Abstract
Sumoylation is a posttranslational modification in which SUMO (small ubiquitin-related modifier) proteins are covalently attached to their substrates. In vertebrates, developmental roles for sumoylation have been studied, but the function of sumoylation during mammalian oocyte growth and maturation is not known. As a prelude to conducting studies on the role of sumoylation during oocyte development, we analyzed the temporal and spatial pattern of expression of UBE2I, a SUMO-conjugating E2 enzyme. Immunocytochemical analysis of UBE2I revealed a punctate nuclear staining pattern, with transcriptionally quiescent, fully grown, GV-intact oocytes having larger UBE2I-containing bodies than transcriptionally active, meiotically incompetent growing oocytes. Inhibiting transcription in incompetent oocytes resulted in an increase in the size of the UBE2I-containing bodies. Overexpression of either wild-type UBE2I or catalytically inactive UBE2I resulted in an increase in the size of the UBE2I-containing bodies but also an increase in BrUTP incorporation, suggesting that transcriptional activation by UBE2I is independent of its catalytic activity. Although UBE2I-containing bodies did not completely colocalize with SUMO1 or SUMO2 and SUMO3, which were localized mainly on the nuclear membrane and in the nucleoplasm, UBE2I strikingly colocalized with SFRS2, which is a component of nuclear speckles and critical for mRNA processing. These results suggest a novel function for UBE2I and therefore sumoylation in gene expression..
Keywords: nuclear speckle, oocyte development, oogenesis, SUMO, transcription, UBE2I
Localization of the SUMO-conjugating enzyme UBE2I within nuclear speckles in mouse oocytes suggests a novel function in RNA processing.
INTRODUCTION
Sumoylation is a posttranslational modification in which SUMO (small ubiquitin-related modifier) proteins are covalently attached to their substrates. Like ubiquitination, sumoylation is ATP dependent and requires the SUMO-activating E1 enzyme AOA1/UBA2 and the SUMO-conjugating E2 enzyme UBE2I, which can catalyze direct SUMO transfer; a variety of SUMO E3 ligases can also facilitate SUMO transfer [1–3]. Covalent modification with SUMO is achieved by formation of an isopeptide bond between the C-terminal glycine of SUMO and the ε-amino group of a lysine in the target protein. Sumoylation can result in conformational changes of target proteins, and the SUMO-modified lysine can be the same as that which can be ubiquitinated. The consequence of sumoylation is alterations in protein-protein interaction, activity, or stability, which in turn affects various cellular processes, e.g., nucleo-cytoplasmic trafficking, subcellular localization, gene expression, cell cycle progression, chromatin organization, DNA repair, apoptosis, and imprinting [1, 2].
Developmental roles of sumoylation during vertebrate development are poorly understood. Loss of Ube2i function in a chick cell line causes polynucleated and cell cycle-independent apoptosis [4]. In zebrafish embryos, expressing a dominant negative UBE2I leads to early embryonic apoptosis, and antisense morpholino oligonucleotides cause developmental defects in organogenesis [5]. In mouse embryos, loss of Ube2i causes chromosome mis-segregation and loss of nuclear integrity, and Ube2i-deficient embryos die at the early postimplantation stage [6]. The function of sumoylation during oocyte growth and maturation is even less well understood.
During its growth phase the mouse oocyte, which is arrested in diplotene of the first meiotic prophase, increases in diameter from ∼10–15 μm in a primordial resting follicle to ∼80 μm in a preovulatory antral follicle [7]. During the growth phase, oocytes sequentially acquire both meiotic competence (i.e., the ability to resume meiosis and progress to and arrest at metaphase II) [8–10] and developmental competence (i.e., the ability to be fertilized and develop to term) [11]. Accumulated organelles and macromolecules are end-products of transcription that occur during oocyte growth. Nevertheless, a progressive decrease in transcription initiates around mid-growth, the time of antrum formation, such that a fully grown oocyte is essentially transcriptionally quiescent [12, 13].
The molecular basis for the decrease in transcription is poorly defined but likely due to changes in both chromatin structure and activity of the transcription machinery. Although chromatin condensation occurs during oocyte growth [10] and could clearly contribute to the global decrease in transcription, the situation is likely far more complex. For example, oocytes deficient in nucleoplasmin 2 (Npm2) do not undergo the growth-associated changes in chromatin condensation but nevertheless undergo the decrease in transcription. Reciprocally, inducing histone hyperacetylation with trichostatin A results in chromatin decondensation in euchromatin regions—centromeric heterochromain associated with the nucleolus shows only a partial response—without any obvious increase in transcription [14].
Transcription factors/coactivators are a main target for sumoylation that usually leads to repression by a poorly understood mechanism [15, 16]. Thus, sumoylation could contribute to the global decrease in transcription that accompanies oocyte growth. As discussed above, transfer of SUMO to target proteins is similar to that for ubiquitination, except sumoylation requires only a single E2-conjugating enzyme UBE2I, making it an ideal candidate to study the function of sumoylation in oocyte development.
