Abstract
Pseudomonas aeruginosa is an opportunistic human pathogen that causes infections in a variety of animal and plant hosts. Caenorhabditis elegans is a simple model with which one can identify bacterial virulence genes. Previous studies with C. elegans have shown that depending on the growth medium, P. aeruginosa provokes different pathologies: slow or fast killing, lethal paralysis and red death. In this study, we developed a high-throughput semi-automated liquid-based assay such that an entire genome can readily be scanned for virulence genes in a short time period. We screened a 2,200-member STM mutant library generated in a cystic fibrosis airway P. aeruginosa isolate, TBCF10839. Twelve mutants were isolated each showing at least 70% attenuation in C. elegans killing. The selected mutants had insertions in regulatory genes, such as a histidine kinase sensor of two-component systems and a member of the AraC family, or in genes involved in adherence or chemotaxis. One mutant had an insertion in a cheB gene homologue, encoding a methylesterase involved in chemotaxis (CheB2). The cheB2 mutant was tested in a murine lung infection model and found to have a highly attenuated virulence. The cheB2 gene is part of the chemotactic gene cluster II, which was shown to be required for an optimal mobility in vitro. In P. aeruginosa, the main player in chemotaxis and mobility is the chemotactic gene cluster I, including cheB1. We show that, in contrast to the cheB2 mutant, a cheB1 mutant is not attenuated for virulence in C. elegans whereas in vitro motility and chemotaxis are severely impaired. We conclude that the virulence defect of the cheB2 mutant is not linked with a global motility defect but that instead the cheB2 gene is involved in a specific chemotactic response, which takes place during infection and is required for P. aeruginosa pathogenicity.
Author Summary
The increase in hospital acquired and multi-drug resistant bacterial infections calls for an urgent development of new antimicrobials. As such, the identification and characterization of novel molecular targets involved in bacterial virulence has become a common goal for researchers. The use of non-mammalian hosts, such as the nematode Caenorhabditis elegans, is useful to accelerate this process. In our study, we developed a high-throughput screening method, which further facilitates the use of C. elegans, and allows the rapid screening of a large collection of bacterial mutants at the genomic scale. We have used Pseudomonas aeruginosa, a potent opportunistic pathogen, to perform this study. The screening of more than 2,000 mutant strains allowed the characterization of a mutant affected in the cheB2 gene. Importantly, this mutant was shown to be impaired in a mouse model of infection, supporting that our new screen is a good model to identify virulence genes relevant for infection in mammals. The cheB2 gene encodes a component of a chemotaxis pathway, which is likely involved in the perception of stimuli during the infection process, and allows an appropriate adaptive response for a successful infection. Our method could be applied to other bacterial pathogens and will help researchers discover candidate genes leading to the design of novel antimicrobials.
Introduction
The ubiquitous Gram-negative bacterium Pseudomonas aeruginosa is an opportunistic pathogen able to infect a broad range of animals and plants hosts including humans. In the course of infection, P. aeruginosa adapts to changing environmental conditions and coordinates the production of molecular determinants involved in host colonization and virulence [1]. Among these are pili and flagella, which are required for attachment and spreading on surfaces [2],[3]. Also necessary are protein secretion systems [4] and toxins required for cytotoxicity and survival within the host. One powerful approach to dissect the interaction between pathogen and host is the use of simple infection models. It has been demonstrated that the nematode Caenorhabditis elegans can be an effective model for studying virulence mechanisms used by a variety of bacterial pathogens [5],[6]. P. aeruginosa is capable of killing C. elegans in several distinct ways. When the P. aeruginosa strain PA14 is cultured on a high-osmolarity peptone-glucose-sorbitol medium (PGS), worms succumb to intoxication termed “fast killing”, as the exposed worms die within hours [7]. Nematodes exposed to PA14 grown on nematode growth media (NGM), succumb to “slow killing”. In this case the bacteria colonize the gut and the infected worms die over a number of days rather than hours [8]. The P. aeruginosa isolate PAO1 cultured on Brain Heart Infusion (BHI) agar induces worm paralysis and death within hours [9]. Finally, C. elegans death, called red death, is observed in response to PAO1 grown on phosphate-depleted medium in conjunction with physiological stress on the nematodes [10].
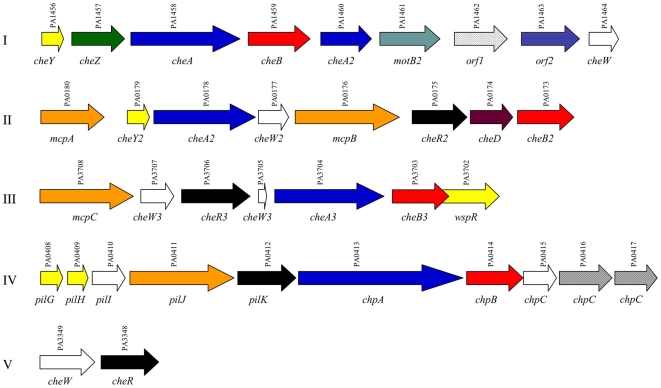
It is known that the P. aeruginosa isolates PA14 and PAO1 show genomic diversity. Strains cultured in vitro for years are known to undergo changes in gene expression. Individual genes and even entire genomic islands have been lost from laboratory isolates when compared to pathogenic strains [11],[12],[13]. In this study, we used the P. aeruginosa strain TBCF10839 (TB), which was isolated from a Cystic Fibrosis (CF) patient [14]. The TB strain belongs to an abundant clonal complex in the P. aeruginosa population [15]. It has a high resistance to detergents [16] and reactive oxygen intermediates [17] and is able to grow within polymorphonuclear leukocytes [18]. We developed a high-throughput killing assay using C. elegans as a host, which will be appropriate for a systematic screen of mutant libraries of any P. aeruginosa isolate of interest. Increasing throughput in an assay such that an entire genome can readily be scanned in a short time period is an important advance. Our protocol is based on a standard killing assay and makes use of a Biosorter to distribute nematodes into the wells of microtitre plates in a fully automated manner. We screened a TB STM (signature tagged mutagenesis) mutant library of 2,200 non-redundant clones [19],[20] and selected a small group of mutants, significantly attenuated for virulence in C. elegans. By testing these mutants in additional phenotypic assays, including adherence to epithelial cells and virulence in a mouse model, we identified a gene, cheB2, necessary for in vivo P. aeruginosa virulence. The cheB2 gene belongs to the P. aeruginosa chemotaxis che2 gene cluster (cluster II, PA0180-PA0173) (Figure 1) [21]. P. aeruginosa has multiple copies of chemotaxis genes arranged in five clusters (Figure 1) [22]. The chemotaxis gene cluster I (PA1456-PA1464) and cluster V (PA3349-PA3348) have previously been shown to be essential for chemotaxis and flagellar mobility in P. aeruginosa [23],[24]. The wsp genes contained in cluster III (PA3708-PA3702) have been proposed to contribute to bacterial biofilm formation [25], whereas the cluster IV (Pil-Chp system, PA0408-PA0417) has been shown to be involved in twitching motility [26],[27]. The che2 genes from cluster II, and more particularly the cheB2 gene, were initially identified as required for an optimal chemotactic response in P. aeruginosa. The CheB proteins are essential components of the chemotactic response and are responsible for demethylation of glutamate residues in methyl-accepting chemotaxis proteins (MCPs) and also deamidation of glutamine residues to form methyl-accepting glutamates [28]. MCPs sense the chemical stimuli that initiate the chemotactic activity [29]. In Escherichia coli, the loss of CheB leads decreased receptor sensitivity, due to an inability to reset the MCP's [30]. This modification leaves the chemotactic pathway in active states causing clockwise rotation of the flagella and continuous tumbling instead of runs and tumbles. It has been reported previously in Salmonella typhimurium that a loss of proper chemotaxis control through a cheB mutation, leads to a ‘tumbly’ swimming phenotype and a strong reduction in the isolates ability to invade human epithelial HEP2 cells as well as a reduction in infectivity in a mouse ligated-loop model [31].
Figure 1. Chemotaxis Clusters in P. aeruginosa PAO1.
Cluster numbers are indicated to the left. Gene names are indicated below each representative arrow bar and PA numbers (http://pseudomonas.com) are indicated above the arrow bars. Homologues are indicated by shared color.
In our study we further compare the phenotypes of cheB2 and cheB1 mutants and propose that the cheB2 gene has a specific role during infection, which is essential for pathogenesis.
Results
High Throughput Assay for Nematode Killing by P. aeruginosa
It was reported earlier that relatively high-throughput screening procedures performed on plates could be used to identify virulence factors in various pathogens such as Staphylococcus aureus [32], P. aeruginosa [8] or Serratia marcescens [33]. Here, we developed a high throughput screening method, in which C. elegans killing is assessed in a liquid assay and which allows a quick selection of attenuated candidates within large transposon libraries of P. aeruginosa. We prepared a bacterial culture medium that both allowed the worms to thrive feeding on the non-pathogenic E. coli OP50, and bacterial isolates being tested to have normal (see Materials and Methods). We filled 96 well plates with the medium and inoculated the bacterial strains to be tested. The virulence of each isolate was assessed in a second microtitre plate, into which a specific population of nematodes had been deposited. Evaluation of virulence was based upon the number of live worms recovered after 24 h exposure to bacteria. Under our assay conditions, more than 90% of the worms grown on E. coli were still alive after 24 h. In contrast, more than 50% of the worms were killed within 24 h when cultured with any one of three isolates of P. aeruginosa, PA14, PAO1 and TB (data not shown). In order to confirm that this assay would allow the recovery of P. aeruginosa mutants with an attenuated virulence, we tested previously characterized mutants. We used a gacA mutant from the PA14 transposon library (http://ausubellab.mgh.harvard.edu/cgi-bin/pa14/home.cgi) [34], which is affected in a gene encoding a response regulator involved in virulence [8]. C. elegans infection using this mutant in the liquid assay resulted in only 40% killing at 24 h, whereas 95% killing was observed with the parental PA14 strain (data not shown).
