Abstract
Repeated daily dosing of rats with the occupational chemical 4-vinylcyclohexene diepoxide (VCD) depletes the ovary of primordial and primary follicles through an increase in the natural process of atresia. Additionally, in vitro exposure of Postnatal Day 4 (PND 4) rat ovaries to VCD causes similar follicular depletion. This study was designed to investigate survival signaling pathways that may be associated with VCD-induced ovotoxicity in small preantral follicles. Female Fischer 344 rats (PND 28) were dosed daily (80 mg/kg/day VCD i.p.; 12 days in vivo), and PND 4 ovaries were cultured (VCD 20 or 30 μM; 8 days in vitro). Microarray analysis identified a subset of 14 genes whose expression was increased or decreased by VCD in both experiments (i.e., via both exposure routes). Particularly, the analysis showed that relative to controls, VCD did not affect mRNA expression of growth and differentiation factor 9 (Gdf9), whereas there were decreases in mRNA encoding bone morphogenic protein receptor 1a (Bmpr1a) and Kit. To confirm findings from microarray, the genes Gdf9, Bmpr1a, and Kit were further examined. When growth factors associated with these pathways were added to ovarian cultures during VCD exposure, GDF9 and BMP4 had no effect on VCD-induced ovotoxicity; however, KITL attenuated this follicle loss. Additionally, there was a decrease in Kit and an increase in Kitl expression (mRNA and protein) following VCD exposure, relative to control. These results support that VCD compromises KIT/KITL signaling, which is critical for follicular survival in primordial and primary follicles.
Keywords: Kit, Kitl, ovary, ovotoxicity, preantral follicles
VCD-induced ovotoxicity in rats is mediated through compromised KIT/KITL signaling, which is critical for follicular survival
INTRODUCTION
At birth, the mammalian ovary contains a finite number of follicles. In an ongoing process, primordial follicles are selected for activation. However, only a few (<1%) of the oocytes contained in these follicles develop to ovulation for possible fertilization [1]. Rather, the vast majority become activated and degrade at some stage of growth by the natural process of atresia. Chemicals that destroy the primordial follicle population can cause permanent infertility (early ovarian failure), because once destroyed these follicles cannot be replaced [2].
Repeated daily dosing of rats with the occupational chemical 4-vinylcyclohexene diepoxide (VCD) selectively destroys primordial and primary ovarian follicles [3–5]. In addition, in vitro VCD exposure in a neonatal rat whole ovarian culture system involves a similar pattern of selective ovarian follicle loss [6]. Although the exact mechanism of follicle depletion is unknown, previous studies have shown that VCD induces follicle loss via an increase in the rate of atresia in both in vivo [4, 5] and in vitro systems [6]. Following repeated daily VCD exposure, the involvement of the proapoptotic branches of the BCL2 family of proteins and the mitogen-activated protein kinase (MAPK) family of proteins were examined in Fischer 344 rat ovarian follicles. Expression of the proapoptotic proteins, BAX [7] and BAD [8], was increased in primordial and primary follicles but not in larger nontargeted follicles (secondary and antral) isolated from rats following in vivo VCD dosing. An increase in the BAX:BCL2L1 mitochondrial ratio was also observed [8] in those follicles. In addition, c-Jun N-terminal protein kinase (MAPK8) and MAPK14 activities were increased following repeated in vivo VCD exposure [9]. Each of these studies demonstrated an increase in apoptotic activity in the target follicle population following VCD exposure, supporting that VCD increases the natural process of atresia [5].
A number of cellular survival pathways within the ovary are involved in maintaining the viability of primordial and small preantral follicles undergoing the early stages of folliculogenesis. If one or more of these survival pathways fail to properly maintain the health of the oocytes or supportive granulosa cells, the end result will be follicular degradation via atresia [10–12].
Two TGFB (TGFβ) family growth factors, growth and differentiation factor 9 (GDF9) and bone morphogenetic factor 4 (BMP4), have both been shown to contribute to rat ovarian follicular survival and folliculogenesis by targeting granulosa cells. GDF9 is produced in oocytes and signals through its receptor on granulosa cells to promote follicular growth [13] and is required during early folliculogenesis [14]. Also, GDF9 elicits antiapoptotic effects during folliculogenesis [15], and exposure of neonatal rat ovaries to GDF9 in vitro specifically promotes development of early primary but not primordial follicles [16]. BMP4 is produced in interstitial cells and binds its receptor, BMPR1A, on granulosa cells to influence follicular development [17]. Exposure of BMP4 in vitro to neonatal rat ovaries has been shown to promote survival and development of primordial follicles [18]. Thus, based on these previous studies, GDF9 and BMP4 appear to play critical roles in the maintenance of normal ovarian folliculogenesis working through granulosa cells.
Another growth factor, Kit ligand (KITL; also referred to as stem cell factor, steel factor, and mast cell growth factor), is produced in granulosa cells and signals the oocyte through KIT, a tyrosine kinase plasma membrane receptor [19]. KITL activates the PI3K/AKT signaling pathway within the oocyte of small preantral follicles. Several studies have shown that female mice that have naturally occurring mutations in Kitl or Kit are infertile due to abnormalities in ovarian development [20]. KITL is able to stimulate oocyte growth [21, 22], induce primordial follicle development, and initiate folliculogenesis [23]. In addition, KITL is able to act as an antiapoptotic factor on oocytes in primordial follicles [24]. Therefore, the interaction between KITL and KIT plays a crucial role in the progression of early folliculogenesis and follicular survival.
Because previous studies have shown that VCD contributes to an increase in proapoptotic signaling pathways, it is possible that VCD also affects cellular survival pathways within primordial and primary follicles, thereby compromising follicle survival and contributing to follicular atresia. Therefore, the purpose of the current study was to examine the involvement of cell survival signaling pathways associated with VCD-induced ovotoxicity in primordial and primary follicles. Oligoarray analysis was performed to identify survival pathways affected by both in vivo and in vitro VCD exposure in the Fischer 344 rat ovary.
MATERIALS AND METHODS
Reagents
VCD (mixture of isomers, purity > 99%), collagenase (clostridium histolytic type I), DNase I, bovine serum albumin (BSA), Sigmacote, ascorbic acid (vitamin C), transferrin, d-biotin, Ribonuclease A, and all other unspecified chemicals were purchased from Sigma-Aldrich Inc. (St. Louis, MO). Dulbecco modified Eagle medium/nutrient mixture F-12 (Ham) 1× (DMEM/Ham F-12), medium 199 (M199), Albumax, penicillin/streptomycin (5000 U/ml, 5000 μg/ml), Hanks Balanced Salt Solution (without CaCl2, MgCl2, or MgSO4), Superscript III reverse transcriptase, oligo(dT)20 primers, MgCl2 (25 mM), Cot-1 DNA, and PBS were purchased from Invitrogen (Carlsbad, CA). Recombinant mouse KITL and human BMP4 were obtained from R & D Systems Inc. (Minneapolis, MN). Recombinant human GDF9 was obtained from PeproTech Inc. (Rock Hill, NJ). Forty-eight-well tissue culture plates and 96-well assay plates were obtained from Corning Inc. (Corning, NY). SenseAMP kits for RNA amplification and DyeSaver2 antifade coating for microarrays were obtained from Genisphere Inc. (Hatfield, PA). SYBR Green I Dye, aminoallyl deoxyuridine triphosphates (dUTPs), and Alexa Fluor 555 and 647 dyes were purchased from Molecular Probes (Eugene, OR). Millicell-CM filter inserts were obtained from Millipore (Bedford, MA). Rabbit polyclonal anti-KITL immunoglobulin G (IgG) antibody, as well as the KIT blocking peptide were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-KIT antibody was purchased from DakoCytomation (Carpenteria, CA). The RNeasy Mini Kit, QiaShredder Kit, RNeasy MinElute Kit, RNase-Free DNase Set, Quantitect SYBR Green PCR Kit, MinElute PCR Purification Kit, and QiaQuick PCR Purification Kit were all purchased from Qiagen Inc. (Valencia, CA). Endo-free Reverse Transcription Kit, 10% SDS solution, 20× saline-sodium citrate (SSC) solution, and 0.5 M EDTA (pH 8.0) were purchased from Ambion (Austin, TX). BCA Protein Assay Kit, SuperSignal West Femto Maximum Sensitivity Substrate, and goat anti-rabbit horseradish peroxidase-conjugated antibodies for Western blots were purchased from Pierce Biotechnology (Rockford, IL). Nylon filter screens (250 μm) were purchased from Small Parts Inc. (Miami Lakes, FL). Random hexamers and all primers used for real-time PCR were purchased from Integrated DNA Technologies (Coralville, IA). The ECL plus chemiluminescence detection kit was purchased from GE Healthcare, Amersham.
