Abstract
X-linked reticulate pigmentary disorder with systemic manifestations in males (PDR) is very rare. Affected males are characterized by cutaneous and visceral symptoms suggestive of abnormally regulated inXammation. A genetic linkage study of a large Canadian kindred previously mapped the PDR gene to a greater than 40 Mb interval of Xp22–p21. The aim of this study was to identify the causative gene for PDR. The Canadian pedigree was expanded and additional PDR families recruited. Genetic linkage was performed using newer microsatellite markers. Positional and functional candidate genes were screened by PCR and sequencing of coding exons in affected males. The location of the PDR gene was narrowed to a ~4.9 Mb interval of Xp22.11–p21.3 between markers DXS1052 and DXS1061. All annotated coding exons within this interval were sequenced in one affected male from each of the three multiplex families as well as one singleton, but no causative mutation was identiWed. Sequencing of other X-linked genes outside of the linked interval also failed to identify the cause of PDR but revealed a novel nonsynonymous cSNP in the GRPR gene in the Maltese population. PDR is most likely due to a mutation within the linked interval not affecting currently annotated coding exons.
Introduction
X-linked reticulate pigmentary disorder with systemic manifestations in males (PDR, OMIM 301220) is an exceedingly rare genetic disease. Thus far, only four families with the disorder, three multiplex with affected relative pairs and one singleton, have been reported (Ades et al. 1993; Anderson et al.2005; Megarbane et al. 2005; Partington et al. 1981). The disorder is X-linked dominant with variable expressivity, with males much more severely affected than females. Affected males in the Wrst family described, a large Canadian kindred, showed diffuse reticulate hyper- and hypopigmentation beginning in infancy, recurrent pneumonia, corneal opacification, gastrointestinal inflammation, urethral stricture, failure to thrive, and characteristic upswept hair and flared eyebrows (Partington and Prentice 1989). Female carriers showed only patchy pigmentary skin lesions along the lines of Blaschko, with incomplete penetrance. Gedeon et al. (1994) mapped the PDR gene by linkage analysis of the Canadian pedigree to a greater than 40 cM interval of Xp22–p21 bounded by DXS999 distally and DXS228 proximally. Three other unrelated families in whom one or more individuals were diagnosed with PDR have since been reported (Ades et al. 1993; Anderson et al. 2005; Megarbane et al. 2005). In an attempt to identify the responsible gene, we narrowed the PDR linked interval by repeating linkage analysis with newer microsatellite markers in the expanded Canadian pedigree together with two other nuclear families. We also sequenced all annotated coding exons in the refined linkage interval as well as other selected Xp genes in one affected male from each of the three multiplex families and one singleton.
Methods
DNA collection
This study was approved by the UT Southwestern Institutional Review Board. DNAs were obtained previously from members of the Canadian family (Gedeon et al. 1994). Informed consent and blood or DNA samples were obtained from additional members of this family and members of other reported PDR families. Lymphoblasts were immortalized from one or more affected males from each of the Canadian, Texas, and Australian PDR families. DNA was extracted from blood or lymphoblastoid cells using standard techniques.
Genotyping
ABI Linkage V2.5 microsatellite markers for the short arm of the X chromosome and additional custom markers DXS365, DXS443, DXS1052, DXS989, DXS8099, DXS1202, and DXS8192 selected from GDB (http://www.gdb.org) were analyzed using an ABI 3100 capillary electrophoresis instrument and ABI GeneMapper software version 3.7.
Linkage analysis
PDR exhibited a dominant mode of inheritance, and the underlying gene was highly penetrant, particularly in males; therefore we performed multipoint model-based linkage analysis by fitting a liability model with penetrance equal to 0.99 and 0.90 for males and females, respectively. Both disease allele frequency and sporadic rate were set to be 0.0001. To evaluate the significance level of the result, we simulated 10,000 replicates under the null hypothesis of no linkage conditional on pedigree structure and marker informativity. We constructed the most likely haplotype to infer the location of the PDR susceptibility gene by comparing haplotype similarity between affected individuals and dissimilarity between discordant individuals in each family. To confirm the linkage signal irrespective of genetic models, we also performed model-free linkage analysis. All analyses were conducted by using the MINX module of the software MERLIN (Abecasis et al. 2002).
