Abstract
Plant seeds can sense diverse environmental signals and integrate the information to regulate developmental responses, such as dormancy and germination. The circadian clock confers a growth advantage on plants and uses environmental information for entrainment. Here, we show that normal circadian clock gene function is essential for the response to dormancy-breaking signals in seeds. We show that mutations in the clock genes LATE ELONGATED HYPOCOTYL, CIRCADIAN CLOCK ASSOCIATED1 (CCA1), and GIGANTEA (GI) cause germination defects in response to low temperature, alternating temperatures, and dry after-ripening. We demonstrate that the transcriptional clock is arrested in an evening-like state in dry seeds but rapidly entrains to light/dark cycles in ambient temperatures upon imbibition. Consistent with a role for clock genes in seed dormancy control, CCA1 expression is transcriptionally induced in response to dry after-ripening and that after-ripening affects the amplitude of subsequent transcriptional clock gene oscillations. Control of abscisic acid- and gibberellin-related gene expression in seeds requires normal circadian function, and GI and TIMING OF CAB EXPRESSION1 regulate the response to ABA and GA in seeds. We conclude that circadian clock genes play a key role in the integration of environmental signaling controlling dormancy release in plants.
INTRODUCTION
Dormancy is a common attribute among biological organisms and allows the timing of growth to coincide with favorable environmental conditions. Seed germination terminates dispersal by immobilizing the developing plant and hence is important in determining where and when plants grow. The germination of dormant seeds is critically dependent on environmental factors that trigger the adaptive timing of plant growth. In Arabidopsis thaliana, temperature, nitrate, and dry after-ripening are important dormancy breaking signals, and these combine with light to promote the germination of dormant seeds.
After-ripening is a little known process occurring over predictable time scales in low-hydrated seeds that promotes dormancy loss upon imbibition. After-ripening can be likened to a drought response because the role of after-ripening is to promote germination swiftly with the arrival of water after a long period of dryness. Importantly, dry after-ripening and cold can substitute for each other, both promoting the germination of dormant seeds in the presence of light (Holdsworth et al., 2008). A key role of after-ripening is to modulate the sensitivity of seed germination to further dormancy-breaking treatments over time, suggesting that these response pathways are closely linked.
It is now well established that germination is regulated by hormone balance, specifically, the ratio of gibberellin (GA) and abscisic acid (ABA) action in seeds. Environmental signals have been shown to promote germination by regulating the levels of the phytohormones GA and ABA in seeds, and the primary mechanism for this is the transcriptional regulation of a suite of enzymes that control the metabolism of the two hormones. These include enzymes for the synthesis, degradation, and conjugation of ABA and for the activation or deactivation of bioactive GAs (Yamaguchi et al., 1998; Kushiro et al., 2004; Yamauchi et al., 2004; Penfield et al., 2005; Millar et al., 2006; Seo et al., 2006; Holdsworth et al., 2008). The transcript levels of these genes have been shown to be affected by light, temperature, and after-ripening. In particular, but not uniquely, the environmental regulation of GIBBERELLIC ACID 3-OXIDASE (GA3OX) and CYTOCHROME P450 707A2 (CYP707A2) is well established in imbibed seeds (Yamaguchi et al., 1998; Yamauchi et al., 2004; Penfield et al., 2005; Millar et al., 2006). CYP707A2 expression is induced transiently after imbibition in a phytochrome- and after-ripening-dependent manner and is required for ABA breakdown (Kushiro et al., 2004; Millar et al., 2006; Seo et al., 2006). GA3OX catalyzes GA synthesis and is induced by light, cold, and after-ripening in imbibed seeds (Yamaguchi et al., 1998; Yamauchi et al., 2004).
Components of the circadian clock are essential in modulating plant responses to the environment, such as the photoperiodic regulation of flowering time. Furthermore, circadian clocks confer an adaptive advantage on plants through other mechanisms that are less well known but are necessary for optimal growth rates (Dodd et al., 2005). The current model of the circadian clock comprises a series of interlocked transcription feedback loops. A negative feedback loop is formed between an evening-expressed pseudoresponse regulator, TIMING OF CAB EXPRESSION1 (TOC1), and two closely related morning-expressed Myb transcription factors, LATE ELONGATED HYPOCOTYL (LHY) and CIRCADIAN CLOCK ASSOCIATED1 (CCA1). TOC1 forms a second interlocking feedback loop with a predicted gene Y, which corresponds in part to the real gene GIGANTEA (GI; Locke et al., 2005). GI is an evening-expressed gene that encodes a plant-specific, nuclear protein with no clear function deducible from its primary sequence (Fowler et al., 1999). A third interlocking loop is formed between LHY/CCA1 and the TOC1 homologs PSEUDO-RESPONSE REGULATOR7 (PRR7) and PRR9.
Because the circadian clock is also highly sensitive to environmental stimuli and can be entrained or modulated by light, temperature, and even nitrate (McClung, 2006; Gutiérrez et al., 2008), it is therefore responsive to many of the signals that regulate dormancy. Furthermore, the expression of many genes involved in the response to environmental signals is also subject to circadian control (e.g., Gould et al., 2006). Recently it has also been shown that the Arabidopsis circadian clock has a role in the transcriptional regulation of hormone metabolism and the modulation of hormone responses (Covington et al., 2008; Michael et al., 2008). Therefore, it is possible that the clock or its components have direct roles in the transduction of environmental signals regulating hormone metabolism and subsequently development.
The role of plant circadian clock genes in seeds has not been extensively tested. This is despite a study showing that the central seed dormancy regulator ABSCISIC ACID INSENSITIVE3 (ABI3; AT3G24650) interacts in vivo with the TOC1 protein (Kurup et al., 2000). Here, we show that genetic manipulation of the key components of the circadian clock alters seed dormancy and the response to dormancy-breaking signals. We propose that the plant circadian clock is an important signal integrator regulating dormancy release in seeds.
RESULTS
Circadian Clock Mutants Show Environmental Sensing Defects in Seeds
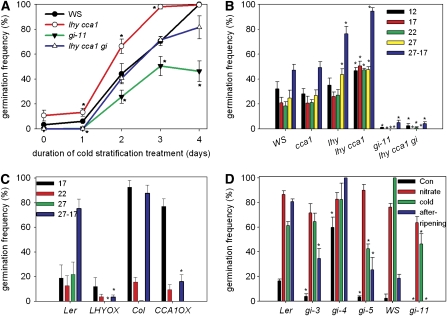
To test whether the circadian clock has a role in seed dormancy regulation, we examined the germination of Arabidopsis mutants deficient in key components of the circadian oscillator. First, we found that the germination of freshly harvested lhy cca1 double mutant seeds (Hall et al., 2003) showed a small but significantly increased germination frequency relative to the wild type and exhibited germination hypersensitive to applied cold treatments (Figure 1A). By contrast, freshly harvested gi-11 seed (Fowler et al., 1999) showed a marked reduced sensitivity to applied cold, reaching a maximum germination frequency of only 50%, while the germination of the lhy cca1 gi triple mutant showed that the two mutants behaved additively. The lhy cca1 double mutant phenotype was dependent on loss of both LHY and CCA1 function (see Supplemental Figure 1 online). Hence, LHY, CCA1, and GI have a role in seed dormancy and germination control. Interestingly, GI has been shown to play a role in other plant responses to cold (Cao et al., 2005; Paltiel et al., 2006), suggesting a common temperature signaling mechanism.