Animals
Female Fischer 344 rats (Postnatal Day 21 [PND 21] or Gestation Day 18) were purchased from Harlan Laboratories (Indianapolis, IN). All animals were housed in plastic cages and maintained in a controlled environment (22°C ± 2°C; 12L:12D cycles). The animals were provided a standard diet with ad libidum access to food and water. All animals were given 1 wk to acclimate to their environment prior to the start of experiments. Pregnant rats were housed one per cage, and the pups were removed on PND 4 for ovary cultures. Postnatal Day 21 rats were housed four per cage during the dosing period. All animal experimentation was approved by the University of Arizona Institutional Animal Care and Use Committee.
Animal Dosing
Postnatal Day 28 female rats were administered an i.p. injection with sesame oil (vehicle control, 2.5 ml/kg) or VCD (80 mg/kg/day, 0.57 mmol/kg, 2.5 ml/kg) for 12 days [4]. The 12-day dosing regime was selected, since previous studies had shown a 50% loss of primordial and primary follicles after 15 days of daily dosing in rats [5]. Thus, the 12-day time point would provide a representation of gene expression changes due to VCD without having lost a great amount of follicles.
Ovarian Follicle Isolations
Rats were asphyxiated using CO2 4 h following the final dose. Ovaries were excised, trimmed of excess tissue, and placed into M199. Eight ovaries from four rats in the same treatment were grouped together for dissociation. Ovaries were minced and gently dissociated in a solution of BSA, DNase I, and collagenase I as previously described [25]. Ovarian digests were filtered through a 250-μm nylon screen to exclude large antral follicles. Using micropipettes, preantral and small antral follicles were hand sorted by size into fraction 1 (25–100 μm; small preantral follicles containing primordial, primary, and some small secondary follicles) and fraction 2 (100–250 μm; large secondary follicles and small antral follicles). A total of four separate pools of fraction 1 and 2 follicles (n = 4) were collected for oligoarray analysis and were designated as control fraction 1 follicles, CF1 (reference sample); control fraction 2 follicles, CF2; VCD fraction 1 follicles, VF1; and VCD fraction 2 follicles, VF2.
Ovarian Cultures
Culturing of ovaries in vitro was performed as previously described [23] with some modifications. Female PND 4 rat pups were killed by CO2 asphyxiation followed by decapitation. Uterus, oviduct, fat, and excess connective tissue were separated from the ovaries in ice-cold Hanks Balanced Salt Solution. Ovarian culture medium (250 μl) consisting of DMEM/Ham F-12, 1 mg/ml BSA, 1 mg/ml Albumax, 50 μg/ml ascorbic acid, 27.5 μg/ml transferrin, 5 U/ml penicillin, and 5 μg/ml streptomycin were allowed to equilibrate in 48-well culture plates in a humidified incubator at 37°C, 5% CO2 in air for at least 1 h prior to ovary collection. Appropriate concentrations of VCD (10, 20, 30, and 40 μM), KITL (25, 50, 100, 200, and 400 ng/ml), GDF9 (50, 100, and 200 ng/ml), or BMP4 (50, 100, and 200 ng/ml) were added to medium prior to equilibration. A small piece of Millicell-CM filter insert was placed on top of the culture medium. With fine forceps, ovaries were placed onto floating filter inserts (one ovary per well). A drop of medium was placed on top of each ovary to prevent drying. Ovaries were cultured for 4, 6, or 8 days, with appropriate treatments and medium changed every 2 days. At the completion of culture, ovaries were processed for oligoarray analyses, histological evaluation, protein isolation, or RNA isolation. Samples for oligoarray analysis were designated as follows: control cultured, OC; VCD cultured, OV.
Ovarian Follicle Counts
Ovaries were processed for histological evaluation by placing them in Bouin fixative solution for 1 h. Each ovary was dehydrated, embedded into a paraffin block, sectioned (4- to 5-μm thickness), and stained with hematoxylin and eosin. Ovarian follicles were classified and counted in every 12th section for cultured ovaries. Only follicles with a distinct oocyte nucleus were counted. The classification scheme for ovarian follicles is as follows: 1) primordial, a single layer of squamous granulosa cells; 2) transient (seen only in Figure 6), an enlarged oocyte with a single layer of squamous granulosa cells (according to Reddy et al. [26]); 3) primary, a single layer of cuboidal granulosa cells; 4) secondary, two or more layers of granulosa cells without the presence of an antral cavity; 5) atretic, oocyte stained pink, follicle structure misshapen, condensed chromatin in oocyte nuclei, or focal contact lost between the oocyte and granulosa cells; and 6) healthy, diffuse chromatin staining in oocyte nuclei and focal contact between oocyte and granulosa cells intact.
FIG. 6.
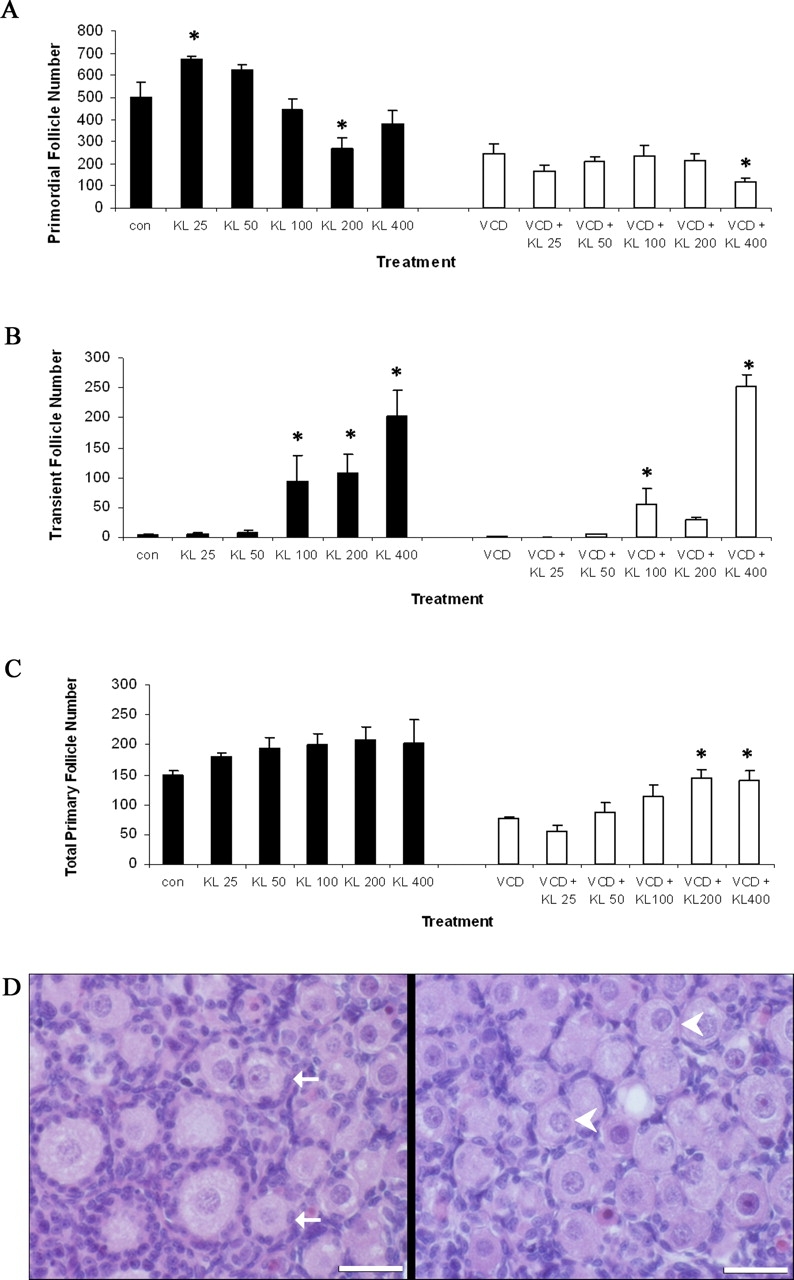
Effect of KITL on VCD-induced follicle loss in specific follicle populations. Postnatal Day 4 Fischer 344 rat ovaries were cultured (8 days) in control medium with or without KITL (25, 50, 100, 200, or 400 ng/ml [labeled KL in figure]) or in culture medium containing VCD (30 μM) ± KITL (25, 50, 100, 200, or 400 ng/ml). On Day 8 of culture, ovaries were collected and processed for histological evaluation as described in Materials and Methods. Healthy follicles in every 12th section were classified and counted. Values indicate the mean number of healthy follicles counted per ovary ± SEM. Primordial (A), transient (B), and primary follicle (C) numbers. D) Micrograph of normal (solid arrows) and transient follicles (400 ng/ml; arrowheads). Bar = 25 μm; original magnification ×40. *Different from respective control (P < 0.05).
RNA Isolation
Total RNA from ovarian follicle fractions or cultured ovaries was isolated with an RNeasy Mini Kit according to the manufacturer's protocol for RNA extraction from tissue. For oligoarray analysis, five cultured ovaries per treatment group for each pool and each group of fraction 1 or 2 follicles were used (n = 4). For real-time PCR analysis, RNA was isolated from 10 cultured ovaries per treatment group. RNA was quantified using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies Inc., Wilmington, DE) and measured for integrity and purity using an Agilent Bioanalyzer 2100 (Agilent Technologies Inc., Santa Clara, CA). RNA was further concentrated for all total RNA samples using an RNeasy MinElute Kit according to the manufacturer's protocol.
