Abstract
The purpose of the study was to understand in more detail the natural history of fetomaternal cell trafficking in healthy pregnant mice. Our goal was to identify the best target organs and days during pregnancy for further mechanistic studies of the role of fetal cells in maternal disease and injury. C57BL/6J wild-type virgin females (n = 54) were mated with congenic enhanced green fluorescent protein (EGFP) transgenic males. During pregnancy and after delivery, female mice were euthanized, and eight organs and blood were analyzed for the presence of fetal GFP+ cells with flow cytometry and real-time quantitative PCR. Maternal lungs, liver, and spleen were also analyzed by fluorescent stereomicroscopy. Fetal GFP+ cells were first found at low frequency at Embryonic Day 11, increased to a maximum at Embryonic Day 19, and decreased rapidly postpartum. These fetal cell dynamics were significantly reproducible among all mice studied. In addition, there was a consistent distribution of fetal cells within maternal organs, with lung, liver, blood, and spleen having the greatest concentrations; these were highly correlated at all time points (P < 0.0001). Maternal lung contained 10- to 100-fold more fetal cells than any other organ, and using all three techniques, the number of fetal cells detected was the most consistent and reproducible in this organ. Stereomicroscopy showed that within the lung, fetal cells were widely and apparently randomly distributed. Using a murine model, our data demonstrate that fetomaternal cellular trafficking occurs in reproducible patterns, is maximal near term delivery, and has predilection for the maternal lung..
Keywords: fetal cell microchimerism, flow cytometry, lung, pregnancy, trafficking
Fetomaternal cellular trafficking is nonrandom, affected by timing in gestation, and has predilection for specific organs, such as the lung, spleen, liver, and blood.
INTRODUCTION
Fetal cell microchimerism, resulting from fetomaternal cell trafficking during pregnancy, labor, and delivery, has been widely described in the human [1–4] and in animal models with hemochorial placentas [5–8]. The clinical significance of fetal cell microchimerism is largely unknown, although an increasing body of literature links it to both “cause” and “cure” of maternal diseases [9]. Using real-time PCR amplification of unique fetal gene sequences, fetomaternal trafficking has been shown to reproducibly exist, but it is relatively rare in healthy organs [10]. In contrast, specific disease states lead to an increased number of fetal cells [11]. Furthermore, fetal cells in the maternal host organ have the apparent capacity to transdifferentiate and assume the phenotype of the maternal tissue [12, 13].
Human studies are limited by a lack of appropriate tissue and/or a complete reproductive history, including that of spontaneous or elective pregnancy loss [14]. Prior animal studies have, in general, limited their focus to a specific organ or time point during pregnancy. In a histologic study of murine placental implantation sites harvested between Days 6 and 19 postcoitum, Vernochet and colleagues [15] demonstrated, using fetomaternal genetic differences, that fetal cells first entered the maternal blood at Embryonic Days 10–12. Fetal cells then entered the maternal decidua in increased amounts as gestation progressed. Other than the placenta, our knowledge of the natural history of fetomaternal trafficking in the organs of healthy pregnant mice is fragmented.
More than a decade ago, Bonney and Matzinger [16] assessed the number of male cells in maternal thymus, spleen, liver, lymph nodes, and peripheral blood in normal mice undergoing their first pregnancy. They detected fetal cells in only a fraction of pregnancies and speculated that the migrating fetal cells were cleared by the maternal immune system. However, they did not have access to real-time quantitative PCR, and they studied only organs related to the immune system. In a previous study, our group added real-time quantitative PCR technology [17], but at the time we focused on gestational weeks (as opposed to days), differences between virgin mice and retired breeders, and the evolution of fetal cell microchimerism after delivery.
More recently, we compared methods (e.g., flow cytometry [FCM] and real-time PCR) to detect rare transgenic fetal cells using a paternally inherited enhanced green fluorescence protein (Gfp) transgene sequence as a fetal marker in the wild-type C57BL/6J pregnant female [18]. The Gfp sequence is transmitted as a hemizygous dominantly expressed gene. All pups that inherit this sequence (approximately 50% of total) will fluorescence green in all of their cells, except for erythrocytes and fur. This allows for the localization of fetal cells in maternal tissues during pregnancy and postpartum. Flow cytometry has the capability to analyze whole organs or sort live cells for further evaluation with the highest sensitivity and specificity [18]. The technique is less time consuming than histochemical evaluation, therefore allowing for the analysis of multiple tissue types at multiple time points. However, real-time PCR allows the study of DNA extracted from entire maternal organs, and it may provide a more complete picture of the total amount of fetal cell microchimerism present. Neither PCR nor FCM, however, give an indication of the physical location of fetal cells within a maternal organ. For this we chose to use fluorescent stereomicroscopy, as this method would allow a general survey without artifacts that might be generated through histochemical analysis using fluorescently conjugated antibodies [19].
We performed the present study to understand in more detail the natural history of fetomaternal cell trafficking in healthy pregnant mice, and to test the hypothesis that such trafficking occurs in reproducible patterns. Our goal was to identify the best target organs and specific days during pregnancy when trafficking would be highest for further mechanistic studies of the role of fetal cells in maternal disease.
