Abstract
Resistance to bacterial speck disease in tomato (Solanum lycopersicum) is activated upon recognition by the host Pto kinase of either one of two sequence-unrelated effector proteins, AvrPto or AvrPtoB, from Pseudomonas syringae pv tomato (Pst). Pto induces Pst immunity by acting in concert with the Prf protein. The recently reported structure of the AvrPto-Pto complex revealed that interaction of AvrPto with Pto appears to relieve an inhibitory effect of Pto, allowing Pto to activate Prf. Here, we present the crystal structure of the Pto binding domain of AvrPtoB (residues 121 to 205) at a resolution of 1.9Å and of the AvrPtoB121-205–Pto complex at a resolution of 3.3 Å. AvrPtoB121-205 exhibits a tertiary fold that is completely different from that of AvrPto, and its conformation remains largely unchanged upon binding to Pto. In common with AvrPto-Pto, the AvrPtoB-Pto complex relies on two interfaces. One of these interfaces is similar in both complexes, although the primary amino acid sequences from the two effector proteins are very different. Amino acid substitutions in Pto at the other interface disrupt the interaction of AvrPtoB-Pto but not that of AvrPto-Pto. Interestingly, substitutions in Pto affecting this unique interface also cause Pto to induce Prf-dependent host cell death independently of either effector protein.
INTRODUCTION
Tomato (Solanum lycopersicum) plants expressing the disease resistance (R) protein Pto (Pseudomonas syringae pv tomato [Pst]), a protein kinase, are resistant to P. s. tomato strains expressing either of the type III effectors AvrPto or AvrPtoB (Pedley and Martin, 2003). Extensive mutational analysis and yeast two-hybrid (Y2H) studies have demonstrated that interaction between Pto and AvrPto or AvrPtoB is the molecular basis for pathogen recognition and activation of the immune response (Scofield et al., 1996; Tang et al., 1996; Kim et al., 2002). Prf, a protein with a nucleotide binding site and leucine-rich repeats, is required for Pto-mediated resistance to Pst (Salmeron et al., 1996), and Pto/Prf have been shown to occur in a complex in the plant cell (Salmeron et al., 1996; Mucyn et al., 2006).
Pto autophosphorylates its Thr-199 residue (Sessa et al., 2000), thereby stabilizing the kinase P+1 loop and allowing interaction with AvrPto (Xing et al., 2007). Interaction between Pto and AvrPto is highly specific; AvrPto does not interact with the Fen kinase, a closely related Pto family member that shares >80% sequence identity with Pto (Scofield et al., 1996; Tang et al., 1996; Jia et al., 1997; Frederick et al., 1998; Chang et al., 2001; Kim et al., 2002). The inability of Fen to interact with AvrPto stems from the fact that three residues, His-49, Val-51, and Thr-204, important for Pto recognition of AvrPto, are not conserved in Fen (Xing et al., 2007). Substitutions at certain amino acid residues clustered around the P+1 loop of Pto cause Pto to induce an AvrPto-independent, but Prf-dependent, host immune response (i.e., a constitutive gain of function (CGF) phenotype). This CGF phenotype suggests that these Pto residues normally play a role in inhibiting Prf-mediated defense signaling (Wu et al., 2004; Xing et al., 2007). Structural studies identified a second loop of Pto that also negatively regulates Prf-mediated defenses in the absence of AvrPto. AvrPto interacts with both of these immunity inhibition loops of Pto (Xing et al., 2007), supporting the hypothesis that AvrPto triggers disease resistance, in part, by disrupting the inhibitory effects of Pto on Prf signaling (Xing et al., 2007).
In vitro studies revealed that interaction with AvrPto inhibits Pto kinase activity. However, this inhibition does not appear to be the signal to trigger the Pto/Prf-mediated defense response, as the mutant PtoH49E/V51D, which is insensitive to AvrPto inhibition, still causes a CGF phenotype (Xing et al., 2007). AvrPto is also known to target mitogen-activated protein kinase signaling to suppress pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) in Arabidopsis thaliana (He et al., 2006). We therefore proposed previously that Pto may act as a decoy by mimicking a kinase that plays a critical role in PTI. Binding of AvrPto by Pto might then interfere with the ability of the effector to disrupt PTI (Xing et al., 2007). Indeed, it was shown recently that AvrPto interacts with the kinase domains of both FLS2/EFR1 from Arabidopsis and inhibits their kinase activity that is needed for PTI (Xiang et al., 2008). Moreover, both the FLS2 kinase domain and Pto bind AvrPto in a competitive manner, supporting structural similarity between FLS2-AvrPto and Pto-AvrPto interaction (Xiang et al., 2008). Interestingly, AvrPto also appears to bind BAK1 (Shan et al., 2008), a protein required for FLS2 and EFR1 function (Chinchilla et al., 2007; Heese et al., 2007).
The second type III effector involved in the tomato–Pst interaction is AvrPtoB, a protein with several discrete domains. The N-terminal domain, AvrPtoB1-307, interacts with Pto and elicits Prf-dependent resistance (Xiao et al., 2007). In tomato lines lacking Pto or Prf, the AvrPtoB1-307 domain exhibits a virulence activity, which promotes ethylene production and enhances growth of Pst (Cohn and Martin, 2005; Xiao et al., 2007). AvrPtoB121-200 has been shown to be necessary and sufficient for this interaction with Pto (Xiao et al., 2007). A longer N-terminal region, AvrPtoB1-387 (but not AvrPtoB1-307), is able to suppress certain PTI responses in Arabidopsis (He et al., 2006) by directly interacting with BAK1 (Shan et al., 2008). The same AvrPtoB fragment was recently shown to interact with Fen, activating Prf-dependent resistance to Pst (Rosebrock et al., 2007). A C-terminal domain of AvrPtoB has evolved to be an E3 ubiquitin ligase that targets Fen for degradation in a proteasome-dependent manner, disrupting Fen-mediated resistance (Abramovitch et al., 2006; Janjusevic et al., 2006; Rosebrock et al., 2007). Elimination of Fen allows the PTI-suppressing ability of AvrPtoB1-387 to function. Thus, much is known about AvrPtoB, and the protein has proven to be an excellent model for use in studying the coevolution between Pst and its host tomato.
Here, we present the crystal structures of AvrPtoB and of the complex of AvrPtoB bound to Pto. These structures reveal that AvrPtoB shares very little structural similarity with AvrPto. In addition, although the AvrPtoB-Pto complex has two interfaces just as AvrPto-Pto does, one of the interfaces is unique. Site-directed mutagenesis confirmed the importance of these two interfaces and also provided further support for a model in which AvrPtoB, like AvrPto, interferes with a negative regulatory function of Pto.
RESULTS
Biochemical Characterization of the AvrPtoB–Pto Interaction
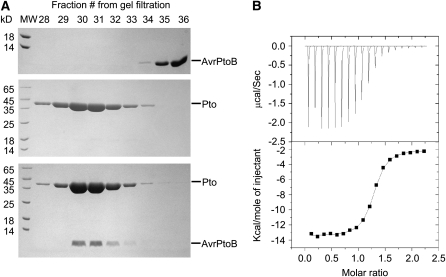
Although genetic and Y2H studies have shown that the interaction between AvrPtoB and Pto is an important event for triggering Pto/Prf-dependent immunity, this interaction has not been verified in vitro using purified proteins. To facilitate the structural study of the AvrPtoB-Pto complex, we purified full-length Pto and AvrPtoB121-205, a region sufficient for interaction with Pto, to homogeneity and then examined their interaction using a gel filtration assay. In agreement with previous results (Xiao et al., 2007), Pto formed a stable complex with AvrPtoB121-205 in solution, as indicated by the comigration of the two proteins (Figure 1A). To further characterize the interaction between Pto and AvrPtoB121-205, we measured their binding affinity using isothermal titration calorimetry (ITC). The ITC results showed that AvrPtoB121-205 interacts with full-length Pto with a dissociation constant of 1.1 μM (Figure 1B) compared with 0.11 μM for AvrPto–Pto interaction quantified by surface plasmon resonance (Xing et al., 2007).
Figure 1.
Characterization of the Interaction between AvrPtoB121-205 and Pto.
(A) A representative gel filtration experiment examining the complex between the Pto binding domain of AvrPtoB (residues 121 to 205) and full-length Pto. Aliquots of the gel filtration fractions were visualized by Coomassie blue staining following SDS-PAGE. The AvrPtoB121-205 fragment was eluted in fractions 35 and 36 when incubated alone and in fractions 30 to 32 when incubated with Pto. MW, molecular weight standards (in kilodaltons) are shown on the left side.
