008) and repressed by SA treatment (Gigolashvili et al., 2008). Thus, BAT5 is a target of the main regulators of aliphatic GS biosynthesis (Figure 1) and appears to integrate the different environmental cues, as has been shown for HAG1/MYB28, HAG2/MYB76, and HAG3/MYB29.
BAT5 Is Involved in the Transport of MTOB and of Chain-Elongated 2-Keto Acids
Besides the observations that BAT5 expression closely correlates with that of other Met-derived GS biosynthetic genes, such as MAM3, CYP79F1, CYP79F2, BCAT4, BCAT3, and St5b, and that BAT5 is a target for three main aliphatic GS regulators HAG1/MYB28, HAG3/MYB29, and HAG2/MYB76 (Table 1, Figure 1), several lines of evidence indicate a role for the plastidic BAT5 in the chain-elongation pathway of Met-derived GS biosynthesis and, particularly, in the transport of short- and long-chain 2-keto acids (Figure 10). First, the reduced levels of aliphatic GSs in the bat5 mutant could not be rescued by feeding with MTOB or Met. By contrast, external MTOB or Met supply to wild-type plants caused a considerable increase in the accumulation of Met-derived GSs (Figures 9B to 9E). The observation that MTOB and Met feeding did not rescue the bat5 mutant chemotype indicates that the import of Met via an amino acid transporter or of MTOB, the BCAT4-mediated transamination product, via a 2-keto acid transporter is impaired in bat5. Assuming that BAT5 represents an amino acid transporter, MTOB feeding should result in wild-type levels of GSs, since MTOB could be taken up, chain-elongated, and exported as chain-elongated 2-keto acid for further conversion into GSs. This is, however, not the case. On the other hand, if BAT5 represents a 2-keto acid transporter affected in bat5, the fed MTOB could be transaminated to Met and imported into chloroplasts for transamination and chain elongation reactions resulting also in aliphatic GS levels similar to those in the wild type. This is also not the case, strongly suggesting that (1) Met is generally only poorly taken up by plastids for GS biosynthesis and (2) that BAT5 most probably represents a transporter for MTOB and other 2-keto acids.
Likewise, because Met, if not transaminated into MTOB, represents only a poor substrate for GS biosynthesis, it could, however, in Met-feeding experiments be transaminated to MTOB and subsequently be taken up and exported as chain-elongated 2-keto acid, thereby resulting in increased accumulation of aliphatic GSs. This actually also does not hold true in bat5, again indicating that BAT5 represents a transporter for MTOB and chain-elongated 2-keto acids. The slight overall increase in aliphatic GSs in the bat5 mutant in feeding experiments with MTOB and Met can be assigned to the residual activity of BAT5 and not to redundant BAT activities because the double bat5 bat4 and the triple bat5 bat4 bat3 mutant phenocopy the single bat5 mutant (Figure 6).
Second, MTOP feeding restored the reduced level of 3MSOP in the bat5 mutant. Likewise, the reduced level of 4MSOB could be rescued by feeding with MTOH and dihomomethionine but not with MTOP, demonstrating that (1) the cytosolic conversion of 2-keto acids into corresponding amino acids proceeds very efficiently and (2) that bypassing the plastidic BAT5 function directly led to the biosynthesis of aliphatic GSs (Figures 9 and 10). Third, feeding with MTOP, MTOH, or dihomomethionine led to elevated levels of the long-chained aliphatic GSs 6MSOH, 7MSOH, 7MTH, and 8MSOO only in wild-type plants but not in the bat5 mutant. This indicates that the chain-elongated 2-keto acids MTOP and MTOH could only be imported into chloroplasts in the wild type but not in the bat5 mutant, again demonstrating a function of BAT5 in the transport of chain-elongated 2-keto acids. Also, the observation that MTOH-supplemented bat5 plants contained lower levels of long-chain aliphatic GSs than dihomomethionine-supplemented plants (Figures 9B to 9I) supports the suggestion that BAT5 is a transporter of 2-keto acids, but not of amino acids. Altogether, the experimental evidence strongly suggests that BAT5 mediates the transport of MTOB and of long-chain 2-keto acids. It is also apparent from these experiments that the fed 2-keto acids and amino acids can enter the long-distance transport pathway, including transport across plasma and plastidic membranes, and that the transport across the plastidic membrane is impaired in the bat5 mutant.