MATERIALS AND METHODS
Mice
The Institutional Animal Care and Use Committee of the Tufts University School of Medicine Division of Laboratory Animal Medicine approved the protocol described here. All institutional guidelines regarding the ethical use of experimental animals were followed. The enhanced green fluorescent protein (EGFP) transgenic C57BL/6-Tg(ACTB-EGFP)10sb/J (CAG; stock no. 003291) and wild-type C57BL/6J mice (stock no. 000664) were purchased from Jackson Laboratories (Bar Harbor, ME). The CAG mice have a C57BL/6J genetic background, with the hemizygously inherited, dominantly expressed Gfp transgene under the control of a ubiquitous chicken beta-actin promoter and a cytomegalovirus enhancer [20]. C57BL/6J virgin female mice (8–10 wk old) were mated to CAG or wild-type (i.e., controls) males. During mating, female and males remained in cages together for 3 days. They were then separated. Each mating group consisted of at least five mating pairs. Day 1 of gestation was assigned based on the presence of a vaginal plug. If, within a given mating group, a vaginal plug was not observed but the female was observed to be pregnant, gestational age was determined by comparing fetal size and developmental characteristics at killing to fetuses from mice in the same mating group that had a vaginal plug. If the fetuses were smaller or larger than the known reference mouse (i.e., with a plug), gestational age was assigned as +1 or −1 day. For the mice at Embryonic Days 10–12 the fetuses were too small to see significant differences, so only mice with vaginal plugs were used. For each pregnant mouse, we recorded the total litter size and the number of GFP+ pups or fetuses using ultraviolet excitation to detect green fluorescence.
Tissue Collection and Single-Cell Suspension
Fifty-four pregnant and postpartum female mice, as well as two nonpregnant wild-type virgin female mice and nine control pregnant or postpartum mice (total 11), were analyzed at various time points in gestation (Table 1). Blood was obtained by cardiac puncture. Heart, lung, thymus, left lobe of liver, spleen, bilateral kidneys, and brain were obtained after gross dissection, and bone marrow cells were isolated from bilateral femoral and tibial bones. Solid tissues were dissected into small pieces in FCM buffer (PBS with 2% fetal bovine serum and 0.1% sodium azide) and mechanically homogenized. The bone marrow cells and homogenates from solid organs were filtered through a 40-μm nylon cell strainer (BD Falcon, Bedford, MA) to remove debris and resuspended in 800 μl FCM buffer as a single-cell suspension. A total of 600 and 200 μl of each single-cell suspension was used for FCM and PCR, respectively.
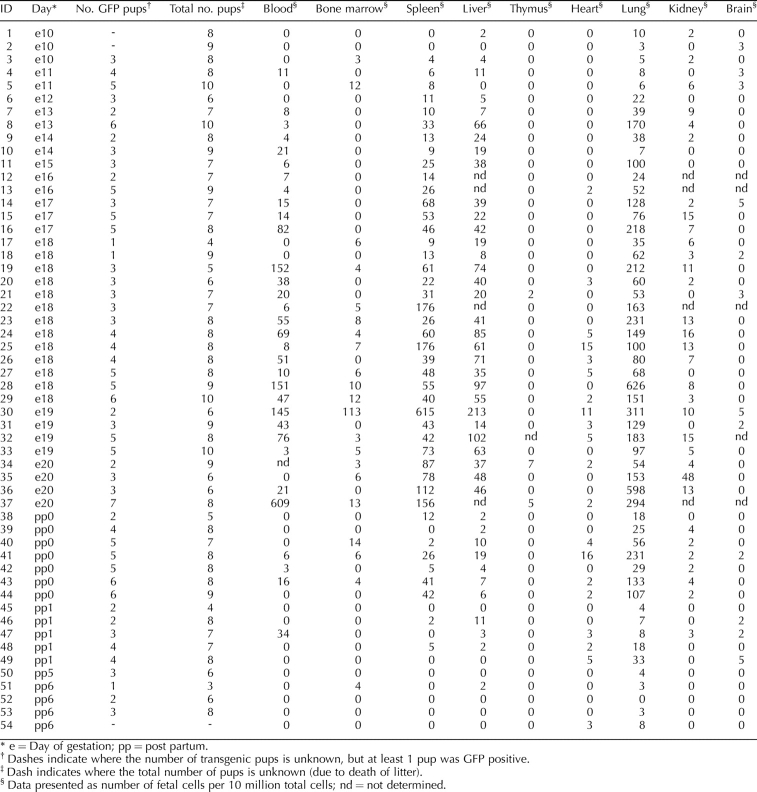
TABLE 1.
Fetal cell count in maternal organs of case mice as determined by flow cytometry (n = 54).
Flow Cytometry
Organs from all mice were analyzed by FCM. Erythrocytes were removed from 600 μl whole blood by lysis (0.15 M NH4Cl, 1.0 mM KHCO3, and 0.1 mM Na2-EDTA). Mononuclear cells (MNCs) in blood samples were washed twice with FCM buffer. Single-cell suspensions from homogenized organs were washed once in FCM buffer. All cells were resuspended in 200 μl FCM buffer with 1 μg/ml 7-amino-actinomycin D (7-AAD; BD Pharmingen, San Diego, CA) to stain dead cells. High-speed FCM analysis was conducted with MoFlo (DAKO, Fort Collins, CO). Data were collected from 7 to 15 million MNCs that were counted using the MoFlo hardware. The 7-AAD and EGFP were excited at 488 nm and measured at 660 nm (FL-3 670/40 filter) and 510 nm (FL-1 530/40 filter), respectively. Abnormally shaped cells were excluded by forward-angle scattered light and side-angle scattered light gating. Autofluorescent noise, which distributes in a linear pattern on FL-2 (570/40 filter) and FL-3, was gated out. The 7-AAD-positive dead cells were also excluded using FL-3 gating. High-speed flow sorting (20 000 to 80 000 cells per second) was conducted for each sample. Summit 4.3 software (DAKO) was used for data analysis. Fetal GFP+ cell counts were determined and shown as the number per 10 million maternal cells.