Figure 1.
The Germination Behavior of lhy cca1 and gi-11 Mutants.
(A) The germination of freshly harvested seed before and after cold stratification at 22°C.
(B) The germination behavior of seed after-ripened for 3 months at the indicated temperatures in degrees Celsius. 27-17 indicates that 27°C was given during the light period and 17°C during the dark period.
(C) The germination of freshly harvested wild-type, LHY-, and CCA1-overexpressing seed at the indicated temperatures. 27-17 indicates that 27°C was given during the light period and 17°C during the dark period.
(D) The germination of wild-type and gi mutant seeds in white light with and without the indicated dormancy breaking treatments at 22°C. Con, control. Values given are means ± se for five to eight independent seed batches. Asterisks indicate germination significantly different from the wild type (P < 0.01).
At constant temperatures, the germination of lines in the Wassilewskija (Ws) background was poor when freshly harvested (see Supplemental Figure 1 online). To determine the role of the circadian clock in dormancy regulation over a wider temperature range, we allowed seed to after-ripen (18 to 22°C in constant darkness) for 3 months. At this time, Ws seed shows incomplete after-ripening and a characteristic temperature response in which elevated germination occurs at 12°C (12°C versus 22°C, P = 0.018; Figure 1B). We also tested the germination response to a temperature shifting regime from 27 to 17°C, as this has been previously reported to break Arabidopsis seed dormancy (Ali-Rachedi et al., 2004). The germination profile of cca1 single mutant seeds closely resembled the wild type, with elevated germination at 12°C and a weak response to a temperature shift (Figure 1B). By contrast, lhy mutant seeds showed increased germination at 27°C compared with the wild type.
Strikingly, the lhy cca1 double mutant showed a strong reduced-dormancy phenotype, and germination that did not vary with temperature. This phenotype is consistent with a role for both LHY and CCA1 in the repression of the after-ripening response (because dry storage has increased the germination of lhy cca1 over the wild type at 22°C; compare Figures 1A to 1B). It is also possible that LHY and CCA1 are necessary for the normal germination response to ambient temperatures; however, we cannot rule out that changes in the temperature regulation of germination are secondary to differences in after-ripening. In addition, we found that both the lhy single mutant and the lhy cca1 double mutant showed a marked hypersensitivity to germination promotion by alternating temperatures, while the germination of both CCA1 overexpressing (CCA1OX; Wang and Tobin, 1998) and LHYOX (Schaffer et al., 1998) seeds were insensitive to germination promotion by alternating temperatures (Figure 1C), despite showing near normal germination at 22°C, and wild-type germination levels after cold stratification (see Supplemental Figure 1 online). We cannot exclude that additional phenotypes could be observed in LHY- and CCA1-overexpressing seeds in a Ws background. We concluded that normal expression of LHY and CCA1 is essential for the response to these alternating temperatures in seeds.
In contrast with lhy cca1, gi-11 seeds showed strong dormancy and poor germination after storage, indicating a failure to after-ripen (Figure 1B). Interestingly, the poor dry after-ripening phenotype of gi-11 mutants was completely epistatic to the increased dry after-ripening phenotype of lhy cca1, demonstrating that LHY and CCA1 require GI function to repress the response to after-ripening. We further analyzed the germination of four gi alleles and their response to dormancy breaking treatments (Figure 1D). The gi-3 and gi-5 mutants too showed an increased dormancy phenotype and slow after-ripening. All gi alleles exhibited a wild-type germination response to nitrate, also demonstrating that GI is not required for the promotion of germination by nitrate or white light (light is a prerequisite for germination promotion by nitrate; Hilhorst and Karssen, 1988). Two alleles, gi-5 and gi-11, also showed reduced germination after a cold stimulus when freshly harvested (Figure 1D). Therefore, we concluded that loss of GI in seeds confers a stronger dormancy and poor after-ripening. In two alleles, this also results in reduced sensitivity to cold. One of the four gi alleles tested, gi-4, did not display any of the above phenotypes but instead displayed a consistent reduced dormancy phenotype. The gi-4 allele has a mutation in the 3′ splice acceptor site of the last intron, intron 12, that is predicted to result in premature termination of translation and elimination of the C terminus (Fowler et al., 1999). These results are interesting because in many ways gi mutants resemble Arabidopsis ecotypes with a strong dormancy, such as CVI (Alonso-Blanco et al., 2003; Ali-Rachedi et al., 2004).
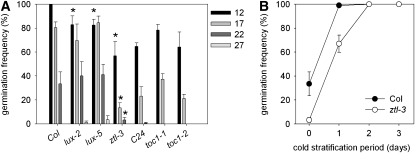
We also noted that mutations in two other circadian clock–associated genes, LUX ARRYTHMO (LUX; Hazen et al., 2005) and ZEITLUPE (ZTL; Somers et al., 2000), also show seed dormancy phenotypes (Figure 2). The two lux mutants tested showed lower germination than the wild type at 12°C. The ztl-3 mutant showed an increased dormancy phenotype at harvest and a reduced response to cold stratification, a phenotype at least superficially resembling gi mutants. Thus, the ztl-3 phenotype could be due to reduced GI protein levels in this mutant (Kim et al., 2007).
Figure 2.
A Role for the Clock-Associated Genes ZTL and LUX in Seed Dormancy and Germination Control.
(A) The germination response of freshly harvested wild type (Col or C24) ztl-3, lux-2, lux-5, and toc1 seed to imbibition at different ambient temperatures.
(B) The response of freshly harvested wild-type and ztl-3 seeds to cold stratification treatment.
Data points in (A) and (B) represent mean and se of data from five to eight replicate seed batches for each genotype.
The fact that ZTL targets TOC1 for degradation and that TOC1 has previously been reported to interact with ABI3 (Kurup et al., 2000) led us to test the germination phenotype of toc1-1. However, we observed no effect of the toc1-1 mutant on germination (Figure 2). TOC1 belongs to a family of pseudoresponse regulators, and it is possible that the lack of phenotype may be due to redundancy within the gene family. Consistent with this hypothesis, we observed phenotypes associated with TOC1 gain of function (see Figure 8). These data, together with a recent report showing that overexpression of a MYB transcription factor alters clock function and germination (Zhang et al., 2007), suggest that modified seed dormancy is a general consequence of the genetic perturbation of Arabidopsis circadian clock–associated genes.
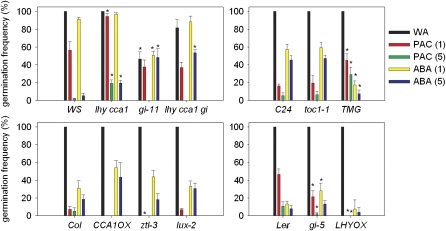
Figure 8.
The Germination of Cold-Stratified Wild-Type and Clock Mutant Seeds in Control Conditions or in Response to Applied ABA and PAC.
Bars represent the mean and se of five replicate seed batches for each genotype. Numbers in brackets indicate the concentration of ABA or PAC in micromoles. TMG refers to the TOC1 minigene (Más et al., 2003), the TOC1 cDNA expressed under its own promoter. WA, water agarose.