In order to screen a large number of bacterial isolates more rapidly, the assay was automated using the COPAS (Complex Object Parametric Analyzer and Sorter) Biosort system (Union Biometrica) (http://www.unionbio.com/). The sorter is able to dispatch systematically a fixed number of worms of a precise developmental stage, into each well of a microtitre plate.
Screening the TB Transposon Library for Mutants Attenuated in Virulence
A signature tagged mutagenesis (STM) library was previously generated in the virulent P. aeruginosa CF isolate, TB, using the plasposon vector pTnModOGm [20],[35]. Auxotrophic mutants were removed from the library by pre-screening post-conjugation TB isolates onto minimal media and selecting only clones, which grew normally. This pre-screening eliminated clones that might have exhibited attenuated virulence based upon reduced growth or viability, rather than loss of a specific virulence function. After this initial segregation, 2,200 clones were picked into a 96-well plate and tested with the C. elegans high throughput-killing assay. Each mutant was tested at least four times using this assay. The mutants selected were segregated into three groups based upon their level of virulence attenuation as compared to the TB parental strain, which killed 85% of worms under the test conditions. In group 1, we included 187 mutants (9% of total library) which allowed more than 50% worm survival, group 2 between 30% and 50% (536 mutants, 25%), and group 3 below 30% (1450 mutants, 65%), approaching the same level of killing as the parental isolate. Within group 1, the 12 most highly attenuated isolates, classified as group 1A (0.4% of the total library), were further studied. When cultured in the presence of these mutants more than 70% of worms survived, representing a near 5-fold reduction in virulence compared to TB.
Characterization of Selected Attenuated Mutants
Genomic DNA was recovered from the 12 group 1A isolates. The transposon insertion site was determined through direct genomic DNA sequencing, using primers located upstream from each transposon inverted repeat (see Materials and Methods). The sequences obtained were compared to the PAO1 genome sequence database (http://www.pseudomonas.com) [22] followed by comparison with the general sequence database. The genes identified are shown in Table 1. Two potential regulatory genes were found. The first, PA2588, encodes an AraC-type regulatory protein, with 48% similarity to the PAO1 PA0831 gene product, OruR [36]. The second matched PA4380, which encodes a protein with 76% similarity to Pseudomonas fluorescens ColS, a histidine kinase sensor from a two-component regulatory system. In P. fluorescens ColS has been implicated in root colonization while in Pseudomonas putida it has been reported to be involved in regulating TN4652 transposition and heavy metal resistance [37],[38],[39],[40]. Another gene was identified as PA4554, which encodes the type 4 fimbrial adhesin PilY1 [41]. We recovered two additional mutants with transposon insertion into genes which have functions related to motility. First, PA4953 encodes a protein with similarity to the E. coli MotB, one of a pair of proteins, which contributes to flagellar rotation [23],[42],[43]. Secondly, we found an insertion in PA0173 (cheB2), one of four cheB gene homologues in the P. aeruginosa genome sequence. PA0173 lies within the chemotactic gene cluster II found in the PAO1 genome (Figure 1) [21]. CheB proteins are responsible for removing methyl groups from MCPs [44]. The adaptation of the chemotaxis system in response to changes in attractant binding is dependent on the methylation state of the MCPs. CheB2 has been proposed to be an essential component for an optimal chemotactic response [21]. In the remaining mutants, one contained a transposon insertion in PA5479, which encodes a protein similar to GltP from E. coli, a glutamate-aspartate carrier protein. Another mutant had an insertion in PA2585 encoding the UvrABC endonuclease subunit UvrC, required for the excision of damaged DNA. The uvrC gene is located downstream from the gacA gene encoding a response regulator known to be involved in P. aeruginosa virulence. Since, in addition to the insertion in PA2585, we isolated a mutant with an insertion in a neighbor gene PA2588, it suggests that this cluster of genes might be required for virulence. Another interrupted gene, PA2478, encodes a probable thiol∶disulfide interchange protein of the DsbD family. Finally, 4 interrupted genes, PA0946, PA3080, PA0260 and PA2769, encode proteins of unknown function.
Table 1. Identification and characterization of selected mutants.
| ORF/STM mutant | Function | Gene name | Slow killing | Cell Adherence | Swimming | Swarming |
| PA0173/TB0173s | probable methylesterase for chemotaxis | cheB2 | − | − | − | − |
| PA0260/TB0260s | hypothetical protein | − | + | − | + | |
| PA0946/TB0946s | hypothetical protein | − | + | + | + | |
| PA2478/TB2478s | probable thiol∶disulfide interchange protein | dsbD2 | + | + | + | + |
| PA2585/TB2585s | endonuclease UvrABC subunit C | uvrC | − | + | + | + |
| PA2588/TB2588s | probable transcriptional regulator | + | − | + | − | |
| PA2769/TB2769s | hypothetical protein | + | − | + | + | |
| PA3080/TB3080s | hypothetical protein | − | − | + | − | |
| PA4380/TB4380s | probable histidine kinase sensor | colS | − | − | + | + |
| PA4554/TB4554s | type 4 fimbrial biogenesis protein | pilY1 | − | +/− | + | + |
| PA4953/TB4953s | flagellar motor protein | motB | + | − | − | − |
| PA5479/TB5479s | proton-glutamate symporter | gltP | + | − | − | + |
ORF PA number, function and gene name annotations were obtained using the Pseudomonas genome database version 2 (http://v2.pseudomonas.com/index.jsp).
+ indicates that the strain exhibited a phenotype not significantly different from the wild type TB isolate, whereas − indicates a reduced phenotype. +/− indicates a reduction at the limit of significance.
In order to compare our liquid assay with the established C. elegans slow killing assay, we compared the lethality of the TB strain with the previously tested PA14 strain. We observed that both strains kill worms with similar efficiency (data not shown). Each of the 12 group 1A mutants derived from TB was further tested in the slow killing assay. The parental TB isolate is able to kill 50% of exposed nematodes in the slow killing assay within 3 days. Seven of the twelve group 1A isolates showed significant virulence attenuation (Table 1). For example, three group 1A mutants TB0173s (cheB2, p<0.001), TB4554s (pilY1, p<0.001) and TB4380s (colS, p<0.001) each required at least one additional day (4 days) in order to reach 50% killing (data not shown). Survival curves are considered significantly different from the control when p-values are <0.05 (see Materials and Methods).
Phenotypic Analysis of Selected TB mutants
Several of the clones recovered were mutated in genes putatively associated with adherence and motility, such as cheB2 (PA0173), motB (PA4953) and pilY1 (PA4554). It has been shown that adherence is an essential first step in the process of P. aeruginosa colonization, allowing the bacteria to persist, expand in numbers and develop a biofilm or establish intimate contact with host cells [45],[46]. We wished to determine if our selected mutants showed a reduction in their capacity to adhere to a biological surface such as the human epithelial cell line 16HBE14o-. In addition to TB4953s (motB, p<0.0001) and TB0173s (cheB2, p<0.001), that presented strongly reduced adherence, five other clones were significantly affected in their adherence to cells when compared to the parental TB isolate (Figure S1 and Table 1). These mutants were TB2588s (araC-like, PA2588, p<0.001), TB4380s (colS-like, PA4380, p<0.001), TB3080s (hypothetical, PA3080, p<0.01), TB2769s (hypothetical, PA2769, p<0.05) and TB5479s (gltP-like, PA5479, p<0.05).
Motility is also an important factor for P. aeruginosa colonization and spread. We evaluated each of the twelve attenuated mutants for both swarming and swimming motility (data not shown, Table 1 and Figure 2). The TB0173s (cheB2) (Figure 2 and Table 1) and TB4953s (motB) (Table 1) mutants showed a reduction in swimming and swarming motility, respectively. Furthermore, the mutants TB0260s (hypothetical, PA0260) and TB5479s (gltP-like) had a significant reduction in swimming motility (Table 1) whereas the mutants TB2588s (araC-like) and TB3080s (hypothetical) had a significant reduction in swarming motility (Table 1).
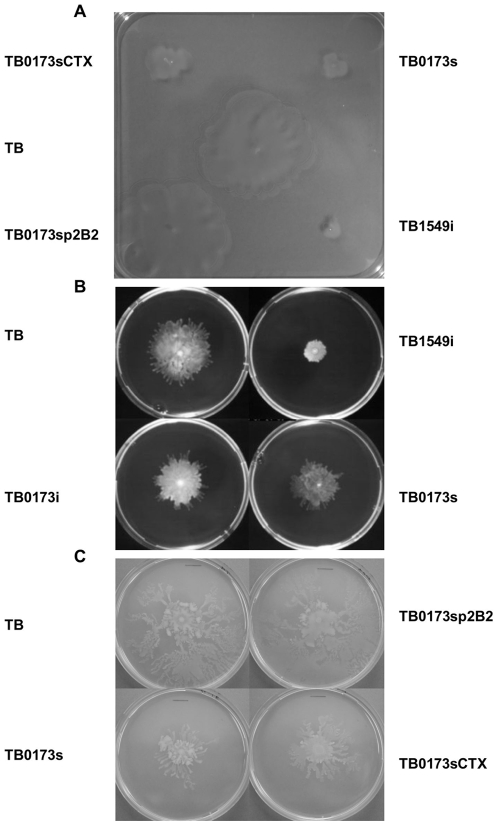
Figure 2. Swimming and Swarming Motility of TB and Isogenic Mutants.