RNA Amplification
Up to 2 μg total RNA was amplified into sense RNA using a SenseAmp Kit. All amplification reactions were carried out according to the manufacturer's protocol. Modifications included using the RT enzyme from an Endo-free kit for cDNA synthesis and using all reverse transcribed cDNA for the in vitro transcription (amplification) reaction. Following cleanup of the sense RNA, the quantity of sense RNA was measured on a NanoDrop ND-1000 spectrophotometer.
Oligoarray Hybridization and Analysis
Oligoarray hybridization and analysis were performed as previously described [27] with some modifications. Samples for oligoarray analysis were: control fraction 1 follicles, CF1 (reference sample); control fraction 2 follicles, CF2; VCD fraction 1 follicles, VF1; VCD fraction 2 follicles, VF2; control cultured, OC; and VCD cultured, OV. Amplified sense RNA (3 μg) was reverse transcribed to antisense cDNA using an Endo-free RT kit according to the manufacturer's protocol with the incorporation of amino allyl dUTPs and dinucleotide triphosphates into the cDNA. RNA was primed using random hexamers, and the RT reaction was maintained at 42°C for 2 h. After incubation, cDNA was denatured and followed by a base hydrolysis. Amino allyl-modified cDNA was purified using a QiaQuick PCR Purification Kit according to the manufacturer's protocol. Purified cDNA was labeled with Alexa 555 or 647 dyes. Labeled cDNA was pooled and purified using a QiaQuick PCR Purification Kit. An equal volume of 2× hybridization buffer (8× SSC, 60% formamide, 0.2% SDS) and 10 μg Cot-1 DNA was added to the purified cDNA. The cDNA was hybridized overnight to an oligoarray slide printed with Operon's 6.1K rat oligo version 1.0 and 1.1 sets (6144 genes in triplicate) using a GeneTac hybridization station (Genomic Solutions Inc., Ann Arbor, MI) at 42°C. The slides were put through a short wash on the hybridization station, removed, rinsed in 0.1× SSC, and dried before being coated with DyeSaver2 solution.
Prior to scanning, slides were stripped of some of the layers of DyeSaver2 with toluene to prevent an uneven distribution of fluorescence. Hybridized slides were scanned using an arrayWoRxe CCD-based scanner from Applied Precision (Issaquah, WA). A bundled spot-finding analysis software (MolecularWare, Irvine, CA) determined the signal intensity for each spot. A multivariate experimental approach based on the fitting of expression data to an additive linear ANOVA model using the custom software package CARMA [28] containing terms for array and dye effects was used to analyze the intensities of spots on the arrays. The reference sample was used to evaluate effects of VCD as well as differences between primordial and primary follicles (F1) isolated from adult rats and primordial and primary follicles (OC) cultured from neonatal rats. Similar effects of VCD in both systems were assumed to be due to direct VCD exposure rather than differences between age and relative purity of samples. CF1 was used as the reference sample for the microarray experiments to determine the differential gene expression of all of the tissue/treatment groups. After the initial analysis, comparisons were made between the VCD-treated samples and their appropriate control samples (i.e., CF1 versus VF1, CF2 versus VF2, and control cultured ovaries versus VCD-treated cultured ovaries).
For oligoarray analysis, CARMA was required to compare the five treatment samples (CF2, VF1, VF2, CO, and VO) to the reference sample CF1, since an “interwoven loop” hybridization scheme was used. In a usual experiment, one would only compare control samples to treatment samples; however, in this case, there was more than one control group (CF1, CF2, and CO). The CF1 group was chosen as the “control” group or reference sample to which all other groups are initially compared, as VCD directly targets this follicle population in the in vivo experiments. A single reference sample is also important for the software to make the necessary corrections for array and dye effects. Using the “interwoven loop” design allows for further comparisons between groups, as samples were hybridized to four of the treatment/tissue groups.
Real-Time Quantitative PCR
Real-time quantitative PCR was performed using the RotorGene RG-3000 (Corbett Research, Sydney, Australia) sequence detection system and the Quantitect SYBR Green PCR Kit as previously described [29]. Briefly, all primers were designed using Primer3 software within 300–500 bp of the 3′ end of the gene. The primer sequences used were Actb (accession no. NM_031144): forward: TCTATCCTGGCCTCACTGTC, reverse: ACGCAGCTCAGTAACAGTCC (product 121 bp); Kit (accession no. NM_022264): forward: CTTTTGCGCAAGCTTTTGT, reverse: ATCCCCCGCTCCAAAGTAT (product 145 bp); and Kitl (accession no. NM_021843): forward: GGCCTACAATGGACAGCAAT, reverse: TCAACTGCCCTTGTAAGACTTG (product 113 bp). Up to 2 μg of total or amplified RNA was reverse transcribed using SuperScript III according to the manufacturer's protocol. The cDNA was diluted to 8 ng/μl using RNase-free water. The PCR reaction contained 0.4 μl of 25 mM MgCl2, 0.35 μl RNase-free water, 0.25 μl SYBR Green I, 5 μl SYBR master mix, 100 pmol forward and reverse primers, and 16 ng cDNA, for a total volume of 10 μl. All reactions were run in triplicate with the following program: 95°C for 15 min, followed by 45 cycles of 95°C for 15 sec, 58°C for 15 sec, and 72°C for 20 sec, finishing with a melt cycle consisting of stepwise increases in temperature from 72°C to 99°C.
Protein Isolation and Western Blot Analysis
Pools of whole ovarian protein homogenates were isolated via homogenization in lysis buffer as previously described [30], with some modifications. Briefly, the homogenized samples were kept on ice for 30 min and centrifuged at 10 000 × g for 15 min at 4°C. Supernatant was centrifuged a second time for 15 min after being removed from the tissue pellet. Supernatant was collected, aliquoted, and stored at −80°C.
The concentration of isolated protein was determined using a standard protocol of a BCA Protein Assay Kit on a 96-well assay plate. Emission absorbance values were detected with a λ = 540-nm excitation on a Synergy HT Multi-Detection Microplate Reader with KC4 software (Bio-Tek Instruments Inc., Winooski, VT). Protein concentrations were calculated from a standard curve using known amounts of BSA protein.
Protein homogenates (10–30 μg) were separated using SDS-PAGE (6%–13%) and transferred to nitrocellulose membranes as previously described [30] with some modifications. Briefly, blots were blocked overnight with agitation in 5% dry milk in Tris-buffered saline with Tween-20 (T-TBS) at 4°C. If KIT blocking peptide was utilized for an experiment, a 5-fold excess of peptide was incubated for 8 h at 4°C with KIT antibody. Then, the blots were incubated in primary antibody (or with antibody-peptide mixture) in 3% dry milk in T-TBS overnight at 4°C (for KIT antibody) or for 1 h at 25°C (for KITL antibody). A 1:200 dilution was used for both anti-KIT and anti-KITL primary antibodies. Blots were washed three times for 10 min each in T-TBS, horseradish peroxidase-conjugated secondary antibody (goat anti-rabbit IgG: 1:1000; donkey anti-goat IgG: 1:10 000) was added for 1 h at 25°C, and then the blots were washed again three times in T-TBS, followed by a single 10-min wash in TBS. Western blots were detected using chemiluminescence (SuperSignal West Femto substrate or ECL plus substrate) and exposed to x-ray film. Densitometry of the appropriate bands (KIT: 120 and 145 kDa; KITL: 23 kDa) was performed using LabWorks software from a UVP Bioimaging System (UVP Inc., Upland, CA).
Immunofluorescence and Confocal Microscopy
Ovaries were fixed in 4% buffered formalin, paraffin embedded, sectioned (5 μM), and deparaffinized. Microwave antigen retrieval in citrate buffer was completed, and tissue sections were blocked with 5% BSA/PBS for 5 min. Anti-KIT (1:400 dilution; DakoCytomation) was applied for 18 h (4°C), followed by biotinylated goat anti-rabbit IgG (Vector) at a 1:75 dilution (1 h). Cy5-streptavidin (Jackson ImmunoResearch Labs) was applied for 1 h at a 1:100 dilution. Sections were treated with Ribonuclease A (50 μg/ml; Sigma) for 1 h, followed by YOYO-1 (Molecular Probes) staining (0.5 μM for 10 min). Slides were repeatedly washed with PBS, coverslipped with aqueous mounting medium, and stored in the dark at 4°C until viewed on a Zeiss (LSM 510 NLO-Meta) confocal microscope with an argon and helium-neon laser projected through the tissue into a photomultiplier at λ = 488 and 633 nm for YOYO-1 (green) and CY-5 (red), respectively. All images were captured using a 40× objective lens. Omission of the primary antibody for an immunonegative section was performed (data not shown).
