Abstract
MicroRNAs (miRNAs) regulate gene expression posttranscriptionally through RNA silencing, a mechanism conserved in eukaryotes. Prevailing models entail most animal miRNAs affecting gene expression by blocking mRNA translation and most plant miRNAs, triggering mRNA cleavage. Here, using polysome fractionation in Arabidopsis thaliana, we found that a portion of mature miRNAs and ARGONAUTE1 (AGO1) is associated with polysomes, likely through their mRNA target. We observed enhanced accumulation of several distinct miRNA targets at both the mRNA and protein levels in an ago1 hypomorphic mutant. By contrast, translational repression, but not cleavage, persisted in transgenic plants expressing the slicing-inhibitor 2b protein from Cucumber mosaic virus. In agreement, we found that the polysome association of miR168 was lost in ago1 but maintained in 2b plants, indicating that translational repression is correlated with the presence of miRNAs and AGO1 in polysomes. This work provides direct biochemical evidence for a translational component in the plant miRNA pathway.
INTRODUCTION
RNA silencing regulates gene expression in many eukaryotic organisms through the activity of distinct classes of endogenous small RNAs (sRNAs). Among these, 19- to 25-bp microRNAs (miRNAs) and small interfering RNAs (siRNAs) are processed from noncoding double-stranded RNA precursors by RNases in the Dicer-like (DCL) family. One strand of the sRNAs duplex is then loaded into an Argonaute (AGO) protein to form a silencing effector complex. These ribonucleoprotein complexes, also called miRNPs or siRNPs, target and postrancriptionally silence mRNAs that are partly or fully complementary to the loaded sRNA. In plants, most known miRNAs interact with fully or near-fully complementary target mRNAs at a single site usually located in the protein coding sequence (Bartel, 2004). Plant miRNAs serve as guides for the AGO1 protein, which cleaves target mRNA owing to its slicer activity (Baumberger and Baulcombe, 2005). By contrast, animal miRNAs typically anneal with nonperfect complementarity to their targets. Animal mRNAs can contain several miRNA recognition elements, often in their 3′-untranslated region, and silencing occurs through slicer-independent mechanisms (Carrington and Ambros, 2003; Pillai et al., 2005). In several models, including Caenorhabditis elegans (Olsen and Ambros, 1999; Seggerson et al., 2002), Drosophila melanogaster (Hammond et al., 2001), and also cultured mammalian cells (Kim et al., 2004; Nelson et al., 2004), miRNAs and their mRNAs targets have been shown to associate with polysomes and repress translation via a slicer-independent mechanism whose nature is debated (Valencia-Sanchez et al., 2006; Pillai et al., 2007). In plants, miRNA-guided translational inhibition has been documented on only a few occasions, and these examples were believed to represent rare exceptions (Aukerman and Sakai, 2003; Chen, 2004; Arteaga-Vazquez et al., 2006; Gandikota et al., 2007). However, a recent study showed that sRNA-guided translational inhibition is in fact widespread in Arabidopsis thaliana and can be genetically uncoupled from slicing (Brodersen et al., 2008). Therefore, plant sRNA action at the posttranscriptional level entails a combination of slicing and translational repression.
Nonetheless, biochemical evidence for interactions between the RNA silencing and translation machinery is still missing in plants. In this work, we show that several miRNAs are associated with polysomes. In addition, we show that AGO1, which is known to confer slicer activity in plants, is also associated with polysomes. By comparative analysis of miRNA target accumulation and translation in an ago1 mutant and in transgenic plants expressing the slicing inhibitor 2b protein from Cucumber mosaic virus (FNY-2b), we reveal that translational repression and miRNA polysomal association can be maintained in 2b plants independently of slicer inhibition. Our results suggest that AGO1 has distinct functions in target cleavage and translational repression and that target mRNAs can be differentially affected by these activities.