(A) Swimming motility. The cheB2 mutant (TB0173s) shows reduced motility as compared to the parental strain (TB) whereas complementation with CTXp2B2 (TB0173sp2B2) fully restores motility. The non-complemented cheB2 mutant strain carrying CTX1 (TB0173sCTX) remains poorly motile. The cheB1 mutant (TB1549i) displays greater motility defect as compared to the cheB2 mutant. Colony area representing the motility zone was calculated using Macnification software (Orbicule BVBA Heverlee, Belgium). TB; 15,92 cm2, TB0173s (cheB2 mutant); 2,58 cm2, TB0173sp2B2 (complemented cheB2 mutant); 14,25 cm2, TB0173CTX (cheB2 mutant); 1,0 cm2, TB1549i (cheB1 mutant); 0,48 cm2. (B) Swarming motility. The cheB2 mutants (TB0173s and TB0173i) display slightly reduced motility as compared to the parental strain (TB), whereas the cheB1 mutant (TB1549i) shows a drastic swarming motility defect. (C) Swarming motility of the cheB2 mutant (TB0173s) is restored to wild-type strain (TB) level upon introduction of the CTXp2B2 (TB0173sp2B2), whereas the cheB2 mutant strain carrying an empty CTX1 (TB0173sCTX) does not display a similar motility restoration.
Characterization of the cheB2 Mutant
We chose to further characterize the TB0173s mutant (cheB2), as this isolate was affected in all phenotypes tested and thus exhibited a general motility and virulence defect (Table 1). To ensure that the cheB2 gene in the TB strain is located in a cluster similar to the che2 cluster described for the PAO1 or PA14 strains, we further analyzed the corresponding TB chromosomal region. A series of 8 PCR reactions were performed using oligonucleotides based on the nucleotide sequence obtained from the PA14 genome sequence (Table S1). The 8 PCR products were meant to cover the totality of the che2 cluster (Figure 1 and Figure S2) starting from the mcpA gene to the PA0171 gene located downstream cheB2 (PA0173). The reactions were performed simultaneously using PA14 or TB genomic DNA as matrix. Each paired reaction set (TB/PA14) resulted in products running at the same migratory rate within the 1.5% agarose gel (data not shown). These results show that there is little difference between the two isolates, TB and PA14, with regards the overall organization of the che2 chromosomal region.
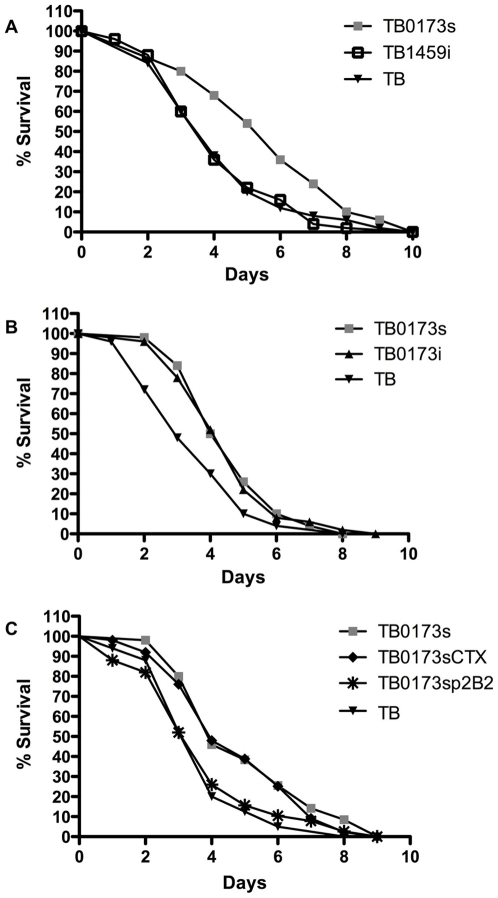
To validate that the loss of virulence and other phenotypic changes observed with the TB strain were a result of the transposon disruption of the cheB2 gene, we generated a new mutant by interrupting the cheB2 gene (see Materials and Methods). As shown in Figure 3A, the newly engineered cheB2 mutant, called TB0173i (p<0.005), showed a delayed C. elegans killing comparable to the original STM cheB2 mutant TB0173s (p<0.004). Both cheB2 mutants, TB0173s and TB0173i, required at least one additional day, as compared to TB, to kill 50% of exposed nematodes. In addition, we engineered a similar cheB2 mutation in the P. aeruginosa PA14 strain, yielding PA140173i (see Materials and Methods), and observed similar virulence attenuation on C. elegans (Figure S3). This further confirmed the virulence phenotype of a cheB2 mutant and indicated that it is not a TB strain-specific trait. Finally, a complementation assay was performed. We reintroduced, in the cheB2 mutant, the wild-type cheB2 gene using an integration-proficient vector, mini-CTX1 [47]. This vector contains the ϕCTX attachment site allowing integration of exogenous DNA fragments at the attB site within the genome of P. aeruginosa. The cheB2 gene was cloned into the mini-CTX1, yielding CTXp2B2 (see Materials and Methods). This construct was introduced into the cheB2 mutant, TB0173s, generating the TB0173sp2B2 strain. The empty vector, mini-CTX1, was also introduced into the TB0173s cheB2 mutant, generating TB0173sCTX. As shown in Figure 3B the virulence was restored in the complemented cheB2 mutant TB0173sp2B2 (p<0.009), whereas the cheB2 mutant strain TB0173sCTX (p<0.005) remained fully attenuated.
Figure 3. C. elegans Slow Killing Assay.
(A) C. elegans survival assay comparison between the parental strain TB, the original cheB2 mutant from the STM library (TB0173s) and the reengineered cheB2 insertion mutant (TB0173i). (B) Comparison between, TB0173s (cheB2 mutant), TB0173sCTX (cheB2 mutant harboring CTX1), TB0173sp2B2 (complemented cheB2 mutant after chromosomal integration of CTXp2B2 carrying the wild type cheB2 gene) and the parental strain (TB). (C) Comparison between, the parental strain TB, the original cheB2 mutant (TB0173s) and the engineered cheB1 mutant (TB1459i). The percent of nematode survival (y axis) is shown with respect to the number of days post-infection (x axis).
A cheB1 Mutant Is not Attenuated for C. elegans Virulence
Several chemotactic gene clusters have been identified in the P. aeruginosa genome [21],[23]. The che1 gene cluster encodes the principle chemotactic device controlling flagellar mobility [23]. We investigated whether the loss of virulence observed in the cheB2 mutant on C. elegans is specific, and results from a loss of chemotaxis under the direction of the che2 chemotactic cluster, rather than being linked with more global chemotactic or motility deficiency. We thus investigated the fate of a cheB1 mutant in a similar assay. The cheB1 mutation was engineered in the TB strain as described in materials and methods, yielding TB1459i. The cheB1 mutant was used to feed C. elegans in the slow killing assay and the survival percentage was compared to those obtained when using the cheB2 mutant or the parental TB strain (Figure 3C). Interestingly, the cheB1 mutant showed no significant reduction in C. elegans killing when compared to the parental TB strain, with only 36% of the worms alive after 4 days post infection. This is in contrast to the loss of virulence in the original cheB2 mutant (TB0173s) and the insertional cheB2 mutant TB0173i.
The mobility of the cheB1 mutant was also analyzed and both swimming and swarming motility appeared to be affected as shown in Figure 2. The swimming or swarming motility defect is, however, much more severe in the cheB1 mutant as compared to the cheB2 mutant. Finally, we used a chemotactic assay using tryptone as chemo-attractant (see Materials and Methods). The parental TB strain was able to move towards the attractant (an increase of slightly more than half a log of bacteria over 45 min) and the TB0173s strain (cheB2 mutant) was also capable of chemotaxis, even though much reduced as compared to TB (Figure S4). However, the TB1459i strain (cheB1 mutant) was no longer able to move towards the attractant, and thus showed a defect in this chemotaxis assay (Figure S4).
The cheB2 Mutant Shows Reduced Virulence in Mice
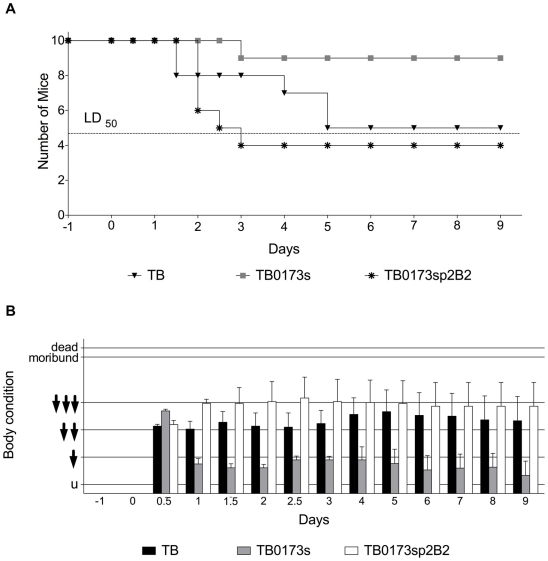
We further tested the virulence of the cheB2 mutant TB0173s in a mouse model of lung infection, utilizing the view-controlled intratracheal infection protocol [48]. Ten female C3H/HeN mice each were inoculated with 7.5×106 CFU equivalent to the LD50 dose of the wild type strain determined in prior experiments (data not shown). Figure 4 displays the representative outcome of one of three independent infection experiments. The cheB2 mutant (TB0173s) showed a significantly attenuated virulence during respiratory tract infection of mice as compared to the parental strain TB and the complemented strain TB0173sp2B2. Infection with the wild type resulted in 50% dead animals (5 of 10) by day 5 compared to 10% deaths for the TB0173s cheB2 mutant (1 of 10; P = 0.036, Fisher's exact test). Infection with the complemented strain TB0173sp2B2 resulted in 60% dead by day 3 (6 of 10; P = 0,0018, Fisher's exact test) (Figure 4A). We have also evaluated the body condition of the mice during bacterial infection (see Materials and Methods). Surviving mice infected with the cheB2 mutant showed a significantly less troubled body condition from days 3 to 6 post infection (P<0.01, Mann-Whitney rank test) than surviving mice infected with the wild type strain indicating that the former group recovered earlier than the latter (Figure 4B).