Dormancy Breaking Signals Regulate Clock Gene Expression in Seeds
Seed imbibition is believed to synchronize and set the phase of the Arabidopsis circadian clock (Zhong et al., 1998); however, it may also be the case that the transcript levels of clock genes do not oscillate in dry seeds. We analyzed the expression of clock genes in dry seeds using available microarray data (Finch-Savage et al., 2007) and found the clock was in an evening-like state, with LHY and CCA1 undetected and transcripts of PRR7, PRR9, GI, TOC1, and LUX present in both dormant and nondormant dry seeds (see Supplemental Table 1 online). To determine whether the transcriptional circadian clock can function in dry seeds, we compared clock gene expression at dawn and dusk in dry seeds after strong entrainment for 3 d in 12-h light/dark cycles (Table 1). We could detect no significant difference in clock gene expression between the two times, confirming that the transcriptional clock is indeed arrested in dry seeds. It is possible that an oscillator could function in the absence of a transcriptional clock, as is observed in dark grown cyanobacteria (Tomita et al., 2005). In fact, it has previously been reported that dry onion seeds exhibit circadian rhythms of respiration (Bryant, 1972). Our data suggest that if circadian oscillations persist in dry Arabidopsis seed, they are uncoupled from the well-characterized transcriptional circadian rhythms and that subsequent imbibition allows the clock to restart or recouple.
Table 1.
The Transcriptional Circadian Clock Is Frozen in Dry Seeds
| Gene | Fold Change Evening/Morning | P Value |
|---|---|---|
| LHY | nd | |
| CCA1 | 1.13 | 0.14 |
| TOC1 | 1.11 | 0.99 |
| GI | 1.16 | 0.13 |
| LUX | 1.20 | 0.42 |
| PRR7 | 1.24 | 0.63 |
| PRR9 | 0.98 | 0.89 |
Gene expression was compared in samples collected 30 min after dawn and 30 min before dusk from dry seeds entrained to 12-h light/dark cycles. Data represent the mean of three biological repeats. No significant difference was observed in the expression of clock-associated genes between the two time points. Nd, not detected in either sample. P values were calculated by two-tailed Student's t test.
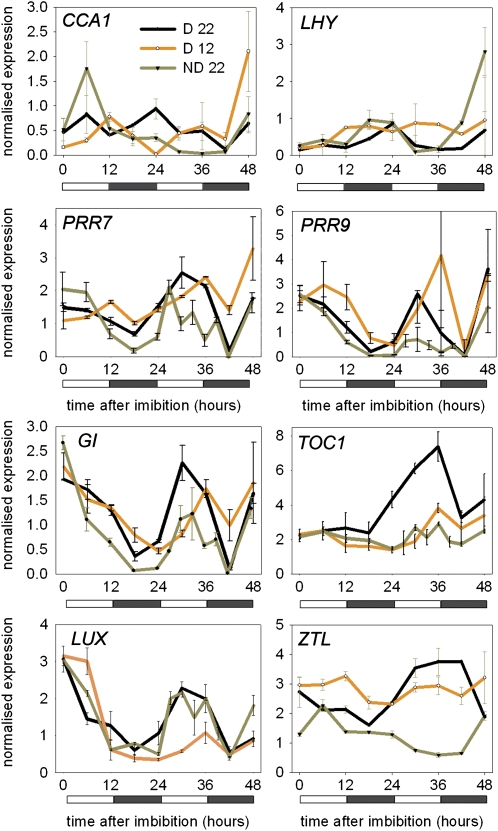
As luciferase functions poorly as a reporter in seeds without first removing the seed coat (Penfield et al., 2004), an act that breaks dormancy in Arabidopsis, we used real-time RT-PCR to compare clock gene expression in dormant and nondormant seeds. For this experiment, we used freshly harvested and after-ripened Columbia-0 (Col-0) seeds at 22°C and also incubated freshly harvested Col seeds at 12°C, a treatment that promotes germination of dormant seeds in this ecotype (see Supplemental Figure 2 online). We analyzed the first 48 h after imbibition, because after this time, nondormant seeds have already germinated; therefore, differences in gene expression in response to dormancy breaking signals with a role in germination control must be manifest before this time. This experiment also confirmed the observations described in Table 1 that in dry seeds (0 h in Figure 3), the circadian clock was frozen in a state in which transcripts for GI, LUX, PRR7, TOC1, and PRR9 were present at relatively high levels and transcripts of the morning expressed genes LHY and CCA1 were present at relatively low levels. Diurnal oscillations in the transcript levels of clock-associated genes were observed in both dormant and nondormant seeds at 22°C during the first 24 h (Figure 3). The key differences between the two states were first that in after-ripened seeds, CCA1 expression was subject to a large induction shortly after imbibition and then assumed a low-level oscillation peaking at dawn as described previously (Wang and Tobin, 1998). The expression of LHY was similar in both 22°C treatments but appeared to be taking longer to entrain to diurnal cycles at 12°C, as did CCA1.
Figure 3.
The Transcript Levels of Circadian Clock-Associated Genes in Dormant and After-Ripened Seeds
The expression of circadian clock–associated genes in imbibed freshly harvested dormant Col-0 seeds at 22°C in 12-h light/dark cycles (D22) and in seeds in which dormancy has been or is being broken by dry after-ripening (ND22) or by imbibition and maintenance at constant 12°C (ND12), a temperature at which primary dormancy is broken. Expression levels were determined by real-time RT-PCR. Data points represent mean ± sd using two biological replicates.
For the evening genes, the expression peaks corresponded to the middle of the day rather than dusk, as had been previously described (Fowler et al., 1999; Strayer et al., 2000). This difference may be due to developmental stage, the fact that the clock is in the process of entrainment, or to differences in the experimental conditions, such as growth medium. Strikingly, dry after-ripening dramatically reduced the amplitude of TOC1, GI, PRR7, and PRR9 gene expression but not that of LUX. By contrast, low temperature treatment also reduced the amplitude of certain gene expression profiles, notably, LUX, GI, and TOC1 gene expression, although most genes were also shifted to a later phase. Together, our data show that the transcript levels of the core clock components in imbibed seeds is responsive to signals that break dormancy and that after-ripening promotes CCA1 expression but has a general amplitude-reducing effect on PRR and GI gene expression. We cannot also rule out small changes in period between the two states that are not detectable with our sampling density or because of entraining signals.