RESULTS
AGO1-miRNPs Are Associated with Active Polysomes in Arabidopsis
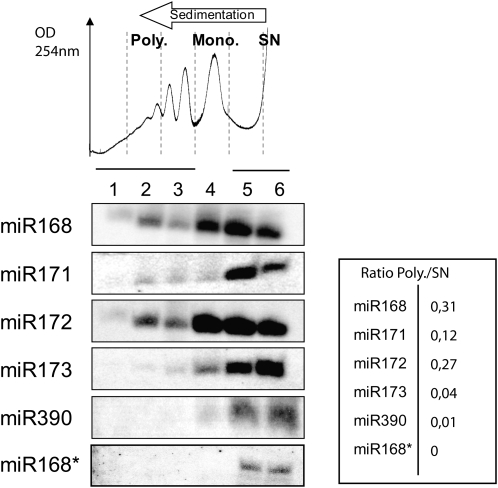
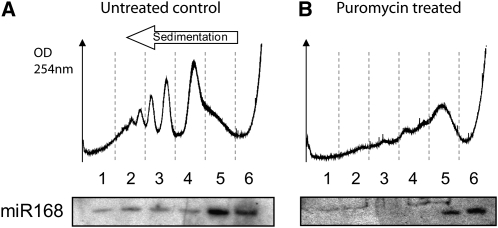
We first investigated the subcellular distribution of several highly expressed miRNAs (Backman et al., 2008). Cytoplasmic extracts were prepared from young Arabidopsis seedlings and fractionated on sucrose gradients (Figure 1). RNA gel blot analysis revealed that a portion of miR168, miR171, and miR172 is present in the polysomal fractions of seedling extracts (fractions 1 to 3, Figure 1). Actively translating polysomes are susceptible to inhibition by the amino acid analog puromycin. We found that, although the level of miRNA association with polysomes versus monosomes varied for different miRNAs, puromycin treatment efficiently removed miR168 from the polysomal fractions. This suggests that miRNAs are associated with active polysomes through association with their cognate mRNA (Figure 2). In order to define whether miRNA association with polysomes correlates with their functional loading into the AGO1 effector complex, we studied the subcellular localization of miR168*, which is the labile complementary (or passenger) strand of miRNA168 in the miR/miR* duplex produced by DCL1. In contrast with miR168, miR168* was only found within light, untranslated fractions (fractions 5 and 6, Figure 1). This suggests that only fully processed miRNAs that are loaded into a functional AGO1 effector complex can associate with polysomes.
Figure 1.
Functional miRNAs Are Associated with Polysomes in Arabidopsis.
In the top panel, absorbance at 254 nm is shown from the heavier (left) to the lighter (right) fractions of the continuous sucrose gradient. Cytoplasmic extracts from young seedlings were separated on gradients and fractionated in six equal fractions (labeled 1 to 6 from the bottom to the top of the gradient). Polysomes (Poly.) are fractions 1 to 3; monosomes (Mono.), fraction 4; and supernatant (SN), fractions 5 and 6. Total RNA was extracted from each fraction and analyzed by RNA gel blots to detect sRNAs. Ratios P/SN were calculated for each miRNA after quantification of signals by phosphor imaging.
Figure 2.
Puromycin Treatment Dissociates miRNAs from Polysomes.
Seedling cytoplasmic extracts were not treated (A) or treated with puromycin (B) before centrifugation on continuous sucrose gradients. In the top panel, absorbance at 254 nm is shown from the heavier (left) to the lighter (right) fractions of the gradient. Total RNA was extracted from each fraction and analyzed by RNA gel blots to detect miR168.