The combined data suggest that BAT5 transports MTOB and chain-elongated 2-keto acids across the chloroplast envelope membrane before, during, and after side chain-elongation of 2-keto acids and is thus an essential component of the aliphatic GS biosynthetic pathway.
METHODS
Plant Material and Growth Conditions
Seeds of Arabidopsis thaliana, ecotype Col-0, and corresponding loss-of-function mutants were plated on soil and cold-treated at 4°C for 3 d in the dark. After stratification, seeds were transferred into a temperature-controlled growth chamber under short-day conditions (8 h light, 16 h dark) at 21 to 22°C and 40% humidity. Transgenic plants were selected by germination on half-strength MS medium containing corresponding antibiotics and were subsequently treated as wild-type plants.
Phylogenetic Analysis
Homologs of human bile acid transporter presented in Figure 2 were identified using BLAST (Altschul et al., 1990). Protein sequences for all putative bile acid transporter proteins (shown in Supplemental Data Set 1 online) were aligned using the ClustalW program with default settings (http://www-ebi.ac.uk/clustalw) and adjusted manually. An unrooted tree was visualized using Treeview software (http:/taxonomy.zoology.gla.ac.uk/rod/treeview.html).
Cotransformation Assays with Cultured Arabidopsis Cells
Cultured Arabidopsis Col-0 cells were maintained in 50 mL of Arabidopsis (AT) medium. The AT medium contained 4.3 g/L MS basal salt media (Duchefa), 1 mg/L 2,4-D, 4 mL of a vitamin B5 mixture (Sigma-Aldrich), and 30 g/L sucrose, pH 5.8. Cells were gently agitated at 160 rpm in the dark at 22°C.
To generate reporter constructs, the promoter regions of the genes of interest (BAT1, BAT2, BAT3, BAT4, BAT5, IPMI1, IPMI2, IMDH1, BCAT3, and MAM1) were amplified from genomic DNA of Arabidopsis plants and cloned into the pTOPO entry or pDONOR-207 vector (Invitrogen Life Technologies). To drive Agrobacterium tumefaciens–mediated expression of the uidA (GUS) reporter gene under control of these promoters, the pGWB3i vector was recombined with the pTOPO entry or pDONOR-207 vector containing corresponding promoters sequences using LR reactions (Invitrogen Life Technologies). As an effector, a previously generated pGWB2 vector containing the full-length clone of HAG1/MYB28, HAG2/MYB76, or HAG3/MYB76 was used (Gigolashvili et al., 2007, 2008). Finally, the reporter and effector constructs were transformed into the supervirulent Agrobacterium strain LBA4404.pBBR1MCS.virGN54D. Transient coexpression assays in cultured cells were performed as described by Berger et al. (2007).
Construction of the GFP Fusion Plasmids and Transfection of Cultured Arabidopsis Cells
To generate GFP fusion constructs, the BAT5, IPMI1, IPMI2, and IPMDH1 coding regions, including the putative chloroplast targeting sequences, were first cloned into the entry pDONOR-207 vector. The obtained entry clones were recombined with the pGWB5 binary vector using LR reactions. Transformation of dark-grown cultured Arabidopsis cells was performed using supervirulent Agrobacterium strains LBA4404.pBBR1MCS.virGN54D each containing one of these constructs as described by Koroleva et al. (2005).
For transient expression of the full-length BAT5 clone in Arabidopsis leaves, the supervirulent Agrobacterium containing the BAT5-GFP construct and the antisilencing Agrobacterium strain 19K were taken from fresh YEB plates, grown overnight, sedimented, resuspended in 10 mM MgCl2 and 10 mM MES, pH 5.6, and adjusted to an OD600 of 0.7 to 0.8. The working solutions for the infiltration of dark-exposed Arabidopsis plants contained a BAT5-GFP suspension together with the Agrobacterium strain 19K in a 1:1 ratio. Acetosyringon was added (0.15 mM, final concentration), and the suspension was incubated for 2 to 4 h at 30°C. Three to five leaves of 5- to 6-week-old Arabidopsis plants were infiltrated with the working solution and sampled after 3 to 5 d of infiltration for microscopy. GFP expression patterns were recorded using a fluorescence microscope (Eclipse E800; Nikon) with a GFP (R)-BP filter (excitation 460 to 500 nm; dichronic mirror 505 nm; barrier filter 510 to 560 nm) or a SP2 confocal laser scanning microscope from Leica. Photomicrographs were taken with Duscus and LCS software.
