Abstract
Embryonic stem (ES) cells are isolated from the inner cell mass (ICM) of blastocysts, whereas epiblast stem cells (EpiSCs) are derived from the post-implantation epiblast and display a restricted developmental potential. Here we characterize pluripotent states in the non-obese diabetic (NOD) mouse strain, which prior to this study was considered “non-permissive” for ES cell derivation. We find that NOD stem cells can be stabilized by providing constitutive expression of Klf4 or c-Myc or small molecules that can replace these factors during in vitro reprogramming. The NOD ES and iPS cells appear “metastable”, as they acquire an alternative EpiSC-like identity after removal of the exogenous factors, while their reintroduction converts the cells back to ICM-like pluripotency. Our findings suggest that stem cells from different genetic backgrounds can assume distinct states of pluripotency in vitro, the stability of which is regulated by endogenous genetic determinants and can be modified by exogenous factors.
Introduction
Mouse embryonic stem cells (ESC), isolated from the inner cell mass (ICM) of blastocysts, can be propagated in vitro in an undifferentiated state and recapitulate all defining features of ICM cells (Jaenisch and Young, 2008). ICM-derived ES cells, when used to generate chimeras, can contribute to all somatic cell lineages and to germ cells, maintain both X chromosome alleles in a reactivated state, but are unable to contribute to the trophectoderm lineages (Rossant, 2008). Mouse ES-like cells derived either by somatic cell nuclear transfer or by direct in vitro reprogramming (termed induced pluripotent stem (iPS) cells) share all of these defining features (Hanna et al., 2008; Takahashi and Yamanaka, 2006; Wakayama et al., 1998; Wernig et al., 2007).
Despite their apparent common origin from the ICM and their ability to propagate in vitro while maintaining pluripotency, human ES cells are phenotypically and functionally distinct from mouse ES cells (Thomson et al., 1998). Human ES cells require different growth conditions and rely on bFGF and ActivinA/TGFbeta signaling to maintain their pluripotent state, whereas mouse ES cells require Lif/Stat3 and Bmp4 signaling (Xu et al., 2005; Ying et al., 2003). Human ES cells differ epigenetically from mouse ES cells by several criteria such as X chromosome inactivation and pluripotency factor promoter occupancy across the genome (Boyer et al., 2005; Silva et al., 2008b; Tesar et al., 2007). Recently pluripotent cells from the epiblast of post-implantation murine embryos termed Epiblast stem cells (EpiSCs) have been isolated and found to recapitulate defining features of human stem cells (Brons et al., 2007; Tesar et al., 2007). EpiSC and human ES cells share the flattened morphology, intolerance to passaging as single cells, dependence on Activin/Nodal signaling, inactivation of the X chromosome in female cell lines, and the ability of some of the isolated EpiSC lines to differentiate into trophectoderm. In contrast to mouse ES cells, EpiSCs are extremely inefficient to generate chimeras and are unable to contribute to the germline (Brons et al., 2007; Guo et al., 2009; Tesar et al., 2007). The similarities between human ES cells and mouse EpiSCs have provoked questions concerning the nature, origin and the in vivo counterpart of human ES cells during normal development (Lovell-Badge, 2007).
Recent advances provided by new genetic and chemical approaches to isolate stem cells have enabled the derivation of iPS cells and ICM- or Epiblast-derived stem cells from different species. For instance, while EpiSCs can be generated from “non-permissive” species such as rat, ES cells cannot be established from the rat ICM under the same conditions used to isolate mouse ES cells (Brons et al., 2007). Moreover, whereas rat ICM-derived ESCs or iPS cells generated via transduction of Oct4, Sox2, Nanog and Lin28 can only be propagated in the presence of glycogen synthase kinase 3 (GSK3) and mitogen-activated protein kinase pathway (ERK) inhibitors (2i conditions) (Li et al., 2008; Li et al., 2009; Liao et al., 2009), rat iPS cells generated with lentiviruses encoding Oct4, Sox2, Klf4 and c-Myc could be propagated like mES cells without these inhibitors (Liao et al., 2009). Moreover, EpiSCs can be converted into ES cells by over expressing KLF4 and growing the cells in 2i conditions (Guo et al., 2009). These findings raise fundamental questions relating to pluripotency: What is the nature of pluripotent states achieved when deriving iPS cells from “permissive” and “non-permissive” strains? What are the molecular determinants involved in defining the epiblast- vs. ICM-like stem cell states? Could such determinants impinge on the ability of certain strains to adapt one pluripotent state but not the other? Would alternative pluripotent states be adapted if key molecular determinants are stabilized or neutralized?
To address these questions we used the non-obese diabetic (NOD) mouse strain as a model to characterize different isolated pluripotent states, as this strain is non-permissive for the derivation of ES cells, but allows the isolation of EpiSCs from day E6.5 embryos (Brons et al., 2007). We found that continuous ectopic expression of Klf4 or c-Myc transcription factors is sufficient for derivation of ICM-like pluripotent NOD iPS and ES cells. Supplementing mouse ES cell growth conditions with small molecules known to replace the function of Klf4 and c-Myc during iPS cell generation can facilitate the derivation of germline competent NOD ES cells. Importantly, upon removal of the exogenous factors that help maintain the ICM-like cell state, the NOD pluripotent cells adapted an alternative Epiblast-like pluripotent state and were functionally and molecularly similar to EpiSCs. Our results provide an example where failure to stabilize ICM-like pluripotency from a non-permissive strain can lead to the attainment of an in vitro acquired alternative pluripotent state. Moreover, these findings support the notion that appropriate growth conditions may have not yet been devised to allow in vitro stabilization of ICM-like pluripotent cells from other species than mouse.
Results
Derivation of transgene-dependent iPS cells from NOD mice
We attempted to derive NOD iPS cells by infecting mouse embryonic fibroblasts (MEFs) with doxycycline (Dox)-inducible lentiviral vectors encoding the four reprogramming factors Oct4, Sox2, Klf4 and c-Myc (see Figure 1A for definitions) and a constitutively active lentivirus encoding the reverse tetracycline transactivator (Ubi-M2rtTA) (Figure 1B). The infected NOD and control 129 MEFs were cultured in the presence of Dox and colonies appeared 12 days after Dox induction, many of which acquired ES-like morphology. However, unlike the 129-derived iPS cells that could be maintained in the absence of Dox after 20–30 days, the NOD ES-like cells differentiated upon Dox withdrawal (Figure 1B). To analyze individual iPS-like clones, colonies were picked from transduced NOD-MEF cultures at day 16 and further passaged in the presence of Dox for up to 45 days. After Dox withdrawal, all 120 Nanog+ NOD-iPS colonies underwent differentiation, while the majority of 129-derived iPS lines remained undifferentiated (Figure 1C). The Dox-dependent NOD-iPS clones had all the characteristics of pluripotent ES cells such as expression of ES cell markers, demethylation of the endogenous Oct4 and Nanog promoters and reactivation of endogenous pluripotency genes (Figure 1D–F). Finally, the Dox-dependent NOD-iPS cells generated differentiated teratomas and adult chimeras (Figure 1G–H). These results suggest that the four factors are capable of inducing a pluripotent ES-like state in NOD somatic cells. However, in contrast to iPS cells from the 129 strain, the NOD pluripotent state was unstable requiring the continuous expression of the reprogramming factors.
Figure 1. Generation of transgene-dependent NOD iPS cells.