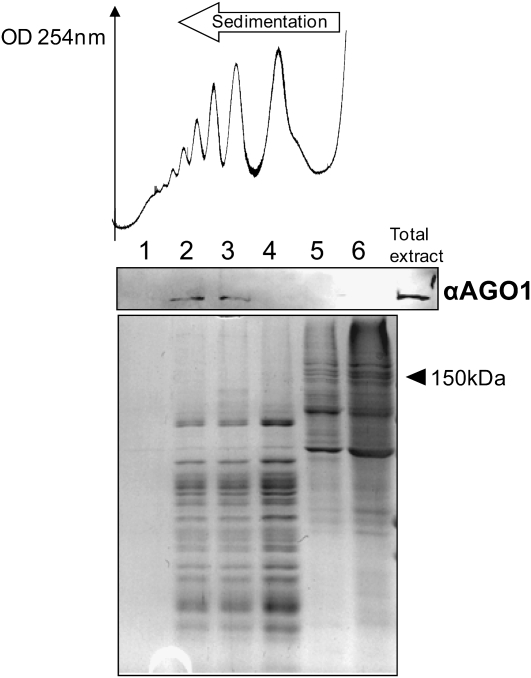
Next, we sought to establish whether the translatability of the target RNA could influence miRNA association with polysomes. The miRNAs miR173 and miR390 target the noncoding (i.e., not translated) Trans-ActingSiRNA precursor 1/2 and TAS3 RNA precursors, respectively, for cleavage, which triggers the subsequent DCL4-dependent phased processing of trans-acting siRNAs (Allen et al., 2005). We found that miR173 is weakly associated with polysomal fractions (Figure 1) and that miR390 is even less strongly associated, with only a faint signal in monosomal fraction 4 (Figure 1). This difference could be due to the fact that miR390, unlike miR173 and indeed most miRNAs, is selectively loaded into AGO7 as opposed to AGO1 (Montgomery et al., 2008). For each miRNA, analysis of the amount associated with high molecular weight polysomes relative to the amount in nonpolysomal fractions suggests a positive correlation between target translatability and the association of miRNA with high molecular weight polysomes. We then asked whether the core component of plant miRNP complexes, the AGO1 protein itself (Baumberger and Baulcombe, 2005), is associated with polysomes. AGO1 associates with many miRNAs and has been implicated in translational inhibition by several miRNAs (Brodersen et al., 2008). Protein gel blot analysis of proteins extracted from six fractions of a sucrose gradient demonstrated that AGO1 is indeed associated with polysomal fractions (Figure 3).
Figure 3.
AGO1 Is Associated with Polysomes.
Absorbance at 254 nm is shown from the heavier (left) to the lighter (right) fractions of the continuous sucrose gradient. Gradients were fractionated in six equal fractions (labeled 1 to 6 from the bottom to the top of the gradient). Proteins extracted from each fraction were separated by SDS-PAGE (Coomassie blue staining is shown in the bottom panel) and analyzed by protein gel blots using an antibody against AGO1. 150 kD is shown to indicate AGO1 position in the gel (116 kD).
The Association of miRNPs with Polysomes Depends on mRNA Binding
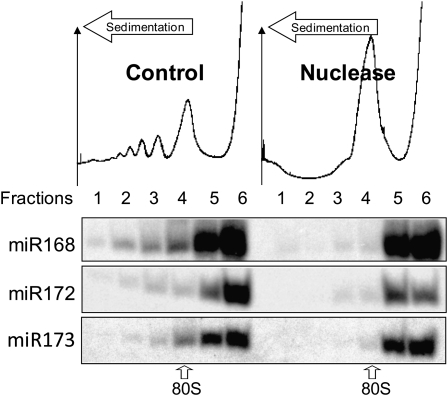
The cosedimentation of miRNPs with polysomes suggests that miRNPs are able to interact with ribosomes or that there is a direct interaction between loaded miRNAs and complementary regions within their mRNA targets. To distinguish between these possibilities, cytoplasmic extracts were digested with micrococcal nuclease. Under these conditions, ribosomes remain intact, but exposed regions of mRNA are degraded (Maroney et al., 2006). Upon micrococcal nuclease treatment, polysomes were almost completely degraded, resulting in a large accumulation of 80S ribosomes (fraction 4, Figure 4). Although ribosome-protected fragments are expected to cosediment with the peak of 80S ribosomes, the sedimentation profile of miRNAs was distinct, as most of these molecules were found near the top of the gradient after nuclease treatment (Figure 4, Nuclease). Therefore, these results suggest that miRNP association with polysomes depends on nuclease sensitive regions of RNA, presumably mRNAs undergoing translation elongation.
Figure 4.
miRNA Association with Polysomes Is RNA Mediated.
Seedling cytoplasmic extracts were prepared before centrifugation on continuous sucrose gradients. Absorbance at 254 nm is shown from the heavier (left) to the lighter (right) fractions of the continuous sucrose gradient. Gradients were fractionated in six equal fractions (labeled 1 to 6 from the bottom to the top of the gradient). Total RNA extracted from each fraction was analyzed by RNA gel blots. Micrococcal nuclease-treated samples are shown in the right panel and a nontreated control sample in the left panel. 80S label indicates the sedimentation level of one assembled ribosome (40S+60S) on an mRNA.