(B) Measurement of binding affinity between Pto and AvrPtoB121-201 by ITC. Top panel: Raw ITC data. Eighteen injections of AvrPtoB121-201 solution were added to the Pto protein solution in the ITC cell. The area of each injection peak corresponds to the total heat released for that injection. Bottom panel: the binding isotherm for AvrPtoB121-205–Pto interaction. The integrated heat is plotted against the molar ratio of AvrPtoB121-205 added to Pto in the cell. Data fitting revealed a binding affinity of 1.1 μM.
Crystal Structure of AvrPtoB121-205
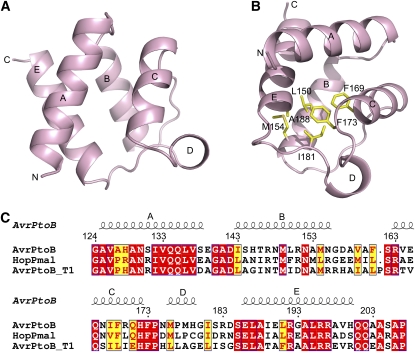
We began by determining the crystal structure of the Pto-interacting domain, AvrPtoB121-205, alone by multiwavelength anomalous dispersion using a single crystal of Se-Met–substituted protein and refined to 1.9-Å resolution (Table 1). The structure of AvrPtoB121-205 is composed primarily of four short α-helices (αA, αB, αC, and αE), forming a globular four-helix bundle with a size of ∼26Å×26Å×23Å (Figure 2A). Located between α-helices C and E is a long rigid loop, with a shorter α-helix (αD) lying within it. Stabilization of this loop in AvrPtoB is achieved mainly through the extensive van der Waals contacts of Ile-181 with residues Leu-150, Met-154, Phe-169, Phe-173, and Ala-188 (Figure 2B). AvrPtoB121-205 has a completely different structure than AvrPto, which consists of an elongated three-helix bundle (Wulf et al., 2004). A structure-based primary sequence alignment showed the amino acids involved in formation of the four-helix bundle in AvrPtoB121-205 are generally conserved among different AvrPtoB homologs (Figure 2C). A Distance mAtrix aLIgnment (DALI) (Holm et al., 2008) search using the refined crystal structure of AvrPtoB121-205 revealed a few structural homologs, with a C-terminal DNA binding domain of the transcriptional pleiotropic repressor CodY (Levdikov et al., 2006) being the most similar one. However, it remains unknown if such similarity is biologically relevant.
Table 1.
Summary of Crystallography Analysis
| AvrPtoB121-205
|
||||
|---|---|---|---|---|
| Se-AvrPtoB B121-205
|
||||
| Peak | Inflection | Remote | AvrPtoB121-205-Pto Complex | |
| Data Set | ||||
| Wavelength (Å) | 0.9789 | 0.9794 | 0.964 | 1.0 |
| Resolution (Å) | 99.0–1.9 | 99.0–1.9 | 99.0–2.0 | 99.0–3.3 |
| Space group | P3121 | P212121 | ||
| Cell dimensions | ||||
| a, b, c (Å) | 77.22, 77.22, 46.72 | 61.07, 104.47, 298.86 | ||
| α, β, γ (°) | 90.0, 90.0, 120.0 | 90.0, 90.0, 90.0 | ||
| Completeness | 99.8% (99.6) | |||
| Rsym | 0.048 (0.35) | 0.051 (0.37) | 0.052 (0.35) | 0.081 (0.530) |
| Data redundancy | 6.9 (6.6) | 6.9 (6.4) | 7.0 (6.1) | 5.6 (5.3) |
| Average I | 22.4 (3.1) | 21.7 (2.8) | 21.4 (2.7) | 22.3 (2.9) |
| Figure of merit (after SOLVE) | 0.77 | |||
| Statistics for refinement | ||||
| Resolution range | 20–1.9 (Å) | 20–3.30 (Å) | ||
| No. of reflections | 12,921 | 28,059 | ||
| Number of atoms | ||||
| Protein | 606 | 11,607 | ||
| Water | 61 | |||
| Completeness | 99.7% | 98.2% | ||
| Rwork (Rfree) | 23.5% (24.9%) | 31.7% (33.1%) | ||
| RMSD bond length (Å) | 0.005 | 0.009 | ||
| RMSD bond angles (°) | 1.119 | 1.320 | ||
Rsym = ΣhΣi|Ih,i − Ih|/ΣhΣiIh,i, where Ih is the mean intensity of the i observations of symmetry related reflections of h. R = Σ|Fobs − Fcalc|/ΣFobs, where Fobs = FP, and Fcalc is the calculated protein structure factor from the atomic model (Rfree was calculated with 5% of the reflections). RMSD in bond lengths and angles are the deviations from ideal values and the root mean square deviation.
Figure 2.
AvrPtoB121-205 Has a Distinct Fold from That Present in AvrPto.
(A) Structure of AvrPtoB121-205, the domain required for interaction with Pto. The five α-helices in AvrPtoB121-205 are labeled as A, B, C, D, and E. N, N terminus; C, C terminus.
(B) Ile-181 in AvrPtoB forms extensive hydrophobic interactions with its neighboring residues for stabilizing the conformation of the rigid coil linking αC and αE. The side chains of certain AvrPtoB residues are shown in yellow.
(C) The structure-based sequence alignment among AvrPtoB homologs (Lin and Martin, 2006). Identical amino acids are boxed in red; similar amino acids are boxed in yellow. The locations of the five α-helices are shown above the alignment. The program ClustalW was used for sequence alignment.
Overall Structure of the AvrPtoB121-205-Pto Complex
To elucidate the structural basis for recognition of AvrPtoB by Pto, we sought to solve the structure of the AvrPtoB121-205-Pto complex. To this end, the two purified proteins were mixed together and subjected to size exclusion chromatography to obtain the correct stoichiometry of the complex. The crystal structure of this complex was determined by molecular replacement using the coordinates of Pto and AvrPtoB121-205 as the searching models and eventually refined to a resolution of 3.3 Å (see Methods and Table 1).
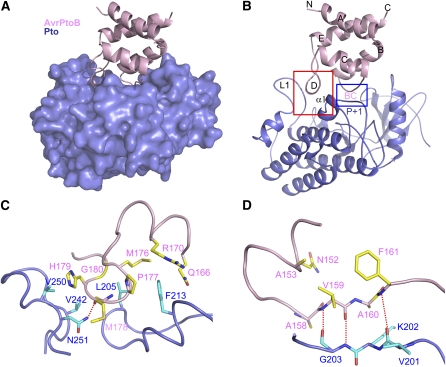
The AvrPtoB121-205-Pto interaction results in a 1:1 complex (Figure 3A) and burial of 1880 Å2 of the surface area that is exposed on the two molecules. The interaction of the two proteins results from two contact surfaces. Interface 1 is created by the interaction of the short helix αD of AvrPtoB121-205 with a surface groove of Pto formed by loop L1 and helix α1 adjacent to the P+1 loop (Figure 3B). Interface 2 involves the loop linking αB and αC in AvrPtoB and the P+1 loop of the Pto kinase (Figure 3B). The P+1 loop of kinases is involved in binding substrates and the interaction of AvrPtoB with this region, suggesting that the effector may compete with certain substrates for binding of Pto.
Figure 3.
Specificity Determinants for Recognition of AvrPtoB by Pto.
(A) Overall structure of the complex between AvrPtoB121-205 and Pto. AvrPtoB121-205 and Pto are colored in pink and slate, respectively. Pto is shown as a surface representation.
(B) Illustration of the overall structure of the complex between AvrPtoB121-201 and Pto with the same orientation as in (A). The red frame highlights interface 1, and the blue frame highlights interface 2 of the complex. BC represents the loop linking helices B and C in AvrPtoB.
(C) The detailed interactions around interface 1 highlighted in the red frame shown in (B). The side chains of AvrPtoB and Pto are shown in yellow (labeled in pink) and cyan (labeled in dark slate), respectively. Relevant amino acid residues are numbered, and a hydrogen bond is shown as a red dashed line.
(D) The detailed interactions around interface 2 highlighted in the blue frame shown in (B). The side chains of AvrPtoB and Pto are shown in yellow and cyan, respectively. Relevant amino acid residues are numbered, and hydrogen bonds are shown as red dashed lines.