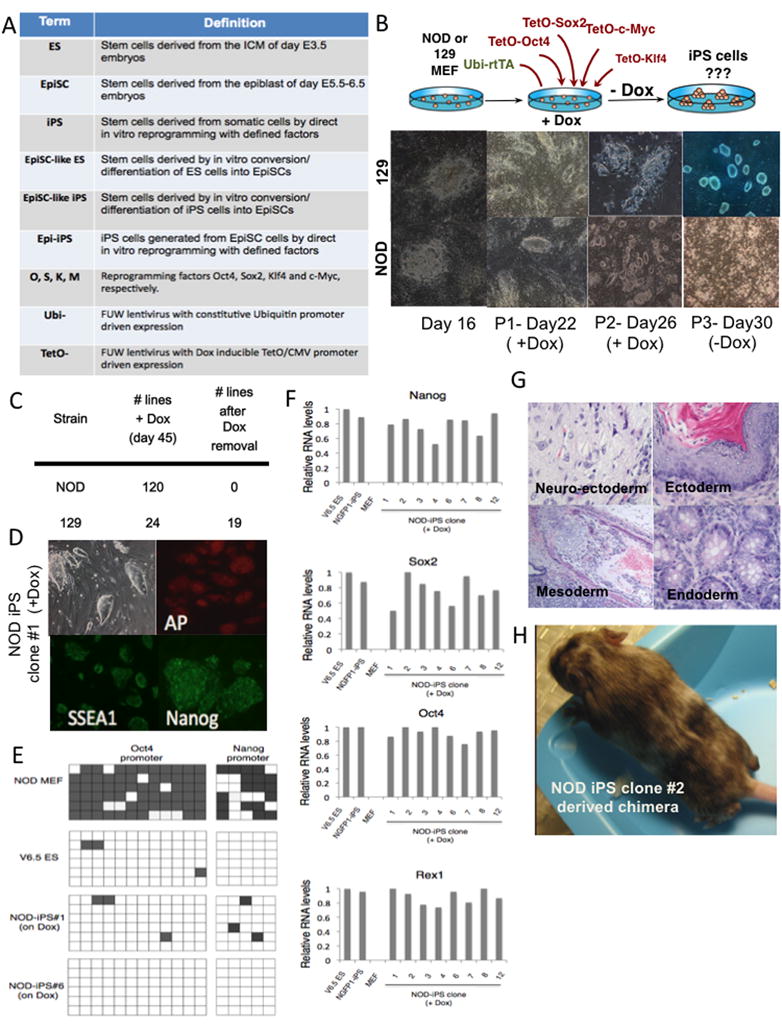
(A) Definitions and terms of reagents and cells used in this study. (B) Strategy used for reprogramming NOD or 129 MEFs into iPS cells by infection with Dox-inducible lentiviruses (TetO) encoding O,S,K,M. Representative images of colonies observed at different stages in the as indicated in the panels (P - passage number). (C) Colonies originally isolated at day 16 after infection from 3 independent experiments from NOD and 129 MEF cultures; only colonies that stained positive for Nanog at day 45 while being maintained with Dox were analyzed. Right column lists the number of iPS lines that remained pluripotent (Nanog and Oct4 positive) without Dox. (D) Immunostaining for pluripotency markers of NOD iPS subclone #1 maintained on Dox. AP: Alkaline Phosphatase. (E) Methylation analysis of Oct4 and Nanog promoters. Open squares indicate unmethylated and filled squares methylated CpG dinucleotides. Shown are 6 representative sequenced clones from NOD MEFs, V6.5 ES cells and three different Dox-dependent NOD iPS cell lines. (F) Quantitative RT-PCR analysis specific for the reactivation of the endogenous indicated pluripotency-related genes in Dox dependent NOD iPS lines. V6.5 ESCs, iPS cells grown in the absence of Dox and MEFs are used as controls. Relative expression levels were normalized to the average expression of control ES line V6.5. (G) Teratoma derived from Dox-dependent NOD-iPS#6 cell line. (H) Chimeric mice derived from Dox-dependent NOD-iPS cells. Chimerism is indicated by the albino and brown coat colors.
Constitutive ectopic expression of Klf4 or c-Myc facilitates derivation of NOD ES and iPS cells
To define the exogenous factors required to stabilize pluripotency in the NOD background, Dox-dependent iPS cells were transduced with constitutively expressed lenti-viruses encoding reprogramming factors and the ability of the cells to propagate was tested after Dox withdrawal (Figure 2A). Constitutive expression of Klf4 or c-Myc, but not Oct4 or Sox2, enabled the derivation of Dox-independent NOD iPS clones (Figure 2B). All Dox-independent NOD iPS cells carried the Ubi-c-Myc and Ubi-Klf4 proviruses and expressed transgene encoded transcripts (Figure 2C–D). The NOD iPS cells stained positive for pluripotency markers and generated teratomas (Figure 2E–F). Factor transduction by retroviral vectors, which are silenced in ES and iPS cells (Jahner et al., 1982), failed to yield any iPS cells from NOD mice, consistent with the requirement for continuous expression of c-Myc and Klf4 (Figure 2G). Conversely, iPS cells were readily isolated from NOD fibroblasts by using a combination of retroviruses encoding OSM factors and a constitutive virus expressing Klf4, or a combination of retroviruses encoding OSK factors and a constitutively expressed lentivirus expressing c-Myc (Figure 2G). When Dox inducible lentiviruses encoding c-Myc or Klf4 were used in the latter combinations instead of the constitutively expressed lentiviruses, NOD iPS lines could only be grown in the presence of Dox (Figure 2G). The derivation of iPS cells from rat fibroblasts required similar conditions (Figure 2G).
Figure 2. Induced pluripotency on NOD background is stabilized by constitutive ectopic expression of Klf4 or c-Myc.

(A) Experimental plan for deriving Dox-independent NOD iPS lines. Constitutive lentiviruses driven by the ubiquitin promoter (abbreviated as Ubi) encoding different transcription factors were used to transduce Dox dependent NOD iPS cells followed by Dox withdrawal. (B) Images of cultures depicting colony growth in the absence of Dox after infection of NOD iPS cells with constitutive lentiviruses encoding the indicated factors. (C) RT-PCR for detection of transgene specific expression of O, S, K, M in NOD-iPS#1 cells infected with c-Myc (M) or Klf4 (K) constitutive lentivirus that were maintained independent of Dox. V6.5 ES cells and parental uninfected pluripotent (on Dox) or differentiated (40 hours after Dox withdrawal) NOD iPS cells in the presence or absence of Dox were used as controls. (D) PCR analysis for the detection of Ubi-c-Myc and Ubi-Klf4 proviruses in genomic DNA of subcloned infected NOD iPS lines. (E) Immunostaining for pluripotency markers of Dox-independent NOD iPS lines that had been infected with Ubi-Klf4 or Ubi-c-Myc lentiviruses. (F) Teratomas derived from NOD iPS#1 infected subclones (G) Summary of infections performed on mouse NOD and 129Sv/Jae MEFs and Rat TTFs with reprogramming factors transduced either by Moloney retroviral backbone vectors (pLib), by constitutively expressed lentiviral vectors (Ubi), or by Dox inducible lentiviral vectors (TetO). Experiments using the three different infection protocols were performed side by side. The ability to derive iPS cells was defined by the detection of Nanog positive clones that were developmentally pluripotent as tested by teratoma formation (129 and NOD cell lines) or EB formation (Rat iPS cell lines) and is indicated by ‘+’. NA: Not applicable.