The Association of miRNAs with Polysomes Is Linked to Translational Repression
We used ago1-25 and 2b transgenic plants with altered AGO1 activity to explore the functional significance of the association of miRNPs with polysomes. ago1-25 is a fertile, hypomorphic mutant impaired in posttranscriptional RNA silencing and virus resistance due to a point mutation in ago1 (Morel et al., 2002). The 2b protein, coded by the FNY strain of Cucumber mosaic virus, inhibits slicing through physical interactions with AGO1 but does not prevent loading of sRNAs (Zhang et al., 2006; Lewsey et al., 2007).
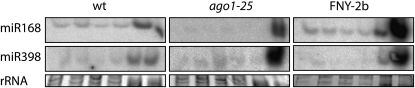
The ago1-25 and 2b transgenic plants were first analyzed for the association of miRNAs with polysomes. We found that miR168 association with polysomes is lost in ago1-25 mutants but persisted in 2b overexpressing plants (Figure 5). These results indicate that the association of miRNAs with polysomes is dependent on AGO1 activities that are differentially affected in ago1-25 and 2b plants. We also found that the proportion of miR398 associated with polysomes was generally less than for miR168. This suggests that the level of miRNA association with polysomes is dependent not only on AGO1 activity but also on the nature of the miRNA itself. In both ago1-25 and 2b plants, the distribution of miR168 and miR398 was found to shift toward low molecular weight fractions, further illustrating a link between RNA silencing and the translational machinery.
Figure 5.
Association of miR168 with Polysomes Is Lost in the ago1-25 Hypomorphic Mutant but Maintained in FNY-2b Plants.
Seedling cytoplasmic extracts were prepared before centrifugation on continuous sucrose gradients. Gradients were fractionated in six equal fractions from the bottom to the top of the gradient. Total RNA was extracted from each fraction and analyzed by RNA gel blots to detect sRNAs. rRNA is shown as a loading control.
Having established the differential behavior of miRNAs in ago1-25 and 2b transgenic plants, we then investigated the mRNA and protein accumulation of three distinct miRNA targets for which antibodies are available: AGO1, CIP4, and CSD2, which are repressed by miR168 (Vaucheret et al., 2004), miR834 (Brodersen et al., 2008), and miR398 (Sunkar et al., 2006), respectively. Mutant analyses recently showed that the CSD2 mRNA undergoes a combination of slicing and translational repression by miR398, while the CIP4 mRNA is inhibited nearly exclusively at the level of translation by Arabidopsis-specific miR834 (Brodersen et al., 2008). We observed a significant increase in the accumulation of AGO1 and CSD2 mRNA in both ago1-25 and 2b plants (Figure 6A), and, as previously reported, there was little variation in CIP4 mRNA levels. This effect at the transcript level was expected because of the impaired AGO1 activity exhibited by both types of plants. The protein levels of all three targets increased in ago1-25 (Figure 6B) but remained similar to wild-type levels in the 2b transgenic plants (Figure 6B). The contrast between an increase in mRNA abundance and low protein accumulation was particularly evident in the case of CSD2. These results indicate that in 2b plants, miRNA-directed repression of target transcripts is maintained at the translational level independently of the slicer activity of AGO1. Our results also show that translational inhibition is a component of miRNA-mediated silencing in wild-type plants.
Figure 6.
Translational Repression of miRNA Targets Is Maintained in FNY-2b Plants.
(A) Total RNA extracted from wild type, FNY-2b, and ago1-25 plants was quantified with real-time RT-PCR using primers surrounding the cleavage sites. Quantifications were normalized to that of ACTIN2 and then to the value of the wild-type plants, which was set to 1 (except for CSD2 mRNA values, which were normalized to ago1-25 because the wild-type value is null). Standard errors were calculated from two independent experiments.
(B) Total proteins were extracted from fresh leaves for each sample and separated by SDS-PAGE. Immunodetection of AGO1, CIP4, and CSD2 is shown in the top panel, and Coomassie blue staining is shown in the bottom panel.