DNA Extraction and Real-Time PCR Amplification
We randomly selected 45 of 54 female mice mated to CAG males and all 11 control mice for PCR analysis. Genomic DNA was extracted from 200 μl whole blood and 200 μl single-cell suspensions from homogenized organs using the QIAamp DNA Mini Kit, as recommended by the manufacturer (Qiagen, Valencia, CA) and stored at −80°C until analysis. Real-time PCR amplification of Gfp and apolipoprotein B (Apob) as an internal control (i.e., to determine the concentration of total genomic DNA) was performed using an ABI 7900 Sequence Detection System with SDS v2.2 software (Applied Biosystems, Foster City, CA) as previously described [21]. All PCR experiments were performed in triplicate, with results reported as mean values. The amount of DNA used in each PCR well for Gfp was 200 000 pg for blood, liver, thymus, heart, and brain and 600 000 pg for bone marrow, spleen, lung, and kidney. DNA concentrations were determined by Apob PCR [21]. We have previously reported a PCR assay sensitivity of approximately two to four microchimeric transgenic cells in a background of 100 000 wild-type cells [22] using the genome conversion factor for the C57BL/6J background strain (6.25 pg DNA per cell of C57BL/6J genome) [23]. Although the transgene copy number has not been determined be gene mapping, it is present at a uniform copy number across animals. Thus, the relative calculations of Gfp quantity were made based on an assumption that the transgene was present at one copy per cell. Calculations of Apob quantity were based on the gene being present at two copies per cell, as this is a naturally occurring housekeeping gene. To be able to compare to the FCM data generated in the present study, we converted amplified product to the equivalent number of microchimeric cells in 10 million total cells.
Whole-Organ Imaging by Fluorescent Stereomicroscopy
Based on the FCM and PCR results, we then examined whole fresh organs (lung, spleen, and liver) of 12 mice (six female mice mated to CAG males and six controls) at Embryonic Day 18 of gestation with fluorescent stereomicroscopy (Model MZ-FLIII; Leica Microsystems, Wetzlar, Germany) equipped with a color digital CCD camera (SPOT RTKE Model 7.4.1; Diagnostic Instruments Inc., Sterling Heights, MI). Imaging studies were performed using GFP2 filter sets with excitation filter 480 ± 40 nm and barrier filter 510 nm. The number of GFP+ foci observed on the surface of organs (both anterior and posterior sides) were counted from the live image and recorded. SPOTS advanced software version 4.1.3 (Diagnostic Instruments) was used to capture images.
Statistical Analyses
Quantities of fetal cells were presented as medians and interquartile ranges (25th, 75th percentiles) for each organ. Spearman correlation analysis was carried out to detect the concordance between fetal cell counts obtained by PCR versus FCM in each organ, as well as between the number of fetal cells and the number of GFP+ pups. Statistical significance was assigned at P < 0.05. All statistical analyses were performed using SAS/STAT software (SAS Institute Inc., Cary, NC). To identify a function that best describes the relationship between fetal cell concentration and time in gestation, quadratic, cubic, restricted cubic spline, and two-slope linear models were fitted to the log-transformed fetal cell count for each organ using the Design library of the R package (R: A language and environment for statistical computing. R Foundation for Statistics. http://www.R-project.org ).
RESULTS
Flow Cytometry
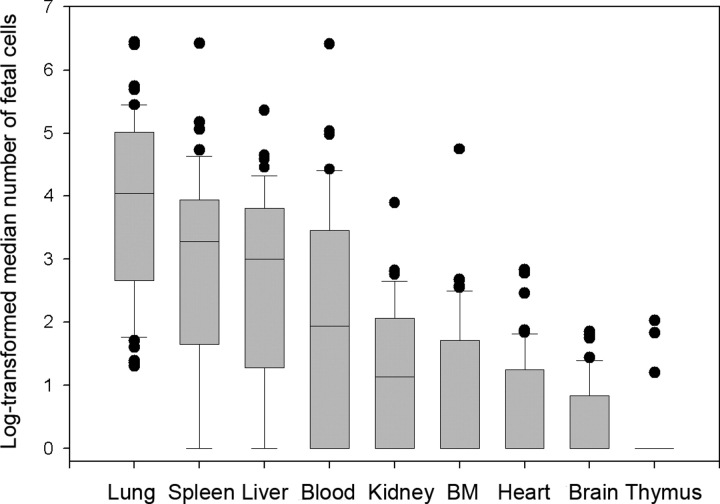
Figure 1 and Table 1 show the numbers of fetal cells analyzed by FCM present in each organ and blood from all female mice mated to CAG males, as log-transformed medians and absolute counts, respectively. Control pregnant mice that carried no GFP+ fetuses always had five or fewer apparently GFP+ cells per 10 million total cells (data not shown); therefore, we used a cutoff of five cells as our limit of detection. The highest median number of fetal cells in female mice mated to CAG males was found in the lung, followed by the spleen, liver, blood, and kidney (Fig. 1). Although fetal cells were found at levels above five fetal cells per 10 million total cells in heart, bone marrow, and thymus at various time points, these organs had an overall median value of 0. The number of fetal cells was at or below the limit of sensitivity at all time points in brain tissue.
FIG. 1.
Graph showing the log-transformed median numbers of fetal cells in all maternal organs and blood analyzed by flow cytometry. Horizontal lines represent median values, boxes represent 25th and 75th percentiles, whiskers represent 10th and 90th percentiles, and filled circles represent outliers. BM, Bone marrow.