Candidate gene sequencing
Primers were designed flanking the splice sites and boundaries of coding exons for all annotated protein-coding genes in the linked region (UCSC Genome Browser, hg36 assembly) and several genes beyond the boundaries. Standard PCR amplification was performed and amplified products were sequences using ABI Big Dye terminator chemistry. Sequences were examined using SeqMan II version 5.05 (DNASTAR, Madison, WI). Variations from the reference sequence were checked against dbSNP. Unannotated nonsynonymous SNPs were evaluated for potential effect on protein structure and function using PolyPhen (Sunyaev et al. 2001).
Expression studies
Total RNA was extracted from lymphoblastoid cell lines using Trizol reagent (InVitrogen, Carlsbad, CA). RT-PCR primers spanning introns were designed for ACOT9, SAT1, GRPR and PRDX4. Primers flanking the entire coding region were included if transcript size allowed, otherwise the product was separated into amplifiable sizes. ACOT9 was amplified both as the entire coding region and as two separate products using the primer pairs ACOT9.FL AGGTTGGCTCATTGCTCTTTCTTT, TTCTTTCTCCCCTCAGCCCCATCC; ACOT9.1 GGCTCCCGGGCTGTCCTCA, ATGCCGGCCCTTTATTTTC; ACOT9.2 AGCTTGGGAGTTCTTATTTGTTAC, AGGGAGGCCACTTCACTG. The full SAT1 coding region was amplified as well as an alternate transcript of SAT1 including an alternate exon 3 and 3’UTR using the primer pairs SAT1.FL GACTGGTGTTTATCCGTACTC, AGAATCAAACAGAAACTCTAAGTACCA; SAT1AE3 TGGTGTTTATCCGTCACTCG, CGGGTCTCCACAGCACTTAT. PRDX4 was amplified into two segments using primer pairs PRDX4.1 GCGCCAAGGGACGTGTTTCTG, CTTGTCTTCGAGGGGGTATTA; PRDX4.2 GTTGATTCACAGTTTACCCATTTG, AACCGTGAACTTTATTGAGAACTT. GRPR was amplified in four segments using primer pairs GRPR.1 TCTGTTAAGCTAGGTAGGAACTGC, GCACTGTGACTGGAGATGTTG; GRPR.2 GACTGTTTCCTTCTGAACTTGGA, GGATTCAATCTGCTTCTTGAC; GRPR.3 ATTCCACTGTCGATCATCTCTG, CACATCAGAAGAAACGTTCACAA; GRPR.4 TTGAAAGAAGCCATCAAGTCTTA, AAAGGATTGCTCTTCTATGGTG.
GRPR RFLP analysis
Primers CAAAGAGCCCGGCATAGAT and GTGAGTGTGAAGACAGACACCC were used to generate a 500 bp PCR product from genomic DNA. This product is cleaved at three sites by restriction enzyme HpyCH4III (New England Biolabs) to give products of 194, 128, 121 and 57 bp. The c.17G→C SNP eliminates one of these sites, resulting in products of 194, 178 and 128 bp. Restriction fragments were resolved by electrophoresis using 4% agarose gels.