We tested whether constitutive expression of c-Myc and/or Klf4 would allow derivation of ES cells from NOD ICMs. NOD blastocysts failed to yield any ES cells under conditions routinely used for ES cell derivation (Figure 3A). While NOD-ICM outgrowths were obtained after plating on feeders, the cells did not survive passaging (Figure 3B). However, when NOD ICM outgrowths were infected with constitutive lenti-viruses encoding Klf4 or c-Myc the cultures could be dissociated and maintained in mouse ES cell conditions (defined as mESM: DMEM supplemented with fetal bovine serum (FBS) and Lif grown on irradiated feeders) resulting in the generation of independent NOD ES cell lines carrying either c-Myc or Klf4 proviruses (Figures 1A, 3E). Southern blot and RT-PCR analysis verified proviral integration and transgene-specific transcripts in the isolated lines (Figure 3D–E). The NOD-ES lines had a normal karyotype and a cell cycle pattern identical to control V6.5 ES cell, expressed pluripotency markers and generated adult chimeras with germ-line contribution (Figure 3F–I and Table S1). Tumor formation was observed in some of the c-Myc transgenic NOD ES line derive chimeras and offspring (data not shown) probably as a result of the ectopic expression of the c-Myc oncogenic transgene (Okita et al., 2007).
Figure 3. Generation of Klf4 and c-Myc transgenic NOD ES lines.

(A) Summary of ESC derivation efficiency from NOD and permissive strain control blastocysts. (B) Representative images for NOD blastocyst at embryonic day 3.5 and ICM outgrowths after plating the blastocysts on feeder cells. The cells were infected with the indicated viruses and passaged in mESM. (C) Representative images of NOD colonies after infection with c-Myc and Klf4 viruses at passage two (P2) and stable established NOD ES lines at P5. NOD ES 1M was derived after infections with Ubi-c-Myc and three lines were obtained following Ubi-Klf4 infection (NOD ES 2K is shown). (D) Transgene expression by RT-PCR in various NOD ES lines. MEFs infected with O, S, K, M viruses were used as positive controls. (E) Southern analysis of c-Myc and Klf4 viral integrations in representative NOD ES cell lines derived following viral infection. NOD MEFs are used as background controls. (F–G) Karyotype and cell cycle analysis of transgenic NOD ES lines. (H) Immuno-fluorescent staining of NOD ES lines for pluripotency markers. (I) NOD ES cell derived chimeras from NOD ES 1M and 2K cell lines were mated with NOD mice with albino offspring indicating germline transmission.
Generation of genetically unmodified germline competent NOD ES cells
Small molecules have been identified to replace some of the reprogramming factors in iPS derivation (Huangfu et al., 2008; Shi et al., 2008). We have found that Wnt signaling promotes the derivation of iPS cells in the absence of c-Myc (Marson et al., 2008) and have identified Kenpaullone, a GSK3b and CDK1/cyclin B inhibitor, as a small molecule that replaces Klf4 during iPS reprogramming (Lyssiotis et al., 2009). We tested whether these compounds could replace constitutive Klf4 or c-Myc expression in propagating NOD iPS and ES cells. Dox-dependent NOD-iPS cells were grown in mESM lacking Dox but supplemented with: (1) Wnt3a, (2) the glycogen synthase kinase 3 inhibitor CHIR99021 (CH), or (3) Kenpaullone (KP). All three conditions supported Dox-independent growth of pluripotent NOD-iPS cells (Figure 4A and S1). Notably, it was recently reported that the “2i” culture conditions, using the ERK-cascade inhibitor PD184352 (PD) and the GSK3 inhibitor CH or 6-bromo-indirubin-3′-oxime (BIO) facilitate the derivation of rat ES cells (Buehr et al., 2008; Li et al., 2008). Consistent with this observation, NOD iPS cells were readily propagated independently of Dox in the presence of 2i, although at reduced single-cell cloning efficiency compared to mESM conditions supplemented with both KP and CH (KP/CH) (Figure 4B). In a similar manner to KP, the combination of PD/CH replaced the requirement for ectopic expression of the Klf4 transgene during iPS generation (Figure S2). Rat iPS cells could also be propagated in mESM with KP/CH (Figure S3). Importantly, KP/CH did not inhibit ERK phosphorylation, suggesting that stabilization of pluripotency can occur in the absence of ERK inhibition (Figure S4).
Figure 4. Generation of genetically unmodified germline competent NOD ES cells.
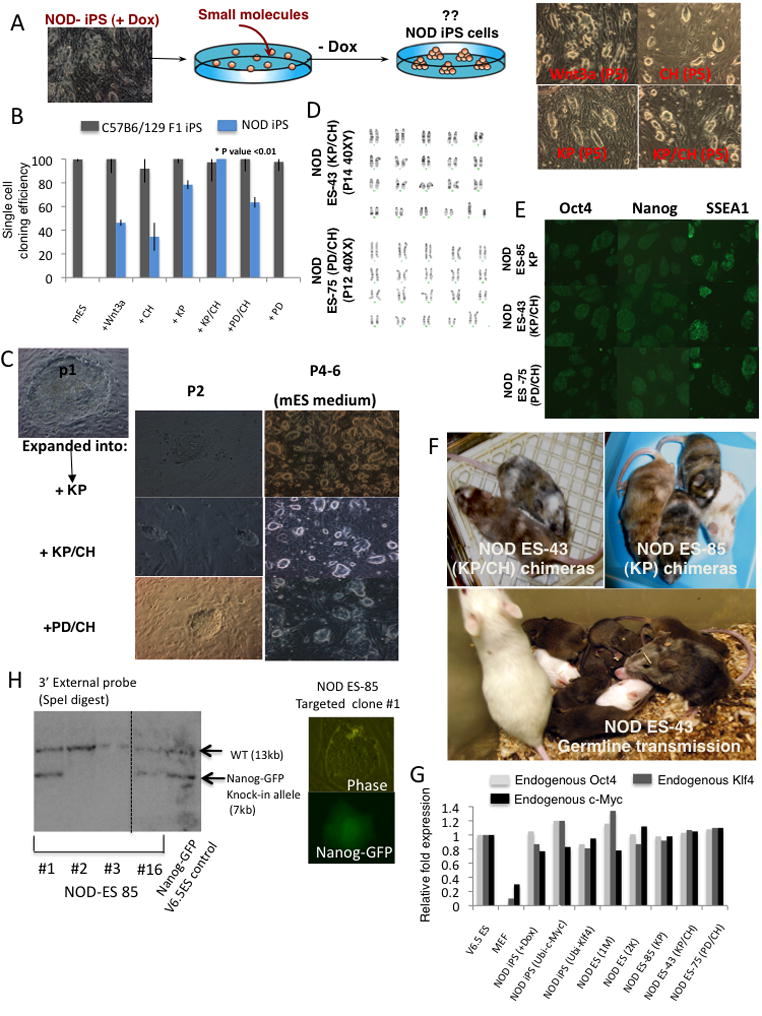
(A) Experimental outline for derivation of iPS lines that grow in the presence of small molecules and in the absence of Dox. Representative images of NOD iPS lines growing in mESM supplemented with the indicated small molecules or growth factors. (B) Single-cell cloning efficiency of NOD iPS cells in different growth conditions. WT or NOD-iPS cells grown in mESM and KP were sorted in 96 well plates and cultured in mESM supplemented with small molecules. The number of wells containing Nanog+ colonies was counted after 6 days. Efficiencies were normalized to that of C57B6/129F1-iPS cells (“permissive” controls) plated in mESM and defined as 100%. SD for average efficiencies from 2 experiments are shown. Student ttest P value compares KP/CH to PD/CH mESM conditions for NOD iPS cells. (C) Expansion of day E3.5 ICM derived NOD ES cells in the presence of KP, KP/CH or PD/CH and derivation of stable ES lines. Images of initial colonies observed at passage 2 after embryo dissociation are shown together with representative images at later passages. (D) Karyotype of NOD ES lines. (E) Immunostaining of NOD ES lines for pluripotency markers. Inhibitors used during cell line derivation and propagation are indicated. (F) Chimerism in adult mice generated from the indicated cell lines is evident by the presence of agouti (brown) and albino coat color originating from the NOD background. Lower panel demonstrates germline transmission obtained from a male NOD ES #43 derived chimera that was mated to an NOD female. (G) Real-Time PCR analysis for endogenous expressed Oct4, c-Myc and Klf4 genes in NOD iPS and ES cells grown in the indicated conditions. Relative expression levels are normalized to levels detected in control 129 ES cells. (H) Southern analysis indicating correct targeting of the endogenous Nanog locus in NOD ES cells with Nanog-GFP knock-in targeting construct.