Statistical Analysis
In quantitative real-time PCR experiments, threshold numbers (Ct values) were set within the exponential phase of the reaction and were used to calculate the relative expression for each gene normalized to Actb RNA in each sample. The ratio of gene expression in VCD-treated samples compared with control samples was then calculated. At each time point, t-test to determine significant differences in gene expression between control and VCD-treated ovaries was carried out. Samples were analyzed separately between sample groups but calculated relative (normalized) to control (1) for the purposes of reporting. For Western blotting, control and VCD-treated protein levels were compared, and t-test was performed. Samples were analyzed separately between sample groups but calculated relative (normalized) to control (100%) for the purposes of reporting.
Comparisons between two means were analyzed using a Student t-test. Comparisons of multiple treatment groups or means were analyzed using one-way analysis of variance (ANOVA) followed by either Fisher protected least-significant difference (PLSD; for three means) or Tukey/Kramer (for more than three means) posthoc tests. For all tests, significance was determined by P < 0.05, with a trend for significance assigned at P < 0.1.
RESULTS
Ovarian In Vitro VCD Exposure
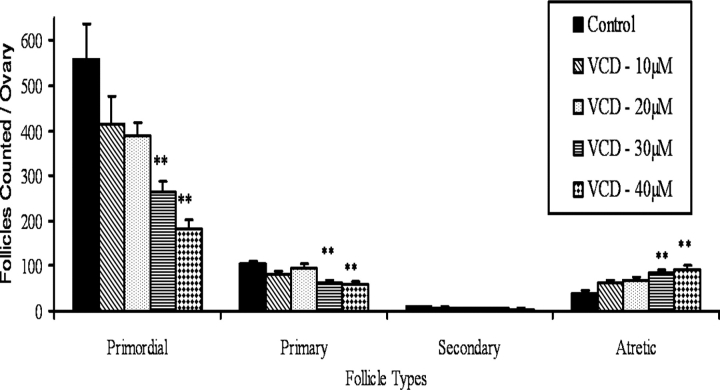
From previous studies, in vivo daily dosing of rats for 12 days with VCD (80 mg/kg/day) was determined to be a time point just prior to 50% loss (P < 0.05) of ovarian follicles [4, 5]. Additionally, in a previous study, 50% follicle loss (P < 0.05) in PND 4 rat ovaries cultured with VCD was seen on Day 8 with 30 μM VCD [6]. In accordance with those findings, in the present study there was nonsignificant primordial and primary follicle loss after 12 days of VCD dosing (primordial follicle number: 75% of control; primary follicle number: 72% of control; P > 0.05). Thus, analogous to follicle loss at the in vivo time point, it was undertaken to identify a VCD concentration at the Day 8 time point in which impending follicle loss was observed just prior to significant loss. Postnatal Day 4 ovaries were cultured for 8 days in the presence of increasing concentrations of VCD (Fig. 1). Decreases (P < 0.05) in healthy ovarian primordial and primary follicles (target populations) were observed only at the highest (30 and 40 μM) VCD exposures compared with ovaries cultured in control medium. Increases (P < 0.05) in ovarian atretic (unhealthy) follicles were also observed only in the highest VCD exposure groups. Based on these data, the concentration determined for impending follicle loss (20 μM VCD) was used to investigate gene expression in follicles in which VCD had initiated its effects, because once follicle loss had occurred, (30 μM VCD), fewer VCD-affected follicles could be recovered for measurement of mRNA. Conversely, in the experiment evaluating effects of growth factors on VCD-induced follicle loss, 30 μM VCD was used to increase the potential for observing a reversal (P < 0.05) of follicle loss. Figure 2 is a micrograph of ovaries collected on Day 0 (PND 4) or on Day 8 following incubation in control medium or medium containing VCD (20 μM or 30 μM). Follicle counts on Day 0 ovaries were: primordial, 481.5 ± 61.5; and small primary, 49.3 ± 8.1 (n = 4). There were no large primary or secondary follicles at that time.
FIG. 1.
Ovarian follicle counts: in vitro VCD exposure dose response. Postnatal Day 4 Fischer 344 rat ovaries were cultured (8 days) in the presence of 0 (control), 10, 20, 30, or 40 μM VCD. Ovaries were prepared for histological evaluation as described in Materials and Methods. Values are mean healthy follicles counted per ovary ± SEM; n = 7–8 ovaries per treatment; **P < 0.05, different from ovaries cultured in control medium.
FIG. 2.
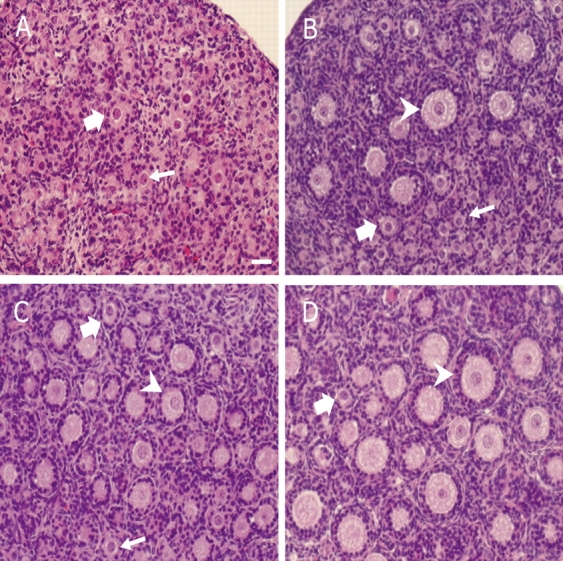
Effect of culture and VCD on PND 4 ovaries. Ovaries were collected from PND 4 Fischer 344 rats on Day 0 (A) or cultured (8 days) in control medium (B) or medium containing 20 μM (C) or 30 μM (D) VCD. At the time of collection, ovaries were processed for histological evaluation as described in Materials and Methods. Small arrow, primordial follicles; thick arrow, primary follicles; arrowhead, secondary follicles. Bar = 25 μm; original magnification ×40.
Oligoarray Analysis—Effect of VCD: In Vivo Versus In Vitro
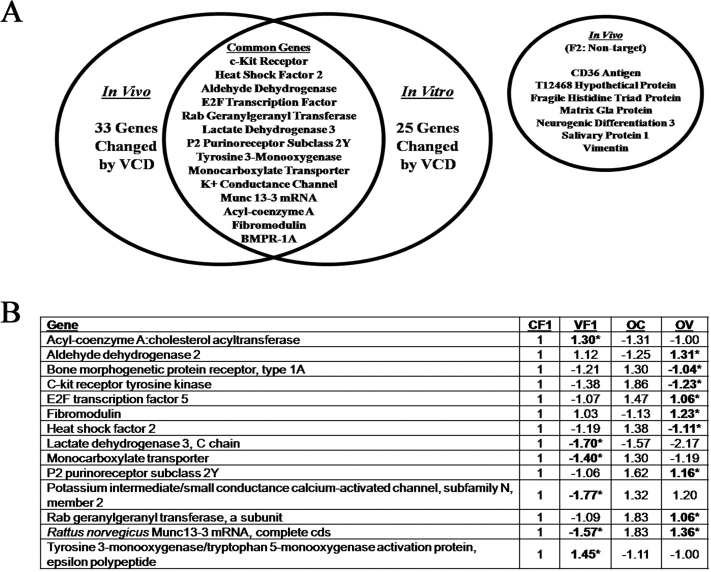
Each rat oligoarray was probed with two labeled antisense RNA samples. After complete analysis of all hybridized arrays, the CARMA software generated a list of selected genes altered in comparison to the reference sample CF1. Relative to CF1, 361 genes were significantly changed in at least one of the other five tissue/treatment groups (CF2, VF1, VF2, OC, and OV). Further analysis made comparisons within the sample types (control sample = CF1, CF2, or OC) for genes that changed significantly in response to VCD exposure within that specific sample type. By that assessment, 33 genes were significantly different between CF1 and VF1 follicles, whereas 25 genes were significantly different between OC versus OV ovaries as determined by a t-test comparison (Fig. 3A). Fourteen genes that followed the same pattern of expression in response to VCD exposure in both systems are shown in the intersect region (Fig. 3A). Conversely, mRNA expression of only seven genes was significantly changed by VCD in nontarget fraction 2 follicles (Fig. 3A). None of those genes were affected in VCD-targeted tissues (fraction 1 follicles and cultured ovaries). The fold-changes in genes affected in both systems by VCD exposure in target follicles relative to their respective controls are listed in Figure 3B.
FIG. 3.
Effect of VCD exposure summary. A) The Venn diagram displays the results of the common genes targeted by VCD exposure in both the in vivo and in vitro systems as measured using oligoarray analysis as described in Materials and Methods. The in vivo VCD-exposed rats had 33 genes change in fraction 1 (VF1) targeted follicles and seven genes change in the fraction 2 (VF2) nontargeted follicles. The in vitro-exposed ovaries from the neonatal ovary culture system had 25 genes that changed in response to VCD exposure (OV). The common genes that changed in both systems in the target population are listed within the intersect of the diagram, with the genes from the nontarget population listed in a separate circle. B) Fold-changes (P < 0.05) in gene expression affected by VCD exposure relative to respective controls (positive values, >CF1; negative values, <CF1). Asterisk (*) and bolded items indicate different from respective control, P < 0.05.