Results
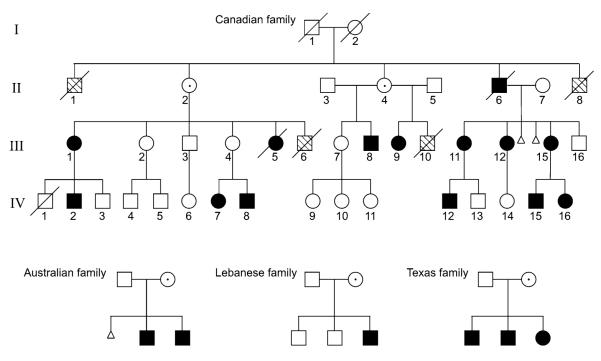
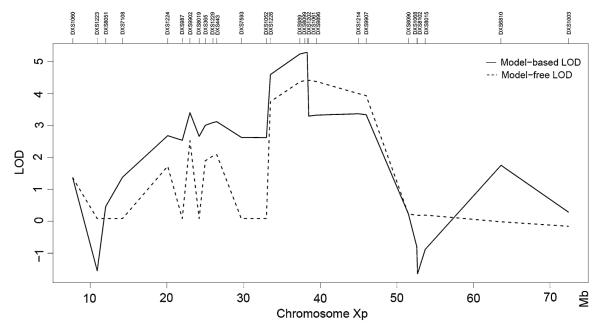
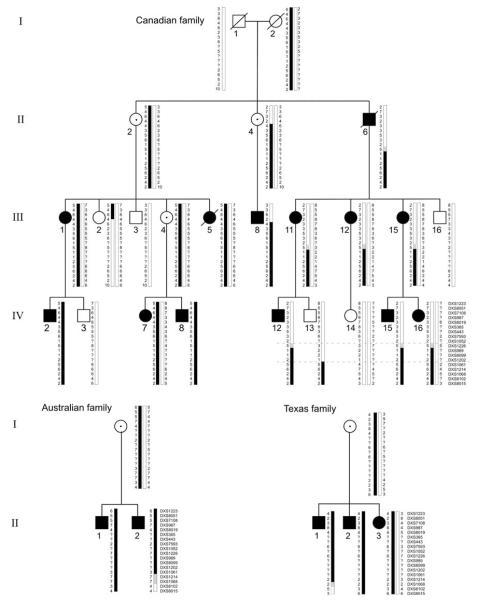
All PDR families in this study have been previously reported (Ades et al. 1993; Anderson et al. 2005; Megarbane et al. 2005; Partington et al. 1981). We obtained stored DNA samples from previously studied members of the Canadian family (Fig. 1) and repeated linkage analysis using newer microsatellite markers for the short arm of the X chromosome. We then expanded the pedigree, obtaining samples from IV-13, IV-14, IV-15 and IV-16, who were born since the original report. The additional family members were then genotyped with the same markers and the combined results analyzed for linkage. Based on the results, selected family members were genotyped for additional microsatellite markers DXS365, DXS443, DXS1052, DXS989, DXS8099, DXS1202, and DXS8192 to narrow the location of meiotic recombination events. The refined PDR linkage interval of ~4.9 Mb was bounded distally by DXS1052 and proximally by DXS1061 (Fig. 2). The proximal boundary was determined by an observed recombination in IV-13 between DXS1202 and DXS1061. The distal boundary was determined by an inferred recombination in II-6 between DXS1052 or DXS1226 and DXS989. DXS1052 and DXS1226 are ~0.5 Mb apart, with no annotated genes in between, and we did not attempt to further refine the distal boundary of the PDR linkage interval. Based on these results, the other multiplex families were genotyped for selected Xp markers. Within each family, all affected members shared common haplotypes, although the mutations in the different families arose independently on different haplotypes, as expected for a highly penetrant deleterious gene (Fig. 3). Australian family member II-2 showed a recombination between DXS1061 and DXS8102, and Texas family member II-1 showed a recombination between DXS1214 and DXS8102, but both these recombinations were proximal to the PDR interval defined by the recombination in Canadian family member IV-13 (Fig. 3). The peak LOD score with model-based linkage is 5.279 in the interval between DXS1226 (slightly more informative than DXS1052) and DXS1061. With model-free linkage analysis, the peak LOD score was 4.41, but the interval is larger, with a proximal boundary of DXS8090. To be conservative, a multipoint model-based lod score of 5.279 corresponds to a P value of 5.3 × 10−6 (Xing et al. 2007), and none of the 10,000 replicates generated a greater LOD score.
Fig. 1.
PDR pedigrees in this study. Canadian family members are numbered according to previous reports (Gedeon et al.1994; Partington et al. 1981)
Fig. 2.
Model-based and model-free LOD score plots for Xp markers genotyped in PDR families
Fig. 3.
Partial pedigrees showing haplotypes defining PDR interval. Black bars denote chromosomes or regions carrying disease allele. Grey bars denote regions of uncertainty. Dashed lines indicate minimal linkage interval for PDR gene
We then sequenced all annotated coding exons and surrounding splice sequences in the DXS1052–DXS1061 interval in one proband from each of the four reported PDR families. Table 1 summarizes the results.
Table 1.