To derive NOD ES cells, E3.5 NOD blastocysts were plated on mouse feeders and grown under the optimized culture conditions supplemented with the different inhibitors (Figure 4C). We isolated 16 lines in mESM containing KP (NOD-ES#85), KP/CH (NOD-ES#43), or PD/CH (NOD-ES#75). The ESCs remained stable in culture but required the continuous presence of the defined inhibitors, expressed pluripotency markers and retained a normal karyotype (Figure 4D–E and S1–2). The cells generated adult chimeras (Figure 4F) and contributed to the germ line (Figure 4F and Table S1). Importantly, the NOD ES or iPS cells maintained with exogenous factors expressed endogenous levels of Klf4 and c-Myc similar to those observed in control 129 ES cells (Figure 4G). This argues that the inability of NOD cells to stabilize pluripotency in the absence of the exogenous factors is not due to failure to reactivate the endogenous Klf4 or c-Myc genes. Finally, we tested whether NOD ES cells could be targeted by homologous recombination. NOD-ES#85 cells were electroporated with a linearized Nanog-GFP knock-in targeting construct (Hatano et al., 2005), followed by puromycin selection. Resistant colonies were picked after 10 days of drug selection and 2 out of 16 colonies analyzed demonstrated correct targeting of the Nanog locus (Figure 4H).
Destabilized NOD ICM-like stem cells adapt an alternative Epiblast-like pluripotent State
The NOD stem cells stabilized by ectopic expression of Klf4 or c-Myc or the presence of small molecules displayed ICM-like pluripotency as evident by their ability to generate chimeras with germline contribution. Upon plating these cells after withdrawal of the exogenous stimuli (Dox or inhibitors), we noticed distinct colonies with flat morphology (~0.3% of plated cells) (Figure 5A). These colonies could not be propagated following dissociation into single cells (by trypsinization), typically used to passage mouse ES cells, but could be stably propagated by passage of smaller clumps using collagenase or mechanical dissociation in the presence of bFGF (DMEM/F12 supplemented with FBS and bFGF, referred to as epiESM). The flat colonies were morphologically distinct from mouse ES cells and had a similar morphology to EpiSC cells and were termed EpiSC-like iPS cells (Figure 1A). Moreover, the NOD EpiSC-like iPS line #1 and the Dox dependent NOD-iPS #1 cells from which the former line was derived, carried an identical Sox2 genetic integration pattern, thus excluding contamination as a source for the EpiSC-like cells (Figure 5B).
Figure 5. Generation of NOD EpiSC-like ES and iPS cells.

(A) Dox dependent NOD-iPS lines were trypsinized and plated on MEF feeders and grown in mESM without Dox. Representative image of colonies typically observed 5–8 days after plating that can be manually passaged using collagenase and stably propagated in epiESM conditions. (B) Southern blot analysis indicating identical integration pattern for Sox2 transgene in NOD-iPS #1 and its derived cell line NOD EpiSC-like iPS #1. Black triangle indicate endogenous band, white triangle indicates transgenic band. (C) Derivation of Epiblast-like ES cells from NOD blastocysts by plating the embryos in EpiSC derivation medium and passaging ICM outgrowths after 5–8 days in epiESM conditions. Continued culture in epiESM and manual dissociation supported the growth of colonies with flat morphology-like colonies. (D) Immunofluorescent staining for pluripotency markers in EpiSC, EpiSC-like NOD iPS and ES lines. (E) Teratoma formation of NOD Epiblast-like NOD ES and iPS cell lines. (F) Evaluation of Oct4 distal enhancer (DE) and proximal enhancer (PE) reporter gene activity in the indicated pluripotent lines. Baseline activity was analyzed by infecting with an empty vector. (G) Cell samples were equally divided and plated on feeders in the indicated growth medium, and 24 hours later the wells were supplemented with either JAKi or ALKi or were kept without any inhibitor. At Day 6 the wells were stained for Oct4 to determine the relative percentage of pluripotent colonies. Values indicated are normalized to internal control sample (Growth medium without inhibitors – Gray Bars) in which each cell line were not exposed to inhibitors and defined as 100%. Relative percentage lower than 30% was defined as “sensitivity” to the presence of the inhibitor. (H) Whole genome cluster analysis of transcripts from pluripotent cell lines analyzed. Top 5000 differentially expressed probes were selected by cross-array standard deviations of the normalized expression values and included in unbiased cluster analysis. Growth conditions and passage number for the cell lines are indicated.
NOD EpiSCs have been previously derived from the epiblast of day E6.5 developing embryos (Brons et al., 2007). This prompted us to test whether EpiSC-like cells can also be isolated from NOD ICM in the absence of exogenous factors. NOD day E3.5 blastocysts were explanted and ICM outgrowths were manually dissociated after plating in epiESM derivation medium (see experimental procedures) and propagated in epiESM, resulting in stable lines termed EpiSC-like ES cells (Figure 1A, 5C and S6). NOD EpiSC-like iPS and NOD EpiSC-like ES cell lines (see Figure 1A) derived by the different approaches expressed pluripotency markers, were not capable of generating chimeric mice, but were pluripotent as evident by their ability to generate teratomas (Figure 5D–E and Table S1) (Brons et al., 2007; Guo et al., 2009; Tesar et al., 2007). In contrast, when the same derivation protocol was used on 129 embryos, typical murine ES cells were generated that could be passaged by trypsinization, were dependent on Lif/Stat3 signaling for their self renewal and were capable of generating adult chimeras with germline transmission (Figure S7 and Table S2).