In complex with AvrPtoB121-205, Pto adopts a typical kinase fold with a smaller N-terminal lobe consisting mostly of β-sheets with one helix and a larger, predominantly helical C-terminal lobe (Figure 3B). The conformation of AvrPtoB121-205 in complex with Pto remains essentially unchanged compared with that of its free form with a root mean square deviation of 0.18 Å. This is in contrast with AvrPto whose CD loop is flexible in its isolated form but becomes stabilized following interaction with Pto (Xing et al., 2007).
Specific Recognition of AvrPtoB121-205 by Pto
In interface 1 of the complex, Pto Phe-213 makes extensive hydrophobic contacts with AvrPtoB residues Met-176 and Pro-177 (side chain and Cα atom) and the aliphatic portion of Gln-166 and forms π-charge interactions with AvrPtoB Arg-170 (Figure 3C). Further reinforcement of the interactions involved in interface 1 result from Leu-205 in Pto forming hydrophobic contacts with Cα atoms of AvrPtoB residues Gly-180 and Met-176 as well as the side chain of Met-176. In addition, Asn-251 in Pto makes a hydrogen bond with the carbonyl oxygen of AvrPtoB Met-178 (Figure 3C). Val-242 and Val-250 in Pto also appear to contribute to interaction with AvrPtoB by making hydrophobic contact the Cα atom and side chain, respectively, of His-179 in AvrPtoB.
Interface 2 is similar to that of the AvrPto-Pto complex, where the Pto P+1 loop forms hydrogen bonds with AvrPto (Xing et al., 2007). In the AvrPtoB-Pto complex, the Pto P+1 loop makes three backbone hydrogen bonds with the AvrPtoB loop linking αB and αC (we refer to this as the BC loop; Figure 3D).
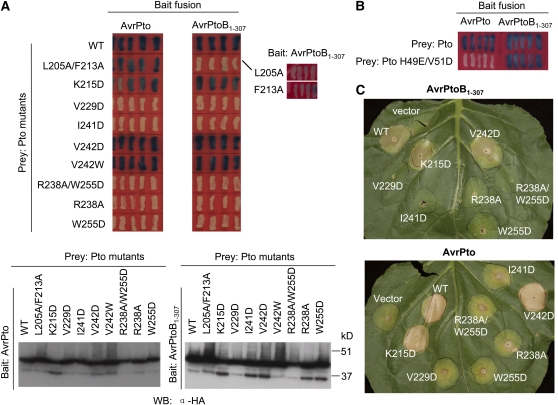
Structure-Based Mutagenesis of AvrPtoB Residues Involved in the Interaction with Pto
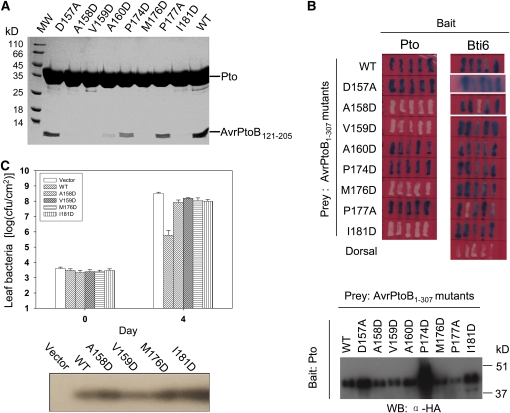
To experimentally test the two interfaces observed in the crystal structure of the AvrPtoB121-205-Pto complex, we made the following substitutions in AvrPtoB121-205: M176D, P177A, and I181D (all involved in interface 1) and A158D, V159D, and A160D (involved in interface 2). We also mutated two AvrPtoB residues Pro-174 and Asp-157 that are not involved in interaction with Pto. Each protein was purified to homogeneity, and its interaction with Pto was investigated using a gel filtration assay. As shown in Figure 4A, wild-type AvrPtoB121-205 formed a stable complex with Pto in this assay, whereas two of the substitutions that were predicted to disrupt interface 1 abolished the interaction. Specifically, an Asp substitution in AvrPtoB at residue Met-176, which makes van der Waals contacts with Leu-205/Phe-213 in Pto, and the substitution I181D, which is expected to destabilize helix αD, both abolished AvrPtoB interaction with Pto. The substitution in interface 1, P177A, affected binding to Pto only slightly (based on the gel filtration experiment) probably because Ala can still form hydrophobic contact with Pto Phe-213. The mutation P174D in AvrPtoB appeared to slightly reduce interaction with Pto in the gel filtration assay, which could result from increased conformational freedom around αD caused by this mutation.
Figure 4.
Pto-Interacting Residues of AvrPtoB Are Important for Its Avirulence Activity.
(A) Effects of substitutions in AvrPtoB121-205 on the interaction with Pto as determined by gel filtration. Each AvrPtoB protein was mixed with full-length Pto and subjected to a gel filtration assay. Aliquots of the fraction corresponding to the peak of AvrPtoB121-205–Pto complex were visualized by Coomassie staining following SDS-PAGE.
(B) Effects of substitutions in AvrPtoB1-307 on the interaction with Pto as determined by a Y2H assay. Blue patches indicate positive interactions. Protein gel blotting with anti-HA antibody in the bottom panel shows similar expression in yeast of wild-type AvrPtoB1-307 and the mutant proteins.
(C) Effects of point mutations in AvrPtoB1-307 on its avirulence activity. Pst DC3000ΔavrPtoΔavrPtoB strains expressing wild-type AvrPtoB1-307, AvrPtoB1-307 mutants, or carrying an empty vector were vacuum inoculated into leaves of tomato RG-PtoR plants (Xiao et al., 2007). Bacterial populations in leaves were determined 0 and 4 d after inoculation. Experiments were repeated twice with similar results. Protein gel blotting with anti-AvrPtoB antibody in the bottom panel shows that all of the proteins were secreted at similar levels from Pst. cfu/cm2 = colony-forming units per centimeter2.
Three of the four AvrPtoB substitutions predicted to affect interface 2 greatly affected the interaction with Pto. Specifically, Asp substitutions at AvrPtoB residues Ala-158 and Ala-160, which would generate steric hindrance with Asp-164 and Lys-202, respectively, in Pto (Figure 3D), abolished or greatly impaired interaction with Pto. In addition, an AvrPtoB Val-159 substitution also disrupted association with Pto (Figure 4A). Val-159 is not directly involved in interaction with Pto, but it appears to play an important role in stabilizing the loop that forms hydrogen bonds with the Pto P+1 loop (Figure 3B). As a control, the D157A substitution did not affect the interaction with Pto as it is located outside the two interfaces.
Although AvrPtoB121-205 is sufficient for binding Pto, a longer region, AvrPtoB1-307, is necessary for triggering Pto-mediated immunity in tomato leaves (Xiao et al., 2007). To examine residues in the two interfaces more fully, we incorporated the substitutions described above into AvrPtoB1-307 and evaluated their effects on interaction with Pto using a Y2H assay. All but one of the substitutions in AvrPtoB121-205 that disrupted interface 1 or 2 with Pto in the gel filtration assays also led to loss of AvrPtoB1-307 binding to Pto in the Y2H system (Figure 4B, top panel; note however, that AvrPtoBA160D interacted very poorly in the gel filtration assay). Normal accumulation of each of the altered AvrPtoB proteins was demonstrated by protein gel blotting (Figure 4B, bottom panel). In addition, each of the AvrPtoB proteins was shown to interact normally with a previously isolated AvrPtoB tomato-interacting (Bti) protein Bti6 (Bti6 is a putative lactate dehydrogenase; Xiao et al., 2007), indicating that these substitutions did not change the overall conformation of AvrPtoB1-307 but specifically disrupted the interaction with Pto.
To investigate the effects of these substitutions on AvrPtoB avirulence activity, the four AvrPtoB1-307 substitutions that were unable to interact with Pto in yeast (M176D and I181D [interface 1] and A158D and A159D [interface 2]) were introduced individually into the broad-host-plasmid pCPP45 under the control of its hrp promoter, and the resulting constructs were transformed into DC3000ΔavrPtoΔavrPtoB (Xiao et al., 2007). After each protein was shown to be expressed and secreted normally from Pst, the strains were inoculated into leaves of Pto-expressing tomato plants, RG-PtoR (Figure 4C). Bacterial population assays revealed that each of the AvrPtoB substitutions that disrupted interaction with Pto also abolished AvrPtoB avirulence activity. Collectively, these results are consistent with previous observations that the amino acids of AvrPtoB required for interaction with Pto are important for eliciting Pto-mediated immunity in tomato.