Generation of Transgenic Pro35S:amiBAT5 Lines
To construct an amiRNA against BAT5, an artificial microRNA fragment was designed as described at http://wmd.weigelworld.org and cloned into the Gateway TOPO Entry vector. For amplification of the miR319a precursor, the RS300 plasmid, kindly provided by D. Weigel (Max Planck Institute for Developmental Biology, Tuebingen, Germany), was used. The obtained pTOPO Entry clone containing the amiBAT5 sequence was recombined into the Gateway destination pGWB2 vector.
To generate Pro35S:amiBAT5 Arabidopsis knockdown plants and corresponding cells, Pro35S:amiBAT5:GWB2 was transformed into the Agrobacterium strain GV3101 and LBA4404.pBBR1MCS.virGN54D by electroporation and further into Arabidopsis plants and cultured cells. Plant transformants were selected with kanamycin (50 μg per mL), and the BAT5 transcript level was monitored by real-time PCR analysis.
Generation of the Estradiol-Inducible ProG10-90:BAT5:GUS Construct for the Complementation Analysis
To rescue BAT5 function, an estrogen receptor-based chemical-inducible system (Zuo et al., 2000) modified for gateway cloning by I. Somssich (Max Planck Institute for Plant Breeding Research, Cologne, Germany) was used. To generate an expression clone, the coding sequence of BAT5 was first amplified by RT-PCR and cloned into the TOPO Entry vector (Invitrogen Life Technologies) before being recombined into the destination pMD-GWY-St vector using the LR reaction. The final ProG10-90:BAT5:GUS construct within the pMD-GWY-St vector was transformed into Agrobacterium cells by electroporation and into Arabidopsis plants by vacuum infiltration (Bechtold et al., 1993). All transformants were selected using BASTA, and the uidA transcript level was monitored by histochemical GUS staining.
Histochemical Analysis of Transgenic Plants Expressing the ProBAT5:uidA Fusion Construct
The promoter region of the BAT5 gene was amplified from genomic DNA of Arabidopsis plants and cloned into the pTOPO Entry vector (Invitrogen Life Technologies). To drive expression of the uidA gene under control of the BAT5 promoter, the binary Gateway-compatible plant transformation vector pGWB3 was recombined with the entry clone using an LR reaction. The obtained ProBAT5:uidA construct was transformed into the GV3101 Agrobacterium strain and Arabidopsis wild-type plants.
Histochemical localization of GUS in several independent transgenic lines harboring the ProBAT5:uidA construct was performed as described previously (Gigolashvili et al., 2007).
Preparation of Methanolic Extracts and HPLC Analysis of GSs
Fifty milligrams of leaves were placed into a 2-mL reaction tube and frozen in liquid nitrogen. Frozen leaf samples were lyophilized and homogenized in a mill (Qiagen). GSs were extracted in 80% methanol after addition of 20 μL of 5 mM of benzyl GS as an internal standard (www.glucosinolates.com). Extracts were applied onto a DEAE Sephadex A25 column (0.1 g powder equilibrated in 0.5 M acetic acid/NaOH, pH 5). GSs were converted to desulfoglucosinolates by overnight incubation with a purified sulfatase (E.C. 3.1.6.1) designated type H-1, from Helix pomatia, 16,400 units/g solid (Sigma-Aldrich). For analysis of desulfoglucosinolates, samples were subjected to UPLC analysis (Waters) with a diode array detector, using an Acquity UPLC column (BEH Schield RP18, 150 × 2.1 mm, 1.7 μm). Peaks were quantified by the peak area at 229 nm relative to the area of the internal standard peak.
Reverse Transcriptase–Mediated First-Strand Synthesis and Real-Time PCR Analysis
Total RNA was extracted from rosette leaves of adult plants from different mutant lines using the TRIsure buffer (Biolone) followed by treatment with RNase-free DNase (Roth) to remove genomic DNA contaminants. Seven-to-ten micrograms of total RNA was reverse transcribed with the first-strand cDNA synthesis SSII kit (Invitrogen) according to the manufacturer's instructions.