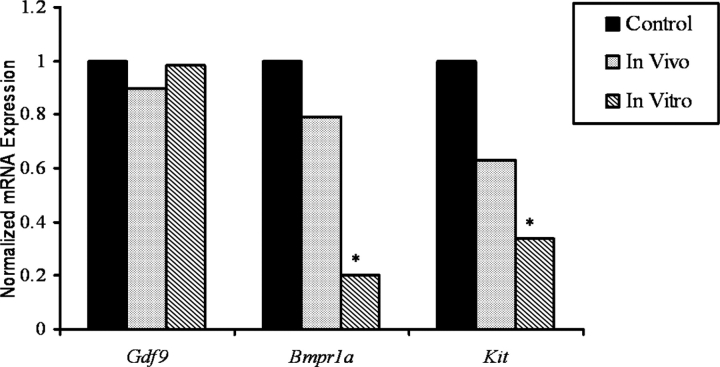
Comparisons of the effect of VCD on mRNA expression were made between two genes (Bmpr1a and Kit) identified by oligoarray analysis in the in vivo as well as the in vitro groups that are known to be involved in survival signaling pathways in small preantral follicles. Additionally, another gene (Gdf9) that was not differentially expressed in the oligoarray experiment but known to be involved in folliculogenesis was examined. Normalized mRNA expression of Gdf9, Bmpr1a, and Kit for the in vivo and in vitro target groups is shown in Figure 4. No changes in expression of mRNA encoding Gdf9 were caused by VCD exposure in either system. However, decreases in expression of mRNA encoding Bmpr1a (BMP4 receptor) and Kit (KITL receptor) were observed in response to VCD exposure in vivo (P > 0.05) and in vitro (P < 0.05).
FIG. 4.
The mRNA expression for genes involved in survival signaling. The effect of VCD exposure on mRNA encoding Gdf9, Bmpr1a, and Kit in both the in vivo and in vitro systems was measured using oligoarray analysis as described in Materials and Methods. Values were normalized to reference sample CF1 as described in Materials and Methods. Values represent the ratio of treatment to control; n = 4 for each exposure system; **P < 0.05, different from control.
Effect of Exogenous GDF9, BMP4, and KITL on Ovarian Follicle Numbers During VCD Exposure
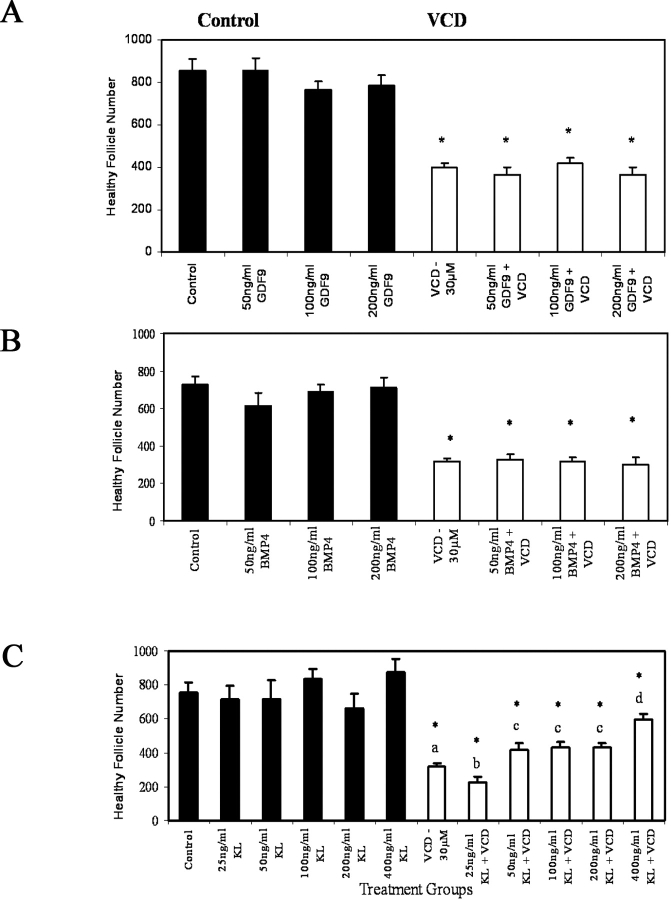
Increasing concentrations of recombinant GDF9 (50, 100, and 200 ng/ml), BMP4 (50, 100, and 200 ng/ml), or KITL (25, 50, 100, 200, and 400 ng/ml) alone in neonatal rat ovary cultures (8 days) had no effect (P > 0.05) on the total number of healthy follicles compared with ovaries cultured in control medium (Fig. 5, A–C). VCD (30 μM) in the medium reduced (P < 0.05) the number of total healthy follicles to approximately 50% of control (Fig. 5, A–C). Addition of GDF9 or BMP4 to the media containing VCD (30 μM) did not prevent (P > 0.05) the VCD-induced decrease in the number of total healthy follicles (Fig. 5, A and B). However, in the presence of KITL, there was a dose-dependent attenuation (P < 0.05) of VCD-induced follicle loss (Fig. 5C).
FIG. 5.
Effect of exogenous GDF9, BMP4, or KITL on VCD-induced follicle loss. Postnatal Day 4 Fischer 344 rat ovaries were cultured (8 days) in control medium with or without recombinant human GDF9 (50, 100, or 200 ng/ml), recombinant human BMP4 (50, 100, or 200 ng/ml), or recombinant mouse KITL (25, 50, 100, 200, or 400 ng/ml [labeled KL in figure]); or in culture medium containing 30 μM VCD with or without GDF9 (50, 100, or 200 ng/ml), BMP4 (50, 100, or 200 ng/ml), or KITL (25, 50, 100, 200, or 400 ng/ml). On Day 8 of culture, ovaries were collected and processed for histological evaluation as described in Materials and Methods. Healthy follicles in every 12th section were counted. Values indicate the mean number of healthy follicles counted per ovary ± SEM after (A) GDF9 or GDF9 + VCD, (B) BMP4 or BMP4 + VCD, and (C) KITL or KITL + VCD; n = 4–11 ovaries per treatment group (A–C); *P < 0.05 different only from ovaries cultured in control medium (C); and different letters indicate different (P < 0.05) follicle numbers within VCD-containing groups.
Effect of Exogenous KITL on Individual Follicle Types During VCD Exposure
Further investigation of the effect of exogenous KITL on specific follicle populations was made (Fig. 6). Increasing concentrations of KITL (25, 50, 100, 200, and 400 ng/ml) alone in neonatal rat ovaries (8 days) did not show a dose-dependent effect on primordial follicle number (Fig. 6A). At two concentrations of KITL, 25 ng/ml and 200 ng/ml, there were increased (P < 0.05) and decreased (P < 0.05) primordial follicle numbers, respectively. Relative to control, exogenous KITL alone had no effect on primary follicle numbers (Fig. 6C). Interestingly, when specific follicle types were classified, a transient follicle classification was apparent with exogenous KITL at higher concentrations (100, 200, and 400 ng/ml; Fig. 6B). These follicles contained an enlarged oocyte, with few surrounding flattened granulosa cells. Figure 6D shows the appearance of transient follicles in response to exogenous KITL in which the oocyte has grown; however, the surrounding granulosa cells appear flattened and have not become activated. Relative to VCD alone, addition of exogenous KITL at the highest concentration (400 ng/ml) in the presence of VCD (30 μM) showed a reduction (P < 0.05) in primordial follicle number. In contrast to exogenous KITL alone, VCD plus KITL (200 ng/ml; 400 ng/ml) resulted in increased (P < 0.05) primary follicle numbers compared with VCD treatment alone. Transient follicles appeared in the KITL- as well as VCD plus KITL-treated ovaries which, relative to VCD alone, were increased (P < 0.05) at 100 ng/ml and 400 ng/ml KITL. Furthermore, the increase in the numbers of transient follicles at 400 ng/ml KITL in VCD-exposed ovaries was 4-fold higher than that in ovaries treated with KITL alone.