Structure-Based Mutagenesis of Pto Residues Involved in the Interaction with AvrPtoB
To further examine the interface surfaces between AvrPtoB121-205 and Pto, we made amino acid substitutions in Pto and evaluated their effects on the interaction with AvrPtoB1-307 using the Y2H system. Protein gel blotting confirmed that each of the altered Pto proteins accumulated normally in yeast (Figure 5A). Consistent with our structural observations of interface 1, individual substitutions to Ala at Pto Leu-205 or Phe-213, each of which makes hydrophobic contacts with AvrPtoB121-205, greatly impaired the Pto interaction with AvrPtoB1-307. A Pto protein with both substitutions was completely unable to interact with AvrPtoB1-307. Notably, the PtoL205A/F213A protein retained the ability to interact with AvrPto (Figure 5A). This observation is consistent with these two residues being involved in a unique interface with AvrPtoB compared with AvrPto. An Asp substitution at Pto Lys-215, which is distantly located from either of the interfaces, caused no detectable effect on interaction with either AvrPto or AvrPtoB1-307 (Figure 5A, top panel). Pto substitutions V242D or V242W that would appear to disturb the hydrophobic contact of this Val with the Cα atom of AvrPtoB H179 nevertheless had no effect on interaction with AvrPtoB1-307 in the Y2H assay, suggesting this contact is not critical (Figure 5A). Based on the complex structure, two residues of Pto, His-49 and Val-51, which are important for the interaction of the kinase with AvrPto (Xing et al., 2007), are not involved in the interaction with AvrPtoB. Indeed, a Pto protein with these two substitutions interacts normally with AvrPtoB1-307 but does not associate with AvrPto (Figure 5B).
Figure 5.
Pto Substitutions L205A/F213A Disrupt Interaction of Pto with AvrPtoB but Not AvrPto.
(A) Effects of substitutions in Pto on its interaction with AvrPtoB1-307 and AvrPto as determined by a Y2H assay. Blue patches indicate positive interactions. Protein gel blotting with anti-HA antibody in the bottom panel shows equal expression of wild-type Pto and derived mutants in yeast.
(B) Effects of Pto substitutions H49E/V51D on Pto interaction with AvrPtoB1-307 or AvrPto as determined by a Y2H assay.
(C) Effect of Pto mutations on effector-elicited cell death in N. benthamiana leaves. Agrobacterium-mediated transient coexpression of wild-type Pto or derived mutants together with AvrPtoB1-307 or AvrPto in N. benthamiana leaves. AvrPto alone causes weak cell death in this assay (see vector control), but much stronger cell death is apparent when AvrPto is coexpressed with wild-type Pto or the Pto K215D and V242D proteins. Note that the wild-type Pto, Pto K215D, or Pto V242D proteins, when expressed alone (without an effector protein), do not cause cell death (see Figure 6A).
The structure also shows that loop L1 of Pto is involved in the interaction with AvrPtoB121-205 (Figure 3B). To test the functional significance of this interaction, we made substitutions in Pto residues that are involved in stabilizing the conformation of this loop (e.g., R238A, I241D, W255D, and R238A/W255D). Each of these substitutions resulted in insoluble Pto when expressed in Escherichia coli, and they were unable to be analyzed by gel filtration. The proteins did appear to accumulate in yeast, and none of them interacted with AvrPtoB1-307 or AvrPto in a Y2H assay (Figure 5A).
To test whether these Pto substitutions affected AvrPtoB-triggered plant immunity, we coexpressed the various Pto mutants with AvrPtoB1-307 or AvrPto in leaves of Nicotiana benthamiana using Agrobacterium tumefaciens–mediated transient expression. As shown in Figure 5C, coexpression of AvrPtoB1-307 or AvrPto with the proteins PtoK215D or PtoV242D (PtoV242W was not tested), both of which retained interaction with AvrPtoB, resulted in rapid death of the inoculated tissue. By contrast, none of the Pto proteins with substitutions that abolished interaction with AvrPtoB caused cell death when coexpressed with this effector protein. The same result was obtained with AvrPto, although this assay is less clear because AvrPto alone causes weak cell death when expressed in N. benthamiana leaves (Figure 5C; He, et al. 2004).
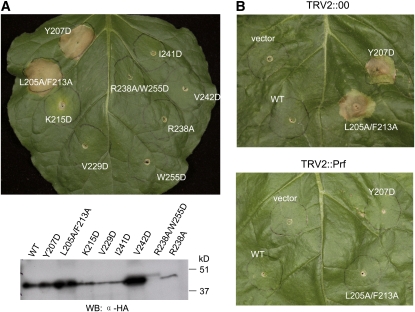
PtoL205A/F213A Interacts with AvrPto but Not AvrPtoB and Possesses CGF Activity
Previously, substitutions at several Pto residues that are involved in the interaction with AvrPto were shown to both disrupt the AvrPto–Pto interaction and to cause a CGF phenotype in the N. benthamiana cell death assay (Wu et al., 2004; Xing et al., 2007). This CGF phenotype is postulated to be due to interference in the negative regulation that Pto exerts on Prf. We tested whether substitutions at Pto Leu-205 or Phe-213, which completely abolish interaction with AvrPtoB but retain the ability to interact with AvrPto, also cause Pto CGF activity by again using the N. benthamiana agroinoculation assay. As anticipated, the PtoL205A/F213A protein caused cell death in this assay in the absence of either AvrPtoB1-307 or AvrPto (Figure 6A). The CGF activity of PtoL205A/F213A is Prf dependent because expression of this protein did not cause cell death on Prf-silenced N. benthamiana leaves (Figure 6B). It was shown previously that single substitutions of L205D or F213D in Pto abolished interaction with AvrPto and AvrPtoB and caused a CGF phenotype (Wu et al., 2004). The reason for this conflicting result is likely due to the greater impact of the charged Asp substitutions on hydrophobic contacts with AvrPto compared with our Ala substitutions.
Figure 6.
The PtoL205A/F213A Protein Has Prf-Dependent CGF Activity.
(A) Agrobacterium-mediated transient expression of PtoL205A/F213A in N. benthamiana leaves caused rapid cell death independent of AvrPto or AvrPtoB. Leaves were inoculated with Agrobacterium strains that deliver T-DNAs encoding the Pto variant proteins with the amino acid substitutions indicated. Expression of the Pto variant proteins in N. benthamiana leaves was confirmed by protein blots as shown at the bottom using an anti-HA antibody.
(B) Prf is required for hypersensitive response triggered by PtoL205A/F213A. N. benthamiana plants were subjected to virus-induced gene silencing using a tobacco rattle virus (TRV2) construct carrying a fragment of Prf. The top panel shows a control leaf from a plant infected with TRV alone. The bottom panel shows a leaf silenced for the Prf gene.
Transient expression in N. benthamiana leaves of other Pto mutants that probably destabilize the L1 loop did not trigger a CGF phenotype (Figure 6A). However, it is important to note that, with the exception of PtoL205A/F213A, the expression level in plants of these altered proteins was much lower compared with wild-type Pto and the other proteins that are still able to interact with AvrPtoB (e.g., Pto Lys-215 and V242D; Figure 6). However, this is not likely the reason they did not have CGF activity because Pto (Y207D), a previously identified CGF mutant (Rathjen et al., 1999), was also expressed at a similarly low level but caused strong cell death (Figure 6A). Taken together with previous studies (Wu et al., 2004, Xing et al., 2007), these results support the hypothesis that binding of Pto by AvrPto or AvrPtoB interferes with different inhibitory residues of Pto to trigger Prf-dependent immune responses.
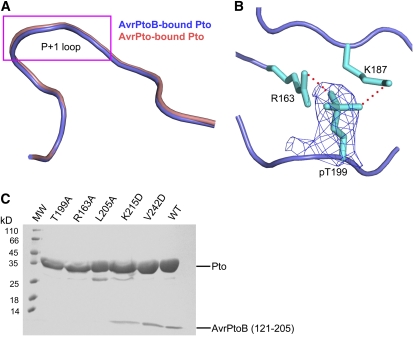
The Active Conformation of Pto Is Required for Interaction with AvrPtoB121-205
The Pto kinase must be in its active conformation to interact with AvrPto (Xing et al., 2007). Structural comparisons revealed that the P+1 loop of Pto has a nearly identical conformation whether it is forming a complex with AvrPto or with AvrPtoB121-205 (Figure 7A), indicating that Pto is also in the active conformation in the AvrPtoB121-205-Pto structure. Indeed, Pto Thr-199, whose autophosphorylation is important for maintaining the active conformation of the Pto P+1 loop, is phosphorylated in our structure as shown by the electron density and previously by mass spectrometry (Figure 7B; Xing et al., 2007). To test whether phosphorylation of this residue is also required for interaction with AvrPtoB, we made the substitution of T199A in Pto, purified the protein to homogeneity, and characterized its interaction with AvrPtoB121-205. As expected, the PtoT199A protein was unable to interact with AvrPtoB121-205 in the gel filtration assay, whereas the control wild-type Pto formed a stable complex with the same AvrPtoB fragment (Figure 7C). Similarly, substitutions at other Pto residues (e.g., Arg-163 and Leu-205) that are important for the active conformation of the kinase (Xing et al., 2007) also abolished their interaction with AvrPtoB121-205 (Figure 7C). These results indicate that, as with AvrPto, the active conformation of Pto is required for its interaction with AvrPtoB121-205.