The expression of genes was analyzed in three independent replicates by real-time RT-PCR analysis using the fluorescent intercalating dye SYBR-Green in a GeneAmp 7300 sequence detection system (Applied Biosystems). The Arabidopsis ACTIN2 gene was used as a standard. Real-time PCR was performed using the SYBR-Green master kit system (Applied Biosystems) according to the manufacturer's instructions. The Ct, defined as the PCR cycle at which a statistically significant increase of reporter fluorescence is detected, is used as a measure for the starting copy number of the target gene. Relative quantification of expression levels was performed using relative quantification software based on the comparative Ct method (Applied Biosystems). The calculated relative expression values are normalized to the wild-type expression level, wild type = 1. The efficiency of each primer pair was tested using wild-type (Col-0) cDNA as a standard template, and the RT-PCR data were normalized dependent on the relative efficiency of each primer pair.
Plant Hormone Induction and Wounding Experiments
Arabidopsis wild-type seedlings (Col-0) were grown on half-strength MS media with 0.5% agar and 0.5% sucrose for 10 d in a growth chamber at 21°C under constant light. Afterwards, roots and portions of the first leaves were immersed in half-strength MS liquid media containing MeJA or SA (10 μM) and incubated for 0, 2, 5, 15, or 120 min, or for 24 h. Subsequently, the seedlings were placed into 2-mL reaction tubes, frozen in liquid nitrogen, and used for RNA isolation. Three independent sets of plants induced by this elicitor were used for real-time PCR analysis.
For wounding experiments, inflorescences of Col-0 plants were slightly cut with a scalpel or blade. Samples were collected 0, 1, 5 10, 30, and 60 min after treatment, immediately frozen in liquid nitrogen, and used for RNA isolation and real-time PCR analysis.
Feeding Experiment with 2-Keto Acids and Amino Acids
Seedlings of the wild type (Col-0) and bat5 mutant were grown on half-strength MS media with 1% agar and 0.5% sucrose for 14 d in a growth chamber at 21°C under short-day conditions. Afterwards, both Col-0 and bat5 were carefully taken out of plates and transferred into six-well plates containing half-strength MS supplemented with 0.2 mM MTOB, MTOP, MTOH, Met, or dihomomethionine, respectively. Plants were left for 14 additional days under the same conditions, followed by analyses of GS contents using UPLC.
MTOB and Met were obtained from Sigma-Aldrich. MTOP and MTOH were synthesized by Applichem. Dihomomethionine was synthesized as described by Dawson et al. (1993). In short, sodium methoxide (0.033 mol) was treated with metyhlmercaptane, and 3-bromovaleronitrile was subsequently added to yield 5-methylthiopentanenitrile. This was converted to 5-methylthiopentanal by treatment with diisobutylaluminium hydride. The 5-methylthiopentanal was first converted into the corresponding aminonitrile and subsequently hydrolyzed to dihomomethionine, which was further purified by anion exchange chromatography.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: BAT5 (At4g12030.2, representative gene model; http://Arabidopsis.org), BAT4 (At4g22840), BAT3 (At3g25410), BAT2 (At1g78560), BAT1 (At2g26900), IPMI1 (At3g58990), IPMI2 (At2g43100), and IPMDH (At1g31180).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Histochemical GUS Staining in Various Tissues of ProBAT1-4:GUS Plants.
Supplemental Figure 2. Plastidic Localization of BAT1, BAT2, BAT3, and BAT4 Full-Length GFP Constructs in BY Tobacco Protoplasts.
Supplemental Figure 3. Transcript Levels of Known Glucosinolate Biosynthesis Regulators in Leaves of bat5 and Pro35S:amiBAT5 Plants in Comparison with the Wild Type (Col-0).
Supplemental Figure 4. Content of the Indolic Glucosinolate Indol-3-Ylmethyl-GS in bat5 Mutant Fed with 2-Keto Acids or Amino Acids.
Supplemental Data Set 1. Protein Sequences Used to Generate the Phylogenetic Tree Presented in Figure 2.