Figure 4. Mouse Lung Infection with P. aeruginosa TB and Mutant cheB2.
Panels A–B show the outcome of a representative single infection experiment with groups of 10 mice each. The impact on virulence of the parental TB, the cheB2 mutant TB0173s or the complemented cheB2 mutant TB0173sp2B2 isolate in a mouse lung infection model was tested. (A) The TB parental strain showed a maximal killing rate 5 days post-infection while the cheB2 mutant strain TB0173s is severely impaired for virulence. The complemented strain, TB0173sp2B2, like the parental TB isolate, is fully virulent and reaches a maximal killing by day 3. Dashed line indicates the LD50. (B) Mouse body condition and behavior was evaluated following infection. While the mice infected with the cheB2 mutant TB0173s, showed a generally untroubled condition throughout the experiment, the mice infected with the parental strain or the complemented cheB2 mutant strain (TB0173sp2B2) experienced much more severe dysfunction in condition and behavior.
Pathohistological Analysis of P. aeruginosa Infection
To further evaluate the change in virulence between the parental TB isolate and the TB0173s cheB2 mutant, mice were sacrificed 2 days after bacterial challenge and the lungs were formalin fixed and paraffin embedded. Lungs from infected mice challenged with the TB strain as well as the complemented TB0173sp2B2 strain had a pronounced polymorphonuclear neutrophil (PMN) infiltration as well as strong peribronchiolar and perivascular inflammation (Figure 5A). In contrast, the TB0173s cheB2 mutant had caused markedly reduced inflammation and very little PMN infiltration (Figure 5B).
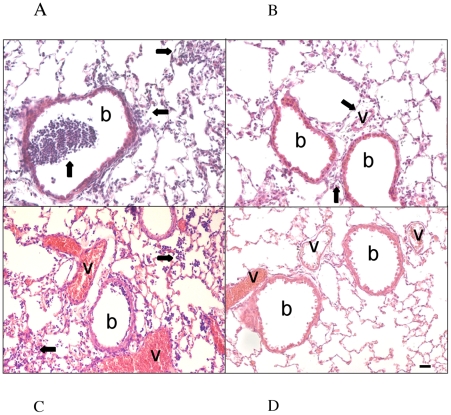
Figure 5. Pathohistological Signs of Inflammation in Murine Lungs.
Two days after intratracheal infection with P. aeruginosa TB or the cheB2 mutant TB0173s, mice were sacrificed. Inflammatory infiltrates are marked by arrows; b: bronchus, v: vessel. (A) TB shows a strong purulent inflammation with intra- and peribronchiolar infiltrates of leucocytes. (B) Slight peribronchiolar and perivascular inflammation is seen with the cheB2 mutant TB0173s. (C) The complemented strain TB0173sp2B2, like the wild type TB isolate, shows a strong purulent inflammation with intra- and peribronchiolar infiltrates of leucocytes. (D) The vehicle control was instilled with 30 µl PBS. Magnification×200.
Discussion
C. elegans is a convenient model for studying bacterial virulence mechanisms [7],[49] and it was shown that virulence factors important in the killing of C. elegans are relevant for virulence in mammalian hosts [50],[51]. Growth conditions and host-pathogen interactions affect the physiology of P. aeruginosa via complex regulatory networks, which in turn control an arsenal of virulence factors [52],[53],[54],[55]. We developed a liquid-based high throughput C. elegans killing assay to speed the screening process of bacterial libraries and to perform multiple screens on virulent clinical isolates. This method was automated using the COPAS Biosort worm sorting system, which greatly reduced the time required to perform the assay. Under slow killing conditions (Figure 3) the TB strain requires approximately three days to kill 50% of a given worm population, and up to 8 days to kill all the worms in a population of 50 nematodes. The liquid killing assay by contrast, is read after overnight feeding of the nematodes with the bacterial strains. It is worth noting that a mutant affected in the gacA gene, previously identified in the slow killing assay, appeared to be attenuated in our liquid-based killing assay.
We used our killing assay to screen a STM library of the P. aeruginosa CF isolate TB [20]. We identified several factors, not yet associated with virulence in P. aeruginosa. Several of the genes identified relate to regulatory, motility or unknown functions. We tested additional virulence and motility related phenotypes including C. elegans slow killing, swarming and swimming motility, or adherence to human cells (Table 1). Most of our selected mutants were impaired in at least one of the additional tested phenotypes.
One of the identified mutants showed reductions in all aspects of virulence we tested. This mutant, named TB0173s, has a transposon insertion in PA0173, cheB2, one of four cheB genes found in P. aeruginosa. CheB protein functions involve interactions with MCPs. As many as 26 MCPs have been identified in P. aeruginosa [56], and it has been proposed that the CheB2 protein functions at least with McpB [57].
In the high throughput C. elegans liquid-based killing assay and the slow killing assay, the cheB2 mutant showed significant virulence attenuation. The cheB2 mutant was also found to have reduced adhesive capabilities to bronchial epithelial cells (Figure S1). Finally, the reductions seen in the motility assays, swimming and swarming, are in agreement with previously reported results revealing a partial loss of motility capabilities [21]. Most importantly in a mouse lung model of infection, the cheB2 mutant was highly attenuated and failed to induce strong inflammatory response in the infected mice lungs. Interestingly, we observed that a mutation in the cheB1 gene yields different phenotypes compared to the cheB2 mutation. The cheB1 mutant virulence is not attenuated in the slow killing assay. Moreover, it shows more severe motility defect than the cheB2 mutant. Finally, in chemotaxis assays using tryptone as chemo-attractant, we observed that, while the cheB1 mutant was strongly affected, the cheB2 mutant had a mild phenotype. These observations suggest that the attenuation in virulence of the cheB2 mutant is specific and not due to a global effect on motility or chemotaxis.
As cluster I of che genes plays the dominant role in P. aeruginosa chemotaxis and flagellar mobility, cluster II genes may be induced under very specific conditions and function in fine-tuning the bacterial response to conditions encountered within a host. Schuster and collaborators have reported that the cluster II genes are induced during stationary phase, are regulated by quorum sensing and that RpoS, the stationary phase RNA polymerase sigma factor, plays a role in controlling cheB2 gene expression [58]. Moreover, Burrowes and collaborators reported that RsmA exerted control over cheB2 with a 10-fold reduction in expression of cheB2 in an rsmA mutant [59]. RsmA works in conjunction with small non-coding RNA to regulate the expression of multiple virulence genes, including the quorum sensing lasI and rhlI genes [60]. Finally, using fluorescent protein-tagged CheY and CheA, it was shown that the Che1 proteins (cluster I) localized to the flagellated pole throughout growth [61]. While the Che1 proteins are still found as bacterial cells entered stationary phase, a patch of Che2 proteins begins to co-localize with the Che1 proteins at that stage. This might indicate that during stationary phase, the chemotactic response will be different than the one observed with exponentially growing cells and the function of the Che2 cluster, even though currently unknown, might be to respond to particular stimuli encountered at that stage.
It has been shown that a cheW mutant in Helicobacter pylori is unable to establish a normal long-term infection in mice, remaining only in one portion of the stomach. The bacterial pathogen may lose its ability to perceive its niche due to the absence of chemotaxis [62]. It is a possibility that within a host, the cluster II che genes of P. aeruginosa might play a similar role in sensing particular conditions during infection.
As previously mentioned, even though the cheB2 mutant is impaired in motility, it is likely that the cheB2 defect in virulence is not linked to this phenotype but to additional traits of virulence and attachment that have not yet been determined. We have shown that the cheB1 mutant is affected in motility but still is virulent. Furthermore, in our study, we isolated a second mutant with reduced motility TB4953s (motB mutant) (Table 1), and while this isolate was attenuated in the high throughput assay, it did not show a reduction in killing in the slow assay (data not shown), further suggesting that the loss of virulence seen in the cheB2 mutant strain (TB0173s) is not solely due to a reduction or change in motility.
While some bacteria require host-derived factors for their growth and are difficult to grow in vitro, others, such as S. marcescens during its infection of C. elegans [33], grow at a much lower rate in the host compared to in vitro. Our observations support the hypothesis that the che2 gene cluster plays a major role during host colonization. For example, we may hypothesize that within the gut of the nematode, or the lungs of the mice, bacteria might reach high cell density and develop as biofilm. This could be a condition to set the function of the Che2 proteins and allow non-growing cells to sense and respond to the environment in a different manner as compared to fast-growing cells. While the idea that the Che2 system might be a specialized chemotaxis system required during host infection is appealing, further experiments are required to understand the chain of command, which switches on the Che2 system and directs the physiological response of P. aeruginosa to Che2 sensing.
Materials and Methods
Bacterial strains and growth conditions
Bacterial strains and plasmids used are listed in Table 2. The P. aeruginosa TB STM transposon library was constructed with the plasposon pTnModOGm [35] carrying variable signature tags as described previously [20]. Plasmids were introduced into P. aeruginosa strains by mating using pRK2013 as a helper or through electroporation. The transformants were selected on Pseudomonas isolation agar containing antibiotics. Plasmids were maintained by adding ampicillin for E. coli (50 µg/ml) and carbenicillin, tetracycline and gentamycin for P. aeruginosa (300, 200 and 10 µg/ml, respectively).