The Mechanism Underlying the Phenotypes of Clock Gene Mutants Is the Aberrant Regulation of Hormone Metabolism
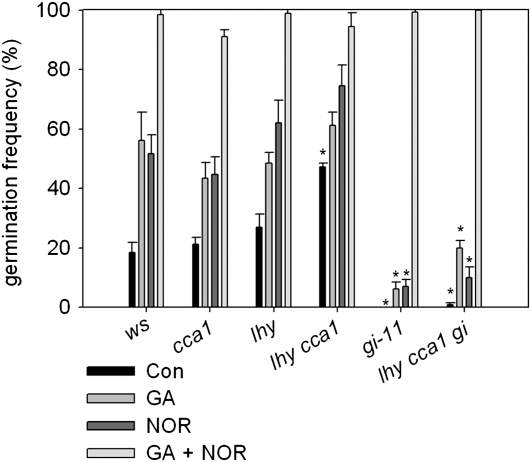
To begin to examine the relationship between LHY, CCA1, and GI activity and the hormonal regulation of dormancy, we analyzed seed germination of seed stored for 3 months in the presence of GA, the ABA biosynthetic inhibitor norfluorazon (NOR), or the combination of both treatments (Figure 4). Wild-type seeds required both GA and NOR application to promote high germination frequencies, and the same was true for the lhy cca1 double mutants. Therefore, we concluded that low dormancy in lhy cca1 was not attributable to perturbation of either the GA or ABA pathways alone or to the seed's sensitivity to either of these pathways, but of both. Similarly, increased dormancy in gi-11 still required both only GA and ABA loss for breakage. This experiment shows that the increased dormancy of gi-11 mutants is not due to altered action of either GA or ABA but that the mutant is likely affected in the metabolism of, or response to, both hormones.
Figure 4.
Seed Germination in Response to Paclobutrazol and Norfluorazon.
The germination of Ws, cca1, lhy, gi-11, and double and triple mutant seeds (as indicated) under control conditions (water agarose; Con) or in response to applied GA (100 μM), NOR (50 μM), or both (GA+NOR) at 22°C. Data points represent the mean and se of data from five to eight independent seed batches. Asterisks indicate germination significantly different from the wild type (P < 0.01).
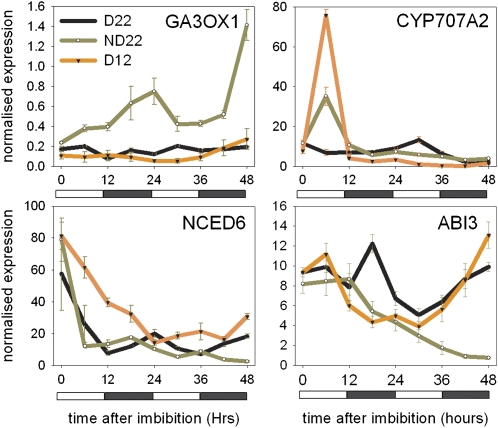
The germination of seeds is triggered by the effects of phytohormones on protein levels of DELLA, ABI3, and ABI5 (Lopez-Molina et al., 2001; Penfield et al., 2006a). Previously, it has been shown that environmental signals promote germination through the regulation of GA and ABA metabolic gene expression in seeds. ABA levels are controlled by cytochrome P450 CYP707A2 (AT2G29090) playing a key role in the catabolism of ABA and NCED6 (AT3G24220) and NCED9 (AT1G78390) essential for ABA synthesis. In turn, GA levels are promoted by the regulation of the mRNA levels of GA biosynthetic enzymes, principally GA3OX1 (Yamaguchi et al., 1998; Yamauchi et al., 2004), the last committed step in active GA synthesis. To test a role for clock genes in this regulation, we first analyzed the expression of key genes in dormant and nondormant seeds in an entraining system over the first 48 h of imbibition and their response to low temperature and dry after-ripening treatments (Figure 5). GA3OX1 was expressed only in after-ripened seeds and expression is coincident with dawn (Figure 5).
Figure 5.
The Expression of Selected Genes Central to Hormone Metabolism Associated with Germination Control in Dormant and After-Ripened Seeds.
The expression of ABA- and GA-related genes in freshly harvested imbibed Col-0 seeds at 22°C (D22), in dry after-ripened seeds at 22°C (ND22), and in freshly harvested seeds maintained at 12°C (ND12), a temperature at which primary dormancy is broken. Data points represent mean ± sd of data from two biological replicates.
We also analyzed key genes in ABA metabolism and signaling. The expression of NCED6 (Lefebvre et al., 2006) in dormant seeds at 22°C peaked at dawn, but this expression pattern was not observed in after-ripened seeds or in dormant seeds at 12°C. Transcript levels of the key ABA-catabolic gene CYP707A2 (Kushiro et al., 2004) were increased by after-ripening as described previously (Millar et al., 2006) and also strikingly by low temperature treatment. Interestingly, under our conditions, ABI3 expression initially declined after imbibition but increased to a peak during the first morning in dormant seeds at 22°C. In nondormant seeds, ABI3 expression declines with increasing imbibition times, as is evident in previous analyses.
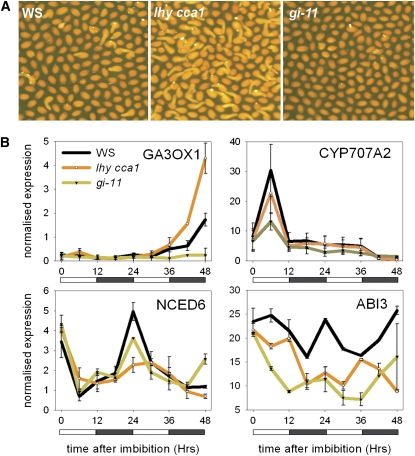
As dormancy-breaking treatments caused altered regulation of CYP707A2, GA3OX, and ABI3 expression, we tested their expression in Ws, lhy cca1, and gi-11 mutant imbibed seeds stored for 3 months at 22°C (Figure 6): under these conditions, all three genotypes show easily distinguishable germination frequencies (Figures 1B and 6A), and the Ws seeds have after-ripened to a state most similar to the dormant Col-0 states analyzed in Figure 3 (see Supplemental Figure 2 online). All dormancy breaking treatments cause an increase in GA3OX1 expression in imbibed seeds. We found that the expression of GA3OX1 was markedly increased in lhy cca1 compared with Ws and was abolished in gi-11 (Figure 6B). This is consistent with the increased germination of lhy cca1 and the absence of germination of gi-11. gi-11 seeds were also found to be defective in GA3OX1 expression following cold stratification (see Supplemental Figure 4 online). CYP707A2 is induced in the presence of red light in after-ripened or cold-treated seeds (Millar et al., 2006; Seo et al., 2006; Figure 5) and is required for ABA breakdown (Kushiro et al., 2004). In gi-11 mutants, the early peak in CYP707A2 expression was also strikingly reduced. This peak in expression is promoted by after-ripening and low temperature (Figure 5), two treatments whereby gi mutants were found to show reduced responsiveness (Figures 1B and 1D). Hence, a critical function of GI is to permit the normal promotion of CYP707A2 expression in nondormant seeds (Kushiro et al., 2004).
Figure 6.
The Expression of ABA- and GA-Related Genes in 3-Month after-Ripened lhy cca1 and gi-11 Seeds during the first 48 h of Imbibition.
(A) Visual illustration of the germination of Ws, lhy cca1, and gi-11 seeds after 48 h of imbibition at 22°C.
(B) The expression of GA3OX1, NCED6, ABI3, and CYP707A2 in Ws, lhy cca1, and gi-11. Data points represent means and sd of data from two biological replicates.