Figure 2 shows fetal cell dynamics across gestation in a subset of maternal organs that were analyzed (liver, lung, and spleen). The restricted cubic spline curve was the best-fitting function to describe log-transformed fetal cell counts. According to this model, the variances in cell number explained by time in gestation, R2, were 79%, 73%, and 76% for liver, lung, and spleen, respectively. With the exception of one lung specimen at Embryonic Day 10, a small number of fetal cells were first observed in most organs at Embryonic Day 11. The frequency of fetal cells continued to increase with advancing gestational age. The maximum number of fetal cells was found in organs obtained at Embryonic Day 19, which corresponds to full-term pregnant mice just before delivery (Fig. 2). After delivery, fetal cell count decreased rapidly in all organs. Fetal cells were still observed at Day 0 postpartum (i.e., 6–12 h after delivery), but decreased by Day 1 postpartum and returned to background level (five or fewer cells per 10 million total cells) at Days 5–6 postpartum. However, restricted cubic spline modeling suggests that there may be a subsequent increase in fetal cell numbers following this decrease after delivery, specifically in liver tissue (Fig. 2A). Nevertheless, the timing of the appearance, increase, and rapid decease following delivery in the frequency of fetal cells was consistent among all organs analyzed. The only difference was in the overall quantity of fetal cells detected in each organ (Fig. 2 and Table 1).
FIG. 2.
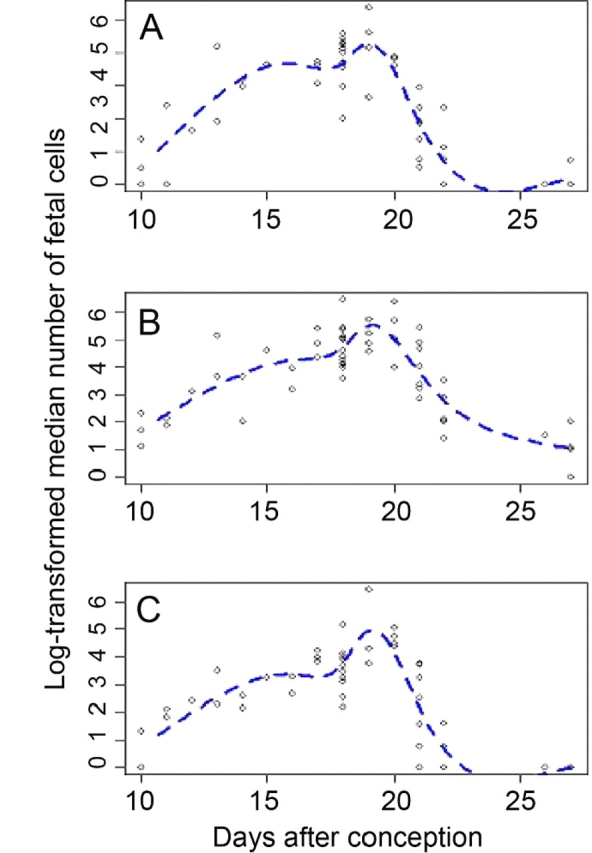
Fetal cell quantitative dynamics as represented by log-transformed median fetal cell numbers in maternal organs during pregnancy and postpartum as measured by flow cytometry. Days 21, 22, 26, and 27 represent Days 0, 1, 5, and 6 postpartum, respectively. Dashed lines show the six-knot restricted cubic spline curves that are the best-fitting functions to describe these data. A) Liver. B) Lung. C) Spleen.
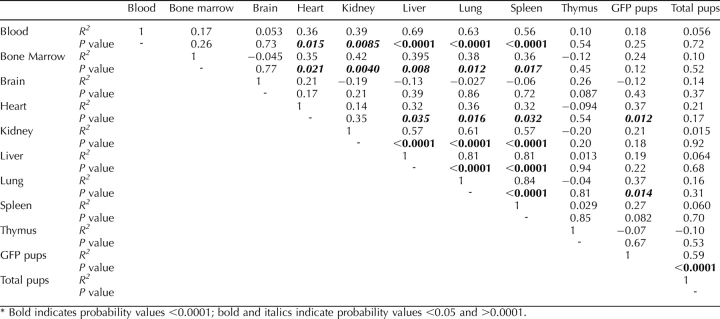
These fetal cell dynamics were significantly reproducible among all organs studied based on correlation analysis (Table 2). For example, the number of fetal cells in lung, liver, spleen, and blood was highly correlated at all time points (P < 0.0001), and significantly correlated among other organs, such as kidney and bone marrow (0.0001 < P < 0.05). The exceptions were brain and thymus tissue, in which the numbers of fetal cells were at or below the limit of sensitivity and were therefore not correlated (P > 0.05).
TABLE 2.
Correlation analysis of flow cytometry data between organs, number of GFP+ pups, and total number pups.*
Effect of Litter Size
In spleen, liver, kidney, blood, bone marrow, thymus, and brain, the number of GFP+ pups per litter did not have an effect on the number of fetal cells found in maternal organs (Table 2). The only exceptions were lung (P = 0.014) and heart (P = 0.012) tissue, in which there was a positive correlation between the number of GFP+ pups per litter and the frequency of fetal cells across all time points. The number of fetal cells was not significantly affected by the number of GFP+ pups per litter in all other organs (Table 2).