Altogether, these results highlight the contrasting effects of the 2b protein and ago1-25 mutation on miRNA activities by suggesting that the translational repression activity of the miRNA/AGO1 effector complex is more active in 2b transgenic plants than in the ago1-25 hypomorphic mutants. In addition, our data suggest that the balance between RNA slicing and translational repression in wild-type plants is tuned for each mRNA/miRNA pair.
DISCUSSION
We have shown that a fraction of several miRNAs is associated with polysomes in Arabidopsis cell culture and seedlings. This association appears to be specific to mature miRNAs because the passenger strand miR168* was only found within nonpolysomal, untranslated fractions (Figure 1). Moreover, we found that the association of miRNAs with polysomes may depend on the translatability of the target RNA. For example, the AGO7/miR390 complex, dedicated for slicing TAS3 noncoding RNA precursors, is not associated with polysomes. We did find weak polysomal association for miR173, which coordinates the phased processing of TAS1/2 noncoding precursors (Figure 1). This weak association could be because TAS1/2 precursors entering the cytoplasm are capped and contain an AUG codon and therefore might be transiently recognized by the translational machinery.
We found that the majority of AGO1 protein is within high molecular weight polysomal fractions (Figure 3). This suggests that miRNA that sediment in low molecular weight fractions of the gradient are not associated with AGO1 or else that they somehow engage with a small AGO1 fraction in an alternative mode of regulation. In fact, we found that a substantial portion of several miRNA target transcripts partition to untranslated fractions in the same manner as the miRNA themselves (see Supplemental Figure 1 online). The copartitioning of miRNAs and targets raises the possibility that the targets may be deployed into multiple effector complexes for distinct types of outputs, possibly allowing subcellular-level regulation of mRNA stability and translation. Indeed, the cytoplasm contains several compartments that are not surrounded by membranes and are composed of ribonucleoprotein aggregates, including processing (P)-bodies and stress granules. In animals, some of these complexes are associated with components of the RNA silencing machinery (Leung et al., 2006), and there is evidence that Drosophila P-bodies are involved in miRNA-directed target decay through deadenylation and decapping, whereas translational repression is P-body independent (Eulalio et al., 2007). In plants, components of the RNA decapping machinery were found involved in RNA silencing (Gazzani et al., 2004; Gy et al., 2007), and the existence of P-bodies was recently demonstrated (Xu et al., 2006). Thus, it will be interesting to investigate which type of proteins, such as other members of the large AGO family, are linked to miRNAs and their mRNA targets in nonpolysomal fractions. Intriguingly, translationally repressed yeast mRNAs sequestered in P-bodies were found to sediment within light sucrose gradient fractions (Brengues et al., 2005).
The contrasting results with 2b transgenic and ago1-25 mutant plants echo those of Brodersen et al. (2008) who identified Arabidopsis mutations that uncouple slicing from translational repression by miRNAs. This raises important issues, including how slicing might be prevented during AGO1-mediated translational repression and why some miRNA targets appear more prone to one specific type of regulation.
METHODS
Plant Material
Arabidopsis thaliana mutant ago1-25 and FNY-2b–overexpressing plants were described previously (Morel et al., 2002; Lewsey et al., 2007). In both cases, Columbia-0 ecotype was used for the wild type. Line 3.13F of FNY-2b–overexpressing plants was used in our experiments. Plants were grown under a short-day photoperiod (8 h of light).
Preparation of Arabidopsis Cytoplasmic Extracts and Sucrose Density Gradient Analysis
Ten-day-old seedlings were used for polysome experiments. After harvesting, fresh material was frozen and ground in liquid nitrogen. To prepare cytoplasmic extracts, 100 to 500 mg of powder were combined with 1 mL of chilled polysome buffer (100 mM Tris-HCl, pH 8.4, 50 mM KCl, 25 mM MgCl2, 5 mM EGTA, 15.4 units·mL−1 heparin, 18 μM cycloheximide, 15.5 μM chloramphenicol, and 0.5% [v/v] Nonidet P-40). Nuclei, cell wall, and membranes were removed by centrifugation at 1200g for 5 min at 4°C. Aliquots of the resultant supernatant were loaded on 20 to 50% (w/w) continuous sucrose gradients and centrifuged at 32,000 rpm for 160 min in a Beckman SW41 rotor at 4°C. After centrifugation, fractions were collected from the bottom to the top of the gradient with continuous monitoring of 254-nm absorbance to monitor the distribution of ribosomes. Six equal fractions were collected after centrifugation. Fractions were numbered 1 to 6 from the bottom to the top of the gradient. Fractions 1 to 3 correspond to polysomes, fraction 4 to monosomes, and fractions 5 and 6 to supernatant. RNAs extracted from each fraction were loaded on a polyacrylamide gel and blotted without prior quantification.