Table 2. Strains and plasmids used in this study.
| Strains/Plasmids | Relevant characteristics* | Reference/origin |
| Escherichia coli | ||
| TG1 | supE (lac-proAB)thi hsdR 5(F′ traD36 proA+B+lacI qZM15) | Lab. collection |
| TOP10F′ | F′ [ lacI qTn 10 (Tet r)] mrcA (mrr-hsdRMS-mcrBC) 80 lacZ M15 lacX74 recA1 araD139 (ara-leu)7697 galU galK rpsL (Str r) endA1 nupG | Invitrogen |
| S17 λpir | thi pro hsdR hsdM+recA RP4-2-Tc::Mu-Km::Tn λpir | Lab. collection |
| OP50 | Uracil auxotroph E. coli B | CGC |
| Pseudomonas aeruginosa | ||
| PA14 | Wild type | Lab. collection |
| TBCF10839 (TB) | CF airway wild type, serotype 4 | Lab. collection |
| TB5479s | Tn mutant from the TB STM library, gltP, Gmr | [19],[20] |
| TB4953s | Tn mutant from the TB STM library, motB, Gmr | [19],[20] |
| TB4554s | Tn mutant from the TB STM library, pilY1, Gmr | [19],[20] |
| TB4380s | Tn mutant from the TB STM library, colS, Gmr | [19],[20] |
| TB3080s | Tn mutant from the TB STM library, PA3080, Gmr | [19],[20] |
| TB2769s | Tn mutant from the TB STM library, PA2769, Gmr | [19],[20] |
| TB2588s | Tn mutant from the TB STM library, PA2588, Gmr | [19],[20] |
| TB2585s | Tn mutant from the TB STM library, uvrC, Gmr | [19],[20] |
| TB2478s | Tn mutant from the TB STM library, dsbD2, Gmr | [19],[20] |
| TB0946s | Tn mutant from the TB STM library, PA0946, Gmr | [19],[20] |
| TB0260s | Tn mutant from the TB STM library, PA0260, Gmr | [19],[20] |
| TB0173s | Tn mutant from the TB STM library, cheB2, Gmr | [19],[20] |
| TB0173i | cheB2 chromosomal insertion mutant derived from TBCF10839. Insertion of PCR2.1 with internal cheB2 DNA fragment of 667 bp, Cbr | This study |
| TB1459i | cheB1 chromosomal insertion mutant derived from TBCF10839. Insertion of PCR2.1 with internal cheB1 DNA fragment of 613 bp, Cbr | This study |
| TB0173sCTX | TB0173s harboring CTX1 on the chromosome, Tcr | This study |
| TB0173sp2B2 | TB0173s harboring CTXp2B2 on the chromosome. CTXp2B2 encodes cheB2 under control of the mcpA promoter, Tcr | This study |
| PA140173i | cheB2 chromosomal insertion mutant derived from PA14. Insertion of PCR2.1 with internal cheB2 DNA fragment of 667 bp, Cbr | This study |
| Plasmids | ||
| pCR2.1 | Apr, ColE1 f1 ori | Invitrogen |
| pCR0173 | pCR2.1 vector with cheB2 DNA fragment of 667 bp | This study |
| pCR1459 | pCR2.1 vector with cheB1 DNA fragment of 612 bp | This study |
| Mini-CTX1 | Tcr, oriT, modified CTX int gene, CTX attachment site | [47] |
| Mini-CTXp2B2 | Tcr, Mini-CTX1 with cheB2 under control of the mcpA promoter region | This study |
Apr, Ampicillin resistance; Cbr, Carbenicillin resistance; Gmr, Gentamycin resistance; Tcr, Tetracycline resistance.
Nematode culture
Nematode strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health National Center for Research Resources. C. elegans strains, wild type N2 (Bristol) and fer-15-(b26) were cultured as described previously [63]. Eggs were isolated by hypochlorite treatment of gravid adults. fer-15 eggs were hatched on NGM at 25°C to obtain synchronized sterile worms for use in the slow killing assays. N2 worms were used for all other killing assays.
Liquid worm assay
Individual clones from the P. aeruginosa STM TB library were grown in 96 well plates in liquid assay media (4.0 g NaCl, 2.5 g peptone, 5.0 g tryptone, 2.5 g yeast extract, dH2O to 1 liter, 1 ml cholesterol (5 mg/ml stock in ethanol) and 7.5 ml glycerol). Each well was filled with 100 µl of media, inoculated with an individual bacterial clone and incubated 24 h at 37°C. The assay was semi-automated using the Copas Biosort (Union Biometrica). This machine is able to sort worms based on extinction (optical density) and time of flight (length) [64]. Using these two parameters we sorted a population of fifty L4 stage worms into the wells of a new 96-well plate. Then 50 µl of the individual P. aeruginosa mutant cultures was transferred to each worm-containing well. Plates were incubated at 25°C overnight with moderate shaking and each well was examined at 24 h with a microscope for surviving worms.
C. elegans slow killing assay
The slow killing assay was performed as described previously [33]. Each independent assay consisted of three replicates. E. coli was used as a control. L4 stage C. elegans were picked onto plates containing overnight growth of each bacterial strain, and on a daily basis worms were evaluated for viability. Animal survival was plotted using the PRISM 5.00 computer program. Survival curves are considered significantly different from the control when P-values are <0.05. Prism calculates survival fractions using the product limit (Kaplan-Meier) method. Prism compares survival curves by two methods: the log-rank test (also called the Mantel-Cox test) and the Gehan-Breslow-Wilcoxon test.
Determination of motility
The media used for the swimming assay was tryptone broth (10 g/l tryptone and 5 g/l NaCl) that contained 0.3% (wt/vol) agarose. Swim plates were inoculated with freshly streaked bacteria from an LB agar plate with a sterile toothpick. The plates were placed in a humidified box and incubated at 37°C for 24 h. Swarm agar was based on M9 minimal medium without NH4Cl, supplemented with MgSO4 (1 mM), glucose (0.2%), and Casamino Acids (0.5%), and solidified with agar (0.5%). Bacteria were spot inoculated on the plates as aliquots taken directly from overnight cultures. Plates were incubated for 16 h at 37°C and incubated an additional 32 h at room temperature [65],[66].
Adherence assays
16HBE14o- human bronchial epithelial cells were grown in Minimal Essential Media (MEM) supplemented with Fetal Calf Serum (FCS), L-glutamine antibiotics and fungisone. Before infection they were seeded in a 24 well plate with a glass coverslip in each well and incubated overnight at 37°C. At the start of the infection, the MEM was replaced with RPMI 1640 media with 25 mM HEPES without phenol red or FCS for 4 h. They were then infected at an MOI of 40 with one of the bacterial isolates for 1 h at 37°C with appropriate antibiotics. After 1 h non-adherent bacteria were removed from each well of the test plate by repeated washings with phosphate buffered saline (PBS). Each well was fixed using 4% paraformaldehyde and the coverslips mounted for observation. The number of crystal violet stained bacteria adhering to cells was determined by visual evaluation using a Leica microscope. At least 30 cells were examined for each isolate. Statistical analysis was performed using GraphPad 4 software, using a non-parametric t- test.
Chemotaxis assays
Chemotaxis assays were performed following a modified version of a previously described assay [67]. Bacterial cultures were grown with shaking overnight in LB at 37°C and subcultured into mineral salts media (MSM) [68] supplemented with 0.4% succinate and 7.6 mM ammonium sulfate. The subcultures were incubated for three hours at 37°C to an OD600 of approximately 1. The cultures were centrifuged and washed twice with Bushnell-Haas media (BHB) and then tested for chemotaxis using BHB and BHB supplemented with 0.1% tryptone as a chemo-attractant. A 1-ml tuberculin syringe with a disposable 23-gauge needle was filled with 100 µl of (BH) mineral salts medium (Difco) containing 0.1% tryptone as a chemoattractant. A 100 µl sample of the washed bacterial suspension was drawn into a 200 µl pipette filter tip. The syringe was then linked and tightly fit into the tip with 3 mm of the needle inserted into the cell suspension. Duplicate apparatus were incubated at 37°C for 45 min, and the content of the syringe was then diluted in 25 mM PBS and plated onto TSA plates for total CFU.
DNA manipulations
Cloning, restriction enzyme analysis, and transformation of E. coli were performed essentially as described by Sambrook et al. [69]. Plasmid DNA was isolated with the QIAprep Spin Miniprep kit, and DNA fragments were purified from agarose gels by using the QIAquick gel extraction kit. Total chromosomal DNA from P. aeruginosa was purified with the BioRad AquaPure genomic DNA kit. Direct genomic sequencing was performed by MilleGen Biotechnologies. Primers used for sequencing were IR1f and IR1r (Table 3). Sequences obtained were subjected to BLAST analysis against the PAO1 genome database (http://www.pseudomonas.com) and general databases.