We characterized NOD derived ES and EpiSC-like cells in a series of assays: (i) Cells were transfected with a luciferase reporter construct under the control of either the distal or the proximal enhancers that control expression of the Oct4 gene in the mouse ICM and Epiblast, respectively (Tesar et al., 2007). The NOD EpiSC-like ES and iPS lines preferentially utilized the Oct4 proximal enhancer similar to EpiSCs (Figure 5F). In contrast, NOD ES and iPS cells grown in mESM with KP/CH, as well as controls including 129 mES or iPS cells derived and grown in mESM or epiESM conditions, utilized the distal enhancer consistent with the notion that these cells resemble mouse ES cells (Tesar et al., 2007). (ii) Lif-dependent self-renewal: Mouse ES cells are dependent on Lif signaling and readily differentiate when exposed to JAK inhibitor (JAKi) that blocks Stat3 phosphorylation. In contrast, mouse EpiSCs or human ESCs rely on activin A (Inhb)/Nodal signaling to maintain pluripotency and rapidly differentiate in the presence of ALKi, an inhibitor of type I activin receptor-like kinases (Tesar et al., 2007). We exposed ESCs and EpiSCs to either ALKi or JAKi to test which of these pathways controls their self-renewal. NOD ICM derived ESCs maintained in mESM KP/CH conditions were dependent on Lif/Stat3 signaling in contrast to NOD EpiSC-like cells, which required Activin A/Nodal signaling (Figure 5G). (iii) Gene expression: EpiSCs and ESCs display different developmental potentials and several key pluripotency genes are differentially expressed in the two cell types (Tesar et al., 2007). Unbiased clustering of global gene expression profiles demonstrated that the NOD EpiSC-like ES and iPS lines clustered closely with EpiSCs, and were distinct from 129 ICM derived ES grown in mESM or epiESM conditions (Figure 5H and S9). Our results support the notion that, in spite of the presence of Lif and other growth factors present in the serum or provided by the feeders, NOD stem cells in vitro adapt a pluripotent state that highly resembles EpiSCs when the exogenous components that maintain their ICM-like pluripotent state are removed. In contrast, 129 ES cells maintained their ICM-like pluripotency in epiESM conditions and were dependent on Lif/Stat3 pathway stimulated by signals originating from the FBS and feeders used in these growth conditions. Consistent with this, differentiating 129 ES cells into EpiSC-like state was achieved only upon removal of FBS and Lif, and required prolonged culturing of the cells in serum free N2B27 defined medium supplemented with high levels of bFGF and Activin A (data not shown, Figure S7 and (Guo et al., 2009)).
Pluripotent states of NOD stem cells are unstable and are affected by exogenous factors
We asked whether the exogenous stimuli defined in this study could interconvert the two distinct pluripotent states. We first tested whether infection of previously characterized E5.5 derived 129 EpiSCs (Tesar et al., 2007) with a Klf4 or c-Myc Dox inducible vectors could convert the EpiSCs to an ICM-like pluripotent state. Consistent with other reports, EpiSCs did not spontaneously convert into ES-like cells upon culturing in mESM, but rather differentiated (Guo et al., 2009; Tesar et al., 2007). Infected EpiSC cultures maintained in mESM with Dox gave rise to distinguishable small round colonies (Figure 6A). When trypsinized into single cells and propagated on MEFs, they eventually acquired typical mouse ES-like morphology. Dox was withdrawn after 7–12 days and clonal lines termed Epi-iPS cells (Figure 1A and (Guo et al., 2009)) were stable and morphologically indistinguishable from mouse ES cells (Figure 6A). Southern analysis verified the presence of c-Myc or Klf4 proviral integrations (Figure S10). Moreover, unlike their donor EpiSCs, Epi-iPS cells grew stably in mESM conditions and their pluripotency was disrupted by inhibition of Stat3, but not of activin/nodal pathway (Figure 6B). Epi-iPS cells showed preferential utilization of the Oct4 distal enhancer similarly to mESCs (Figure 6C). Epi-iPS cells were transcriptionally indistinguishable from 129 ICM-like ES cells and did not cluster with the EpiSCs (Figure 5H and S9). High contribution chimeras were derived from Epi-iPS cells, verifying that ICM-like pluripotency had been reestablished (Figure 6D). A recent report (Guo et al. 2009) showed that Klf4 over-expression as well as growth in PD/CH was required to convert EpiSCs into iPS cells. In contrast, we found that such conversion can be accomplished either by over expression of the Klf4 or the c-Myc transcription factor, or by supplementing the medium with KP or KP/CH or PD/CH, consistent with the ability of these small molecules to replace Klf4 or c-Myc during iPS cell generation (Figure 6A–D, S2, S10–11). It should be emphasized that only a small fraction of Klf4 or c-Myc transduced EpiSC cells (up to 2%) can be reprogrammed back to ICM-like pluripotency (Figure 6E and (Guo et al., 2009)). However, the efficiency of converting EpiSCs into iPS cells was within the same range as reprogramming efficiencies observed of hematopoietic cells at various differentiation stages (Figure 6E). These findings indicate that expression of OSKM transcription factors can induce ICM-like pluripotency in different somatic cells and that the efficiency of reprogramming is not dictated by the differentiation state of the donor cell but rather depends on additional parameters such as cytokine stimulation, cell cycle state, enhancement by additional reprogramming factors (e.g. C/EBPα for mature B cells reprogramming efficiency) (Figure 6E and (Hanna et al., 2008)).
Figure 6. Resetting the identity of pluripotent states.
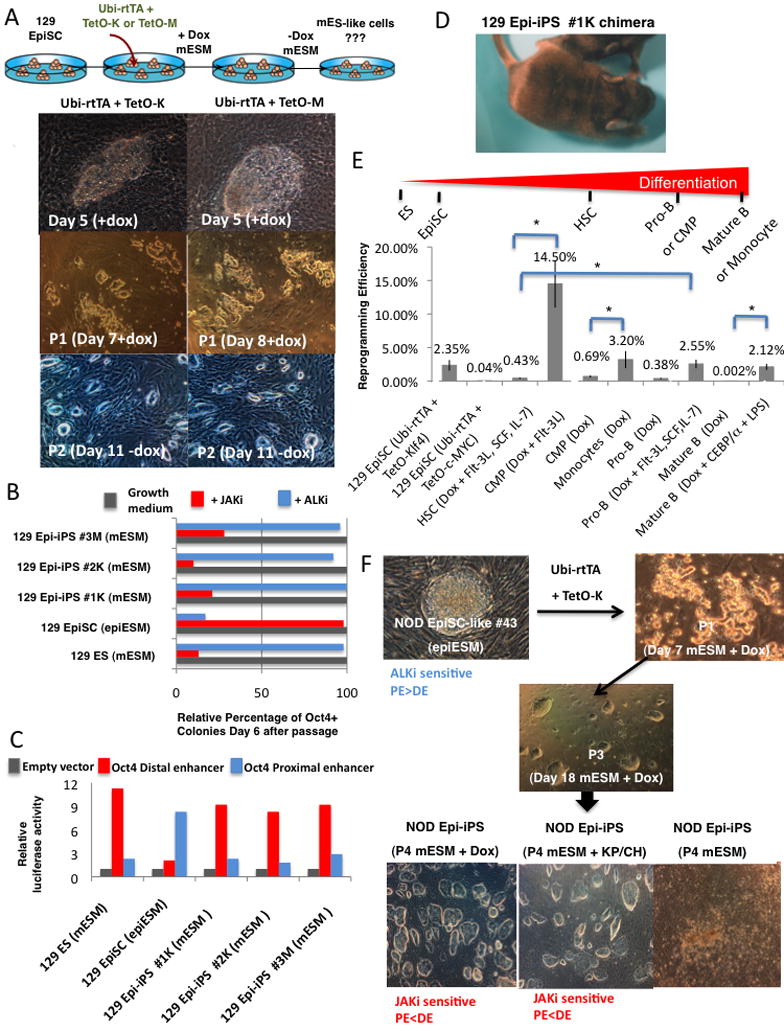
(A) 129 EpiSC cells derived from the epiblast of day E5.5 mouse embryos were reprogrammed into ICM-like cells after infection with TetO-Klf4 or c-Myc lentiviruses and culture in the mESM + Dox. Images on the bottom show representative colony formation observed 4–6 days after infection, which were passaged on MEF feeders. The cell lines were termed Epi-iPS. (B–C) Evaluation of Oct4 enhancer activity and sensitivity to JAKi and ALKi on the converted cell lines was performed as indicated in (Figure 4E–F). (D) Chimeric mice generated from the indicated Epi-iPS cells lines as evident by the agouti coat color. (E) Efficiencies of iPS derivation from EpiSC and somatic cells: 129 EpiSCs were infected with Teto-Klf4-2A-mOrange or Teto-cMyc-2A-mOrange lentiviruses and after 3 days of Dox inductions mOrange+ infected cells were single cell sorted in 96 well plates. Different hematopoietic cells from transgenic reprogrammable mice carrying the Dox inducible OSKM transgenes were single cell sorted in 96 well plates. HSC- hematopoietic stem cell enriched fraction, CMP- common myeloid progenitors. Efficiency for Nanog+ cells stable in mESM was determined at day 25 for all samples. Average results from 2 independent experiments are shown. *P value < 0.05 for the outlined efficiency comparisons. (F) Converting NOD EpiSC-like ES cells into transgene dependent ICM-like cells. EpiSC like cells were generated from NOD-ES line #43 after withdrawal of KP/CH and growing the cells in epiESM. As in (A) the line was infected with TetO-K and within 2 passages in mESM in the presence of Dox. Stable lines termed Epi-iPS were derived. The lower pictures indicate that the cell line required Dox or the presence of KP/CH to remain stable in culture. Factors and growth conditions utilized to facilitate the identity conversion at each step are indicated. The pluripotency state was evaluated and defined based on Oct4 enhancer activity and ALKi or JAKi sensitivity (indicated in blue or red).