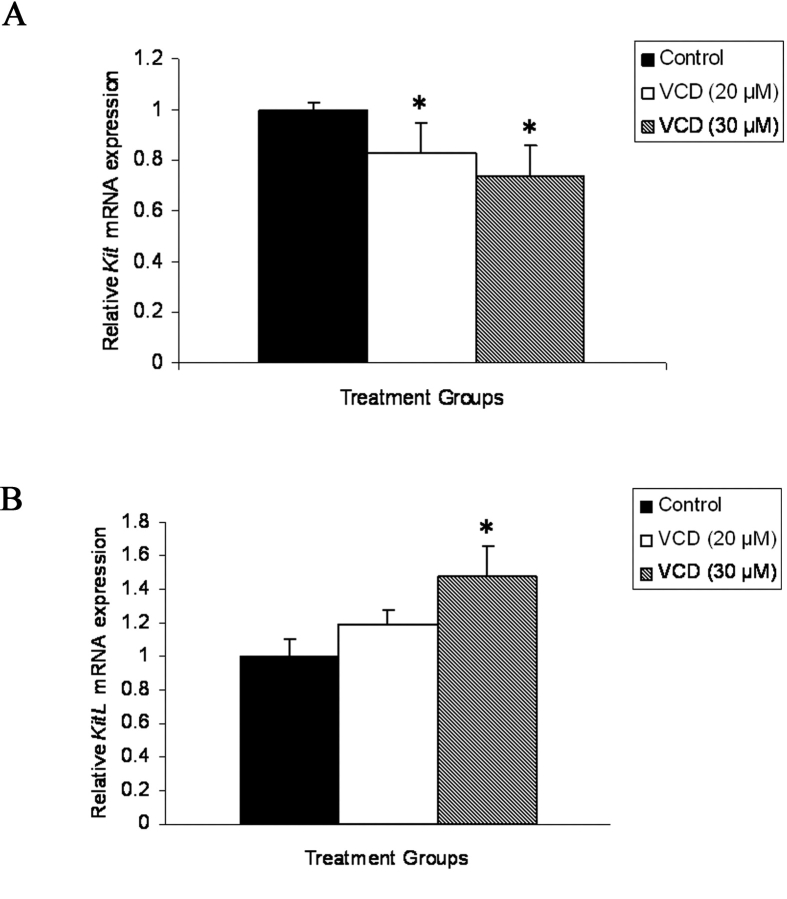
Ovarian Kit and Kitl mRNA Expression Following VCD Exposure
The effect of VCD (20 and 30 μM) on Kit and Kitl mRNA expression was measured using real-time PCR in cultured PND 4 ovaries (Fig. 7). The relative abundance of Kit and Kitl mRNA on Day 8 following both 20 and 30 μM VCD exposure is shown in Figure 7A (Kit) and Figure 7B (Kitl). During a time course of 20 μM VCD exposure, the relative abundance of Kit mRNA was first decreased on Day 8 of culture (0.83 ± 0.12 VCD/control; P < 0.05), whereas only a trend increase in Kitl mRNA was observed on Day 8 of culture (1.19 ± 0.09 VCD/control; P = 0.09). During a time course of 30 μM VCD exposure, the relative abundance of Kit mRNA was first decreased on Day 4 of culture (Day 4: 0.88 ± 0.07 VCD/control; Day 6: 0.75 ± 0.12 VCD/control; Day 8: 0.74 ± 0.07 VCD/control; P < 0.05), whereas Kitl mRNA was increased beginning on Day 6 of culture (Day 6: 1.25 ± 0.05 VCD/control; Day 8: 1.48 ± 0.18 VCD/control; P < 0.05). Thus, for both concentrations of VCD, the decrease in Kit mRNA preceded an increase in Kitl mRNA.
FIG. 7.
Effect of VCD on expression of mRNA encoding Kit and Kitl. Postnatal Day 4 Fischer 344 rat ovaries were cultured (8 days) in control medium or medium containing 20 or 30 μM VCD. Total RNA was extracted, and relative amounts of mRNA encoding (A) Kit or (B) Kitl were determined using quantitative real-time PCR as described in Materials and Methods. Values indicate the relative mRNA expression (normalized to Actb) ± SEM; n = 3 (10 ovaries/pool); *P < 0.05, different from control medium.
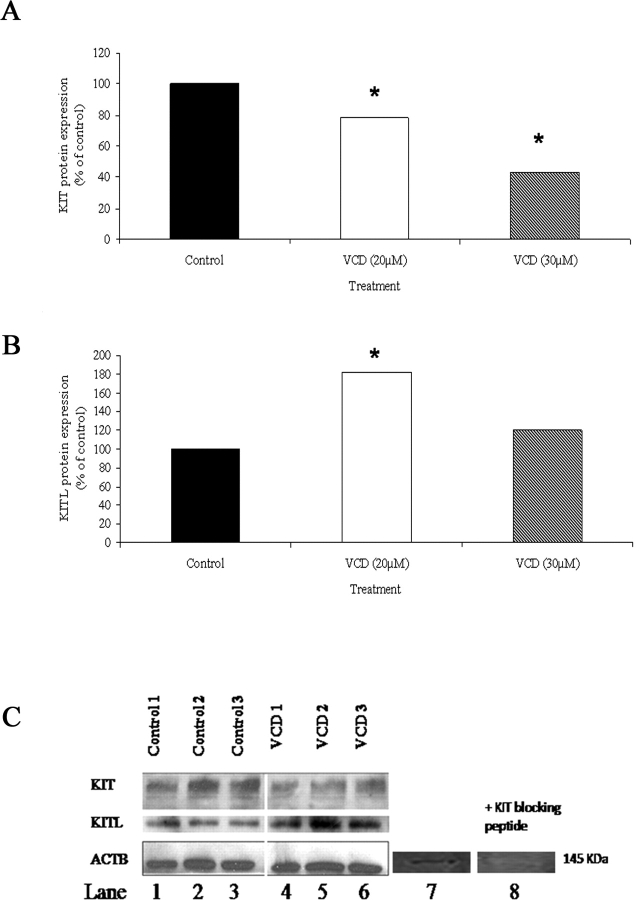
Ovarian KIT and KITL Protein Expression Following VCD Exposure
Expression of ovarian KIT and KITL protein expression was measured using Western blotting followed by densitometric analysis from PND 4 rat ovaries cultured 8 days in control medium or in 20 or 30 μM VCD-containing medium (Fig. 8). Two forms of KIT protein were detected in all control and VCD-treated groups (Fig. 8C). These two protein bands have been noted by Santa Cruz Biotechnology to be KIT precursor (120 kDa) and mature protein (145 kDa). Although the 120-kDa band was not visible in the adult rat ovary, a KIT blocking peptide prevented the appearance of the 145-kDa band for the KIT protein, indicating specificity of the antibody (Fig. 8C). Densitometric analysis of the 120 + 145-kDa band revealed that 20 and 30 μM VCD caused a dose-dependent decrease (P < 0.05) in KIT ovarian protein after 8 days in culture (Fig. 8A). KITL protein (23 kDa) was detected in all control and 20 μM VCD-treated groups (Fig. 8B). Densitometric analysis of bands revealed that 20 μM VCD caused an increase (P < 0.05) in KITL ovarian protein after 8 days in culture (Fig. 8B). However, relative to control, at 30 μM VCD, KL protein was not different (P > 0.05) from control.
FIG. 8.
Effect of VCD on ovarian KIT and KITL protein. Postnatal Day 4 Fischer 344 rat ovaries were cultured (8 days) in control medium or medium containing 20 or 30 μM VCD. Total protein was extracted and analyzed as described in Materials and Methods. A) Densitometric analysis of KIT (120 + 145 kDa) protein expression on Day 8 in response to VCD (20 and 30 μM). B) Densitometric analysis of KITL protein expression on Day 8 in response to VCD (20 and 30 μM). Values represent the % of control; n = 3 (10 ovaries/pool); *P < 0.05, different from control. C) Representative Western blots of KIT, KITL, and ACTB. Lanes 1–3, control groups; lanes 4–6, VCD-exposed ovaries; lanes 7 and 8, KIT protein in adult rat ovary with or without blocking peptide.
KIT protein in ovaries incubated with VCD (Day 8; 30 μM) was visualized by immunofluorescence and confocal microscopy (Fig. 9). Specific staining for KIT was selectively localized within oocytes, with the greatest intensity at the plasma membrane. Additionally, staining intensity was greater in primordial and primary relative to secondary follicles (Fig. 9C). There was a visible decrease in staining intensity in oocytes in ovaries incubated with VCD (Fig. 9B). Relative to control, VCD (30 μM) exposure resulted in fewer total numbers of primordial (45.5% of control) and primary (48.7% of control) follicles; however, of those remaining, many did not stain for KIT. KIT staining was unaffected by VCD exposure in secondary follicle nontargets (n = 5; Fig. 9, B and D).
FIG. 9.