Real-Time PCR Amplification
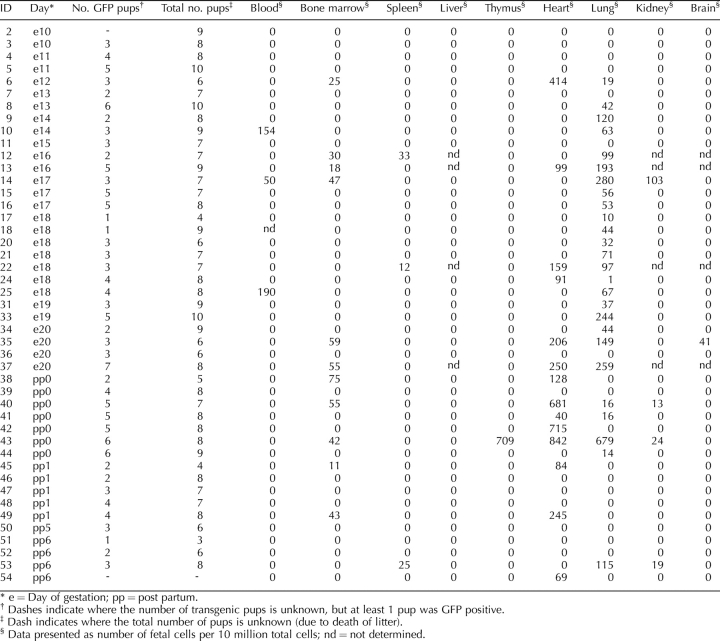
Real-time PCR data confirmed the results of FCM in the organs that had the greatest quantity of fetal cells (i.e., lung; Table 3). In the lung, fetal Gfp sequence was first detected at Embryonic Day 12 of gestation and increased with gestation. Although fetal cells were still found at Day 0 postpartum, their level became undetectable by Day 1 postpartum. The PCR results showed a generally similar trend to the FCM results as a result of advancing gestation, but there was an overall lower sensitivity in the detection of fetal cells. Fetal cells were also found in heart. Fetal cells were not detected in almost all spleens or livers with PCR. Among negative control PCR results, only one thymus sample from one mouse (Embryonic Day 14) showed amplification of Gfp (data not shown).
TABLE 3.
Fetal cell count in maternal organs of case mice as determined by PCR (n = 45).
Fluorescent Stereomicroscopy
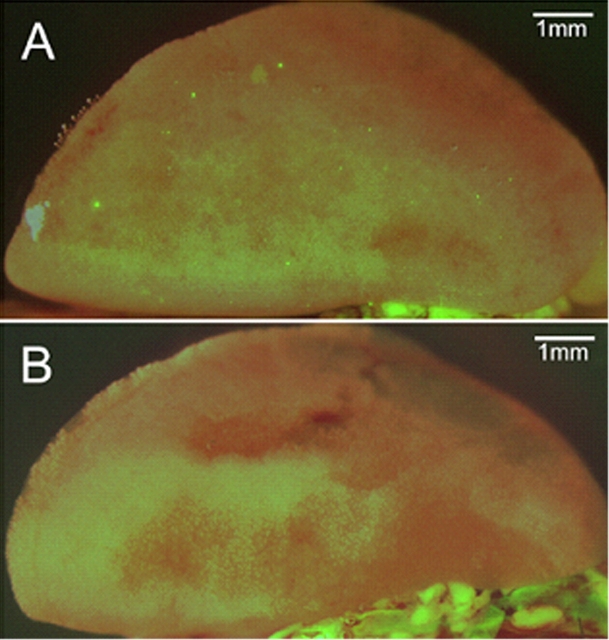
Analysis by fluorescent stereomicroscopy showed that fetal cells, observed as discrete green fluorescent foci, were not grouped in specific locations, but rather distributed widely in lung tissue (Fig. 3A). On average, a total of 100 distinct GFP+ foci (range: 54–140) were observed in whole-lung specimens from all six wild-type female mice (100%) at Embryonic Day 18 of gestation following mating with GFP+ males. In contrast, there were no fluorescent foci in any lungs from control mice (P < 0.0001; Fig. 3B). In addition, no fluorescent foci were seen in any liver specimens or from control spleen. A single green focus was observed in one spleen specimen from a female mouse mated to a CAG male.
FIG. 3.
Photomicrographs of (A) GFP+ fetal cells (foci) in wild-type maternal lung tissue at Day 18 of gestation following mating with a GFP+ male and (B) absence of GFP signal in wild-type maternal tissue at Day 18 of gestation following mating with a wild-type male.
Comparison of Techniques
Table 4 shows the categorization of results for each maternal organ analyzed by each of three techniques (FCM, PCR, and stereomicroscopy). For FCM, categories (per 10 million total cells) include strongly positive (+++; >100 mean fetal cells), moderately positive (++; 10–100 mean fetal cells), weakly positive (+; five to nine mean fetal cells), and negative (−; fewer than five mean fetal cells) results. For PCR, categories (per 10 million total cells) include strongly positive (+++; >50 mean fetal cells), moderately positive (++; 10–50 mean fetal cells), weakly positive (+; 1–10 mean fetal cells), and negative (−; fewer than one mean fetal cell) results. For stereomicroscopy, categories include strongly positive (+++; >100 mean fluorescent foci), moderately positive (++; 10–100 mean fluorescent foci), weakly positive (+; 1–10 mean fluorescent foci), and negative (−; 0 mean fluorescent foci) results. In some cases (e.g., blood, bone marrow, thymus, lung, and brain), the results of FCM and PCR analyses are correlated. In others (e.g., spleen, liver, heart, and kidney), the results of FCM and PCR analyses are not correlated. However, by statistical analysis, the numbers of fetal cells measured by PCR and FCM across all time points were significantly positively correlated only in lung tissue (P = 0.0004). Fluorescent stereomicroscopy was only performed on spleen, liver, and lung specimens. In these cases, the results of stereomicroscopy analysis correlated to FCM and PCR analyses of lung specimens and roughly correlated to PCR analyses of spleen and liver specimens.
TABLE 4.
Fetal cell microchimerism at Day 18 of gestation.*
DISCUSSION
Using a murine model, our data demonstrate that fetomaternal cellular trafficking occurs in reproducible patterns, is maximal near term delivery, and has a predilection for the maternal lung.