Supplementary Material
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft. We thank A. Berkessel (University of Cologne, Germany) for providing facilities for the synthesis of dihomomethionine I.E. Somssich (Max Planck Institute for Plant Breeding Research, Cologne, Germany) for providing the ergosterol-inducible gateway construct, and Katja Wester for help with confocal microscopy.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with policy described in the Instruction of Authors (www.plantcell.org) is: Ulf-Ingo Flügge (ui.fluegge@uni-koeln.de).
Online version contains Web-only data.
References
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215 403–410. [DOI] [PubMed] [Google Scholar]
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris Life Sci. 316 1194–1199. [Google Scholar]
- Berger, B., Stracke, R., Yatusevich, R., Weisshaar, B., Flügge, U.I., and Gigolashvili, T. (2007). A simplified method for the analysis of transcription factor-promoter interactions that allows high-throughput data generation. Plant J. 50 911–916. [DOI] [PubMed] [Google Scholar]
- Brader, G., Mikkelsen, M.D., Halkier, B.A., and Palva, E.T. (2006). Altering glucosinolate profiles modulates disease resistance in plants. Plant J. 46 758–767. [DOI] [PubMed] [Google Scholar]
- Brader, G., Tas, E., and Palva, E.T. (2001). Jasmonate-dependent induction of indole glucosinolates in Arabidopsis by culture filtrates of the nonspecific pathogen Erwinia carotovora. Plant Physiol. 126 849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, P.D., Tokuhisa, J.G., Reichelt, M., and Gershenzon, J. (2003). Variation of glucosinolate accumulation among different organs and developmental stages of Arabidopsis thaliana. Phytochemistry 62 471–481. [DOI] [PubMed] [Google Scholar]
- Chen, S.X., Glawischnig, E., Jorgensen, K., Naur, P., Jorgensen, B., Olsen, C.E., Hansen, C.H., Rasmussen, H., Pickett, J.A., and Halkier, B.A. (2003). CYP79F1 and CYP79F2 have distinct functions in the biosynthesis of aliphatic glucosinolates in Arabidopsis. Plant J. 33 923–937. [DOI] [PubMed] [Google Scholar]
- Chung, W.C., Huang, H.C., Chiang, B.T., Huang, H.C., and Huang, J.W. (2005). Inhibition of soil-borne plant pathogens by the treatment of sinigrin and myrosinases released from reconstructed Escherichia coli and Pichia pastoris. Biocontrol Sci. Technol. 15 455–465. [Google Scholar]
- Cipollini, D., Enright, S., Traw, M.B., and Bergelson, J. (2004). Salicylic acid inhibits jasmonic acid-induced resistance of Arabidopsis thaliana to Spodoptera exigua. Mol. Ecol. 13 1643–1653. [DOI] [PubMed] [Google Scholar]
- Dawson, G.W., Hick, A.J., Bennett, R.N., Donald, A., Pickett, J.A., and Wallsgrove, R.M. (1993). Synthesis of glucosinolate precursors and investigations into the biosynthesis of phenylalkyl- and methylthioalkylglucosinolates. J. Biol. Chem. 268 27154–27159. [PubMed] [Google Scholar]
- Devoto, A., and Turner, J.G. (2005). Jasmonate-regulated Arabidopsis stress signalling network. Physiol. Plant. 123 161–172. [Google Scholar]
- Fliege, R., Flügge, U.I., Werdan, K., and Heldt, H.W. (1978). Specific transport of inorganic phosphate, 3-phosphoglycerate and triosephosphates across the inner membrane of the envelope in spinach chloroplasts. Biochim. Biophys. Acta 502 232–247. [DOI] [PubMed] [Google Scholar]
- Giamoustaris, A., and Mithen, R. (1995). The effect of modifying the glucosinolate content of leaves of oilseed rape (Brassica Napus Ssp Oleifera) on its interaction with specialist and generalist pests. Ann. Appl. Biol. 126 347–363. [Google Scholar]
- Gigolashvili, T., Engqvist, M., Yatusevich, R., Müller, C., and Flügge, U.I. (2008). HAG2/MYB76 and HAG3/MYB29 exert a specific and coordinated control on the regulation of aliphatic glucosinolate biosynthesis in Arabidopsis thaliana. New Phytol. 177 627–642. [DOI] [PubMed] [Google Scholar]