Figure 7.
The Active Conformation of Pto Is Important for Pto Interaction with AvrPtoB121-205.
(A) Pto P+1 loop adopts a similar conformation in its complex with AvrPto or AvrPtoB121-205. Superimposition of Pto around the P+1 loop region from AvrPtoB121-205-Pto (slate) and AvrPto-Pto (pink) complexes.
(B) Pto T199 in AvrPtoB121-205-Pto complex is phosphorylated as indicated by electron density. Omit electron density map around PtoT199 (shown at 1.2 sigma). The map was calculated using the CNS program. Hydrogen bonds are represented by red dashed lines. PtopT199 was not used for calculation of the electron density map.
(C) Effects of various Pto substitutions on the interaction of Pto with AvrPtoB121-205. Gel filtration was used to assay the interaction of Pto proteins and AvrPtoB121-205 as described in Figure 1A.
DISCUSSION
We determined the crystal structures of AvrPtoB121-205 alone and in complex with full-length Pto. The structure of AvrPtoB121-205 is mainly α-helical as predicted previously (Xiao et al., 2007). However, in contrast with the elongated three-helix bundle found in AvrPto and that predicted for AvrPtoB121-200 (Wulf et al., 2004; Xiao et al., 2007), the α-helices in AvrPtoB121-205 form a globular four-helix bundle (Figure 2A). Despite this striking structural difference between AvrPtoB121-205 and AvrPto, both effectors interact directly with the Pto kinase with a comparable binding affinity (Figure 1B; Xing et al., 2007).
The structure of the AvrPtoB-Pto complex reveals that the interaction of these two proteins is mediated by two interfaces. Supporting that the P+1 loop of Pto acts as a shared binding site for AvrPtoB and AvrPto, many mutations in this region resulted in loss of Pto interaction with both effector proteins. Interaction between AvrPtoB and Pto at this interface was further verified by the mutations in AvrPtoB (Figures 4A and 4B). Interestingly, Pto employs a distinct surface for recognition of AvrPtoB compared with AvrPto. In agreement with this, mutation of residues such as M176D and I181D in AvrPtoB from the unique interface 1 (Figure 3C) disrupted the AvrPtoB–Pto interaction (Figures 4A and 4B) and abolished AvrPtoB-induced resistance (Figure 4C).
Our structure also sheds light on previously reported mutations in AvrPtoB and Pto that affect their interactions. Five substitutions in AvrPtoB (F173A, E165K, F169S, G180V, and L195H) caused a loss of interaction with Pto (Xiao et al., 2007). Some of these mutations (E165K, F169A, and L195A) may disturb the structural integrity of the Pto-interacting domain and result in loss of interaction with Pto. For example, the inability of G180V and F173A mutants to interact with Pto may stem from the fact that these two residues contribute to stabilizing the conformation of the αD helix.
Leu-205 in Pto makes hydrophobic contacts with the Ca atoms from the αD helix of AvrPtoB (Figure 3C). Mutation of this residue to the smaller residue Ala (Figure 5A) or the charged residue Asp (Wu et al., 2004) greatly impairs its interaction with Pto. By contrast, the substitution L205I still interacted with AvrPtoB (Kim et al., 2002), presumably because Ile has a similar size to Leu. Asn-251 in Pto is involved in interaction with AvrPtoB but not with AvrPto. Consistently, the substitution N251K specifically disrupted interaction with AvrPtoB (see Supplemental Figure 1 online). Substitutions at several buried residues in the L1 loop (e.g., R238A, I241D, W255D, and R238A/W255D) abolished interaction with both effector proteins even though these residues are not directly involved in the protein–protein interaction. The reason for this may be that these mutations have a deleterious effect on folding of Pto kinase. Notably, substitutions at other buried residues (Leu-252, Ala-256, and Val-257) around L1 loop resulted in no expression of Pto in plant cells (Wu et al., 2004), whereas mutations of certain solvent-exposed residues in this region (Glu-248 and Glu-254) had no effect on interaction with either AvrPtoB or AvrPto (Bernal et al., 2005). Our current data support that the buried residues from this region are important for proper folding of Pto as suggested before (Bernal et al., 2005).
Other effectors in the HopAB (AvrPtoB) family are recognized by Pto and elicit an immune response in plant leaves (Lin and Martin, 2006). However, except Met-176, other Pto-interacting residues from AvrPtoB are not conserved in these other HopAB effectors (Figure 2C). How does Pto recognize these other HopAB effectors? Interactions in the AvrPtoB-Pto complex are mainly mediated by main chain contacts (Figures 3C and 3D), which would not be strictly dependent on the identity of the residues involved in binding. This is exemplified by the interaction of the Pto P+1 loop with the CD loop of AvrPto and the BC loop of AvrPtoB, which do not have significant amino acid identity to each other. However, proper conformation of the short helix αD and the BC loop are absolutely required for AvrPtoB binding to Pto because the AvrPtoB substitutions V159D and I181D, which can locally disturb the conformations of the two Pto-interacting regions, resulted in loss of interaction with Pto (Figures 4A and 4B).
The conserved residues within the Pto binding domain of the AvrPtoB homologs are clustered around αA αB, αC, and αE helices (Figure 2C). Although the residues surrounding αB are comparatively variable, all three residues (Ile-143, Leu-150, and Met-154) involved in the four-helix bundle are conserved. These results suggest that the other HopAB effectors can form a similar four-helix bundle, thus presenting the two Pto-interacting regions in similar conformations and allowing their recognition by Pto. Therefore, recognition of the peptide motifs in AvrPtoB and its homologs by Pto is not governed by their primary sequence, but by their tertiary contacts with Pto. Such a strategy might aid Pto in accommodating mutations in Pto-interacting residues of AvrPtoB and its homologs.
A previous structural study suggested that AvrPto triggers Pto/Prf-mediated immune responses by removing the inhibitory effect on Prf signaling imposed by Pto (Xing et al., 2007). Our data indicate that AvrPtoB likely uses a similar mechanism for activation of plant immunity, although these two effector proteins exhibit strikingly different tertiary structures. Residues surrounding the Pto P+1 loop are important for negative regulation of immune signaling by Pto, as many substitutions at this region induced the hypersensitive response (HR) independent of AvrPtoB or AvrPto (Rathjen et al., 1999; Wu et al., 2004; Bernal et al., 2005; Xing et al., 2007). The simultaneous substitutions Leu-205/Phe-213 in Pto around the unique AvrPtoB-interacting interface 1 specifically disrupted interaction with AvrPtoB (Figure 5B) and led to a CGF phenotype (Figure 6A), indicating that the region flanking these two residues also negatively regulates Prf signaling. Collectively, our results indicate the AvrPtoB-interacting residues of Pto play a role in inhibiting Prf activation, which can be masked by the binding of AvrPtoB. The previously proposed model (Xing et al., 2007) for how AvrPto–Pto interaction activates Prf signaling is thus also applicable to AvrPtoB. The direct target of Pto inhibition is likely Prf, based on the fact these two proteins directly interact (Mucyn et al., 2006). This interaction likely involves the interfaces used by Pto for AvrPtoB and AvrPto binding, as mutations at these sites generated CGF Pto mutants presumably through disturbing interaction with Prf. It is also possible, however, that the surfaces of Pto involved in inhibition of Prf signaling simply overlap with those for binding to AvrPtoB or AvrPto.