Sequencing results for annotated genes in PDR linkage interval
| Gene name | RefSeq or ENSEMBL identifer | No. of coding exons |
Variation detected | Comment (see also text) |
|---|---|---|---|---|
| DDX53 | NM_182699 | 1 | ||
| FLJ11556 | ENST00000356867 | 1 | ||
| PTCHD1 | NM_173495 | 3 | ||
| Predicted gene | ENST00000352495 | 1 | ||
| PRDX4 | NM_006406 | 7, ae1 | Regulates NFκB | |
| ACOT9 | NM_001033583 | 15, ae1, ae3 | c.934G→C (E312Q) | |
| SAT1 | NM_002970 | 6, ae4 | Overexpression may cause keratosis follicularis spinulosa decalvans |
|
| APOO | NM_024122 | 9 | ||
| RPL9-like | ENST00000329019 | 1 | Retrotransposed pseudogene? | |
| CXorf58 | NM_152761 | 9 | ||
| KLHL15 | NM_030624 | 2 | ||
| EIF2S3 | NM_001415 | 12 | ||
| ZFX | NM_003410 | 9 | ||
| FAM48B1 | ENST00000327866 | 1 | ||
| PDK3 | NM_005391 | 11 | ||
| PCYT1B | NM_004845 | 8, ae1 | ||
| POLA | NM_016937 | 37 | ||
| SCARNA23 | ENSG00000212474 | 0 | snoRNA ACA12 | |
| LOC644820 | NM_001089876 | 1 | ||
| ARX | NM_139058 | 5 | ||
| Predicted gene | ENST00000351630 | 0 | Noncoding RNA | |
| Predicted gene | ENST00000366348 | 8 | ||
| RANBP1-like | ENST00000304245 | 1 | Retrotransposed pseudogene? | |
| MAGEB18 | NM_173699 | 3 | ||
| MAGEB6 | NM_173523 | 2 | ||
| MAGEB5 | ENST00000379029 | 1 |
ae Alternate exon
Only one variation was identiWed, a c.934G→C transversion in the ACOT9 gene in the proband from the Australian family. This variation, which was not present in dbSNP, alters the first nucleotide of ACOT9 exon 12, changing glutamic acid 312 to glutamine. Polyphen predicted that this substitution is benign. Although G is the preferred base at this exon position, it is not invariant among spliced mRNAs (Zhang 1998). To test whether the mutation abrogated efficient ACOT9 splicing, we performed RT-PCR, using RNA from EBV-immortalized lymphoblasts and primers from flanking exons. We obtained an RT-PCR product of the expected size that was not qualitatively different in abundance in the Australian proband versus normal controls, suggesting that the mutation did not prevent efficient splicing (data not shown). Quantitative RT-PCR also showed no systematic difference in ACOT9 mRNA abundance in immortalized lymphoblasts from any of the PDR probands compared to controls (data not shown). Given that there were no ACOT9 coding mutations in the other three unrelated PDR probands and no evidence of abnormal ACOT9 expression in any proband tested, we decided that the ACOT9 c.934G→C mutation in exon 12 is a rare variation unrelated to PDR.
Several biologically plausible candidate genes outside of the reduced linked interval were also sequenced in some or all of the probands before results from the analysis of the expanded pedigree were obtained (Table 2). One of these was the spermine synthase gene SMS. SMS is in the same polyamine synthetic pathway as spermine acyl transferase, encoded by SAT1, one of the candidate genes in the critical region. An Xp duplication that includes SAT1 has been associated with keratosis follicularis spinulosa decalvans (Gimelli et al. 2002), a disease with skin and cornea involvement, like PDR. Furthermore, transgenic overexpression of SAT1 in the skin of mice resulted in permanent hair loss and dermal cysts (Pietila et al. 2005). While loss of function mutations in SMS cause X-linked mental retardation (Cason et al. 2003), we surmised that a gain of function mutation could result in a disease phenotype similar to KFSD and might cause PDR. However, no SMS coding mutations were identified, and quantitative RT-PCR studies did not show any increase in SAT1 expression in lymphoblastoid cells of PDR probands compared to controls (data not shown).
Table 2.