Table 3. Oligonucleotides used in this study.
| Oligonucleotide name | Sequence# | Position* |
| IR1f | 5′-GCTGCGTTCGGTCAAGGTTC-3′ | 130 bp downstream from the IR upstream Gmr in pTnMod-OGm |
| IR1r | 5′-CATTTTTTGTGATGCTCGTCAGGG-3′ | 100 bp upstream of the IR upstream oriR in pTnMod-OGm |
| CheIN1 | 5′-GACCTGATCAAGCAGCACGC-3′ | 120 bp downstream from the cheB2 ATG |
| CheIN2 | 5′-CGAACATCACGTCCACTGCC-3′ | 260 bp upstream from the cheB2 stop codon |
| ChebUP | 5′-AGCTGAAGAACGACACCGTGC-3′ | 100 bp upstream of the cheB2 ATG |
| ChebDW | 5′-TGCGGGGGGGCGAAAAAGC-3′ | 24 bp downstream the cheB2 stop codon |
| Che1INU | 5′-GCTGGCCCTGAGACCCGA-3′ | 131 bp downstream from the cheB1 ATG |
| Che1INR | 5′-GATGTCGCCGTCCTCTGCT-3′ | 363 bp upstream from the cheB1 stop codon |
| Che1UP | 5′-AAGCCACTCGGCAAGATGTTG-3′ | 154 bp upstream of the cheB1 ATG |
| Che1DW | 5′-GCGACGAACGCCAGGATG-3′ | 115 bp downstream the cheB1 stop codon |
| B2CTX1 | 5′-CGTAAGCTTGCAGTACGCCTATGCCTACC-3′ | 393 bp upstream of the mcpA (PA0180) ATG |
| B2CTX2 | 5′-CGTGAATTCCAGCTTCAGCAGCAGGGGC-3′ | 35 bp downstream of the mcpA ATG |
| B2CTX3 | 5′-TAACGAATTCTCCTGGTGAAGTCCCTGGTGG-3′ | 130 bp upstream of the cheB2 ATG |
| B2CTX4 | 5′-TAACAAGCTTACACAATGATGGCGAGCAGGC-3′ | 100 bp downstream of the cheB2 stop codon |
*: Gmr: Gentamycin resistance cassette.
restriction sites within oligonucleotides are with italics.
Comparison of the che2 genomic regions of the TB and PA14 strains
PCR analyses were performed with eight sets of oligonucleotides covering the che2 cluster-containing region in the PA14 genome (Table S1). These oligonucleotides were used to amplify similar regions within the PA14 and TB genomes. The PA14 and TB genomic DNAs were purified with the BioRad AquaPure genomic DNA kit. PCR reaction products were separated on a 1.5% agarose gel.
Construction of the P. aeruginosa cheB2 and cheB1 mutants
An internal DNA fragment of cheB2 was PCR amplified using the primer pair CheIN1 and CheIN2 (Table 3). The resulting 667 bp PCR product was cloned into pCR2.1 yielding pCR0173. The construct was used as a suicide plasmid and introduced into P. aeruginosa TB and PA14 strains by electroporation. One selected clone for each strain, TB0173i and PA140173i, respectively, was confirmed for the chromosomal integration event into the cheB2 gene by PCR using the primer pair ChebUP and ChebDW (Table 3). An internal DNA fragment of cheB1 was PCR amplified using the primer pair Che1INU and Che1INR (Table 3). The resulting 613 bp product was cloned into pCR2.1 yielding pCR1459. The construct was used as a suicide plasmid and introduced into P. aeruginosa TB strain by electroporation. One selected clone (TB1459i) was confirmed for the chromosomal integration event into the cheB1 gene by PCR using the primer pair Che1UP and Che1DW (Table 3).
Complementation of cheB2 mutant
The cheB2 gene along with the putative promoter region for the che2 chemotaxis cluster located upstream of the mcpA gene (PA0180) were cloned into the mini-CTX1 vector [64], yielding mini-CTXp2B2. These DNA fragments were PCR amplified using B2CTX1, harboring a HindIII site, and B2CTX2, harboring an EcoR1 site, to amplify a 463 bp-long mcpA upstream region, or B2CTX3, harboring an EcoR1 site, and B2CTX4, harboring a HindIII site, to amplify the 1300 bp-long cheB2-containing coding region. These two fragments were cloned in the same orientation, with the promoter region preceding the cheB2 gene. The cloning was done in two steps. Firstly, the promoter region was cloned into the CTX1 using EcoR1 and HindIII. Secondly, this construct was cut using EcoR1 and BamH1, the cheB2-containing coding region was cut with the same restriction enzymes, taking advantage of a BamH1 site following the coding region by 4 bp. The vector and insert were ligated together generating CTXp2B2. This vector was mated into the cheB2 mutant TB0173s generating TB0173sp2B2. The empty CTX1 vector was also mated into the cheB2 mutant TB0173s generating TB0173sCTX. The recombinant clones containing the mini-CTX inserted at the attB locus on the P. aeruginosa genome were selected on tetracycline-containing PIA plates.
Murine respiratory tract infection
Prior to animal experiments, bacteria were grown in LB at 37°C with shaking until stationary phase growth was reached. Bacteria were pelleted by centrifugation (5000 g, 10 min), washed twice with PBS. The intended number of CFU was extrapolated from a standard growth curve and appropriate dilutions with sterile PBS were made to prepare the inoculum for the mice. Ten to twelve week old female mice of the inbred strain C3H/HeN were inoculated via view controlled intratracheal instillation with a bacterial suspension containing 7.5×106 CFU [48]. This noninvasive application technique via catheter allows controlled delivery of bacteria to the lungs. During the experiments mice were maintained in micro-isolator cages with filter top lids at 21±2°C, 50±5% humidity and 12 h light/dark cycle. They were fed with autoclaved acidulated water and standard mouse food (SSniff).
To characterize the course of the bacterial infection the weight and rectal temperature of the mice were measured daily and their body condition was determined using a self-developed score. For this score mice behavior was observed and evaluated for the parameters: vocalization, piloerection, attitude, locomotion, breathing, curiosity, nasal secretion, grooming and dehydration. Dysfunctions in single behavioral parameters were assessed by zero, one or two points, respectively. The body condition of the mice was determined by adding the points resulting in the following score: untroubled 0–1 (u); slightly troubled 2–4 ( ); moderately troubled 5–7 (
); moderately troubled 5–7 (
 ); profoundly troubled 8–10 (
); profoundly troubled 8–10 (

 ); moribund≥11; death≥16. The outcome of the infection experiments was assessed for significant differences between the inoculation of wild type strain and mutant by Fisher's exact test (survival) and Mann-Whitney tests (body condition score).
); moribund≥11; death≥16. The outcome of the infection experiments was assessed for significant differences between the inoculation of wild type strain and mutant by Fisher's exact test (survival) and Mann-Whitney tests (body condition score).
For pathohistological signs of inflammation, lungs were fixed in 4% formalin and embedded in paraffin. Tissue sections (5-µm) were stained by haematoxylin and eosin and evaluated using a Zeiss light microscope.
Supporting Information
Adherence capabilities of TB and selected isogenic mutants on epithelial cells. Examination of the binding capacity to 16HBE14o- human airway epithelial cells of the parental TB isolate and mutant clones. Cells were infected at approximately ten to one for one hour with each isolate. Numbers of bacteria per cell were visually quantified by counting numbers of adherent bacteria on at least 30 cells per isolate. (A) Results obtained for each strain were compared to TB strain in unpaired t tests using Graph Pad Prism 4 software. The * indicates the level of significance, ns indicates : not significant. Data shown are mean {plus minus} standard error from three experiments. (B) representative images showing the typical binding of the TB strain and the cheB2 mutant TB0173s to airway epithelial cells.
(3.00 MB TIF)
Comparison of chemotaxis cluster 2 in P. aeruginosa PA14 and TB by PCR analysis. (A) Gene names and PA numbers within cluster 2 are indicated below each representative arrow bar. Oligonucleotide identity is indicated above the cluster, whereas the line indicates their position within the cluster. (B) Bar diagram indicating the overlap between the eight oligonucleotide pairs used.
(3.00 MB TIF)
C. elegans slow killing assay. C. elegans survival assay comparison between the parental strain PA14 and the cheB2 insertion mutant (PA140173i). The percent of nematode survival (y axis) is shown with respect to the number of days post-infection (x axis).
(3.00 MB TIF)
Chemotaxis assays. Bacterial cultures were grown in LB at 37°C and subcultured into mineral salts media (MSM) supplemented with succinate and ammonium sulfate. The subcultures were grown to an OD600 of approximately 1. The cultures were centrifuged, washed with Bushnell-Haas media (BHB) and tested for chemotaxis using BHB and BHB supplemented with 0.1% tryptone as a chemo-attractant where indicated as 〈〈trypt〉〉. TB is the parental strain, TB1459i is the cheB1 mutant, and TB0173s is the cheB2 mutant. Data are mean±standard error of triplicate cultures from three experiments.
(3.00 MB TIF)
Oligonucleotides used for che2 region mapping.
(0.04 MB DOC)
Acknowledgments
We thank A. Blanc for help in worm sorting, carried-out at the C. elegans functional genomics platform of the Marseille-Nice Génopole, supported by the Réseau Nationale des Génopoles.
Footnotes
The authors have declared that no competing interests exist.