Most strikingly, however, LHY, CCA1, and GI were found to be important in the regulation of ABI3 expression, a central germination repressor previously shown to interact with TOC1 (Kurup et al., 2000). ABI3 expression was low even in dry lhy cca1 double mutant seeds and was lower than wild type at all nine time points tested, suggesting that lhy cca1 double mutants are unable to maintain high ABI3 expression, even in seeds that do not germinate (germination frequency is only 50%; Figure 2B). As ABI3 is required for dormancy, reduced ABI3 expression coupled with low NCED expression and increased GA3OX expression underlies the reduced dormancy phenotypes of lhy cca1 double mutant seeds.
After-Ripening Promotes a Light-Independent Increase in CCA1 Transcript Levels upon Imbibition
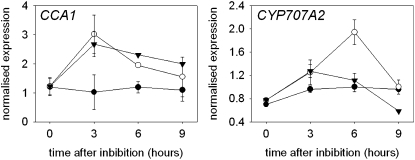
Two genes, CCA1 and CYP707A2, show strong transient expression 6 h after imbibition in after-ripened, but not dormant, seeds (Figures 3 and 5). The transcriptional induction of CYP707A2 in seeds is red/far-red reversible, suggesting regulation by the phytochromes (Seo et al., 2006). As CCA1 expression can also be induced by phytochrome-dependent light signaling (Wang and Tobin, 1998), we analyzed whether the increase in expression of CCA1 in after-ripened seeds required light (Figure 7). In agreement with previous studies (Millar et al., 2006; Seo et al., 2006; Figure 3), we found that CYP707A2 expression was induced only by a combination of after-ripening and light. By contrast, CCA1 expression was increased in after-ripened seeds whether in white light or maintained in the dark, when compared with freshly harvested dormant seeds in the light. Therefore, we conclude that after-ripening precipitates a light-independent promotion of CCA1 expression upon imbibition.
Figure 7.
The Transcriptional Induction of CCA1 Expression by after-Ripening Is Independent of the Light Signal.
After-ripened Landsberg erecta wild-type seeds were imbibed for 9 h in the light or dark, and the expression of CCA1 and of the after-ripening and light-induced CYP707A2 gene was compared with the expression in freshly harvested seeds. Closed circles, freshly harvested seeds in white light; closed triangles, 3-month after-ripened seed maintained in darkness; open circles, 3-month after-ripened seeds in white light. This treatment only resulted in germination at high frequency. Data points represent the mean and sd of data from two biological replicates.
Clock Genes Are Required for Normal ABA and GA Responses in Seeds
We noted that ABI3 expression was consistently reduced in gi-11 mutants compared with the wild type (Figure 6B). This result was surprising given that these mutants do not germinate under these conditions and led us to test the ABA sensitivity of seed germination in lhy cca1 and gi-11 mutants (Figure 8). The lhy cca1 double mutant showed germination weakly resistant to ABA or the GA biosynthesis inhibitor paclobutrazol (PAC), consistent with its increased germination potential. However, gi-11 seeds whose germination could be promoted by cold were specifically insensitive to applied ABA. This effect was most striking in lhy cca1 gi, where the increased germination under controlled conditions attributable to loss of LHY and CCA1 was ABA sensitive, but the remainder showed strong ABA insensitivity. Hence, this experiment uncovers a duality in GI function during germination: first, a role in germination promotion required for the perception of after-ripening that is epistatic to lhy cca1; and second, a role in germination inhibition that is closely linked to ABA action and that functions additively to LHY and CCA1.
Because of this strong effect, we analyzed the ABA and PAC sensitivity of germination in further clock gene mutants (Figure 8). Of those that showed significant phenotypes, we found that gi-5 exhibited a weak ABA-resistant germination phenotype but was slightly more sensitive to PAC (at 1 μM, P = 0.002). We also analyzed the germination sensitivity of toc1-1 and the TOC1 minigene gain-of-function line (Más et al., 2003; this line expresses the TOC1 cDNA fused yellow fluorescent protein under the control of the TOC1 promoter). While toc1-1 exhibited a wild-type response to PAC and ABA, TMG seeds showed ABA hypersensitivity, coupled by PAC hyposensitivity of germination. This is opposite to, and consistent with, the phenotype of gi mutants, which accumulate lower levels of TOC1 protein than the wild type (Kim et al., 2007). Any differences from the wild type in other lines tested were not significantly different, other than a very slight increased PAC-resistant germination phenotype for ztl-3 and LHY-OX seeds. Interestingly, it appears to be lines in which TOC1 protein levels are altered but rhythmicity is preserved (gi and TMG) that have these ABA and GA response phenotypes, whereas those lines with disrupted TOC1 protein rhythms (e.g., CCA1OX and ztl-3) or complete loss of TOC1 are unaffected. Together, these results suggest that altering TOC1 protein levels in the evening, or altering the amplitude of TOC1 oscillations, can affect hormone signaling in seeds.
DISCUSSION
Our data show that the plant circadian clock genes play a central role in the regulation of seed dormancy and germination. Lesions in at least five independent clock gene loci result in seed dormancy phenotypes, implicating LHY, CCA1, GI, ZTL, and LUX in dormancy control. At least one other protein associated with the circadian oscillator, TOC1, interacts in vivo with ABI3, a central regulator of dormancy control (Kurup et al., 2000), and while loss of TOC1 does not on its own affect dormancy, we showed that TOC1 gain of function affects the seed germination response to ABA and GA (Figure 8). Our data also demonstrate that clock genes are essential for the regulation of the transcription of enzymes required for GA and ABA metabolism in seeds, the action of which has been previously shown to be essential for normal dormancy and germination. Some of these, such as CYP707A2 and NCED genes, have also been recently shown to be circadian regulated in seedlings (Michael et al., 2008). Therefore, we show a role for circadian clock genes in plant dormancy regulation.
In the after-ripening response, GI action is epistatic to LHY and CCA1 in a manner strongly reminiscent of the photoperiodic control of flowering time (Mizoguchi et al., 2005). In this case, the epistasis results from the role for GI in controlling the critical output of the clock, the regulation of CONSTANS transcription. GI has an obvious role in light signaling (Mizoguchi et al., 2005; Oliverio et al., 2007), and light is essential for germination. This may in part explain the poor germination of gi mutants in the absence of dormancy-breaking signals, particularly as genes such as CYP707A2, which are light, clock, and GI regulated (Seo et al., 2006; Michael et al., 2008; Figure 6). However, the epistatic relationship between gi and lhy cca1 is inconsistent with their genetic relationship in the light signaling pathway in seedlings, where the action of their gene products is additive (Mizoguchi et al., 2005). This suggests that the role of these genes in seeds is subtly different. In some responses, notably the promotion of germination by cold, LHY and CCA1 also have a GI-independent role in germination repression (Figure 1A). This shows that there is germination regulation by clock genes that does not require GI and suggests that different targets are activated by clock genes during the breaking of dormancy by different signals. Equally, although the increased after-ripening speed of lhy cca1 requires GI, it is not caused by high GI expression in the lhy cca1 mutant (see Supplemental Figure 3 online).