Sequencing results for selected candidate genes outside PDR linkage interval
| Gene name | RefSeq or other identifier | No. of coding exons | Variation detected | Comment (see also text) |
|---|---|---|---|---|
| GRPR | NM_005314 | 3 | c.17G→C (C6S) | Gastrin-releasing peptide receptor |
| PPEF1 | NM_006240 | 16 | ||
| SH3KBP1 | NM_031892 | 18 | Enhances TNF-mediated apoptosis | |
| EIF1AX | NM_001412 | 7 | ||
| CNKSR2 | NM_014927 | 22 | ||
| MBTPS2 | NM_015884 | 11 | ||
| SMS | NM_004595 | 11 | Encodes enzyme in same pathway as SAT1 | |
| FLJ32742 | AK057304 | 2 | ||
| IL1RAPL1 | NM_014271 | 11 | ||
| MAP3K7IP3 | NM_152787 | 11 | Upstream activator of NFκB |
Having failed to identify a causal mutation for PDR, we took advantage of the International Genetics Of Learning Disability (IGOLD) study (carried out at the Wellcome Trust Sanger Institute, Hinxton, UK) (Raymond and Tarpey 2006; Tarpey et al. 2006, and data not shown) and resequenced probands from all three families in 737 X-chromosome genes annotated by VEGA (Vertebrate Genome Annotation Database; http://vega.sanger.ac.uk/index.html). This large scale re-sequencing identified a nonsynonymous substitution in the Australian proband in the first exon of the GRPR (gastrin-releasing peptide receptor) gene, c.17G→C, that changes conserved cysteine residue seven to serine in the extracellular portion of the protein. We verified this mutation by independent sequencing and sequenced GRPR in our other PDR probands but found no other coding mutations. GRPR was a plausible biological candidate for PDR, as it encodes a G-protein coupled receptor that is highly expressed in lung epithelium. The C7S mutation was not present in any of ~250 predominantly Caucasians with XLMR also sequenced by the Sanger Center.
The Australian PDR family was of Maltese ancestry (Ades et al. 1993). To determine whether the GRPR C7S mutation is a polymorphism in the Maltese population, we assayed cord blood DNA samples from 191 Maltese subjects of unknown sex. The c.17G→C mutation eliminates an HpyCH4III restriction site. PCR/RFLP analysis demonstrated that 14 of the Maltese samples (7.3%) carried at least one mutant allele, 9 heterozygous and 5 either homozygous or hemizygous, demonstrating that it is present at polymorphic frequency in this population and therefore ruling it out as the cause of PDR.
Discussion
Linkage mapping narrowed the location of the gene causing PDR to a ~4.9 Mb interval containing approximately 18 genes. Several of these genes were plausible candidates, based upon the likely inXammatory pathogenesis of the disorder. PRDX4 encodes a thioredoxin peroxidase that has been shown to regulate NFκB activation in cultured cells (Jin et al. 1997). SAT1 encodes spermine synthase, overexpression of which may be responsible for keratosis follicularis spinulosa decalvans, another disorder affecting skin and cornea (Gimelli et al. 2002). However, comprehensive sequencing of the coding exons of these genes did not reveal a mutation in PDR kindreds, nor was there any evidence of aberrant expression in lymphoblastoid cells from affected males. Systematic resequencing of other coding sequences in the critical region also failed to reveal the cause of PDR.
There are several possible explanations for this inconclusive result. First, the linkage interval could be erroneous. The distal boundary is based on an inferred meiotic recombination. The proximal boundary is based on an observed recombination, assuming complete penetrance. PDR clearly shows incomplete penetrance in females, but the literature suggests that the disorder is fully penetrant in males. Sex-specific penetrance is consistent with X-linked inheritance. Linkage analysis could also be erroneous if there is variation in marker order in the Canadian family, e.g., due to a cryptic chromosomal inversion. Finally, there is a small but not zero (<0.0001) probability that a LOD score >5 could occur by chance. However, comprehensive re-sequencing of VEGA-annotated X chromosome coding sequences also failed to reveal a causal PDR mutation.
A more likely explanation for the failure to identify the cause of PDR is that the disorder is due to mutation(s) outside of the exons that were sequenced. There could be a mutation in an unannotated gene, a noncoding region, or an alternative transcript of one of the annotated genes in the interval. It is also possible that mutation of a noncoding RNA gene in the interval causes PDR. We sequenced one annotated noncoding small nucleolar RNA, ACA12, encoded by the SCARNA23 gene within an intron of POLA. The ACA12 RNA is predicted to guide the pseudouridylation of residue U40 of the spliceosomal U6 snRNA (Kiss et al. 2004). We considered this RNA gene to be a plausible PDR candidate because mutations in dyskerin (DKC1), a component of the telomerase complex that associates with H/ACA small nucleolar RNAs (Tollervey and Kiss 1997), cause dyskeratosis congenita, a skin disorder with some similarities to PDR (Mitchell et al. 1999). However, no SCARNA23 mutations were identified in any PDR proband.