Finally, we tested whether similar manipulations could convert the identity of NOD EpiSC-like cells to that of the ICM-like pluripotent state. We derived NOD EpiSC-like ES cells from germline competent NOD-ES#43 by withdrawing KP/CH and growing the cells in epiESM for over 8 passages (Figure 6F). Subsequently, the cells were infected with TetO- Klf4 and grown in mESM and Dox. This treatment readily converted the EpiSC-like into ICM-like cells. However, the ICM-like pluripotent state on the NOD background remained stable only in the presence of Dox inducing the Klf4 transgene or, alternatively, by supplementing mESM with KP/CH (Figure 6F). Identical results were obtained when KP/CH or PD/CH instead of Klf4 and c-Myc transgenes were used to reprogram NOD and 129 EpiSC cells to ICM like pluripotency (Figure S11). In summary, expression of Klf4 or c-Myc converts the EpiSC-like state to ICM-like pluripotency. However, unlike 129 Epi-iPS cell lines, continuous presence of the same exogenous factors is required to stabilize the NOD Epi-iPS cells and to prevent reversion into the EpiSC-like state.
Discussion
Stem cells characterized by different states of pluripotency and developmental potential have been derived under defined growth conditions. For example, growth of explanted mouse epiblasts in conditions containing bFGF and Activin, that are routinely used to isolate human ES cells, generates EpiSC cells that have a restricted ability to contribute to chimeric mice (Brons et al., 2007; Tesar et al., 2007; Thomson et al., 1998). In contrast, “mouse ES conditions” stabilize a pluripotent ES-like state with the potential to generate high-contribution chimeras in “permissive” mouse strains such as 129 (Ying et al., 2003). We found that identical culture conditions failed to induce ICM-like ES cells in the “non-permissive” NOD derived ICM explants. Stable transduction of the NOD ICM explants with c-Myc or Klf4 or addition of small molecules to the medium that replace the action of these factors during iPS derivation was sufficient to generate NOD ES cells that were indistinguishable from 129 ICM derived ES cells. The same stimuli were required to stabilize NOD fibroblast derived iPS cells, supporting the notion that in vitro stability of pluripotent state is dependent on genetic background rather than on the method used to derive the stem cells. However, the pluripotent state of NOD-ES cells was unstable despite the presence of Lif and could be maintained only through continuous expression of Klf4 or c-Myc or in the presence of inhibitors. Silencing of the transcription factors or removal of the inhibitors readily converted the cells to the EpiSC-like state. These cells resembled previously defined EpiSCs by multiple criteria including cellular morphology, signaling requirements and gene expression profiles (Tesar et al., 2007). We hypothesize that the NOD genome lacks or carries genetic determinants that cause instability of the ICM–like ES cell state in vitro unless supported by exogenous factors. It is also important to emphasize that our findings relate to differences in stem cells of different genetic background grown in culture rather than reflecting in vivo differences, as NOD embryos develop normally. Our defined conditions, either with inhibitors or over-expressing transcription factors, enable maintaining fully pluripotent NOD stem cells in vitro, and upon injection of the cells into host blastocysts, the in vivo environment probably substitutes for the requirement of exogenous factors and allows the cells to contribute to chimera formation.
Metastable states of pluripotency
The term “metastability” has been previously used to describe transient changes within ICM-like ES cell populations resulting from oscillations in Nanog or Stella gene expression (Chambers et al., 2007; Hayashi et al., 2008). Here, we apply this term to describe the interconversion between two distinct pluripotent states in NOD and 129 mouse strains. Our results suggest that the ICM and EpiSC pluripotent states may be in a “metastable” equilibrium dictated by the genetic background where exogenous factors can convert one state into another. Thus, one may consider the two states of pluripotency, the ICM/ES cell-like state and the epiblast/EpiSC cell-like state, as two different levels of pluripotency (Figure 7). Exogenous factors such as c-Myc and Klf4 in combination with Oct4 and Sox2 can induce the ICM-ES like state from somatic cells. However, the stability of the ES cell state is determined by the genetic background: while ICM-ES cells or iPS cells derived from a “permissive” genetic background such as 129 or C57BL/6 are stable once established in the presence of Lif, the ES cell like state of iPS cells or of ICM derived pluripotent cells of the ”non-permissive” NOD background remains unstable with the maintenance of the pluripotent state depending on the continuous expression of the exogenous factors in addition to Lif/Stat3 signaling. Inactivation of the transcription factors or removal of the inhibitors causes the ES like NOD cells to assume an EpiSC-like state, characterized by reduced pluripotency. Inter-conversion between these states can be controlled by the absence or presence of the same factors (Figure 7).
Figure 7. Pluripotent states are “metastable”.

Model summarizing the requirements for in vitro stabilization of different pluripotent states in 129 (left half) and NOD mouse strains (right half). “Metastability” pertains to describing a system with two or more equilibrium states (indicated by the stippled grey horizontal lines) that can interconvert by defined signals. The ICM-like state is characterized by a greater developmental potential than the EpiSCs-like state. The factors required for stabilizing the respective pluripotent states in the different genetic backgrounds are indicated in green on the plateau lines for each state. The ICM-like pluripotent state in permissive 129 cells, whether achieved by direct in vitro reprogramming or by ICM explantation is stabilized by Lif/Stat3 signaling, while the bFGF/Activin/Nodal signaling stabilizes the EpiSC-like state. Defined transcription factor or small molecules convert the EpiSC-like cells to the ICM-like ES cell state (highlighted in orange on the left). The 129 and NOD EpiSC-states are indistinguishable in their stability and growth condition requirements and can be reverted into ICM-like pluripotency by expression of Klf4 or c-Myc or by specific inhibitors added to the medium. However, the ICM-ES like state is unstable on the NOD genetic background and requires continuous expression of exogenous factors in addition to Lif (dashed black and red arrows).