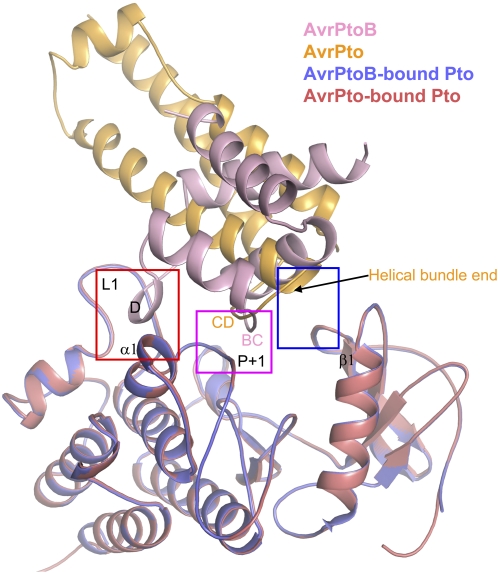
Despite strikingly different structural folds, both AvrPtoB121-205 and AvrPto are recognized by Pto and bind to the P+1 loop of Pto (Figure 8). However, the primary sequence used by them for interaction with Pto P+1 loop is completely different (Figure 8). The P+1 loop is also a site for substrate binding, and multiple autophosphorylation sites of Pto (Sessa et al., 2000) or for phosphorylation of a residue in AvrPtoB (Ntoukakis et al., 2009) have been identified. Thus, the Pto P+1 loop is a shared binding site for many peptides with diverse sequences, suggesting that some effectors could compete with certain substrates of Pto for their virulence activities. Pto does use two different hydrophobic patches to distinguish AvrPtoB from AvrPto (Figure 8). In support of these distinct interaction mechanisms, the mutants L205A/F213A and H49E/V51E of Pto specifically disrupted interaction with AvrPtoB and AvrPto, respectively. Nonetheless, interaction of both effectors with Pto appears to result in the same activation mechanism, relief of Pto inhibition of Prf-mediated signaling, and thus induces the same downstream host responses.
Figure 8.
Structural Comparison of the AvrPtoB121-205-Pto and AvrPto-Pto Complexes.
Superimposition of the AvrPtoB121-201-Pto and AvrPto-Pto complexes. The red and blue frames highlight the two unique interfaces in the complexes, with red indicating the AvrPtoB121-205-Pto interface 1 involving contact of the helix D in AvrPtoB with loop L1 and helix α1 in Pto and blue indicating the AvrPto-Pto unique interface involving H49/V51 from the loop preceding β1 in Pto. The pink frame indicates the shared interface between AvrPtoB-Pto and AvrPto-Pto complexes. “BC” with a sequence AVAF and “CD” with a sequence GINP represent the two loops in AvrPtoB and AvrPto, respectively, that are involved in the interaction with the P+1 loop of Pto. Pto (shown in slate in AvrPtoB121-205-Pto and deep salmon in AvrPto-Pto) from both complexes was used for structural superimposition.
Both AvrPto and AvrPtoB were recently found to interact with BAK1 to inhibit PAMP-triggered immunity in Arabidopsis (Shan et al., 2008). However the AvrPtoB1-307 domain does not interact with BAK1 and still enhances virulence in tomato plants. This suggests that there is another host virulence target of AvrPtoB1-307. Pto does not seem to fill this role, as many mutations that disrupted AvrPtoB1-307 interaction with Pto generated no detectable effect on the virulence activity of this AvrPtoB fragment (Xiao et al., 2007). Furthermore, a tomato line lacking the Pto gene still supports AvrPtoB1-307 virulence activity. The observation that the F173A in AvrPtoB affects both virulence and avirulence activities (Xiao et al., 2007) suggests that the virulence target of AvrPtoB1-307 might be a Pto-related protein, or at least a protein kinase. Indeed, AvrPtoB appears to preferentially interact with kinases for both its avirulence and virulence activities (Rosebrock et al., 2007; Göhre et al., 2008; Shan et al., 2008).
In summary, our study reveals the molecular mechanisms by which Pto recognizes two sequence-divergent effector proteins and also supports the hypothesis that both AvrPtoB and AvrPto induce Pto-dependent immunity by subverting the inhibitory effect of Pto on Prf signaling. However, many questions regarding the mechanism of AvrPtoB-triggered immune responses remain unanswered. For example, how does Pto negatively regulate Prf? In addition to its negative regulatory role, Pto is also required for activation of Prf. Then how does Pto also activate Prf upon binding of AvrPtoB or AvrPto? Finally, do the other domains of AvrPtoB, in addition to AvrPtoB1-307, function completely independently or is there any synergism among these domains? Future studies, in particular biochemical and structural investigations, are needed to address these questions.
METHODS
Protein Expression and Purification
Pto and avrPtoB, wild type or mutants, were subcloned into prokaryotic expression vectors for protein production. AvrPtoB (encoding residues 121 to 205) and the full-length Pto were cloned into pGEX-2T (Pharmacia) and pET30a (Novagen), respectively, and were expressed in Escherichia coli strain BL21(DE3). All the primers and restriction enzymes used are included in Supplemental Table 1 online. Cells expressing AvrPtoB and Pto were induced with 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) for 12 h at room temperature. Cells were collected, pelleted, and then resuspended in buffer A (50 mM Tris, pH 8.0, and 100 mM NaCl, supplemented with protease inhibitors). The cells were lysed by sonication and then centrifuged at 14,000 rpm for 1 h. The soluble fraction of AvrPtoB was purified using GS4B resin (Pharmacia). After removal of GST by Precision protease (GE Healthcare Life Sciences), AvrPtoB was further purified by anion-exchange column (Source-15Q; Pharmacia) and gel filtration chromatography (Superdex200; Pharmacia). Soluble Pto was first purified using Ni2+ resin (Invitrogen) and then subjected to anion-exchange column and gel filtration chromatography. To form the AvrPtoB-Pto complex, purified wild-type AvrPtoB (residues 121 to 205) and Pto were mixed together and then subjected to gel filtration chromatography. Fractions corresponding to the AvrPtoB-Pto complex were pooled for crystallization.
ITC
A direct binding affinity between Pto and AvrPtoB (201 to 205) was measured using ITC. Approximately 0.1 mM AvrPtoB protein was titrated against 10 μM full-length Pto using a VP-ITC microcalorimeter (MicroCal). All proteins were prepared in a buffer containing 25 mM HEPES, pH 8.0, and 150 mM NaCl. The titration data, collected at 25°C, were analyzed using ORIGIN data analysis software (MicroCal Software). The binding parameters are as follows: n = 1.25 ± 0.00362 sites; K= 8.851 × 105 ± 4.92 × 104 M−1; △H = −1.149 × 104 ± 50.50 cal/mol; △S = −13.4 cal/mol/deg.
Crystallization and Data Collection
Crystallization conditions for AvrPtoB (residues 121 to 205) and the AvrPtoB-Pto complex were determined from the sparse matrix screen (Hampton Research). Screening was done using hanging drop vapor diffusion by combining 2 μL of protein solution with an equal volume of well buffer. Native and SeMet crystals of AvrPtoB were grown under the same conditions: 60% (v/v) Tacsimate, pH 7.0. The crystals grew to their maximum size (0.2 × 0.2 × 0.3 mm3) within ∼2 d. The selenium crystals were transferred to the mother liquor, containing 25% glycerol, and then flash cooled in liquid nitrogen. The SAD data set for the selenium crystal to 2.1 Å was collected at the Photon Factory (Tsukuba, Japan) beam line NW12 using a CCD detector and processed using the software Denzo and Scalepack (Otwinowski and Minor, 1997). The crystals belong to space group P3121 with a cell dimension a = b = 77.38 Å, c = 46.73 Å, and contain one AvrPtoB per asymmetric unit. Several conditions from the initial screen generated AvrPtoB-Pto complex crystals, but none of them diffracted x-rays well. Phenol was found to strikingly improve the diffraction ability of crystals grown from 0.1 M tri-sodium citrate dihydrate, 25% (w/v) polyethylene glycol 3350, and 0.1mM Tris-HCl, pH 8.0. The best crystals of the AvrPtoB-Pto complex were grown under the condition of 0.1 M tri-sodium citrate dihydrate, 17.5% (w/v) polyethylene glycol 3350, 0.1 mM Tris-HCl, pH 7.9, and 10.0 mM phenol. The crystals belong to space group P212121 with a cell dimension a = 61.07 Å, b = 104.47 Å, c = 298.86 Å, and contain four AvrPtoB-Pto complex molecules per asymmetric unit.