For puromycin treatment, cytoplasmic extracts were prepared in a modified polysome buffer (100 mM Tris-HCl, pH 8.4, 50 mM KCl, 25 mM MgCl2, and 0.5% [v/v] Nonidet P-40). After centrifugation, supernatant was incubated with 0.25 mg.mL−1 of puromycin dihydrochloride from Streptomyces alboniger (Sigma-Aldrich) for 30 min at 37°C and loaded on a sucrose gradient for sedimentation as previously described. For micrococcal nuclease treatment, cytoplasmic extracts were prepared in a modified polysome buffer (100 mM Tris-HCl, pH 8.4, 50 mM KCl, 25 mM MgCl2, 18 μM cycloheximide, 15.5 μM chloramphenicol, and 0.5% [v/v] Nonidet P-40). After centrifugation, supernantant was incubated with 0.75 units·μL−1 of micrococcal nuclease (Biolabs) for 20 min at room temperature and loaded on a sucrose gradient for sedimentation as previously described.
RNA Analysis
After fractionation, fractions were deproteinized with phenol chloroform/isoamyl alcohol, and RNA was recovered by ethanol precipitation. For miRNA analyses, RNAs extracted from the fractions were separated on a 15% polyacrylamide (w/v) 8 M urea gel and transferred to GeneScreen nylon membranes. DNA oligonucleotides complementary to miRNAs or trans-acting siRNAs were labeled with [γ-32P]ATP using T4 PNK (Promega). Hybridizations were performed at 50°C overnight. Hybridized membranes were exposed to imaging plates that were recorded after 12 h (FLA-5000; Fuji). Total RNA was also extracted from tissues by phenol and chloroform/isoamylic alcohol and was recovered by ethanol precipitation. mRNAs were reverse transcribed with oligo dT(21) by RT-AMV (Promega) and amplified with GoTaq (Promega) during 25 cycles.
The following primers were used for amplification: AGO1, forward 5′-AAGGAGGTCGAGGAGGGTATGG-3′/reverse 5′-GCTGAGAAGACACCGCTTGATAAG-3′; SPL10, forward 5′-GGTGTGGGAGAATGCTCAGGAG-3′/reverse 5′-GAGTGTGTTTGATCCCTTGTGAATCC-3′; ACT2, forward 5′-GCACCCTGTTCTTCTTACCG-3′/reverse 5′-AACCCTCGTAGATTGGCACA-3′; ARF17, forward 5′-AGCACCTGATCCAAGTCCTTCTATG-3′/reverse 5′-TGGTGAATAGCTGGGGAGGATTTC-3′; PPR, forward 5′-CAACTCTCTCATTACTCGCCTTTTCC-3′/reverse 5′-TGCCCCTCTTTCCCATACACATC-3′; CIP4, forward 5′-CAGTGAGTTGACATCTACTCCAGTTAC-3′/reverse 5′-CGTTCACAATTTCTCTTGAAGC-3′; CSD2, forward 5′-ACACGGAGCTCCAGAAGATG-3′/reverse 5′-TCAAGCCAATCACACCACAT-3′. Quantifications were performed on a Bio-Rad IQcycler using the IQ SYBR Green supermix during 40 cycles. Target quantifications were performed with the same primers as above.