Several lines of evidence support the notion that the conversions between the different pluripotency states are due to cells being inefficiently induced to successfully convert from one state to another, rather than due to selection for rare pre-existing cells constantly present in heterogeneous stem cell populations. First, evidence for direct reprogramming of EpiSCs into iPS cells is supported by the observations that EpiSC cells do not convert spontaneously into ES like cells and that all derived Epi-iPS cell lines carried integrated viral transgenes (Figure S11 and (Guo et al., 2009)). Second, the EpiSC to ES cell conversion requires multiple passages in defined media and continuous transgene induction, which is similar to generating iPS cells from somatic cells (Fig. 6, (Jaenisch and Young, 2008)). Third, it is unlikely that NOD ES cultures carry already rare EpiSC-like cells since the NOD iPS or ES lines were passaged routinely by trypsinization, which does not allow propagation of the EpiSC cells. Finally, the NOD EpiSC-like iPS cell line carried an identical Sox2 integration as its parental Dox dependent NOD iPS line indicating a clonal relation (Figure 5B). An important question remains why only a small fraction of the NOD ES cells convert into an EpiSC state. One possibility is that after removal of the exogenous stimuli, the EpiSC state becomes one of several epigenetic states that can be acquired by the NOD ES cells upon differentiation.
Pluripotency in NOD strain as a paradigm for other “non-permissive” species?
It has been established that human and rhesus macaque ES cells resemble the EpiSC rather than the ICM pluripotent state of mouse cells suggesting that ICM-like pluripotent cells might have not been isolated from several species (Brons et al., 2007; Byrne et al., 2007; Lovell-Badge, 2007; Tesar et al., 2007; Thomson et al., 1998). When cultured under standard ES cell growth conditions, the pluripotent state of NOD stem cells isolated from explanted blastocysts or from somatic cells by in vitro reprogramming was the EpiSC-like state. The ICM-ES like state in NOD cells could only be stabilized when exogenous factors such as Klf4 or c-Myc were added. Thus, it is possible that “non-permissive” species such as human, that have yielded only EpiSC-like pluripotent cells, require specific exogenous factors to maintain the ICM-like pluripotent state. Consistent with this notion is that ICM-like iPS cells could be generated from rat fibroblasts under identical culture conditions to those used for the isolation of NOD iPS cells. This raises the question whether the conditions that were successful for the isolation of NOD ICM-like ES cells could be used to maintain ICM-like ES from other “non-permissive” species. It will also be of great interest to define the genetic determinants that affect the in vitro stability of pluripotent states from different genetic backgrounds. Moreover, uncovering how Klf4, c-Myc, Wnt and MAPK pathways might converge and cross-talk in the reprogramming process, and whether they play a similar role in fibroblasts to iPS and EpiSC to iPS conversion, is a fundamental question relevant to understanding the mechanisms of reprogramming (Markoulaki et al., 2009; Marson et al., 2008; Silva et al., 2008a; Ying et al., 2008).
Generation of germline competent NOD ES cells
The inability to derive germline competent embryonic stem cells on the NOD background has posed limitations in generating genetically engineered NOD mice (Bach and Mathis, 1997). The NOD strain has been instrumental for the studying of disease progression and pathology of Insulin Dependent Diabetes Mellitus (IDDM). NOD mice spontaneously develop a form of diabetes that closely resembles human IDDM as a result of an autoimmune process directed against the pancreatic beta cells. IDDM is a polygenic disease with multiple parameters influencing susceptibility disease progression. Until now, the only available approach to generate an NOD knock-out involves crossing of NOD mice with a non-NOD strain carrying the desired allele and subsequently back-crossing to NOD mice for at least 15 generations to ensure re-establishment of the original NOD inbred genetic background harboring all IDDM susceptibility loci. Thus, the generation of germline competent NOD ES cells circumvents an obstacle posed on modeling IDDM in mice, and would establish an efficient platform for achieving direct gene targeting on the NOD background.
Experimental Procedures
Cell culture and viral infections
mESM conditions refer to culturing the cells on irradiated MEFs in DME containing 15% FCS, leukemia inhibiting factor (Lif), 1 mM glutamine (Invitrogen), 1% nonessential amino acids (Invitrogen), 0.1 mM -mercaptoethanol (Sigma)). These cells (ICM-like) were passaged every third day as a singe cell suspension using 0.25% trypsin/EDTA. EpiSC and EpiSC-like cells were maintained in epiESM conditions which include maintaining the cells on mitomycin C inactivated mouse embryonic fibroblast (MEF) feeder layers in epiESM [DMEM/F12 (Invitrogen) supplemented with 15 % FBS (Hyclone), 5% Knockout replacement serum (KSR – Invitrogen), 1 mM glutamine, 1% nonessential amino acids and 4 ng/ml human FGF2 (bFGF) (R&D systems)]. Cultures were passaged every 5 to 7 days either manually or enzymatically with collagenase type IV (Invitrogen; 1.5 mg/ml). The density of feeder cells was crucial to maintaining the EpiSC- and EpiSC-like cells in an undifferentiated state (density of 4 * 10^4 cells per cm2). Lentiviral preparation and infection with Doxycycline inducible lentiviruses encoding Oct4, Klf4, c-Myc and Sox2 cDNA driven by the TetO/CMV promoter or constitutive lentiviruses driven by ubiquitin promoter, were done as previously described (Hanna et al., 2008; Hanna et al., 2007).
Small molecule compounds
ALK inhibitor (ALKi - SB431542 Stemgent technologies) (20μM final concentration); JAK inhibitor I (Calbiochem 420099 - 0.6μM). Kenpaullone (KP) (Sigma –5μM), PD184352 (PD) (Pfizer - 0.8μM); CHIR99021 (Stemgent - 3μM).
Animals and ES derivation
NOD/ShiLtJ were obtained from the Jackson laboratory and bred in specific-pathogen-free animal facility. Control MEFs were made from 129SvJae mice. Non-NOD ES lines derived used in this study as controls were derived from matings between 129SvJae or B6D2F1 mice. To derive ES lines, ICM explants were derived from day 3.5 blastocysts following procedures previously described (Markoulaki et al., 2008). Where applicable, ES derivation medium was also supplemented with the indicated compounds (Table S2). Dissociation of the outgrowths by treatment with Trypsin was performed on day 5 after plating and the cells were further cultured in mESC derivation medium until colonies appeared (typically after 5–7 days). From then on, established ES lines were cultured in mESM condition supplemented with the indicated compounds. NOD EpiSC-like ES lines were derived by similarly plating blastocysts in EpiSC-derivation medium (For 100 ml, we added 15 ml FBS (Hyclone), 5 ml KSR, 5 μl Lif (1× 10^7 U ESGRO/ml; Chemicon), 100 μl Mek1 inhibitor (PD98059; Cell Signaling Technology), hFGF2 (12g/ml), 1 ml non-essential amino acids, 1 ml glutamine solution, and 1ml pen/strep solution into DMEM/F12). ICM outgrowths were manually passaged after 5–7 days and were stably maintained in epiESM conditions. 129 EpiSC cell line used was previously described (Tesar et al., 2007), and were obtained from day E5.5 129SvEv (Taconic) embryos and propagated in epiESM conditions.
Luciferase reporter assay
Constructs encoding the two previously characterized Oct4 enhancer (the Oct3/4 DE- and Oct3/4 PE- SV40-Luc constructs) cloned into the pGL3-Promoter Vector (Promega) were used to determine regulation pattern of Oct4 expression (Tesar et al., 2007). Constructs were transfected into 0.25–0.5*10^6 cells using the Amaxa Nucleofection kit or Biorad along with the pRL-TK vector for normalization. Assays were performed 24–48 hours later using the Dual-Glo Luciferase Assay System (Promega). The basal activity of the empty luciferase vector was set as 1.0.