The Lung Is the Organ with the Most Microchimerism
The highest frequency of fetal cell microchimerism was consistently observed in maternal lung at all gestational ages. This finding was also reproducible with all methods used to detect fetal cells, including FCM, PCR, and fluorescence stereomicroscopy. By direct observation in whole lung, fetal cells in lung tissue were widely distributed. We previously showed that there were more fetal cells in the lungs of pregnant and postpartum mice with real-time PCR [17]. Fetal placental cells (e.g., trophoblasts) were first observed more than 100 years ago in a study of lung tissue from women who died of preeclampsia [24]. Fetal cell microchimerism in the human lung was reported again recently [25]. In the human, trophoblasts have been found in maternal broad ligament veins or inferior vena cava [26]. In the mouse, Bonney and Matzinger [16] found that peripheral blood and (uterine) draining lymph nodes had the highest frequency of detectable fetal cells. These investigators did not study the maternal lungs. Taken together, these studies suggest that microchimeric fetal cells may originate from blood flow through the uterine vein into maternal pulmonary arteries, resulting in the highest frequency of fetal cells being found in the lung. The high frequency of fetal cells in lung tissue may also reflect the high rate of blood flow through lung tissue. Moreover, as certain serious diseases like primary pulmonary hypertension and pulmonary thrombosis are more frequent in pregnant women than in nonpregnant women, we speculate that fetal cells may be in a position to play an active functional (biological) or pathological role in the maternal lung. It is still unclear whether fetal cells actively accumulate in lung or are just trapped there passively. Carey et al. [27] suggested that gender affects the incidence, susceptibility, and severity of several lung diseases in pregnant and nonpregnant women. Therefore, it is possible that fetal cells may be a factor in the observed gender differences of lung disease.
Other Organ-Specific Differences in Microchimerism
Fetal cells were also found in hematopoietic organs, such as spleen and liver, and blood. These organs, particularly the spleen, which functions as a blood filter, may sequester fetal cells and remove them from circulation [28]. The results observed in liver, combined with that of lung, also suggest that those organs that experience high blood flow are more likely to contain fetal cells. Nevertheless, further investigation is needed to determine the role of fetal cells in hematopoietic organs.
Fetal cells were found in low numbers in bone marrow and heart, but were not observed in thymus and brain by FCM. It is possible that the dearth of fetal cells in these organs reflects that no blood filtration occurs at these sites. It is also possible that no specific homing of fetal cells to the brain and thymus occurs, or that brain and thymus are nonconducive microenvironments for the acquisition or maintenance of fetal cells. Our results differ from those of Tan et al. [11], who detected fetal cells in maternal brain following induced injury. In noninjured mice, this group found fetal cells primarily in the olfactory region, an area that we did not isolate for our studies. In contrast, our results do not significantly differ from those of O'Donoghue et al. [2], who found fetal cells present in human maternal bone marrow at a frequency of ∼1 per 100 000 maternal cells, which is no more than an order of magnitude greater than our findings.
Fetal GFP+ cells were not observed by stereomicroscopic examination in spleen and liver, even though fetal cells were detected in these organs by FCM. The reason for this discrepancy is unclear, although it is possible that the higher cell density in these organs restricted GFP fluorescence penetration to the organ surface, thereby shielding the fetal cells from detection by fluorescent microscopy.
Overall, the reproducible patterns of fetal cells among maternal organs suggest that the deposition of these cells may be due to more than one mechanism, including passive (e.g., blood flow and filtration) and active (i.e., organ-specific homing or recruitment) processes. Future experiments, including the phenotypic assessment of fetal cells found in various maternal organs, will help to determine the relative contribution of these different processes to the observed organ-specific distribution of fetal cells.
Fetal Cell Microchimerism Is Maximal Near Term
Fetal cell microchimerism was observed at low frequency at Embryonic Day 11 of gestation in all organs analyzed, and their numbers increased with advancing gestational age. This timing of fetal cell trafficking is similar to that reported by Vernochet and colleagues [15], in which the number of fetal cells increased in placenta with advancing gestation. Fetal cell microchimerism was maximal near term. After delivery, fetal cells were effectively cleared from each maternal organ. Although fetal cells and DNA still remained at low levels at Day 0 postpartum (∼6 h after delivery), their quantities were reduced further by Day 1 postpartum (∼30 h after delivery) and were undetectable by 6 days after delivery. These dynamics are also similar to fetal cells and cell free fetal DNA in human postpartum peripheral blood [29–32].
In the present study, we analyzed eight organs and blood with two or three methods at various time points to comprehensively understand fetal cell microchimerism. We have also applied different statistical models to estimate the functional shape of the relationship between fetal cell count and gestational age. The best-fit model (i.e., six-knot restricted cubic spline) described consistently well the changes in the fetal cell microchimerism observed in different organs.
Effect of Litter Size
At each gestational stage, the number of fetal cells in the maternal organs and blood was widely distributed. One explanation for this is a difference in the number of GFP+ fetuses. The frequency of fetal cell microchimerism may depend on how many GFP+ fetuses were in utero. Because the number of GFP+ fetuses varied in each pregnancy (Table 1), it is possible that the number of fetal cells in maternal organs is dependent on the number of GFP+ fetuses in a litter. Our data show that with the exception of lung and heart, there is no apparent effect of the number of GFP+ fetuses per litter on the subsequent number of fetal cells detected in maternal organs. However, since lung is the organ with the highest number of observed fetal cells, it is possible that the observed effect of litter size is only apparent in organs where a large number of fetal cells are present. Therefore, while our results suggest that litter size may not influence the trafficking of fetal cells into the maternal murine circulation, more work is required to further investigate this issue.
Comparison of Techniques
In certain organs (e.g., bone marrow, heart, lung, thymus, and brain), the results of FCM and PCR analyses are somewhat correlated. These results suggest that in these organs, either no fetal cells were observed by both techniques (i.e., in brain and thymus tissue) or a sufficient number of cells present with sufficient Gfp expression so that the detection of either the transgene sequence (by PCR) or the transgenic protein (by FCM) is above the threshold of sensitivity for both of these techniques (e.g., in lung).