Protein Analysis
Proteins were extracted from each fraction of the gradient, resolved on SDS-PAGE, and stained using Coomassie Brilliant Blue. After electroblotting on a nitrocellulose membrane, protein gel blot analysis was performed using antibodies against AGO1 (1:8000), CIP4 (1:3000), or CSD2 (1:5000). The AGO1, CIP4, and CSD2 antibodies have been described previously (Kliebenstein et al., 1998; Yamamoto et al., 2001; Qi et al., 2005). Detection was performed using goat anti-rabbit IgG (H+L) horseradish peroxidase conjugate secondary antibodies (Promega) (1:2000) and TMB-stabilized substrate for horseradish peroxidase (Promega).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: AGO1, At1g48410; SPL10, At1g27370; ACT2, At3g18780; ARF17, At1g77850; PPR, At1g06580; CIP4, At4g00930; CSD2, At2g28190; DCL1, At1g01040; and SCL6-IV, At4g00150.
Supplemental Data
The following material is available in the online version of this article.
Supplemental Figure 1. miRNA Targets Are Less Associated with Polysomes Than mRNA Not Known to Be Targeted by miRNAs.
Supplementary Material
Acknowledgments
We thank Mathew Lewsey and John Carr for providing 2b overexpressing lines, Hervé Vaucheret for providing ago1-25 seeds, and Bruce Veit and Ben Field for careful correction of the manuscript. E.L. was supported by a Commissariat à l'Energie Atomique-Région Provence Côte d'Azur doctoral fellowship, and R.S. and E.D. were supported by Commissariat à l'Energie Atomique doctoral and postdoctoral fellowships, respectively. This work was supported by Commissariat à l'Energie Atomique, Centre National de la Recherche Scientifique, Aix-Marseille University, and the Provence Alpes Côte d'Azur Région.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Christophe Robaglia (christophe.robaglia@univmed.fr).
Online version contains Web-only data.
References
- Allen, E., Xie, Z., Gustafson, A.M., and Carrington, J.C. (2005). microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121 207–221. [DOI] [PubMed] [Google Scholar]
- Arteaga-Vazquez, M., Caballero-Perez, J., and Vielle-Calzada, J.P. (2006). A family of microRNAs present in plants and animals. Plant Cell 18 3355–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aukerman, M.J., and Sakai, H. (2003). Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15 2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backman, T.W., Sullivan, C.M., Cumbie, J.S., Miller, Z.A., Chapman, E.J., Fahlgren, N., Givan, S.A., Carrington, J.C., and Kasschau, K.D. (2008). Update of ASRP: The Arabidopsis Small RNA Project database. Nucleic Acids Res. 36 D982–D985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel, D.P. (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116 281–297. [DOI] [PubMed] [Google Scholar]
- Baumberger, N., and Baulcombe, D.C. (2005). Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl. Acad. Sci. USA 102 11928–11933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brengues, M., Teixeira, D., and Parker, R. (2005). Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science 310 486–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen, P., Sakvarelidze-Achard, L., Bruun-Rasmussen, M., Dunoyer, P., Yamamoto, Y.Y., Sieburth, L., and Voinnet, O. (2008). Widespread translational inhibition by plant miRNAs and siRNAs. Science 320 1185–1190. [DOI] [PubMed] [Google Scholar]
- Carrington, J.C., and Ambros, V. (2003). Role of microRNAs in plant and animal development. Science 301 336–338. [DOI] [PubMed] [Google Scholar]
- Chen, X. (2004). A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303 2022–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio, A., Rehwinkel, J., Stricker, M., Huntzinger, E., Yang, S.F., Doerks, T., Dorner, S., Bork, P., Boutros, M., and Izaurralde, E. (2007). Target-specific requirements for enhancers of decapping in miRNA-mediated gene silencing. Genes Dev. 21 2558–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandikota, M., Birkenbihl, R.P., Hohmann, S., Cardon, G.H., Saedler, H., and Huijser, P. (2007). The miRNA156/157 recognition element in the 3′ UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J. 49 683–693. [DOI] [PubMed] [Google Scholar]
- Gazzani, S., Lawrenson, T., Woodward, C., Headon, D., and Sablowski, R. (2004). A link between mRNA turnover and RNA interference in Arabidopsis. Science 306 1046–1048. [DOI] [PubMed] [Google Scholar]