We also considered that possibility that PDR could result from copy number variation that could be missed by PCR and sequencing, e.g., a duplication. Incontinentia pigmenti presents in females with a reticular pigmentary abnormality reminiscent of that seen in males with PDR (Partington et al. 1981). Identification of IKKB (NEMO) mutations as the cause of incontinentia pigmenti was hindered by the presence of a nearby pseudogene, recombination of which gives rise to IKKB deletions (Smahi et al. 2000). Interestingly, an Xp duplication including SAT1 has been associated with keratosis follicularis spinulosa decalvans, a disorder also affecting skin and cornea (Gimelli et al. 2002). We therefore performed high resolution array comparative genomic hybridization on an affected male from each of the four PDR families using an X chromosome tiling array with average probe spacing of ~169 bp (Nimblegen), but we did not detect copy number variations anywhere on the X chromosome (data not shown).
The molecular basis of PDR remains unknown. Largescale genomic resequencing of the PDR linkage interval, or if necessary, the entire X chromosome of affected PDR family members should ultimately reveal the genetic basis of this enigmatic disorder.
Acknowledgments
We thank Carrie Soukup, Matt Stevenson, and Bo Luo for contributing to candidate gene sequencing. We also thank Professor Alex Felice and Joseph Borg from the Laboratory of Medical Genetics, University of Malta for providing cord blood samples. This work was supported in part by NIH P30 grant AR041940.
Footnotes
Competing interests None declared.
References
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlinrapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- Ades LC, Rogers M, Sillence DO. An X-linked reticulate pigmentary disorder with systemic manifestations: report of a second family. Pediatr Dermatol. 1993;10:344–351. doi: 10.1111/j.1525-1470.1993.tb00396.x. [DOI] [PubMed] [Google Scholar]
- Anderson RC, Zinn AR, Kim J, Carder KR. X-linked reticulate pigmentary disorder with systemic manifestations: report of a third family and literature review. Pediatr Dermatol. 2005;22:122–126. doi: 10.1111/j.1525-1470.2005.22206.x. [DOI] [PubMed] [Google Scholar]
- Cason AL, Ikeguchi Y, Skinner C, Wood TC, Holden KR, Lubs HA, Martinez F, Simensen RJ, Stevenson RE, Pegg AE, Schwartz CE. X-linked spermine synthase gene (SMS) defect: the first polyamine deficiency syndrome. Eur J Hum Genet. 2003;11:937–944. doi: 10.1038/sj.ejhg.5201072. [DOI] [PubMed] [Google Scholar]
- Gedeon AK, Mulley JC, Kozman H, Donnelly A, Partington MW. Localisation of the gene for X-linked reticulate pigmentary disorder with systemic manifestations (PDR), previously known as X-linked cutaneous amyloidosis. Am J Med Genet. 1994;52:75–78. doi: 10.1002/ajmg.1320520115. [DOI] [PubMed] [Google Scholar]
- Gimelli G, Giglio S, Zuffardi O, Alhonen L, Suppola S, Cusano R, Lo Nigro C, Gatti R, Ravazzolo R, Seri M. Gene dosage of the spermidine/spermine N(1)-acetyltransferase (SSAT) gene with putrescine accumulation in a patient with a Xp21.1p22.12 duplication and keratosis follicularis spinulosa decalvans (KFSD) Hum Genet. 2002;111:235–241. doi: 10.1007/s00439-002-0791-6. [DOI] [PubMed] [Google Scholar]
- Jin DY, Chae HZ, Rhee SG, Jeang KT. Regulatory role for a novel human thioredoxin peroxidase in NF-kappaB activation. J Biol Chem. 1997;272:30952–30961. doi: 10.1074/jbc.272.49.30952. [DOI] [PubMed] [Google Scholar]