- Gigolashvili, T., Berger, B., Mock, H.P., Müller, C., Weisshaar, B., and Flügge, U.I. (2007. a). The transcription factor HIG1/MYB51 regulates indolic glucosinolate biosynthesis in Arabidopsis thaliana. Plant J. 50 886–901. [DOI] [PubMed] [Google Scholar]
- Gigolashvili, T., Yatusevich, R., Berger, B., Müller, C., and Flügge, U.I. (2007. b). The R2R3-MYB transcription factor HAG1/MYB28 is a regulator of methionine-derived glucosinolate biosynthesis in Arabidopsis thaliana. Plant J. 51 247–261. [DOI] [PubMed] [Google Scholar]
- Grubb, C.D., and Abel, S. (2006). Glucosinolate metabolism and its control. Trends Plant Sci. 11 89–100. [DOI] [PubMed] [Google Scholar]
- Grubb, C.D., Zipp, B.J., Ludwig-Muller, J., Masuno, M.N., Molinski, T.F., and Abel, S. (2004). Arabidopsis glucosyltransferase UGT74B1 functions in glucosinolate biosynthesis and auxin homeostasis. Plant J. 40 893–908. [DOI] [PubMed] [Google Scholar]
- Hagenbuch, B., Stieger, B., Foguet, M., Lubbert, H., and Meier, P.J. (1991). Functional expression cloning and characterization of the hepatocyte Na+/bile acid cotransport system. Proc. Natl. Acad. Sci. USA 88 10629–10633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halkier, B.A., and Gershenzon, J. (2006). Biology and biochemistry of glucosinolates. Annu. Rev. Plant Biol. 57 303–333. [DOI] [PubMed] [Google Scholar]
- Hansen, C.H., Wittstock, U., Olsen, C.E., Hick, A.J., Pickett, J.A., and Halkier, B.A. (2001). Cytochrome P450 CYP79F1 from Arabidopsis catalyzes the conversion of dihomomethionine and trihomomethionine to the corresponding aldoximes in the biosynthesis of aliphatic glucosinolates. J. Biol. Chem. 276 11078–11085. [DOI] [PubMed] [Google Scholar]
- Hayes, D., Kelleher, O., and Eggelston, M. (2008). The cancer chemoprotective actions of phytochemicals derived from glucosinolates. Eur. J. Nutr. 47 (Suppl. 2): 73–88. [DOI] [PubMed] [Google Scholar]
- Hemm, M.R., Ruegger, M.O., and Chapple, C. (2003). The Arabidopsis ref2 mutant is defective in the gene encoding CYP83A1 and shows both phenylpropanoid and glucosinolate phenotypes. Plant Cell 15 179–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai, M.Y., et al. (2007). Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc. Natl. Acad. Sci. USA 104 6478–6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein, D.J., Kroymann, J., Brown, P., Figuth, A., Pedersen, D., Gershenzon, J., and Mitchell-Olds, T. (2001). Genetic control of natural variation in Arabidopsis glucosinolate accumulation. Plant Physiol. 126 811–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knill, T., Schuster, J., Reichelt, M., Gershenzon, J., and Binder, S. (2008). Arabidopsis branched-chain aminotransferase 3 functions in both amino acid and glucosinolate biosynthesis. Plant Physiol. 146 1028–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koroleva, O.A., Tomlinson, M.L., Leader, D., Shaw, P., and Doonan, J.H. (2005). High-throughput protein localization in Arabidopsis using Agrobacterium-mediated transient expression of GFP-ORF fusions. Plant J. 41 162–174. [DOI] [PubMed] [Google Scholar]
- Kroymann, J., Textor, S., Tokuhisa, J.G., Falk, K.L., Bartram, S., Gershenzon, J., and Mitchell-Olds, T. (2001). A gene controlling variation in Arabidopsis glucosinolate composition is part of the methionine chain elongation pathway. Plant Physiol. 127 1077–1088. [PMC free article] [PubMed] [Google Scholar]