- Gy, I., Gasciolli, V., Lauressergues, D., Morel, J.B., Gombert, J., Proux, F., Proux, C., Vaucheret, H., and Mallory, A.C. (2007). Arabidopsis FIERY1, XRN2, and XRN3 are endogenous RNA silencing suppressors. Plant Cell 19 3451–3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond, S.M., Boettcher, S., Caudy, A.A., Kobayashi, R., and Hannon, G.J. (2001). Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293 1146–1150. [DOI] [PubMed] [Google Scholar]
- Kim, J., Krichevsky, A., Grad, Y., Hayes, G.D., Kosik, K.S., Church, G.M., and Ruvkun, G. (2004). Identification of many microRNAs that copurify with polyribosomes in mammalian neurons. Proc. Natl. Acad. Sci. USA 101 360–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein, D.J., Monde, R.A., and Last, R.L. (1998). Superoxide dismutase in Arabidopsis: An eclectic enzyme family with disparate regulation and protein localization. Plant Physiol. 118 637–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung, A.K., Calabrese, J.M., and Sharp, P.A. (2006). Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proc. Natl. Acad. Sci. USA 103 18125–18130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewsey, M., Robertson, F.C., Canto, T., Palukaitis, P., and Carr, J.P. (2007). Selective targeting of miRNA-regulated plant development by a viral counter-silencing protein. Plant J. 50 240–252. [DOI] [PubMed] [Google Scholar]
- Maroney, P.A., Yu, Y., Fisher, J., and Nilsen, T.W. (2006). Evidence that microRNAs are associated with translating messenger RNAs in human cells. Nat. Struct. Mol. Biol. 13 1102–1107. [DOI] [PubMed] [Google Scholar]
- Montgomery, T.A., Howell, M.D., Cuperus, J.T., Li, D., Hansen, J.E., Alexander, A.L., Chapman, E.J., Fahlgren, N., Allen, E., and Carrington, J.C. (2008). Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell 133 128–141. [DOI] [PubMed] [Google Scholar]
- Morel, J.B., Godon, C., Mourrain, P., Beclin, C., Boutet, S., Feuerbach, F., Proux, F., and Vaucheret, H. (2002). Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell 14 629–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, P.T., Hatzigeorgiou, A.G., and Mourelatos, Z. (2004). miRNP:mRNA association in polyribosomes in a human neuronal cell line. RNA 10 387–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen, P.H., and Ambros, V. (1999). The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev. Biol. 216 671–680. [DOI] [PubMed] [Google Scholar]
- Pillai, R.S., Bhattacharyya, S.N., Artus, C.G., Zoller, T., Cougot, N., Basyuk, E., Bertrand, E., and Filipowicz, W. (2005). Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science 309 1573–1576. [DOI] [PubMed] [Google Scholar]
- Pillai, R.S., Bhattacharyya, S.N., and Filipowicz, W. (2007). Repression of protein synthesis by miRNAs: How many mechanisms? Trends Cell Biol. 17 118–126. [DOI] [PubMed] [Google Scholar]
- Qi, Y., Denli, A.M., and Hannon, G.J. (2005). Biochemical specialization within Arabidopsis RNA silencing pathways. Mol. Cell 19 421–428. [DOI] [PubMed] [Google Scholar]
- Seggerson, K., Tang, L., and Moss, E.G. (2002). Two genetic circuits repress the Caenorhabditis elegans heterochronic gene lin-28 after translation initiation. Dev. Biol. 243 215–225. [DOI] [PubMed] [Google Scholar]
- Sunkar, R., Kapoor, A., and Zhu, J.K. (2006). Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 18 2051–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia-Sanchez, M.A., Liu, J., Hannon, G.J., and Parker, R. (2006). Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 20 515–524. [DOI] [PubMed] [Google Scholar]
- Vaucheret, H., Vazquez, F., Crete, P., and Bartel, D.P. (2004). The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 18 1187–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J., Yang, J.Y., Niu, Q.W., and Chua, N.H. (2006). Arabidopsis DCP2, DCP1, and VARICOSE form a decapping complex required for postembryonic development. Plant Cell 18 3386–3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, Y.Y., Deng, X., and Matsui, M. (2001). Cip4, a new COP1 target, is a nucleus-localized positive regulator of Arabidopsis photomorphogenesis. Plant Cell 13 399–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., Yuan, Y.R., Pei, Y., Lin, S.S., Tuschl, T., Patel, D.J., and Chua, N.H. (2006). Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev. 20 3255–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.