Research at LISM/CNRS/Marseille is supported by grants from the French Cystic Fibrosis Foundation (VLM), the European Union QLK2CT-2001-01339, the Bettencourt-Schueller Foundation and institutional grants from the CNRS. Research in JJE's laboratory is funded by institutional grants from INSERM and the CNRS, and the French Ministry of Research Programme de Microbiologie. Research in BT's laboratory was supported by grants first provided by the BMBF (PathoGenomics programme, Förderkennzeichen: 0313134) and the European Union QLK2CT-2001-01339 and later by the DFG (SFB 587, A9). AF is supported by the Royal Society. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Lyczak JB, Cannon CL, Pier GB. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect. 2000;2:1051–1060. doi: 10.1016/s1286-4579(00)01259-4. [DOI] [PubMed] [Google Scholar]
- 2.O'Toole GA, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol. 1998;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 3.Vallet I, Olson JW, Lory S, Lazdunski A, Filloux A. The chaperone/usher pathways of Pseudomonas aeruginosa: identification of fimbrial gene clusters (cup) and their involvement in biofilm formation. Proc Natl Acad Sci U S A. 2001;98:6911–6916. doi: 10.1073/pnas.111551898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Filloux A, Hachani A, Bleves S. The bacterial type VI secretion machine: yet another player for protein transport across membranes. Microbiology. 2008;154:1570–1583. doi: 10.1099/mic.0.2008/016840-0. [DOI] [PubMed] [Google Scholar]
- 5.Pradel E, Ewbank JJ. Genetic models in pathogenesis. Annu Rev Genet. 2004;38:347–363. doi: 10.1146/annurev.genet.38.072902.092528. [DOI] [PubMed] [Google Scholar]
- 6.Sifri CD, Begun J, Ausubel FM. The worm has turned–microbial virulence modeled in Caenorhabditis elegans. Trends Microbiol. 2005;13:119–127. doi: 10.1016/j.tim.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 7.Tan MW, Mahajan-Miklos S, Ausubel FM. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc Natl Acad Sci U S A. 1999;96:715–720. doi: 10.1073/pnas.96.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan MW, Rahme LG, Sternberg JA, Tompkins RG, Ausubel FM. Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proc Natl Acad Sci U S A. 1999;96:2408–2413. doi: 10.1073/pnas.96.5.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gallagher LA, Manoil C. Pseudomonas aeruginosa PAO1 kills Caenorhabditis elegans by cyanide poisoning. J Bacteriol. 2001;183:6207–6214. doi: 10.1128/JB.183.21.6207-6214.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zaborin A, Romanowski K, Gerdes S, Holbrook C, Lepine F, et al. Red death in Caenorhabditis elegans caused by Pseudomonas aeruginosa PAO1. Proc Natl Acad Sci U S A. 2009;106:6327–6332. doi: 10.1073/pnas.0813199106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi JY, Sifri CD, Goumnerov BC, Rahme LG, Ausubel FM, et al. Identification of virulence genes in a pathogenic strain of Pseudomonas aeruginosa by representational difference analysis. J Bacteriol. 2002;184:952–961. doi: 10.1128/jb.184.4.952-961.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heurlier K, Denervaud V, Haenni M, Guy L, Krishnapillai V, et al. Quorum-sensing-negative (lasR) mutants of Pseudomonas aeruginosa avoid cell lysis and death. J Bacteriol. 2005;187:4875–4883. doi: 10.1128/JB.187.14.4875-4883.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liang X, Pham XQ, Olson MV, Lory S. Identification of a genomic island present in the majority of pathogenic isolates of Pseudomonas aeruginosa. J Bacteriol. 2001;183:843–853. doi: 10.1128/JB.183.3.843-853.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tümmler B, Koopmann U, Grothues D, Weissbrodt H, Steinkamp G, et al. Nosocomial acquisition of Pseudomonas aeruginosa by cystic fibrosis patients. J Clin Microbiol. 1991;29:1265–1267. doi: 10.1128/jcm.29.6.1265-1267.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wiehlmann L, Wagner G, Cramer N, Siebert B, Gudowius P, et al. Population structure of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2007;104:8101–8106. doi: 10.1073/pnas.0609213104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hagelueken G, Adams TM, Wiehlmann L, Widow U, Kolmar H, et al. The crystal structure of SdsA1, an alkylsulfatase from Pseudomonas aeruginosa, defines a third class of sulfatases. Proc Natl Acad Sci USA. 2006;103:7631–7636. doi: 10.1073/pnas.0510501103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hagelueken G, Wiehlmann L, Adams TM, Kolmar H, Heinz DW, et al. Crystal structure of the electron transfer complex rubredoxin rubredoxin reductase of Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2007;104:12276–12281. doi: 10.1073/pnas.0702919104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tümmler B. Unusual mechanism of pathogenicity of Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Infection. 1987;15:311–312. doi: 10.1007/BF01644144. [DOI] [PubMed] [Google Scholar]
- 19.Rakhimova E, Munder A, Wiehlmann L, Bredenbruch F, Tümmler B. Fitness of isogenic colony morphology variants of Pseudomonas aeruginosa in murine airway infection. PLoS ONE. 2008;3:1685. doi: 10.1371/journal.pone.0001685. doi: 10.1371/journal.pone.0001685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wiehlmann L, Larbig K, Ritzka M, Tümmler B. Signature tagged mutagenesis of Pseudomonas aeruginosa. Genome Lett. 2002;1:131–139. [Google Scholar]
- 21.Ferrandez A, Hawkins AC, Summerfield DT, Harwood CS. Cluster II che genes from Pseudomonas aeruginosa are required for an optimal chemotactic response. J Bacteriol. 2002;184:4374–4383. doi: 10.1128/JB.184.16.4374-4383.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, et al. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 23.Kato J, Nakamura T, Kuroda A, Ohtake H. Cloning and characterization of chemotaxis genes in Pseudomonas aeruginosa. Biosci Biotechnol Biochem. 1999;63:155–161. doi: 10.1271/bbb.63.155. [DOI] [PubMed] [Google Scholar]
- 24.Masduki A, Nakamura J, Ohga T, Umezaki R, Kato J, et al. Isolation and characterization of chemotaxis mutants and genes of Pseudomonas aeruginosa. J Bacteriol. 1995;177:948–952. doi: 10.1128/jb.177.4.948-952.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hickman JW, Tifrea DF, Harwood CS. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci U S A. 2005;102:14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Darzins A. Characterization of a Pseudomonas aeruginosa gene cluster involved in pilus biosynthesis and twitching motility: sequence similarity to the chemotaxis proteins of enterics and the gliding bacterium Myxococcus xanthus. Mol Microbiol. 1994;11:137–153. doi: 10.1111/j.1365-2958.1994.tb00296.x. [DOI] [PubMed] [Google Scholar]
- 27.Kearns DB, Robinson J, Shimkets LJ. Pseudomonas aeruginosa exhibits directed twitching motility up phosphatidylethanolamine gradients. J Bacteriol. 2001;183:763–767. doi: 10.1128/JB.183.2.763-767.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li M, Hazelbauer GL. Adaptational assistance in clusters of bacterial chemoreceptors. Mol Microbiol. 2005;56:1617–1626. doi: 10.1111/j.1365-2958.2005.04641.x. [DOI] [PubMed] [Google Scholar]
- 29.Baker MD, Wolanin PM, Stock JB. Signal transduction in bacterial chemotaxis. Bioessays. 2006;28:9–22. doi: 10.1002/bies.20343. [DOI] [PubMed] [Google Scholar]
- 30.Yonekawa H, Hayashi H, Parkinson JS. Requirement of the CheB function for sensory adaptation in Escherichia coli. J Bacteriol. 1983;156:1228–1235. doi: 10.1128/jb.156.3.1228-1235.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones BD, Lee CA, Falkow S. Invasion by Salmonella typhimurium is affected by the direction of flagellar rotation. Infect Immun. 1992;60:2475–2480. doi: 10.1128/iai.60.6.2475-2480.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Begun J, Sifri CD, Goldman S, Calderwood SB, Ausubel FM. Staphylococcus aureus virulence factors identified by using a high-throughput Caenorhabditis elegans-killing model. Infect Immun. 2005;73:872–877. doi: 10.1128/IAI.73.2.872-877.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kurz CL, Chauvet S, Andres E, Aurouze M, Vallet I, et al. Virulence factors of the human opportunistic pathogen Serratia marcescens identified by in vivo screening. EMBO J. 2003;22:1451–1460. doi: 10.1093/emboj/cdg159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liberati NT, Urbach JM, Miyata S, Lee DG, Drenkard E, et al. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci U S A. 2006;103:2833–2838. doi: 10.1073/pnas.0511100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dennis JJ, Zylstra GJ. Plasposons: modular self-cloning minitransposon derivatives for rapid genetic analysis of gram-negative bacterial genomes. Appl Environ Microbiol. 1998;64:2710–2715. doi: 10.1128/aem.64.7.2710-2715.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hebert MD, Houghton JE. Regulation of ornithine utilization in Pseudomonas aeruginosa (PAO1) is mediated by a transcriptional regulator, OruR. J Bacteriol. 1997;179:7834–7842. doi: 10.1128/jb.179.24.7834-7842.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dekkers LC, Bloemendaal CJ, de Weger LA, Wijffelman CA, Spaink HP, et al. A two-component system plays an important role in the root-colonizing ability of Pseudomonas fluorescens strain WCS365. Mol Plant Microbe Interact. 1998;11:45–56. doi: 10.1094/MPMI.1998.11.1.45. [DOI] [PubMed] [Google Scholar]
- 38.de Weert S, Dekkers LC, Bitter W, Tuinman S, Wijfjes AH, et al. The two-component ColR/S system of Pseudomonas fluorescens WCS365 plays a role in rhizosphere competence through maintaining the structure and function of the outer membrane. FEMS Microbiol Ecol. 2006;58:205–213. doi: 10.1111/j.1574-6941.2006.00158.x. [DOI] [PubMed] [Google Scholar]