- Kiss AM, Jady BE, Bertrand E, Kiss T. Human box H/ACA pseudouridylation guide RNA machinery. Mol Cell Biol. 2004;24:5797–807. doi: 10.1128/MCB.24.13.5797-5807.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megarbane H, Boehm N, Chouery E, Bernard R, Salem N, Halaby E, Levy N, Megarbane A. X-linked reticulate pigmentary layer. Report of a new patient and demonstration of a skewed X-inactivation. Genet Couns. 2005;16:85–89. [PubMed] [Google Scholar]
- Mitchell JR, Wood E, Collins K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature. 1999;402:551–555. doi: 10.1038/990141. [DOI] [PubMed] [Google Scholar]
- Partington MW, Marriott PJ, Prentice RS, Cavaglia A, Simpson NE. Familial cutaneous amyloidosis with systemic manifestations in males. Am J Med Genet. 1981;10:65–75. doi: 10.1002/ajmg.1320100109. [DOI] [PubMed] [Google Scholar]
- Partington MW, Prentice RS. X-linked cutaneous amyloidosis: further clinical and pathological observations. Am J Med Genet. 1989;32:115–119. doi: 10.1002/ajmg.1320320125. [DOI] [PubMed] [Google Scholar]
- Pietila M, Pirinen E, Keskitalo S, Juutinen S, Pasonen-Seppanen S, Keinanen T, Alhonen L, Janne J. Disturbed keratinocyte differentiation in transgenic mice and organotypic keratinocyte cultures as a result of spermidine/spermine N-acetyltransferase overexpression. J Invest Dermatol. 2005;124:596–601. doi: 10.1111/j.0022-202X.2005.23636.x. [DOI] [PubMed] [Google Scholar]
- Raymond FL, Tarpey P. The genetics of mental retardation. Hum Mol Genet. 2006;15:R110–R116. doi: 10.1093/hmg/ddl189. [DOI] [PubMed] [Google Scholar]
- Smahi A, Courtois G, Vabres P, Yamaoka S, Heuertz S, Munnich A, Israel A, Heiss NS, Klauck SM, Kioschis P, Wiemann S, Poustka A, Esposito T, Bardaro T, Gianfrancesco F, Ciccodicola A, D’Urso M, Woffendin H, Jakins T, Donnai D, Stewart H, Kenwrick SJ, Aradhya S, Yamagata T, Levy M, Lewis RA, Nelson DL. Genomic rearrangement in NEMO impairs NF-kappaB activation and is a cause of incontinentia pigmenti. The international incontinentia pigmenti (IP) consortium. Nature. 2000;405:466–472. doi: 10.1038/35013114. [DOI] [PubMed] [Google Scholar]
- Sunyaev S, Ramensky V, Koch I, Lathe W, Kondrashov AS, III, Bork F. Prediction of deleterious human alleles. Hum Mol Genet. 2001;10:591–597. doi: 10.1093/hmg/10.6.591. [DOI] [PubMed] [Google Scholar]
- Tarpey PS, Stevens C, Tcague l, Edkins S, O’Meara S, Avis T, Barthorpe S, Buck G, Butler A, Cole J, Dicks E, Gray K, Halliday K, Harrison R, Hills K, Hinton J, Jones D, Menzies A, Mironenko T, Perry J, Rainc K, Richardson D, Shepherd R, Small A, Tofts C, Varian J, West S, Widaa S, Yates A, Catford R, Butler J, Mallya U, Moon J, Luo Y, Dorkins H, Thompson D, Easton DF, Wooster R, Bobrow M, Carpenter N, Simensen RJ, Schwartz CE, Stevenson RE, Turner G, Partington M, Gecz J, Stratton MR, Futreal PA, Raymond FL. Mutations in the gene encoding the Sigma 2 subunit of the adaptor protein 1 complex, AP1S2, cause X-linked mental retardation. Am J Hum Genet. 2006;79:1119–1124. doi: 10.1086/510137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey D, Kiss T. Function and synthesis of small nucleolar RNAs. Curr Opin Cell Biol. 1997;9:337–342. doi: 10.1016/s0955-0674(97)80005-1. [DOI] [PubMed] [Google Scholar]
- Xing C, Sestak AL, Kelly JA, Nguyen KL, Bruner GR, Harley JB, Gray-McGuire C. Localization and replication of the systemic lupus erythematosus linkage signal at 4p16: interaction with 2p11, 12q24 and 19q13 in European Americans. Hum Genet. 2007;120:623–631. doi: 10.1007/s00439-006-0248-4. [DOI] [PubMed] [Google Scholar]
- Zhang MQ. Statistical features of human exons and their flanking regions. Hum Mol Genet. 1998;7:919–932. doi: 10.1093/hmg/7.5.919. [DOI] [PubMed] [Google Scholar]