The differences in endpoint technique for the detection of rare DNA sequences and/or protein may explain the discrepancies in the literature as to the frequency and distribution of fetal cell microchimerism. We suggest that FCM is the preferred quantitative technique to use for studies of fetal cell microchimerism, as it generates much more reliable and reproducible results than that of PCR. Interestingly, the observed frequency of fetal cells might be underestimated when using our FCM gating strategy, as we can count only those GFP+ cells that were bright enough to be reliably distinguished from autofluorescent maternal cells. Therefore, using FCM we likely underreported dull GFP+ cells (i.e., those with little or no Gfp expression). As GFP+ foci were only observed by stereomicroscopy in lung specimens, we suggest that this technique has limited utility for qualitative analyses but is not useful as a quantitative tool. Indeed, others have also reported statistically significant limitations in fluorescence detection using stereomicroscopy. For example, Cabellero et al. [33] found that 17% of samples were found to be positive by quantitative PCR while observed to be negative by fluorescence microscopy (P < 0.02). Although these investigators suggested that the discrepancy was somewhat negligible, further comparison between techniques is clearly warranted.
In this study we used three different techniques to demonstrate the presence of fetal cells in maternal organs and blood on specific gestational days during pregnancy. All three confirm that the lung is the site of the most fetal cells. The combination of these three techniques has allowed us to evaluate the natural course of fetal cell microchimerism in a thorough and more comprehensive manner than in prior reports.
Further study is necessary for the continued understanding of fetal cell microchimerism and its role in maternal health. This includes the analysis of different immunological background strains of mice to determine the role of histocompatibility on the development of microchimerism and the use of a homozygous transgenic strain to increase the detectable number of fetal cells that can be located based on the presence of transgenic DNA and/or protein. Additional investigation into the dynamics of the long-term persistence of fetal cells following delivery is also clearly warranted, based on the apparent increase after Days 5 and 6 postpartum as suggested by the best-fit modeling (i.e., restricted cubic spline). Nonetheless, the consistent and reproducible results presented here support the validity of this animal model to investigate the mechanisms of fetal cell trafficking and will allow for the testing of new hypotheses regarding the relationship between fetal cell microchimerism and lung disease in mothers, both human and murine.
Acknowledgments
The authors wish to thank Stephen Kwok and Allen Parmelee for flow cytometry expertise, Ina Klebba and Charlotte Kuperwasser for stereomicroscopy expertise, Kai Tao and Gary Sahagian for additional technical advice, and Sicco Scherjon for a constructive evaluation of this manuscript.
Footnotes
1Supported by National Institutes of Health grant R01 HD049469-04 to D.W.B. Y.F. was supported by a grant from the Kanzawa Medical Research Foundation in Nagano, Japan.
REFERENCES
- Bianchi DW, Zickwolf GK, Weil GJ, Sylvester S, DeMaria MA.Male fetal progenitor cells persist in maternal blood for as long as 27 years postpartum. Proc Natl Acad Sci U S A 1996; 93: 705–708.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donoghue K, Chan J, de la Fuente J, Kennea N, Sandison A, Anderson JR, Roberts IA, Fisk NM.Microchimerism in female bone marrow and bone decades after fetal mesenchymal stem cell trafficking in pregnancy. Lancet 2004; 364: 179–182.. [DOI] [PubMed] [Google Scholar]
- Koopmans M, Kremer Hovinga IC, Baelde HJ, Fernandes RJ, de Heer E, Bruijn JA, Bajema IM.Chimerism in kidneys, livers and hearts of normal women: implications for transplantation studies. Am J Transplant 2005; 5: 1495–1502.. [DOI] [PubMed] [Google Scholar]
- Bianchi DW, Fisk NM.Fetomaternal cell trafficking and the stem cell debate: gender matters. JAMA 2007; 297: 1489–1491.. [DOI] [PubMed] [Google Scholar]
- Liégeois A, Escourrou J, Ouvre E, Charreire J.Microchimerism: a stable state of low-ratio proliferation of allogeneic bone marrow. Transplant Proc 1977; 9: 273–276.. [PubMed] [Google Scholar]
- Liégeois A, Gaillard MC, Ouvre E, Lewin D.Microchimerism in pregnant mice. Transplant Proc 1981; 13: 1250–1252.. [PubMed] [Google Scholar]
- Wang Y, Iwatani H, Ito T, Horimoto N, Yamato M, Matsui I, Imai E, Hori M.Fetal cells in mother rats contribute to the remodeling of liver and kidney after injury. Biochem Biophys Res Commun 2004; 325: 961–967.. [DOI] [PubMed] [Google Scholar]
- Jimenez DF, Leapley AC, Lee CI, Ultsch MN, Tarantal AF.Fetal CD34+ cells in the maternal circulation and long-term microchimerism in rhesus monkeys (Macaca mulatta). Transplantation 2005; 79: 142–146.. [DOI] [PubMed] [Google Scholar]
- Johnson KL, Bianchi DW.Fetal cells in maternal tissue following pregnancy: what are the consequences? Hum Reprod Update 2004; 10: 497–502.. [DOI] [PubMed] [Google Scholar]