- Kunst, L., Browse, J., and Somerville, C. (1988). Altered regulation of lipid biosynthesis in a mutant of Arabidopsis deficient in chloroplast glycerol-3-phosphate acyltransferase activity. Proc. Natl. Acad. Sci. USA 85 4143–4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., Hansen, B.G., Ober, J.A., Kliebenstein, D.J., and Halkier, B.A. (2008). Subclade of flavin-monooxygenases involved in aliphatic glucosinolate biosynthesis. Plant Physiol. 148 1721–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loddenkötter, B., Kammerer, B., Fischer, K., and Flügge, U.I. (1993). Expression of the functional mature chloroplast triose phosphate translocator in yeast internal membranes and purification of the histidine-tagged protein by a single metal-affinity chromatography step. Proc. Natl. Acad. Sci. USA 90 2155–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malitsky, S., Blum, E., Less, H., Venger, I., Elbaz, M., Morin, S., Eshed, Y., and Aharoni, A. (2008). The transcript and metabolite networks affected by the two clades of Arabidopsis glucosinolate biosynthesis regulators. Plant Physiol. 148 2021–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manici, L.M., Lazzeri, L., Baruzzi, G., Leoni, O., Galletti, S., and Palmieri, S. (2000). Suppressive activity of some glucosinolate enzyme degradation products on Pythium irregulare and Rhizoctonia solani in sterile soil. Pest Manag. Sci. 56 921–926. [Google Scholar]
- Mari, M., Iori, R., Leoni, O., and Marchi, A. (1996). Bioassays of glucosinolate-derived isothiocyanates against postharvest pear pathogens. Plant Pathol. 45 753–760. [Google Scholar]
- Mewis, I., Appel, H.M., Hom, A., Raina, R., and Schultz, J.C. (2005). Major signaling pathways modulate Arabidopsis glucosinolate accumulation and response to both phloem-feeding and chewing insects. Plant Physiol. 138 1149–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen, M.D., and Halkier, B.A. (2003). Metabolic engineering of valine- and isoleucine-derived glucosinolates in Arabidopsis expressing CYP79D2 from cassava. Plant Physiol. 131 773–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen, M.D., Naur, P., and Halkier, B.A. (2004). Arabidopsis mutants in the C-S lyase of glucosinolate biosynthesis establish a critical role for indole-3-acetaldoxime in auxin homeostasis. Plant J. 37 770–777. [DOI] [PubMed] [Google Scholar]
- Mithen, R.F. (2001). Glucosinolates and their degradation products. Adv. Bot. Res. 35 213–262. [Google Scholar]
- Mugford, S.G., et al. (2009). Disruption of adenosine-5′-phosphosulfate kinase in Arabidopsis reduces levels of sulfated secondary metabolites. Plant Cell 21 910–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naur, P., Petersen, B.L., Mikkelsen, M.D., Bak, S., Rasmussen, H., Olsen, C.E., and Halkier, B.A. (2003). CYP83A1 and CYP83B1, two nonredundant cytochrome P450 enzymes metabolizing oximes in the biosynthesis of glucosinolates in Arabidopsis. Plant Physiol. 133: 63–72. [DOI] [PMC free article] [PubMed]
- Ogier, G., Chantepie, J., Deshayes, C., Chantegrel, B., Charlot, C., Doutheau, A., and Quash, G. (1993). Contribution of 4-methylthio-2-oxobutanoate and its transaminase to the growth of methionine-dependent cells in culture. Effect of transaminase inhibitors. Biochem. Pharmacol. 45 1631–1644. [DOI] [PubMed] [Google Scholar]
- Piotrowski, M., Schemenewitz, A., Lopukhina, A., Müller, A., Janowitz, T., Weiler, E.W., and Oecking, C. (2004). Desulfoglucosinolate sulfotransferases from Arabidopsis thaliana catalyze the final step in the biosynthesis of the glucosinolate core structure. J. Biol. Chem. 279 50717–50725. [DOI] [PubMed] [Google Scholar]
- Price, C.E., Reid, S.J., Driessen, A.J.M., and Abratt, V.R. (2006). The Bifidobacterium longum NCIMB 702259T ctr gene codes for a novel cholate transporter. Appl. Environ. Microbiol. 72 923–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reintanz, B., Lehnen, M., Reichelt, M., Gershenzon, J., Kowalczyk, M., Sandberg, G., Godde, M., Uhl, R., and Palme, K. (2001). bus, a bushy Arabidopsis CYP79F1 knockout mutant with abolished synthesis of short-chain aliphatic glucosinolates. Plant Cell 13 351–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond, P., Bodenhausen, N., Van Poecke, R.M.P., Krishnamurthy, V., Dicke, M., and Farmer, E.E. (2004). A conserved transcript pattern in response to a specialist and a generalist herbivore. Plant Cell 16 3132–3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzewuski, G., and Sauter, M. (2002). The novel rice (Oryza sativa L.) gene OsSbf1 encodes a putative member of the Na+/bile acid symporter family. J. Exp. Bot. 53 1991–1993. [DOI] [PubMed] [Google Scholar]