- 39.Hu N, Zhao B. Key genes involved in heavy-metal resistance in Pseudomonas putida CD2. FEMS Microbiol Lett. 2007;267:17–22. doi: 10.1111/j.1574-6968.2006.00505.x. [DOI] [PubMed] [Google Scholar]
- 40.Horak R, Ilves H, Pruunsild P, Kuljus M, Kivisaar M. The ColR-ColS two-component signal transduction system is involved in regulation of Tn4652 transposition in Pseudomonas putida under starvation conditions. Mol Microbiol. 2004;54:795–807. doi: 10.1111/j.1365-2958.2004.04311.x. [DOI] [PubMed] [Google Scholar]
- 41.Alm RA, Hallinan JP, Watson AA, Mattick JS. Fimbrial biogenesis genes of Pseudomonas aeruginosa: pilW and pilX increase the similarity of type 4 fimbriae to the GSP protein-secretion systems and pilY1 encodes a gonococcal PilC homologue. Mol Microbiol. 1996;22:161–173. doi: 10.1111/j.1365-2958.1996.tb02665.x. [DOI] [PubMed] [Google Scholar]
- 42.Doyle TB, Hawkins AC, McCarter LL. The complex flagellar torque generator of Pseudomonas aeruginosa. J Bacteriol. 2004;186:6341–6350. doi: 10.1128/JB.186.19.6341-6350.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Toutain CM, Zegans ME, O'Toole GA. Evidence for two flagellar stators and their role in the motility of Pseudomonas aeruginosa. J Bacteriol. 2005;187:771–777. doi: 10.1128/JB.187.2.771-777.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stewart RC, Russell CB, Roth AF, Dahlquist FW. Interaction of CheB with chemotaxis signal transduction components in Escherichia coli: modulation of the methylesterase activity and effects on cell swimming behavior. Cold Spring Harb Symp Quant Biol. 1988;53 Pt 1:27–40. doi: 10.1101/sqb.1988.053.01.007. [DOI] [PubMed] [Google Scholar]
- 45.Comolli JC, Waite LL, Mostov KE, Engel JN. Pili binding to asialo-GM1 on epithelial cells can mediate cytotoxicity or bacterial internalization by Pseudomonas aeruginosa. Infect Immun. 1999;67:3207–3214. doi: 10.1128/iai.67.7.3207-3214.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prince A. Adhesins and receptors of Pseudomonas aeruginosa associated with infection of the respiratory tract. Microb Pathog. 1992;13:251–260. doi: 10.1016/0882-4010(92)90035-m. [DOI] [PubMed] [Google Scholar]
- 47.Hoang TT, Kutchma AJ, Becher A, Schweizer HP. Integration-proficient plasmids for Pseudomonas aeruginosa: site-specific integration and use for engineering of reporter and expression strains. Plasmid. 2000;43:59–72. doi: 10.1006/plas.1999.1441. [DOI] [PubMed] [Google Scholar]
- 48.Munder A, Zelmer A, Schmiedl A, Dittmar KE, Rohde M, et al. Murine pulmonary infection with Listeria monocytogenes: differential susceptibility of BALB/c, C57BL/6 and DBA/2 mice. Microbes Infect. 2005;7:600–611. doi: 10.1016/j.micinf.2004.12.021. [DOI] [PubMed] [Google Scholar]
- 49.Kurz CL, Ewbank JJ. Caenorhabditis elegans: an emerging genetic model for the study of innate immunity. Nat Rev Genet. 2003;4:380–390. doi: 10.1038/nrg1067. [DOI] [PubMed] [Google Scholar]
- 50.Ewbank JJ. Tackling both sides of the host-pathogen equation with Caenorhabditis elegans. Microbes Infect. 2002;4:247–256. doi: 10.1016/s1286-4579(01)01531-3. [DOI] [PubMed] [Google Scholar]
- 51.Mahajan-Miklos S, Rahme LG, Ausubel FM. Elucidating the molecular mechanisms of bacterial virulence using non-mammalian hosts. Mol Microbiol. 2000;37:981–988. doi: 10.1046/j.1365-2958.2000.02056.x. [DOI] [PubMed] [Google Scholar]
- 52.Rodrigue A, Quentin Y, Lazdunski A, Méjean V, Foglino M. Two-component systems in Pseudomonas aeruginosa: why so many? Trends Microbiol. 2000;8:498–504. doi: 10.1016/s0966-842x(00)01833-3. [DOI] [PubMed] [Google Scholar]
- 53.Goodman AL, Kulasekara B, Rietsch A, Boyd D, Smith RS, et al. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev Cell. 2004;7:745–754. doi: 10.1016/j.devcel.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 54.Schuster M, Greenberg EP. A network of networks: quorum-sensing gene regulation in Pseudomonas aeruginosa. Int J Med Microbiol. 2006;296:73–81. doi: 10.1016/j.ijmm.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 55.Ventre I, Goodman AL, Vallet-Gely I, Vasseur P, Soscia C, et al. Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc Natl Acad Sci U S A. 2006;103:171–176. doi: 10.1073/pnas.0507407103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kato J, Kim HE, Takiguchi N, Kuroda A, Ohtake H. Pseudomonas aeruginosa as a model microorganism for investigation of chemotactic behaviors in ecosystem. J Biosci Bioeng. 2008;106:1–7. doi: 10.1263/jbb.106.1. [DOI] [PubMed] [Google Scholar]
- 57.Schuster M, Hawkins AC, Harwood CS, Greenberg EP. The Pseudomonas aeruginosa RpoS regulon and its relationship to quorum sensing. Mol Microbiol. 2004;51:973–985. doi: 10.1046/j.1365-2958.2003.03886.x. [DOI] [PubMed] [Google Scholar]
- 58.Schuster M, Lostroh CP, Ogi T, Greenberg EP. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J Bacteriol. 2003;185:2066–2079. doi: 10.1128/JB.185.7.2066-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Burrowes E, Baysse C, Adams C, O'Gara F. Influence of the regulatory protein RsmA on cellular functions in Pseudomonas aeruginosa PAO1, as revealed by transcriptome analysis. Microbiology. 2006;152:405–418. doi: 10.1099/mic.0.28324-0. [DOI] [PubMed] [Google Scholar]
- 60.Pessi G, Williams F, Hindle Z, Heurlier K, Holden MT, et al. The global posttranscriptional regulator RsmA modulates production of virulence determinants and N-acylhomoserine lactones in Pseudomonas aeruginosa. J Bacteriol. 2001;183:6676–6683. doi: 10.1128/JB.183.22.6676-6683.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guvener ZT, Tifrea DF, Harwood CS. Two different Pseudomonas aeruginosa chemosensory signal transduction complexes localize to cell poles and form and remould in stationary phase. Mol Microbiol. 2006;61:106–118. doi: 10.1111/j.1365-2958.2006.05218.x. [DOI] [PubMed] [Google Scholar]
- 62.Terry K, Williams SM, Connolly L, Ottemann KM. Chemotaxis plays multiple roles during Helicobacter pylori animal infection. Infect Immun. 2005;73:803–811. doi: 10.1128/IAI.73.2.803-811.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lewis JA, Fleming JT. Basic culture methods. Methods Cell Biol. 1995;48:3–29. [PubMed] [Google Scholar]
- 64.Couillault C, Pujol N, Reboul J, Sabatier L, Guichou JF, et al. TLR-independent control of innate immunity in Caenorhabditis elegans by the TIR domain adaptor protein TIR-1, an ortholog of human SARM. Nat Immunol. 2004;5:488–494. doi: 10.1038/ni1060. [DOI] [PubMed] [Google Scholar]
- 65.Head NE, Yu H. Cross-sectional analysis of clinical and environmental isolates of Pseudomonas aeruginosa: biofilm formation, virulence, and genome diversity. Infect Immun. 2004;72:133–144. doi: 10.1128/IAI.72.1.133-144.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kohler T, Curty LK, Barja F, van Delden C, Pechere JC. Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J Bacteriol. 2000;182:5990–5996. doi: 10.1128/jb.182.21.5990-5996.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mazumder R, Phelps TJ, Krieg NR, Benoit RE. Determining chemotactic responses by two subsurface microaerophiles using a simplified capillary assay method. J Microbiol Methods. 1999;37:255–263. doi: 10.1016/s0167-7012(99)00072-x. [DOI] [PubMed] [Google Scholar]
- 68.Moulton RC, Montie TC. Chemotaxis by Pseudomonas aeruginosa. J Bacteriol. 1979;137:274–280. doi: 10.1128/jb.137.1.274-280.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sambrook J, Fritsch E, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Adherence capabilities of TB and selected isogenic mutants on epithelial cells. Examination of the binding capacity to 16HBE14o- human airway epithelial cells of the parental TB isolate and mutant clones. Cells were infected at approximately ten to one for one hour with each isolate. Numbers of bacteria per cell were visually quantified by counting numbers of adherent bacteria on at least 30 cells per isolate. (A) Results obtained for each strain were compared to TB strain in unpaired t tests using Graph Pad Prism 4 software. The * indicates the level of significance, ns indicates : not significant. Data shown are mean {plus minus} standard error from three experiments. (B) representative images showing the typical binding of the TB strain and the cheB2 mutant TB0173s to airway epithelial cells.
(3.00 MB TIF)
Comparison of chemotaxis cluster 2 in P. aeruginosa PA14 and TB by PCR analysis. (A) Gene names and PA numbers within cluster 2 are indicated below each representative arrow bar. Oligonucleotide identity is indicated above the cluster, whereas the line indicates their position within the cluster. (B) Bar diagram indicating the overlap between the eight oligonucleotide pairs used.
(3.00 MB TIF)
C. elegans slow killing assay. C. elegans survival assay comparison between the parental strain PA14 and the cheB2 insertion mutant (PA140173i). The percent of nematode survival (y axis) is shown with respect to the number of days post-infection (x axis).
(3.00 MB TIF)
Chemotaxis assays. Bacterial cultures were grown in LB at 37°C and subcultured into mineral salts media (MSM) supplemented with succinate and ammonium sulfate. The subcultures were grown to an OD600 of approximately 1. The cultures were centrifuged, washed with Bushnell-Haas media (BHB) and tested for chemotaxis using BHB and BHB supplemented with 0.1% tryptone as a chemo-attractant where indicated as 〈〈trypt〉〉. TB is the parental strain, TB1459i is the cheB1 mutant, and TB0173s is the cheB2 mutant. Data are mean±standard error of triplicate cultures from three experiments.
(3.00 MB TIF)
Oligonucleotides used for che2 region mapping.
(0.04 MB DOC)