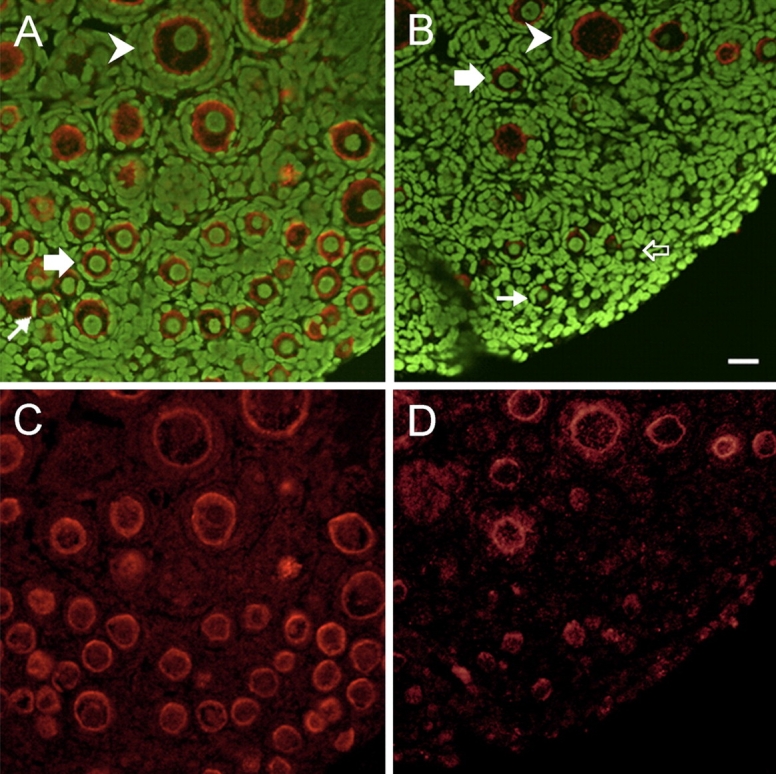
Effect of VCD on KIT protein staining. Postnatal Day 4 Fischer 344 rat ovaries were cultured (8 days) in control medium or medium containing 30 μM VCD. Ovaries were collected and processed for immunofluorescence staining and visualization by confocal microscopy as described in Materials and Methods. Green stain (YOYO 1), genomic DNA; red stain (CY5), anti-KIT antibody. Ovaries incubated in control (A, C) or VCD-containing (B, D) medium. KIT staining is predominantly localized in the oocyte plasma membrane, and it is greatly reduced in VCD-incubated ovaries. Small arrows, KIT-staining primordial follicles; open arrows, KIT-nonstaining primordial follicles; thick arrows, primary follicles; arrowheads, secondary follicles. Bar = 25 μm; original magnification ×40.
DISCUSSION
Repeated in vivo daily i.p. dosing of rats with VCD (80 mg/kg/day) for 12 days was found to be a time point just prior to significant small preantral follicle loss. Therefore, an analogous condition for follicle loss during in vitro VCD exposure was determined by a dose response in the whole ovarian culture system. Postnatal Day 4 rat ovaries exposed to VCD (20 μM) for 8 days showed a nonsignificant trend (P < 0.1) in small preantral follicle loss. This supported these as conditions that produce a similar endpoint with in vivo VCD dosing (80 mg/kg i.p.) for 12 days.
Since many of the genes involved in the mechanism by which VCD induces ovotoxicity are unknown, the use of oligoarray analysis, a large-scale gene screening technique, was used to determine genes involved in signaling pathways that were altered by VCD via both (in vivo versus in vitro) exposure systems. A subset of 14 common genes responded within primordial and primary follicles in a similar pattern during both in vivo and in vitro exposure to VCD. Interestingly, only seven genes in the nontarget fraction 2 follicle populations were altered during VCD exposure. None of those genes were affected by VCD in targeted follicles (fraction 1 of rats treated in vivo or whole cultured neonatal rat ovaries). Furthermore, none of them are identified to be associated with any known follicular signaling pathways. Therefore, this supports that the genes identified in fraction 1 follicles (in vivo) that were common to those identified in vitro are likely altered due to specific ovotoxic effects of VCD in the target population of follicles. Because few genes were changed in nontarget fraction 2 follicles in response to VCD exposure, this further suggests that VCD selectively targets those genes affected in small preantral follicles.
Two families involving cell survival pathways that have been shown to play a key role in the intraovarian regulation of follicular development in small preantral follicles are the TGFB family [31], which includes GDF9 and BMP4, and the PI3K/AKT signaling pathway [10–12, 32], which includes Kit/KITL. Three genes, Gdf9, Bmpr1a, and Kit, associated with these follicle survival pathways were further assessed. In agreement with oligoarray analysis, there was no change in Gdf9 expression; however, expression of mRNA encoding Bmpr1a and Kit was decreased (P > 0.05, in vivo exposure; P < 0.05, in vitro exposure). The reasons that effects were significant in the in vitro system and showed only a trend in the in vivo system probably relate to differences in the isolation of tissue. Isolated follicles (in vivo) contain few primordial as well as a number of secondary follicles. This dilutes the VCD-targeted population. Additionally, the follicle isolation takes several hours, during which reversible signaling pathways may return to a more basal level. Conversely, ovaries in culture are collected immediately upon removal, and follicles need not be sorted, since those ovaries are highly enriched in primordial and primary follicles that are targeted by VCD.
The results of the oligoarray analysis suggest that the survival pathways associated with the Bmpr1a and Kit genes may be disrupted in response to VCD. Further examination into the role of each of these genes in VCD-induced ovotoxicity was explored by addition of exogenous survival growth factors to the culture medium of in vitro VCD-exposed ovaries. Although VCD caused no changes in mRNA encoding Gdf9 in the oligoarray analysis, VCD may have affected its receptor. Thus, it was also further considered in the next experiment to investigate a functional impact of VCD on cell survival pathways.
Addition of the two granulosa cell-associated TGFβ survival factors, GDF9 and BMP4, had no effect on healthy follicle numbers during VCD exposure. However, addition of the oocyte-targeted factor, KITL, caused a dose-dependent attenuation of follicle loss caused by VCD exposure. In particular, this was evident in the attenuation of VCD-induced primary follicle loss. Additionally, the effects of high concentrations of exogenous KITL on oocyte growth were demonstrated by the appearance of transient follicles containing enlarged oocytes without the supporting cuboidal layer of granulosa cells. Undetectable numbers of transient follicles were seen in cultured neonatal ovaries incubated in control medium or media containing VCD, BMP4, and/or GDF9. This points to an effect of KITL on selective activation of oocyte growth [21–23]. Thus, the transient follicles recruited by exogenous KITL are likely from the primordial follicle pool. Follicles with a similar morphological appearance were observed when Pten, a negative repressor of the KIT/KITL signaling pathway, was conditionally turned off in the mouse ovary. Increased recruitment along with premature ovarian failure was seen in the mice lacking Pten expression, presumably due to activation of the PI3 kinase pathway [26]. However, in that study, in the absence of granulosa cell proliferation, these oocytes were destined for atresia. When compared to the respective control (plus KITL), there was a 4-fold increase in numbers of transient follicles formed when VCD was present. These results suggest that VCD accelerates recruitment of primordial oocyte growth and support that KITL can protect the primary follicle pool from the ovotoxic effects of VCD. Collectively, VCD appears to interact with the KIT/KITL and not the GDF9 or BMP4 signaling pathway of survival.
After 8 days in culture, both mRNA and protein expressions of Kit were decreased in response to VCD exposure (20 and 30 μM). At a concentration of 20 μM VCD, follicle loss has not yet begun, confirming that the reduced expression of Kit is not simply due to lower availability of oocytes contained in small preantral follicles. The effect on KIT protein was confirmed by immunofluorescent staining, which was intensified at the plasma membrane and diffused into the cytoplasm of the oocyte in primordial, primary, and secondary follicles. The staining was reduced by VCD (30 μM) in remaining primordial and primary but not secondary follicles, providing visual evidence of the selectivity of VCD for primordial and primary follicles. Expression of Kitl mRNA and protein was increased following 8 days of culture with VCD. There was a dose-dependent increase in mRNA (20 and 30 μM). However, protein was increased at 20 μM but not 30 μM. The reason for the apparent loss of responsiveness with protein at 30 μM VCD could relate to the fact that follicle loss resulted in less KITL-expressing cells for measurement of protein at the concentration. However, because mRNA was increased at that concentration, there may be increased protein degradation in the face of elevated mRNA.
Measurement of the relative abundance of Kit and Kitl mRNA after shorter VCD exposure periods revealed that the decrease in oocyte-derived Kit mRNA preceded an increase in granulosa cell-derived Kitl mRNA. Therefore, the oocyte might be directly affected initially in response to VCD, whereas granulosa cells produce more KITL in a feedback response to the reduced survival signaling within the oocyte. However, it cannot be ruled out that VCD causes direct effects on granulosa cells that compromise their ability to support oocyte survival, thus indirectly affecting KIT. At any rate, the changes in Kit and Kitl expression suggest that VCD is causing a disruption in the associated cellular survival signaling pathway, thereby contributing to follicular atresia. Further studies are necessary to examine the involvement of other signaling components within the PI3K/AKT signaling pathway.
Because the effect of VCD in the target population of follicles was selective for the KIT survival pathway and did not appear to affect the GDF9 or BMP4 pathways, this suggests that VCD is acting directly via signaling pathways and not nonspecifically compromising follicle viability. These results support that either the oocyte is a direct target for VCD or the effects in the oocyte result from indirect targeting of granulosa cells.
In summary, the current studies revealed that KITL is able to attenuate VCD-induced ovotoxicity, whereas GDF9 and BMP4 do not appear to be involved. Also, VCD caused a decrease in Kit and an increase in Kitl expression. These results support that in addition to activation of proapoptotic pathways (BCL2 and MAPK families), VCD also compromises the KIT/KITL signaling pathway, which is a critical survival pathway in primordial and small preantral follicles. This disruption can explain, in part, the selective sensitivity of primordial and primary follicles to VCD and represents one mechanism by which VCD is ovotoxic.