We showed that the transcriptional response of CCA1 to dry after-ripening is independent of the light condition (Figure 7); therefore, we conclude that at least CCA1, and (from their mutant phenotypes) possibly LHY and GI, have a light-independent role in the after-ripening response. That this induction of CCA1 expression is light independent is consistent with the observation that there is no simultaneous induction of LHY expression in after-ripened seeds, itself a light-inducible gene (Schaffer et al., 1998). Furthermore, nitrate, another dormancy breaking signal (Hilhorst and Karssen, 1988), also specifically affects CCA1 expression in seedlings, but not LHY (Gutiérrez et al., 2008), suggesting that this may be a common and necessary attribute of a dormancy breaking signal. However, the relatively normal dormancy of CCA1-overexpressing seeds (Figure 1C), coupled with the counterintuitive increased after-ripening of lhy cca1 double mutants (Figure 1B), show that a simple induction of CCA1 expression is not in itself sufficient for dormancy loss. Given that plants with altered clocks show altered water use efficiency (Dodd et al., 2005), this response to dryness in seeds is further evidence that the circadian clock has a role in the normal plant response to water availability.
In addition to the direct action of after-ripening on CCA1 expression, after-ripening reduces the amplitude of subsequent downstream clock gene expression, especially that of GI and the PRR genes (Figure 3). This may be a consequence of the induction of CCA1 expression by after-ripening or subject to a common cause. However, as overexpression of CCA1 does not in itself lower dormancy (see Supplemental Figure 1B online), and the lhy cca1 mutant has lower dormancy than the wild type (Figure 1), it seems reasonable to exclude the activation of CCA1 expression in after-ripened seeds as the direct cause of dormancy loss. One possibility is that after-ripening acts on GI/PRR proteins present at imbibition and that this mediates the induction of CCA1 expression. This would rationalize the requirement of GI for germination. That lhy cca1 has a reduced dormancy phenotype may indicate that these genes are important when CCA1 expression is lower in the nondormant state than in the dormant state, such as at dawn on the second day when GA3OXs and NCEDs are expressed. Because of the complexity of the gene networks involved, a computational approach is probably required to test these hypotheses.
A recent study suggests one mechanism through which these changes in amplitude might affect germination control. Covington et al. (2008) show that >40% of ABA- and GA-regulated gene expression is under circadian control, with ABA downregulated and GA upregulated genes commonly expressed around dusk and ABA-activated and GA-repressed genes peaking on average close to dawn. Thus, a shift to high amplitude dawn expression and low amplitude dusk expression of clock genes in response to after-ripening can be expected to have a significant impact on the GA and ABA signaling pathways. Genetic evidence for this comes from the analysis of ABA and PAC sensitivity of germination in gi mutants (which have low amplitude TOC1 protein oscillations; Kim et al., 2007) and TMG seeds, which show TOC1 gain-of-function phenotypes (and are expected to have higher amplitude TOC1 oscillations; Más et al., 2003; Figure 8). gi mutant seeds show a strong trend to ABA insensitivity, while TMG seeds show hypersensitivity.
This suggests that changing the amplitude of TOC1 protein levels can affect hormone signaling in seeds, either through hormone levels or signaling. However, toc1-1 mutants show a wild-type phenotype, suggesting that there may be redundant action of PRR genes. Seed dormancy is controlled by hormone balance, with the ratio of GA and ABA action determining dormancy status (Wareing and Saunders, 1971). In this context, the finding that TMG seeds combine ABA hyposensitivity with significant PAC-resistant germination is interesting. Similarly, the combination of low germination coupled with ABA-resistant germination found in gi-11 is unexpected. The gi-5 mutant is also weakly PAC hypersensitive. Together, these results suggest the perturbation of GI and PRR proteins can alter the signaling balance between GA and ABA in seeds in a manner not previously seen in Arabidopsis seed dormancy-affected mutants.
It has been shown that the ability of a far-red pulse to promote germination varies over a 24-h period, suggesting circadian input into the germination program (Oliverio et al., 2007). As clock genes control the response to both light and after-ripening in seeds and are controlling the expression of outputs known to be both necessary for regulation of germination and responsive to these signals, the clock genes are integrating signaling data from both environmental input pathways.
METHODS
Plant Material
gi-11 (Ws background) was isolated in a screen of T-DNA insertion lines and has been described previously, as have gi-3, gi-4, and gi-5 each of which are ethyl methanesulfonate–induced point mutations (Fowler et al., 1999). Single and double cca1-11 lhy-21 complete loss-of-function mutants have been described previously (Hall et al., 2003; ecotype Ws). The LHY OX lines (Landsberg erecta background) were described by Schaffer et al. (1998) and the CCA1 OX line 038 (Col background) by Wang and Tobin (1998). Lux (Col background; Hazen et al., 2005) TMG, toc1-1, toc1-2 (all C24 background), and ztl-3 (Col background) mutant seeds (Millar et al., 1995; Somers et al., 2000; Más et al., 2003) were a gift from Steve Kay to A.H.
Seed Germination Experiments
Plants for seed production were grown in a glasshouse supplemented with artificial light to maintain a photoperiod of 16 h light (minimum 150 μmol m−2 s−1) and 8 h darkness. Before germination, each seed batch was size fractionated to exclude poorly filled seeds, with only those failing to fall through a 200-μm mesh sieve (Fisher Scientific) used for experiments. Seeds were surface sterilized by rinsing in 100% ethanol and air-drying before sowing on 0.9% water-agarose plates. Cold stratification treatments were in constant darkness at 4°C. GA3 (Sigma-Aldrich) was added at 100 μM NOR (Greyhound Chromatography) at 50 μM and potassium nitrate (Sigma-Aldrich) at 60 mM where indicated. PAC was obtained from Greyhound Chromatography. Germination experiments used five to eight independent seed batches for each genotype and were scored 7 d after imbibition (or stratification release) as radicle emergence. The germination conditions were 12 h at 75 μmol m−2 s−1 white light and 12 h darkness at the indicated temperatures. Oscillating temperature experiments applied 27°C during the 12-h light period and 17°C during the 12-h dark period. After-ripening was achieved by storing at 18 to 22°C in constant darkness. Germination significantly different from controls was determined using the Student's t test. Red light treatments (10 μmol m−2 s−1) were performed on water agar plates using seed imbibed in the dark for 1 h prior to light exposure, using an LED light source (peak 660 nm). Seeds were subsequently incubated in the dark for 7 d at 22°C before scoring germination. For experiments where darkness is required, extreme care was taken not to expose imbibed seeds to light, especially during sterilization (rinse in 100% ethanol for 10 min before drying) or sowing.