- Sasaki-Sekimoto, Y., et al. (2005). Coordinated activation of metabolic pathways for antioxidants and defence compounds by jasmonates and their roles in stress tolerance in Arabidopsis. Plant J. 44 653–668. [DOI] [PubMed] [Google Scholar]
- Schuster, J., Knill, T., Reichelt, M., Gershenzon, J., and Binder, S. (2006). BRANCHED-CHAIN AMINOTRANSFERASE4 is part of the chain elongation pathway in the biosynthesis of methionine-derived glucosinolates in Arabidopsis. Plant Cell 18 2664–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab, R., Ossowski, S., Riester, M., Warthmann, N., and Weigel, D. (2006). Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18 1121–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacke, R., Schneider, A., van der Graaff, E., Fischer, K., Catoni, E., Desimone, M., Frommer, W.B., Flügge, U.I., and Kunze, R. (2003). ARAMEMNON: A novel database for Arabidopsis thaliana integral membrane proteins. Plant Physiol. 131 16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sønderby, I.E., Hansen, B.G., Bjarnholt, N., Ticconi, C., Halkier, B.A., and Kliebenstein, D.J. (2007). A systems biology approach identifies a R2R3 MYB gene subfamily with distinct and overlapping functions in regulation of aliphatic glucosinolates. PloS One 2 e1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talalay, P., and Fahey, J.W. (2001). Phytochemicals from cruciferous plants protect against cancer by modulating carcinogen metabolism. J. Nutr. 131 3027S–3033S. [DOI] [PubMed] [Google Scholar]
- Tantikanjana, T., Mikkelsen, M.D., Hussain, M., Halkier, B.A., and Sundaresan, V. (2004). Functional analysis of the tandem-duplicated P450 genes SPS/BUS/CYP79F1 and CYP79F2 in glucosinolate biosynthesis and plant development by Ds transposition-generated double mutants. Plant Physiol. 135 840–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Textor, S., Bartram, S., Kroymann, J., Falk, K.L., Hick, A., Pickett, J.A., and Gershenzon, J. (2004). Biosynthesis of methionine-derived glucosinolates in Arabidopsis thaliana: Recombinant expression and characterization of methylthioalkylmalate synthase, the condensing enzyme of the chain-elongation cycle. Planta 218 1026–1035. [DOI] [PubMed] [Google Scholar]
- Textor, S., de Kraker, J.W., Hause, B., Gershenzon, J., and Tokuhisa, J.G. (2007). MAM3 catalyzes the formation of all aliphatic glucosinolate chain lengths in Arabidopsis. Plant Physiol. 144 60–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trauner, M., and Boyer, J.L. (2003). Bile salt transporters: molecular characterization, function, and regulation. Physiol. Rev. 83 633–671. [DOI] [PubMed] [Google Scholar]
- Traw, M.B., Kim, J., Enright, S., Cipollini, D.F., and Bergelson, J. (2003). Negative cross-talk between salicylate- and jasmonate-mediated pathways in the Wassilewskija ecotype of Arabidopsis thaliana. Mol. Ecol. 12 1125–1135. [DOI] [PubMed] [Google Scholar]
- Wong, M.H., Oelkers, P., Craddock, A.L., Dawson, P.A. (1994). Expression cloning and characterization of the hamster ileal sodium-dependent bile acid transporter. J. Biol. Chem. 269 1340–1347. [PubMed] [Google Scholar]
- Zuo, J., Niu, Q.W., and Chua, N.H. (2000). Technical advance: An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24 265–273. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.