- Lambert NC, Lo YM, Erickson TD, Tylee TS, Cuthrie KA, Furst DE, Nelson JL.Male microchimerism in healthy women and women with scleroderma: cells or circulating DNA? A quantitative answer. Blood 2002; 100: 2845–2851.. [DOI] [PubMed] [Google Scholar]
- Tan XW, Liao H, Sun L, Okabe M, Xiao ZC, Dawe GS.Fetal microchimerism in the maternal mouse brain: a novel population of fetal progenitor or stem cells able to cross the blood-brain barrier? Stem Cells 2005; 23: 1443–1452.. [DOI] [PubMed] [Google Scholar]
- Khosrotehrani K, Johnson KL, Cha DH, Salomon RN, Bianchi DW.Transfer of fetal cells with multi-lineage potential to maternal tissue. JAMA 2004; 292: 75–80.. [DOI] [PubMed] [Google Scholar]
- Stevens AM, McDonnell WM, Mullarkey ME, Pang JM, Leisenring W, Nelson JL.Liver biopsies from human females contain male hepatocytes in the absence of transplantation. Lab Invest 2004; 84: 1603–1609.. [DOI] [PubMed] [Google Scholar]
- Khosrotehrani K, Johnson KL, Lau J, Dupuy A, Cha DH, Bianchi DW.The influence of fetal loss on the presence of fetal cell microchimerism: a systematic review. Arthritis Rheum 2003; 48: 3237–3241.. [DOI] [PubMed] [Google Scholar]
- Vernochet C, Caucheteux SM, Kanellopoulos-Langevin C.Bi-directional cell trafficking between mother and fetus in mouse placenta. Placenta 2007; 28: 639–649.. [DOI] [PubMed] [Google Scholar]
- Bonney EA, Matzinger P.The maternal immune systems' interaction with circulating fetal cells. J Immunol 1997; 158: 40–47.. [PubMed] [Google Scholar]
- Khosrotehrani K, Johnson KL, Guégan S, Stroh H, Bianchi DW.Natural history of fetal cell microchimerism during and following murine pregnancy. J Reprod Immunol 2005; 66: 1–12.. [DOI] [PubMed] [Google Scholar]
- Fujiki Y, Tao K, Bianchi DW, Giel-Maloney M, Leiter AB, Johnson KL.Quantification of green fluorescence protein by in vivo imaging, PCR and flow cytometry: comparison of transgenic strains and relevance for fetal cell microchimerism. Cytometry 2008; 73A: 111–118.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri TR, Mountz JM, Rogers BE, Partridge EE, Zinn KR.Light-based imaging of green fluorescent protein-positive ovarian cancer xenografts during therapy. Gynecol Oncol 2001; 82: 581–589.. [DOI] [PubMed] [Google Scholar]
- Okabe M, Ikawa M, Kominami K, Nakanishi T, Nishimune Y.‘Green mice' as a source of ubiquitous green cells. FEBS Lett 1997; 407: 313–319.. [DOI] [PubMed] [Google Scholar]
- Khosrotehrani K, Wataganara T, Bianchi DW, Johnson KL.Fetal cell-free DNA circulates in the plasma of pregnant mice: relevance for animal models of fetomaternal trafficking. Hum Reprod 2004; 19: 2460–2464.. [DOI] [PubMed] [Google Scholar]
- Su EC, Johnson KL, Tighiouart H, Bianchi DW.Murine maternal cell microchimerism: analysis using real-time PCR and in vivo imaging. Biol Reprod 2008; 78: 883–887.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capparelli R, Cottone C, D'Apice L, Viscardi L, Colantonio L, Lucretti L, Ianelli D.DNA content differences in laboratory mouse strains determined by flow cytometry. Cytometry 1997; 29: 261–266.. [DOI] [PubMed] [Google Scholar]
- Lapaire O, Holzgreve W, Oosterwijk JC, Brinkhaus R, Bianchi DW.Georg Schmorl on trophoblasts in the maternal circulation. Placenta 2007; 28: 1–5.. [DOI] [PubMed] [Google Scholar]
- O'Donoghue K, Sultan HA, Al-Allaf FA, Anderson JR, Wyatt-Ashmead J, Fisk NM.Microchimeric fetal cells cluster at sites of tissue injury in lung decades after pregnancy. Reprod Biomed Online 2008; 16: 382–390.. [DOI] [PubMed] [Google Scholar]
- Douglas GW, Thomas L, Carr M, Cullen NM, Morris R.Trophoblast in the circulating blood during pregnancy. Am J Obstet Gynecol 1959; 78: 960–973.. [DOI] [PubMed] [Google Scholar]
- Carey MA, Card JW, Voltz JW, Arbes SL, Jr, Germolec DR, Korach KS, Zeldin DC.It's all about sex: gender, lung development and lung disease. Trends Endocrinol Metab 2007; 18: 308–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KL, Nelson JL, Furst DE, McSweeney PA, Robert DJ, Zhen DK, Bianchi DW.Fetal cell microchimerism in tissue from multiple sites in women with systemic sclerosis. Arthritis Rheum 2001; 44: 1848–1854.. [DOI] [PubMed] [Google Scholar]
- Hamada H, Arinami T, Kubo T, Hamaguchi H, Iwasaki H.Fetal nucleated cells in maternal peripheral blood: frequency and relationship to gestational age. Hum Genet 1993; 91: 427–432.. [DOI] [PubMed] [Google Scholar]
- Hamada H, Arinami T, Hamaguchi H, Kubo T.Fetal nucleated cells in maternal peripheral blood after delivery. Am J Obstet Gynecol 1994; 170: 1188–1193.. [DOI] [PubMed] [Google Scholar]
- Lo YM, Tein MS, Lau TK, Haines CJ, Leung TN, Poon PM, Wainscoat JS, Johnson PJ, Chang AM, Hjelm NM.Quantitative analysis of fetal DNA in maternal plasma and serum: implications for noninvasive prenatal diagnosis. Am J Hum Genet 1998; 62: 768–775.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo YM, Zhang J, Leung TN, Lau TK, Chang AM, Hjelm NM.Rapid clearance of fetal DNA from maternal plasma. Am J Hum Genet 1999; 64: 218–224.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabellero M, Lightfoot HM, LaPaglia M, Pleasant A, Hatada S, Cairns BA, Fair JH.Detection and characterization of hepatic engraftment of embryonic stem derived cells by fluorescent stereomicroscopy. J Surg Res 2007; 141: 134–140.. [DOI] [PMC free article] [PubMed] [Google Scholar]