Structure Determination and Refinement
The AvrPtoB (residues 121 to 205) crystal structure was determined by single-wavelength anomalous dispersion. The ordered selenium sites were positioned by SOLVE (Terwilliger and Berendzen, 1999), and the phases calculated from the initial sites were further refined by RESOLVE (Terwilliger, 2000). The experimental electron density was sufficient for manual model building under the program O (Jones et al., 1991). Residues 121 to 205 of AvrPtoB were built into the electron density (note residues 202 to 205 were excluded because they lacked clear electron density). Structure refinement of AvrPtoB was performed with the program CNS (Brünger et al., 1998). The final atomic model of AvrPtoB was refined to crystallographic Rwork 23.5% and Rfree 24.9 to 1.9 Å. The crystal structure of the AvrPtoB-Pto complex was determined by molecular replacement (MolRep, included in CCP4) using the coordinates of AvrPtoB and Pto as the searching models. Refmac5 in CCP4 was used for the structure refinement of AvrPtoB-Pto complex. Pto purified using the above method was phosphorylated at the residue Thr-199 as shown before (Xing et al., 2007) and further confirmed by the electron density in structure of AvrPtoB-Pto complex. The final atomic model of AvrPtoB-Pto was refined to crystallographic Rwork 31.7% and Rfree 33.1% to 3.3 Å.
Gel Filtration Assay for Testing Protein–Protein Interaction
Pto and AvrPtoB proteins purified by affinity chromatography and on an anion-exchange column (Source-15Q; Pharmacia) were used for gel filtration interaction assays. To examine the interaction between Pto (wild type or mutants) and AvrPtoB (wild type or mutants) (residues 121 to 205), size exclusion chromatography (Superdex200 column; Pharmacia Biotech) was employed. In all test runs, AvrPtoB (residues 121 to 205) and Pto were mixed together and incubated at 4°C for 1 h. Buffer containing 25.0 mM Tris, pH 8.0, 100.0 mM NaCl, and 3 mM DTT was used for gel filtration assays, with a flow rate of 0.5 mL/min. Aliquots of the peak fraction corresponding to the position of AvrPtoB-Pto complex were subjected to SDS-PAGE. The proteins were visualized by Coomassie Brilliant Blue staining.
Y2H Assays
A LexA Y2H system was used to test protein interactions (Xiao et al., 2007). All bait proteins, wild-type Pto, AvrPtoB1-307, and Bti6 were expressed from the bait vector pEG202, except AvrPto, which was expressed from pNLexA (Tang et al., 1996). Mutants derived from Pto or AvrPtoB1-307 were generated in the prey vector pJG4-5 by site-directed mutagenesis (Xiao et al., 2007) using the primers described in Supplemental Table 2 online. Yeast cells containing both prey and bait proteins were streaked on X-Gal plates with galactose to analyze the interactions and photographed 2 d after incubation at 30°C. For protein gel blot, yeast colonies were directly harvested from the X-Gal plates, washed with ice-cold water, lysed with ice-cold 2 n NaOH + 8% 2-mercaptoethanol. The lysate was precipitated with 5% trichloracetic acid and washed with ice-cold acetone, followed by SDS-PAGE separation, and protein gel blot against anti-HA antibody (2000-fold dilution; Roche Applied Science). Detection of proteins was performed using horseradish peroxidase–conjugated anti-rat secondary antibody and the ECL plus detection system (Amersham-Pharmacia).
Agrobacterium tumefaciens–Mediated Transient Assay
Pto mutant alleles were PCR amplified using the primer set 5′-ACGGTACCATGGGAAGCAAGTATTCTAAGGCAACA-3′ and 5′-TTTCTAGACACTACTGTGCTAAGATGGGTTTGATC-3′ and cloned into the KpnI and XbaI sites of binary vector pBTEX followed by electroporation into Agrobacterium strain GV2260. The Agrobacterium-mediated transient expression assay on Nicotiana benthamiana leaves was performed as described previously (Sessa et al., 2000). The SGT1-silenced and control N. benthamiana plants were previously described (Liu et al., 2002). GV2260 strains containing AvrPto or AvrPtoB1-307 (Xiao et al., 2007) were inoculated by syringe infiltration at a concentration of OD600 = 0.05, whereas GV2260 strains harboring Pto or derived mutant alleles were inoculated at a concentration of OD600 = 0.2. Photographs were taken 7 d after inoculation.
Pseudomonas Protein Secretion and Protein Gel Blotting Assays
AvrPtoB1-307 mutant alleles were PCR amplified from pJG4-5 constructs using primer set 5′-ATGGCGGGTATCAATAGAGCGGGAC-3′ and 5′-CACTGCAGTCAGGGGACTATTCTAAAAGCATACTTGGC-3′, digested with BamHI and PstI, and cloned into the broad-host-range plasmid pCPP45 at BamHI and PstI sites by replacing the wild-type AvrPtoB1-307 allele in pCPP45:AvrPtoB1-307, in which AvrPtoB1-307 is under the control of the hrp promoter (Xiao et al., 2007). The resulting constructs were confirmed by sequencing, transformed into DC3000ΔavrPtoΔavrPtoB by electroporation, and analyzed for protein secretion as described previously (Lin and Martin, 2006). Protein gel blotting was performed using a rat anti-HA antibody (2000-fold dilution; Roche Applied Science) or anti-AvrPtoB rabbit antibody (Lin and Martin, 2007). Detection of proteins was performed using horseradish peroxidase–conjugated secondary antibodies (anti-rat and anti-rabbit) and the ECL plus detection system (Amersham-Pharmacia).
Measurement of Bacterial Populations in Tomato Leaves
Details of preparation of Pseudomonas syringae inoculum have been described (Anderson et al., 2006). Six-week-old greenhouse-grown tomato (Solanum lycopersicum) plants containing functional Pto and Prf genes were vacuum inoculated with Pseudomonas strains containing AvrPtoB1-307 wild-type or mutant alleles or empty vector at an inoculum of 2 × 104 colony-forming units/mL (Xiao et al., 2007). For consistent results, after inoculation with Pseudomonas bacteria, tomato plants were kept in a climate-controlled growth chamber under optimized conditions (24°C during the day and 20°C at night, with 75% humidity and 16-h days; for details, see Anderson et al., 2006). Bacterial populations were recovered from plant leaves and quantitated at 4 d after inoculation following methods described by Anderson et al. (2006).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: Pto (DQ019170), AvrPtoB (Ay074795), HopPmal (Q8RP04), AvrPtoB_T1 (NZ_ABSM01000023), and ArPto (EU024545). The coordinates of AvrPtoB and its complex with Pto have been deposited in the Protein Data Bank with the accession numbers 3HGL and 3HGK, respectively.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Pto N251K Interacts with AvrPto but Not AvrPtoB as Determined by a Yeast Two-Hybrid Assay.
Supplemental Table 1. Primers Used for Expression of AvrPtoB and Pto.
Supplemental Table 2. Primers Used for Yeast Two-Hybrid Assays.
Supplementary Material
Acknowledgments
We thank Yusuke Yamada at the Photon Factory of Japan for assistance with data collection. This research was funded by the Chinese Ministry of Science and Technology “863” Grant 2008-AA022305 and “973” Grant 2006CB806704 to J.C. and National Institutes of Health Grant R01GM078021 and National Science Foundation Grant DBI-0605059 to G.B.M.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) are: Jijie Chai (chaijijie@nibs.ac.cn) about structural biology and Gregory B. Martin (gbm7@cornell.edu) about functional analyses.
Online version contains Web-only data.