Gene array expression analysis and RT-PCR analysis
RNA was isolated from MEF depleted mouse ES cells or from mechanically separated EpiSC and EpiSC-like cells lines using the RNeasy Mini Kit (Qiagen). Sample processing is described in supplementary Methods section. Heatmap and trees were visualized by Java Treeview. Microarray data are available at the NCBI Gene Expression Omnibus database (GSE15603). For RT-PCR analysis, 1 microgram of DNase I-treated RNA was reverse transcribed using a First Strand Synthesis kit (Invitrogen) and ultimately resuspended in 100ul of water. Quantitative PCR analysis was performed in triplicate using 1/50 of the reverse transcription reaction in an ABI Prism 7000 (Applied Biosystems).
Supplementary Material
Acknowledgments
We would like to thank D. Mathis, C. Benoist, M. Wilson and members of the Jaenisch lab for discussions. We thank Peter Andrews for discussing the concept of metastability of pluripotent states. We thank R. Mckay for EpiSC cells. We thank T. Di Cesare for graphics. R.J. is supported by grants from the NIH: RO1-HDO45022, R37-CA084198, RO1-CA087869. J.H. is a Helen Hay Whitney fellow, J.S is a HFSP fellow.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bach JF, Mathis D. The NOD mouse. Res Immunol. 1997;148:285–286. doi: 10.1016/s0923-2494(97)87235-5. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- Buehr M, Meek S, Blair K, Yang J, Ure J, Silva J, McLay R, Hall J, Ying QL, Smith A. Capture of authentic embryonic stem cells from rat blastocysts. Cell. 2008;135:1287–1298. doi: 10.1016/j.cell.2008.12.007. [DOI] [PubMed] [Google Scholar]
- Byrne JA, Pedersen DA, Clepper LL, Nelson M, Sanger WG, Gokhale S, Wolf DP, Mitalipov SM. Producing primate embryonic stem cells by somatic cell nuclear transfer. Nature. 2007;450:497–502. doi: 10.1038/nature06357. [DOI] [PubMed] [Google Scholar]
- Chambers I, Silva J, Colby D, Nichols J, Nijmeijer B, Robertson M, Vrana J, Jones K, Grotewold L, Smith A. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- Guo G, Yang J, Nichols J, Hall JS, Eyres I, Mansfield W, Smith A. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development. 2009 doi: 10.1242/dev.030957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Markoulaki S, Schorderet P, Carey BW, Beard C, Wernig M, Creyghton MP, Steine EJ, Cassady JP, Foreman R, et al. Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell. 2008;133:250–264. doi: 10.1016/j.cell.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Wernig M, Markoulaki S, Sun CW, Meissner A, Cassady JP, Beard C, Brambrink T, Wu LC, Townes TM, et al. Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science. 2007;318:1920–1923. doi: 10.1126/science.1152092. [DOI] [PubMed] [Google Scholar]
- Hatano SY, Tada M, Kimura H, Yamaguchi S, Kono T, Nakano T, Suemori H, Nakatsuji N, Tada T. Pluripotential competence of cells associated with Nanog activity. Mech Dev. 2005;122:67–79. doi: 10.1016/j.mod.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Hayashi K, Lopes SM, Tang F, Surani MA. Dynamic equilibrium and heterogeneity of mouse pluripotent stem cells with distinct functional and epigenetic states. Cell Stem Cell. 2008;3:391–401. doi: 10.1016/j.stem.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Osafune K, Maehr R, Guo W, Eijkelenboom A, Chen S, Muhlestein W, Melton DA. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat Biotechnol. 2008;26:1269–1275. doi: 10.1038/nbt.1502. [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132:567–582. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahner D, Stuhlmann H, Stewart CL, Harbers K, Lohler J, Simon I, Jaenisch R. De novo methylation and expression of retroviral genomes during mouse embryogenesis. Nature. 1982;298:623–628. doi: 10.1038/298623a0. [DOI] [PubMed] [Google Scholar]
- Li P, Tong C, Mehrian-Shai R, Jia L, Wu N, Yan Y, Maxson RE, Schulze EN, Song H, Hsieh CL, et al. Germline competent embryonic stem cells derived from rat blastocysts. Cell. 2008;135:1299–1310. doi: 10.1016/j.cell.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Wei W, Zhu S, Zhu J, Shi Y, Lin T, Hao E, Hayek A, Deng H, Ding S. Generation of rat and human induced pluripotent stem cells by combining genetic reprogramming and chemical inhibitors. Cell Stem Cell. 2009;4:16–19. doi: 10.1016/j.stem.2008.11.014. [DOI] [PubMed] [Google Scholar]
- Liao J, Cui C, Chen S, Ren J, Chen J, Gao Y, Li H, Jia N, Cheng L, Xiao H, et al. Generation of induced pluripotent stem cell lines from adult rat cells. Cell Stem Cell. 2009;4:11–15. doi: 10.1016/j.stem.2008.11.013. [DOI] [PubMed] [Google Scholar]
- Lovell-Badge R. Many ways to pluripotency. Nat Biotechnol. 2007;25:1114–1116. doi: 10.1038/nbt1007-1114. [DOI] [PubMed] [Google Scholar]
- Lyssiotis CA, Foreman R, Staerk J, Garcia M, Mathur D, Maroulaki S, Hanna J, et al. Reprogramming of murine fibroblasts to iPS cells: chemical complementation of Klf4. 2009 doi: 10.1073/pnas.0903860106. (Submitted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markoulaki S, Hanna J, Beard C, Carey BW, Cheng AW, Lengner CJ, Dausman JA, Fu D, Gao Q, Wu S, et al. Transgenic mice with defined combinations of drug-inducible reprogramming factors. Nat Biotechnol. 2009;27:169–171. doi: 10.1038/nbt.1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markoulaki S, Meissner A, Jaenisch R. Somatic cell nuclear transfer and derivation of embryonic stem cells in the mouse. Methods. 2008;45:101–114. doi: 10.1016/j.ymeth.2008.04.002. [DOI] [PubMed] [Google Scholar]
- Marson A, Foreman R, Chevalier B, Bilodeau S, Kahn M, Young RA, Jaenisch R. Wnt signaling promotes reprogramming of somatic cells to pluripotency. Cell Stem Cell. 2008;3:132–135. doi: 10.1016/j.stem.2008.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- Rossant J. Stem cells and early lineage development. Cell. 2008;132:527–531. doi: 10.1016/j.cell.2008.01.039. [DOI] [PubMed] [Google Scholar]
- Shi Y, Desponts C, Do JT, Hahm HS, Scholer HR, Ding S. Induction of pluripotent stem cells from mouse embryonic fibroblasts by Oct4 and Klf4 with small-molecule compounds. Cell Stem Cell. 2008;3:568–574. doi: 10.1016/j.stem.2008.10.004. [DOI] [PubMed] [Google Scholar]
- Silva J, Barrandon O, Nichols J, Kawaguchi J, Theunissen TW, Smith A. Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol. 2008a;6:e253. doi: 10.1371/journal.pbio.0060253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva SS, Rowntree RK, Mekhoubad S, Lee JT. X-chromosome inactivation and epigenetic fluidity in human embryonic stem cells. Proc Natl Acad Sci U S A. 2008b;105:4820–4825. doi: 10.1073/pnas.0712136105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RD. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Wakayama T, Perry AC, Zuccotti M, Johnson KR, Yanagimachi R. Full-term development of mice from enucleated oocytes injected with cumulus cell nuclei. Nature. 1998;394:369–374. doi: 10.1038/28615. [DOI] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- Xu RH, Peck RM, Li DS, Feng X, Ludwig T, Thomson JA. Basic FGF and suppression of BMP signaling sustain undifferentiated proliferation of human ES cells. Nat Methods. 2005;2:185–190. doi: 10.1038/nmeth744. [DOI] [PubMed] [Google Scholar]
- Ying QL, Nichols J, Chambers I, Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115:281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