Acknowledgments
The authors wish to thank Andrea Grantham and Doug Cromey from the Histology Core Facility, and Adam Hoying from the Genomics Research Laboratory for all of their technical assistance.
Footnotes
1Supported by grant R01 ES09246, Center Grant ES06694, and Training Grant ES007091.
REFERENCES
- Hirshfield AN.Development of follicles in the mammalian ovary. Int Rev Cytol 1991; 124: 43–101. [DOI] [PubMed] [Google Scholar]
- Hoyer PB, Sipes IG.Assessment of follicle destruction in chemical-induced ovarian toxicity. Annu Rev Pharmacol Toxicol 1996; 36: 307–331. [DOI] [PubMed] [Google Scholar]
- Flaws JA, Doerr JK, Sipes IG, Hoyer PB.Destruction of preantral follicles in adult rats by 4-vinyl-1-cyclohexene diepoxide. Reprod Toxicol 1994; 8: 509–514. [DOI] [PubMed] [Google Scholar]
- Kao SW, Sipes IG, Hoyer PB.Early effects of ovotoxicity induced by 4-vinylcyclohexene diepoxide in rats and mice. Reprod Toxicol 1999; 13: 67–75. [DOI] [PubMed] [Google Scholar]
- Springer LN, McAsey ME, Flaws JA, Tilly JL, Sipes IG, Hoyer PB.Involvement of apoptosis in 4-vinylcyclohexene diepoxide-induced ovotoxicity in rats. Toxicol Appl Pharmacol 1996; 139: 394–401. [DOI] [PubMed] [Google Scholar]
- Devine PJ, Sipes IG, Skinner MK, Hoyer PB.Characterization of a rat in vitro ovarian culture system to study the ovarian toxicant 4-vinylcyclohexene diepoxide. Toxicol Appl Pharmacol 2002; 184: 107–115. [PubMed] [Google Scholar]
- Springer LN, Tilly JL, Sipes IG, Hoyer PB.Enhanced expression of bax in small preantral follicles during 4-vinylcyclohexene diepoxide-induced ovotoxicity in the rat. Toxicol Appl Pharmacol 1996; 139: 402–410. [DOI] [PubMed] [Google Scholar]
- Hu X, Christian P, Sipes IG, Hoyer PB.Expression and redistribution of cellular Bad, Bax, and Bcl-X(L) protein is associated with VCD-induced ovotoxicity in rats. Biol Reprod 2001; 65: 1489–1495. [DOI] [PubMed] [Google Scholar]
- Hu X, Flaws JA, Sipes IG, Hoyer PB.Activation of mitogen-activated protein kinases and AP-1 transcription factor in ovotoxicity induced by 4-vinylcyclohexene diepoxide in rats. Biol Reprod 2002; 67: 718–724. [DOI] [PubMed] [Google Scholar]
- Johnson AL.Intracellular mechanisms regulating cell survival in ovarian follicles. Anim Reprod Sci 2003; 78: 185–201. [DOI] [PubMed] [Google Scholar]
- Markstrom E, Svensson E, Shao R, Svanberg B, Billig H.Survival factors regulating ovarian apoptosis–dependence on follicle differentiation. Reproduction 2002; 123: 23–30. [DOI] [PubMed] [Google Scholar]
- Quirk SM, Cowan RG, Harman RM, Hu CL, Porter DA.Ovarian follicular growth and atresia: the relationship between cell proliferation and survival. J Anim Sci 2004; 82: E40–E52. [DOI] [PubMed] [Google Scholar]
- Su YQ, Wu X, O'Brien MJ, Pendola FL, Denegre JN, Matzuk MM, Eppig JJ.Synergistic roles of BMP15 and GDF9 in the development and function of the oocyte-cumulus cell complex in mice: genetic evidence for an oocyte-granulosa cell regulatory loop. Dev Biol 2004; 276: 64–73. [DOI] [PubMed] [Google Scholar]
- Dong J, Albertini DF, Nishimori K, Kumar TR, Lu N, Matzuk MM.Growth differentiation factor-9 is required during early ovarian folliculogenesis. Nature 1996; 383: 531–535. [DOI] [PubMed] [Google Scholar]
- Orisaka M, Orisaka S, Jiang JY, Craig J, Wang Y, Kotsuji F, Tsang BK.Growth differentiation factor 9 is antiapoptotic during follicular development from preantral to early antral stage. Mol Endocrinol 2006; 20: 2456–2468. [DOI] [PubMed] [Google Scholar]
- Nilsson EE, Skinner MK.Growth and differentiation factor-9 stimulates progression of early primary but not primordial rat ovarian follicle development. Biol Reprod 2002; 67: 1018–1024. [DOI] [PubMed] [Google Scholar]
- Shimasaki S, Moore RK, Otsuka F, Erickson GF.The bone morphogenetic protein system in mammalian reproduction. Endocr Rev 2004; 25: 72–101. [DOI] [PubMed] [Google Scholar]
- Nilsson EE, Skinner MK.Bone morphogenetic protein-4 acts as an ovarian follicle survival factor and promotes primordial follicle development. Biol Reprod 2003; 69: 1265–1272. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Takakura N, Kataoka H, Kunisada T, Okamura H, Nishikawa SI.Stepwise requirement of c-kit tyrosine kinase in mouse ovarian follicle development. Dev Biol 1997; 184: 122–137. [DOI] [PubMed] [Google Scholar]
- Driancourt MA, Reynaud K, Cortvrindt R, Smitz J.Roles of KIT and KIT LIGAND in ovarian function. Rev Reprod 2000; 5: 143–152. [DOI] [PubMed] [Google Scholar]
- Manova K, Huang EJ, Angeles M, De Leon V, Sanchez S, Pronovost SM, Besmer P, Bachvarova RF.The expression pattern of the c-kit ligand in gonads of mice supports a role for the c-kit receptor in oocyte growth and in proliferation of spermatogonia. Dev Biol 1993; 157: 85–99. [DOI] [PubMed] [Google Scholar]
- Packer AI, Hsu YC, Besmer P, Bachvarova RF.The ligand of the c-kit receptor promotes oocyte growth. Dev Biol 1994; 161: 194–205. [DOI] [PubMed] [Google Scholar]
- Parrott JA, Skinner MK.Kit-ligand/stem cell factor induces primordial follicle development and initiates folliculogenesis. Endocrinology 1999; 140: 4262–4271. [DOI] [PubMed] [Google Scholar]
- Jin X, Han CS, Yu FQ, Wei P, Hu ZY, Liu YX.Anti-apoptotic action of stem cell factor on oocytes in primordial follicles and its signal transduction. Mol Reprod Dev 2005; 70: 82–90. [DOI] [PubMed] [Google Scholar]
- Springer LN, Flaws JA, Sipes IG, Hoyer PB.Follicular mechanisms associated with 4-vinylcyclohexene diepoxide-induced ovotoxicity in rats. Reprod Toxicol 1996; 10: 137–143. [DOI] [PubMed] [Google Scholar]
- Reddy P, Liu L, Adhikari D, Jagarlamudi K, Rajareddy S, Shen Y, Du C, Tang W, Hamalainen T, Peng SL, Lan ZJ, Cooney AJ, et al. Oocyte-specific deletion of Pten causes premature activation of the primordial follicle pool. Science 2008; 319: 611–613. [DOI] [PubMed] [Google Scholar]
- Schwartz MA, Stone AL, Greer KA, Hoying JB, Williams SK.Gene expression in tissue associated with extracellular matrix modified ePTFE. J Biomed Mater Res A 2005; 73: 30–38. [DOI] [PubMed] [Google Scholar]
- Greer KA, McReynolds MR, Brooks HL, Hoying JB.CARMA: a platform for analyzing microarray datasets that incorporate replicate measures. BMC Bioinformatics 2006; 7: 149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McReynolds MR, Taylor-Garcia KM, Greer KA, Hoying JB, Brooks HL.Renal medullary gene expression in aquaporin-1 null mice. Am J Physiol Renal Physiol 2005; 288: F315–F321. [DOI] [PubMed] [Google Scholar]
- Thompson KE, Bourguet SM, Christian PJ, Benedict JC, Sipes IG, Flaws JA, Hoyer PB.Differences between rats and mice in the involvement of the aryl hydrocarbon receptor in 4-vinylcyclohexene diepoxide-induced ovarian follicle loss. Toxicol Appl Pharmacol 2005; 203: 114–123. [DOI] [PubMed] [Google Scholar]
- Juengel JL, McNatty KP.The role of proteins of the transforming growth factor-beta superfamily in the intraovarian regulation of follicular development. Hum Reprod Update 2005; 11: 143–160. [DOI] [PubMed] [Google Scholar]
- Liu K, Rajareddy S, Liu L, Jagarlamudi K, Boman K, Selstam G, Reddy P.Control of mammalian oocyte growth and early follicular development by the oocyte PI3 kinase pathway: new roles for an old timer. Dev Biol 2006; 299: 1–11. [DOI] [PubMed] [Google Scholar]