References
- Abramovitch, R.B., Anderson, J.C., and Martin, G.B. (2006). Bacterial elicitation and evasion of plant innate immunity. Nat. Rev. Mol. Cell Biol. 7 601–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, J.C., Pascuzzi, P.E., Xiao, F., Sessa, G., and Martin, G.B. (2006). Host-mediated phosphorylation of type III effector AvrPto promotes Pseudomonas virulence and avirulence in tomato. Plant Cell 18 502–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal, A.J., Pan, Q., Pollack, J., Rose, L., Kozik, A., Willits, N., Luo, Y., Guittet, M., Kochetkova, E., and Michelmore, R.W. (2005). Functional analysis of the plant disease resistance gene Pto using DNA shuffling. J. Biol. Chem. 280 23073–23083. [DOI] [PubMed] [Google Scholar]
- Brünger, A.T., et al. (1998). Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 50 905–921. [DOI] [PubMed] [Google Scholar]
- Chang, J.H., Tobias, C.M., Staskawicz, B.J., and Michelmore, R.W. (2001). Functional studies of the bacterial avirulence protein AvrPto by mutational analysis. Mol. Plant Microbe Interact. 14 451–459. [DOI] [PubMed] [Google Scholar]
- Chinchilla, D., Zipfel, C., Robatzek, S., Kemmerling, B., Nürnberger, T., Jones, J.D., Felix, G., and Boller, T.A. (2007). Flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 48 497–500. [DOI] [PubMed] [Google Scholar]
- Cohn, J.R., and Martin, G.B. (2005). Pseudomonas syringae pv. tomato type III effectors AvrPto and AvrPtoB promote ethylene-dependent cell death in tomato. Plant J. 44 139–154. [DOI] [PubMed] [Google Scholar]
- Frederick, R.D., Thilmony, R.L., Sessa, G., and Martin, G.B. (1998). Recognition specificity for the bacterial avirulence protein AvrPto is determined by Thr-204 in the activation loop of the tomato Pto kinase. Mol. Cell 2 241–245. [DOI] [PubMed] [Google Scholar]
- Göhre, V., Spallek, T., Häweker, H., Mersmann, S., Mentzel, T., Boller, T., de Torres, M., Mansfield, J.W., and Robatzek, S. (2008). Plant pattern-recognition receptor FLS2 is directed for degradation by the bacterial ubiquitin ligase AvrPtoB. Curr. Biol. 18 1824–1832. [DOI] [PubMed] [Google Scholar]
- He, P., Shan, L., Lin, N.C., Martin, G.B., Kemmerling, B., Nürnberger, T., and Sheen, J. (2006). Specific bacterial suppressors of MAMP signaling upstream of MAPKKK in Arabidopsis innate immunity. Cell 125 563–575. [DOI] [PubMed] [Google Scholar]
- He, X., Anderson, J.C., del Pozo, O., Gu, Y.Q., Tang, X., and Martin, G.B. (2004). Silencing of subfamily I of protein phosphatase 2A catalytic subunits results in activation of plant defense responses and localized cell death. Plant J. 38 563–577. [DOI] [PubMed] [Google Scholar]
- Heese, A., Hann, D.R., Gimenez-Ibanez, S., Jones, A.M., He, K., Li, J., Schroeder, J.I., Peck, S.C., and Rathjen, J.P. (2007). The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl. Acad. Sci. USA 104 12217–12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm, L., Kaariainen, S., Rosenstrom, P., and Schenkel, A. (2008). Searching protein structure databases with DaliLite v.3. Bioinformatics 24 2780–2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janjusevic, R., Abramovitch, R.B., Martin, G.B., and Stebbins, C.E. (2006). A bacterial inhibitor of host programmed cell death defenses is an E3 ubiquitin ligase. Science 311 222–226. [DOI] [PubMed] [Google Scholar]
- Jia, Y., Loh, Y.T., Zhou, J., and Martin, G.B. (1997). Alleles of Pto and Fen occur in bacterial speck-susceptible and fenthion-insensitive tomato cultivars and encode active protein kinases. Plant Cell 9 61–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, T.A., Zou, J.Y., Cowan, S.W., and Kjeldgaard, M. (1991). Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47 110–119. [DOI] [PubMed] [Google Scholar]
- Kim, Y.J., Lin, N.C., and Martin, G.B. (2002). Two distinct Pseudomonas effector proteins interact with the Pto kinase and activate plant immunity. Cell 109 589–598. [DOI] [PubMed] [Google Scholar]
- Levdikov, V.M., Blagova, E., Joseph, P., Sonenshein, A.L., and Wilkinson, A.J. (2006). The structure of CodY, a GTP- and isoleucine-responsive regulator of stationary phase and virulence in gram-positive bacteria. J. Biol. Chem. 281 11366–11373. [DOI] [PubMed] [Google Scholar]
- Lin, N.-C., and Martin, G.B. (2006). Diverse AvrPtoB homologs from several Pseudomonas syringae pathovars elicit Pto-dependent resistance and have similar virulence activities. Appl. Environ. Microbiol. 72 702–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, N.-C., and Martin, G.B. (2007). Pto- and Prf-mediated recognition of AvrPto and AvrPtoB restricts the ability of diverse pseudomonas syringae pathovars to infect tomato. Mol. Plant Microbe Interact. 20 806–815. [DOI] [PubMed] [Google Scholar]
- Liu, Y., Schiff, M., Serino, G., Deng, X.W., and Dinesh-Kumar, S.P. (2002). Role of SCF ubiquitin-ligase and the COP9 signalosome in the N gene-mediated resistance response to Tobacco mosaic virus. Plant Cell 14 1483–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucyn, T.S., Clemente, A., Andriotis, V.M., Balmuth, A.L., Oldroyd, G.E., Staskawicz, B.J., and Rathjen, J.P. (2006). The tomato NBARC-LRR protein Prf interacts with Pto kinase in vivo to regulate specific plant immunity. Plant Cell 18 2792–2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntoukakis, V., Mucyn, T.S., Gimenez-Ibanez, S., Chapman, H.C., Gutierrez, J.R., Balmuth, A.L., Jones, A.M., and Rathjen, J.P. (2009). Host inhibition of a bacterial virulence effector triggers immunity to infection. Science 324 784–787. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z., and Minor, W. (1997). Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276 307–326. [DOI] [PubMed] [Google Scholar]
- Pedley, K.F., and Martin, G.B. (2003). Molecular basis of Pto-mediated resistance to bacterial speck disease in tomato. Annu. Rev. Phytopathol. 41 215–243. [DOI] [PubMed] [Google Scholar]
- Rathjen, J.P., Chang, J.H., Staskawicz, B.J., and Michelmore, R.W. (1999). Constitutively active Pto induces a Prf-dependent hypersensitive response in the absence of avrPto. EMBO J. 18 3232–3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosebrock, T.R., Zeng, L., Brady, J.J., Abramovitch, R.B., Xiao, F., and Martin, G.B. (2007). A bacterial E3 ubiquitin ligase targets a host protein kinase to disrupt plant immunity. Nature 448 370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmeron, J.M., Oldroyd, G.E., Rommens, C.M., Scofield, S.R., Kim, H.S., Lavelle, D.T., Dahlbeck, D., and Staskawicz, B.J. (1996). Tomato Prf is a member of the leucine-rich repeat class of plant disease resistance genes and lies embedded within the Pto kinase gene cluster. Cell 86 123–133. [DOI] [PubMed] [Google Scholar]
- Scofield, S.R., Tobias, C.M., Rathjen, J.P., Chang, J.H., Lavelle, D.T., Michelmore, R.W., and Staskawic, B.J. (1996). Molecular basis of gene-for-gene specificity in bacterial speck disease of tomato. Science 274 2063–2065. [DOI] [PubMed] [Google Scholar]
- Sessa, G., D'Ascenzo, M., and Martin, G.B. (2000). Thr38 and Ser198 are Pto autophosphorylation sites required for the AvrPto-Pto-mediated hypersensitive response. EMBO J. 19 2257–2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan, L., He, P., Li, J., Heese, A., Peck, S.C., Nürnberger, T., Martin, G.B., and Sheen, J. (2008). Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe 4 17–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, X., Frederick, R.D., Zhou, J., Halterman, D.A., Jia, Y., and Martin, G.B. (1996). Initiation of plant disease resistance by physical interaction of AvrPto and Pto kinase. Science 274 2060–2063. [DOI] [PubMed] [Google Scholar]
- Terwilliger, T.C. (2000). Maximum likelihood density modification. Acta Crystallogr. D Biol. Crystallogr. 56 965–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger, T.C., and Berendzen, J. (1999). Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 55 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, A.J., Andriotis, V.M., Durrant, M.C., and Rathjen, J.P. (2004). A patch of surface-exposed residues mediates negative regulation of immune signaling by tomato Pto kinase. Plant Cell 16 2809–2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulf, J., Pascuzzi, P.E., Fahmy, A., Martin, G.B., and Nicholson, L.K. (2004). The solution structure of type III effector protein AvrPto reveals conformational and dynamic features important for plant pathogenesis. Structure 12 1257–1268. [DOI] [PubMed] [Google Scholar]
- Xiang, T., Zong, N., Zou, Y., Wu, Y., Zhang, J., Xing, W., Li, Y., Tang, X., Zhu, L., Chai, J., and Zhou, J.M. (2008). Pseudomonas syringae effector AvrPto blocks innate immunity by targeting receptor kinases. Curr. Biol. 18 74–80. [DOI] [PubMed] [Google Scholar]
- Xiao, F., He, P., Abramovitch, R.B., Dawson, J.E., Nicholson, L.K., Sheen, J., and Martin, G.B. (2007). The N-terminal region of Pseudomonas type III effector AvrPtoB elicits Pto-dependent immunity and has two distinct virulence determinants. Plant J. 52 595–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing, W., et al. (2007). The structural basis for activation of plant immunity by bacterial effector protein AvrPto. Nature 449 243–247. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.