RNA Extraction and Real-Time RT-PCR
Seeds for RNA extraction were treated as for the germination experiments above. RNA was extracted from 75 to 90 mg of dry or imbibed seeds and cDNA synthesized as described previously (Penfield et al., 2005). cDNA was diluted 1:30 with distilled water and used for real-time RT-PCR with SYBR green detection using an ABI Prism 7300 thermocycler (Applied Biosystems). Transcript levels in two biological replicates for each sample were quantified using a standard curve derived from one sample set as a reference sample with an arbitrary value set to one. (Reference samples were an ND22 sample 48 h after imbibition for data shown in Figures 3 and 5, a Ws sample 48 h after imbibition for data shown in Figure 6 and Supplemental Figures 3 and 4, or a wild-type dry seed sample for data shown in Figure 7.) Transcripts were then normalized to those of the housekeeping gene CITRATE SYNTHASE3 (CSY3; Pracharoenwattana et al., 2005). Analysis of publicly available Affymetrix array data shows that this is one of only a few noncircadian regulated genes expressed stably throughout the transition from dry seed to seedling establishment (S. Penfield, unpublished data). Each experiment was repeated twice, and data for the two biological replicates are presented. PCR primers used for GA3OX1, GA3OX2, ABI3, CCA1, LHY, TOC1, LUX, and GI have been described previously (Penfield et al., 2005, 2006b; Edwards et al., 2006; Gould et al., 2006). Novel primers were used were as follows: CYP707A2F, 5′-AAAACGCAACGGCTTAAGTGA-3′; CYP707A2R, 5′-GGTGCGGCGAATATAACAC-3′; NCED6F, 5′-GGAATGCGTGGGAAGAGAGA-3′; NCED6R, 5′-ATACATGACCCGATTACGACGAT-3′; CSY3F, 5′-AGCGCTTTATGGTCCACTTCA-3′; CSY3R, 5′-CAACAGTCCCAATCTCTGACAA-3′.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: TOC1, At5g61380, BT005816; LHY, At1g01060; CCA1, At2g46830, BT001096; GI, At1g22770; LUX, At3g46640, BT006425; ZTL, At5g57360, BT008772; PRR7, At5g02810, AY142560; PRR9, At2g46790, AY128856; GA3OX1, At1g15550, BT005827; CYP707A2, At2g29090; ABI3, At3g24650, X68141; NCED6, At3g24220; CSY3, At2g42790.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. The Germination after 7 d of Freshly Harvested Seeds in Response to Cold Stratification.
Supplemental Figure 2. The Germination of Freshly Harvested and 3 Months after-Ripened Wild-Type Seeds from 12 to 27°C.
Supplemental Figure 3. The Expression of Genes Encoding Arabidopsis Circadian Clock Components under Diurnal Light/Dark Cycles in Imbibed 3 Month after-Ripened Seeds in Ws, gi-11, and lhy cca1 during the first 48 h of Imbibition.
Supplemental Figure 4. Real-Time RT-PCR to Show the Expression of GA3OX1 after the Release of Freshly Harvested Wild-Type Ws and the Cold Stratification Nonresponsive gi-11 Mutant Seeds into Ambient Temperatures from 3 d Cold Stratification at 4°C.
Supplemental Table 1. Microarray Data Showing Expression of Arabidopsis Circadian Clock Genes in Dry Seeds, Taken from Finch-Savage et al. (2007).
Supplementary Material
Acknowledgments
S.P. thanks Susannah Bird for technical assistance. S.P. was supported by a Royal Society University Research Fellowship and by the University of York.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Steven Penfield (sdp5@york.ac.uk).
Online version contains Web-only data.
References
- Ali-Rachedi, S., Bouinot, D., Wagner, M.H., Bonnet, M., Sotta, B., Grappin, P., and Jullien, M. (2004). Changes in endogenous abscisic acid levels during dormancy release and maintenance of mature seeds: Studies with the Cape Verde Islands ecotype, the dormant model of Arabidopsis thaliana. Planta 219 479–488. [DOI] [PubMed] [Google Scholar]
- Alonso-Blanco, C., Bentsink, L., Hanhart, C.J., Blankestijn-de Vries, H., and Koornneef, M. (2003). Analysis of natural allelic variation at seed dormancy loci of Arabidopsis thaliana. Genetics 164 711–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant, T.R. (1972). Gas exchange in dry seeds: Circadian rhythmicity in the absence of DNA replication, transcription, and translation. Science 178 634–636. [DOI] [PubMed] [Google Scholar]
- Cao, S., Ye, M., and Jiang, S. (2005). Involvement of GIGANTEA gene in the regulation of the cold stress response in Arabidopsis. Plant Cell Rep. 24 683–690. [DOI] [PubMed] [Google Scholar]
- Covington, M.F., Maloof, J.N., Straume, M., Kay, S.A., and Harmer, S.L. (2008). Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 9 R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd, A.N., Salathia, N., Hall, A., Kévei, E., Tóth, R., Nagy, F., Hibberd, J.M., Millar, A.J., and Webb, A.A. (2005). Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309 630–633. [DOI] [PubMed] [Google Scholar]
- Edwards, K.D., Anderson, P.E., Hall, A., Salathia, N.S., Locke, J.C., Lynn, J.R., Straume, M., Smith, J.Q., and Millar, A.J. (2006). FLOWERING LOCUS C mediates natural variation in the high-temperature response of the Arabidopsis circadian clock. Plant Cell 18 639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch-Savage, W.E., Cadman, C.S., Toorop, P.E., Lynn, J.R., and Hilhorst, H.W. (2007). Seed dormancy release in Arabidopsis Cvi by dry after-ripening, low temperature, nitrate and light shows common quantitative patterns of gene expression directed by environmentally specific sensing. Plant J. 51 60–78. [DOI] [PubMed] [Google Scholar]
- Fowler, S., Lee, K., Onouchi, H., Samach, A., Richardson, K., Morris, B., Coupland, G., and Putterill, J. (1999). GIGANTEA: A circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 18 4679–4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould, P.D., Locke, J.C., Larue, C., Southern, M.M., Davis, S.J., Hanano, S., Moyle, R., Milich, R., Putterill, J., Millar, A.J., and Hall, A. (2006). The molecular basis of temperature compensation in the Arabidopsis circadian clock. Plant Cell 18 1177–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez, R.A., Stokes, T.L., Thum, K., Xu, X., Obertello, M., Katari, M.S., Tanurdzic, M., Dean, A., Nero, D.C., McClung, C.R., and Coruzzi, G.M. (2008). Systems approach identifies an organic nitrogen-responsive gene network that is regulated by the master clock control gene CCA1. Proc. Natl. Acad. Sci. USA 105 4939–4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, A., Bastow, R.M., Davis, S.J., Hanano, S., McWatters, H.G., Hibberd, V., Doyle, M.R., Sung, S., Halliday, K.J., Amasino, R.M., and Millar, A.J. (2003). The TIME FOR COFFEE gene maintains the amplitude and timing of Arabidopsis circadian clocks. Plant Cell 15 2719–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen, S.P., Schultz, T.F., Pruneda-Paz, J.L., Borevitz, J.O., Ecker, J.R., and Kay, S.A. (2005). LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc. Natl. Acad. Sci. USA 102 10387–10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilhorst, H.W., and Karssen, C.M. (1988). Dual effect of light on the gibberellin- and nitrate-stimulated seed germination of Sisymbrium officinale and Arabidopsis thaliana. Plant Physiol. 86 591–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdsworth, M.J., Bentsink, L., and Soppe, W.J. (2008). Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol. 179 33–54. [DOI] [PubMed] [Google Scholar]
- Kim, W.Y., Fujiwara, S., Suh, S.S., Kim, J., Kim, Y., Han, L., David, K., Putterill, J., Nam, H.G., and Somers, D.E. (2007). ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 449 356–360. [DOI] [PubMed] [Google Scholar]
- Kurup, S., Jones, H.D., and Holdsworth, M.J. (2000). Interactions of the developmental regulator ABI3 with proteins identified from developing Arabidopsis seeds. Plant J. 21 143–155. [DOI] [PubMed] [Google Scholar]
- Kushiro, T., Okamoto, M., Nakabayashi, K., Yamagishi, K., Kitamura, S., Asami, T., Hirai, N., Koshiba, T., Kamiya, Y., and Nambara, E. (2004). The Arabidopsis cytochrome P450 CYP707A encodes ABA 8'-hydroxylases: key enzymes in ABA catabolism. EMBO J. 23 1647–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre, V., North, H., Frey, A., Sotta, B., Seo, M., Okamoto, M., Nambara, E., and Marion-Poll, A. (2006). Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J. 45 309–319. [DOI] [PubMed] [Google Scholar]
- Locke, J.C., Southern, M.M., Kozma-Bognár, L., Hibberd, V., Brown, P.E., Turner, M.S., and Millar, A.J. (June 28, 2005). Extension of a genetic network model by iterative experimentation and mathematical analysis. Mol. Syst. Biol. (online), doi/10.1038/msb4100018. [DOI] [PMC free article] [PubMed]
- Lopez-Molina, L., Mongrand, S., and Chua, N.H. (2001). A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. USA 98 4782–4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Más, P., Alabadí, D., Yanovsky, M.J., Oyama, T., and Kay, S.A. (2003). Dual role of TOC1 in the control of circadian and photomorphogenic responses in Arabidopsis. Plant Cell 15 223–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung, C.R. (2006). Plant circadian rhythms. Plant Cell 18 792–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael, T.P., Breton, G., Hazen, S.P., Priest, H., Mockler, T.C., Kay, S.A., and Chory, J. (2008). A morning-specific phytohormone gene expression program underlying rhythmic plant growth. PLoS Biol. 6 e225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar, A.A., Jacobsen, J.V., Ross, J.J., Helliwell, C.A., Poole, A.T., Scofield, G., Reid, J.B., and Gubler, F. (2006). Seed dormancy and ABA metabolism in Arabidopsis and barley: The role of ABA 8'-hydroxylase. Plant J. 45 942–954. [DOI] [PubMed] [Google Scholar]
- Millar, A.J., Carré, I.A., Strayer, C.A., Chua, N.H., Kay, S.A. (1995). Circadian clock mutants in Arabidopsis identified by luciferase imaging. Science 267 1161–1163. [DOI] [PubMed] [Google Scholar]
- Mizoguchi, T., Wright, L., Fujiwara, S., Cremer, F., Lee, K., Onouchi, H., Mouradov, A., Fowler, S., Kamada, H., Putterill, J., and Coupland, G. (2005). Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis. Plant Cell 17 2255–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliverio, K.A., Crepy, M., Martin-Tryon, E.L., Milich, R., Harmer, S.L., Putterill, J., Yanovsky, M.J., and Casal, J.J. (2007). GIGANTEA regulates phytochrome A-mediated photomorphogenesis independently of its role in the circadian clock. Plant Physiol. 144 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paltiel, J., Amin, R., Gover, A., Ori, N., and Samach, A. (2006). Novel roles for GIGANTEA revealed under environmental conditions that modify its expression in Arabidopsis and Medicago truncatula. Planta 224 1255–1268. [DOI] [PubMed] [Google Scholar]
- Penfield, S., Gilday, A.D., Halliday, K.J., and Graham, I.A. (2006. a). DELLA-mediated cotyledon expansion breaks coat-imposed seed dormancy. Curr. Biol. 16 2366–2370. [DOI] [PubMed] [Google Scholar]
- Penfield, S., Josse, E.M., Kannangara, R., Gilday, A.D., Halliday, K.J., and Graham, I.A. (2005). Cold and light control seed germination through the bHLH transcription factor SPATULA. Curr. Biol. 15 1998–2006. [DOI] [PubMed] [Google Scholar]
- Penfield, S., Li, Y., Gilday, A.D., Graham, S., and Graham, I.A. (2006. b). Arabidopsis ABA INSENSITIVE4 regulates lipid mobilization in the embryo and reveals repression of seed germination by the endosperm. Plant Cell 18 1887–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield, S., Rylott, E.L., Gilday, A.D., Graham, S., Larson, T.R., and Graham, I.A. (2004). Reserve mobilization in the Arabidopsis endosperm fuels hypocotyl elongation in the dark, is independent of abscisic acid, and requires PHOSPHOENOLPYRUVATE CARBOXYKINASE1. Plant Cell 16 2705–2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pracharoenwattana, I., Cornah, J.E., and Smith, S.M. (2005). Arabidopsis peroxisomal citrate synthase is required for fatty acid respiration and seed germination. Plant Cell 17 2037–2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer, R., Ramsay, N., Samach, A., Corden, S., Putterill, J., Carré, I.A., and Coupland, G. (1998). The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93 1219–1229. [DOI] [PubMed] [Google Scholar]
- Seo, M., Hanada, A., Kuwahara, A., Endo, A., Okamoto, M., Yamauchi, Y., North, H., Marion-Poll, A., Sun, T.P., Koshiba, T., Kamiya, Y., Yamaguchi, S., and Nambara, E. (2006). Regulation of hormone metabolism in Arabidopsis seeds: Phytochrome regulation of abscisic acid metabolism and abscisic acid regulation of gibberellin metabolism. Plant J. 48 3543–3566. [DOI] [PubMed] [Google Scholar]
- Somers, D.E., Schultz, T.F., Milnamow, M., and Kay, S.A. (2000). ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 101 319–329. [DOI] [PubMed] [Google Scholar]
- Strayer, C., Oyama, T., Schultz, T.F., Raman, R., Somers, D.E., Más, P., Panda, S., Kreps, J.A., and Kay, S.A. (2000). Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289 768–771. [DOI] [PubMed] [Google Scholar]
- Tomita, J., Nakajima, M., Kondo, T., and Iwasaki, H. (2005). No transcription-translation feedback in circadian rhythm of KaiC phosphorylation. Science 307 251–254. [DOI] [PubMed] [Google Scholar]
- Wang, Z.Y., and Tobin, E.M. (1998). Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93 1207–1217. [DOI] [PubMed] [Google Scholar]
- Wareing, P.F., and Saunders, P.F. (1971). Hormones and dormancy. Annu. Rev. Plant Physiol. 22 261–288. [Google Scholar]
- Yamaguchi, S., Smith, M.W., Brown, R.G., Kamiya, Y., and Sun, T. (1998). Phytochrome regulation and differential expression of gibberellin 3beta-hydroxylase genes in germinating Arabidopsis seeds. Plant Cell 10 2115–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi, Y., Ogawa, M., Kuwahara, A., Hanada, A., Kamiya, Y., and Yamaguchi, S. (2004). Activation of gibberellin biosynthesis and response pathways by low temperature during imbibition of Arabidopsis thaliana seeds. Plant Cell 16 367–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., Chen, Y., Wang, Z.Y., Chen, Z., Gu, H., and Qu, L.J. (2007). Constitutive expression of CIR1 (RVE2) affects several circadian-regulated processes and seed germination in Arabidopsis. Plant J. 51 512–525. [DOI] [PubMed] [Google Scholar]
- Zhong, H.H., Painter, J.E., Salomé, P.A., Straume, M., and McClung, C.R. (1998). Imbibition, but not release from stratification, sets the circadian clock in Arabidopsis seedlings. Plant Cell 10 2005–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.