As an initial foray into ascertaining the role(s) of sumoylation during oocyte development, we analyzed the temporal and spatial pattern of expression of UBE2I. We report that UBE2I-containing structures present in the nucleus accumulate and increase in size during oocyte growth, and inhibiting global transcription in meiotically incompetent oocytes results in a dramatic increase in the size of the UBE2I-containing structures. In contrast, BrUTP incorporation is increased following overexpression of either wild-type or catalytically inactive UBE2I. Although the localization of SUMO1, and SUMO2 and SUMO3, changes during oocyte growth, the UBE2I-containing bodies do not completely colocalize with either SUMO1 or SUMO2 and SUMO3. Strikingly and unexpectedly, UBE2I colocalizes with SFRS2 (SC35), which is a component of nuclear speckles and critical for mRNA processing [17].
MATERIALS AND METHODS
Oocyte and Embryo Collection, Culture, and Microinjection
Collection and culture of meiotically incompetent oocytes, fully grown oocytes, and premplantation embryos and microinjection were performed as previously described [18, 19]. Briefly, fully grown germinal vesicle (GV)-intact oocytes were collected from 6- to 8-wk-old CF1 female mice 46 h after eCG injection (5 IU). Embryos were obtained from CF1 female mice mated to B6D2F1/J males. Females were superovulated by eCG (5 IU) followed by an injection of 5 IU hCG 48 h later. Meiotically incompetent oocytes were obtained from 13-day-old female CF1 mice by incubating pieces of ovarian tissue in Ca2+- and Mg2+-free CZB medium [20] containing 1 mg/ml collagenase and 0.2 mg/ml DNase I at 37°C for up to 120 min. Oocytes and embryos were cultured in CZB medium or potassium simplex optimized medium [21] in an atmosphere of 5% CO2/5% O2/90% N2, respectively. Oocytes were microinjected in bicarbonate-free Whitten [22] medium containing 25 mM HEPES (pH 7.2) and 0.01% PVA as previously described [23]. All animal experiments were approved by the Institutional Animal Use and Care Committee and were consistent with National Institutes of Health guidelines. HeLa S3 cells were cultured in Dulbecco modified Eagle medium containing 10% fetal calf serum. Other materials were from commercial sources.
Antibodies
Antibodies were purchased from the following companies: anti-UBE2I antibody (Abcam, Inc., Cambridge, MA), anti-SFRS2 antibody (Sigma-Aldrich, Inc., St. Louis, MO), anti-HA antibody (Covance Laboratories, Inc., Richmond, CA), anti-SUMO1 antibody (Zymed Laboratories, Inc., San Francisco, CA), anti-SUMO2 and SUMO3 antibody (Chemicon International, Inc., Temecula, CA), anti-TUBB antibody (Sigma-Aldrich), anti-RANGAP1 antibody (Santa Cruz Biotechnology, Santa Cruz, CA), and anti-BrdU antibody (Boehringer Mannheim Biochemicals, Indianapolis, IN).
Plasmid Construction
Standard recombinant DNA techniques were used to construct the following plasmids: pIVT/non-tagged Ube2i wild type, pIVT/non-tagged Ube2i C93S, pIVT/Egfp-Ube2i, pIVT/Ube2i-Egfp, pIVT/HA-Ube2i, and pIVT/Ube2i-HA.
In Vitro Transcription
Polyadenylated capped mRNA for injections was generated using mMESSAGE mMACHINE T7 Kit and Poly(A) Tailing Kit (Ambion, Inc., Austin, TX). The template for in vitro transcription was a pIVT-plasmid containing full-length mouse Ube2i cDNA or point-mutated Ube2i cDNA, each digested with NdeI.
Quantitative Real-Time PCR
Total RNA from 20 to 25 oocytes, eggs, or embryos was isolated using an Absolutely RNA Microprep Kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions. Complementary DNA was prepared by reverse transcription of total RNA with Superscript II enzyme and random hexamer primers. One oocyte equivalent of the resulting cDNA was amplified using TaqMan probes and the ABI Prism Sequence Detection System 7000 (Applied Biosystems, Foster City, CA) according to the manufacturer's protocols. PCR conditions were 40 cycles at 95°C for 15 sec and 60°C for 60 sec. Three replicates were used for each real-time (RT) PCR reaction; a minus RT and a minus template served as controls. Quantification was normalized to endogenous Hist2h2a or external Egfp mRNA added prior to RNA isolation. The TaqMan gene expression assays used were: Mm00504077_m1 (Hist2h2a). For Ube2i, the custom TaqMan Gene Expression Assay used had the following primers and probes: forward primer 5′-CAGGTGAGAGCCAAGGACAAA-3′, reverse primer 5′-GGCCCACTGTACAGCTAACA-3′, probe 5′-CTGGCCTGCATTGATC-3′. For Egfp, the custom TaqMan Gene Expression Assay used had the following primers and probes: forward primer 5′-GCTACCCCGACCACATGAAG-3′, reverse primer 5′-CGGGCATGGCGGACTT-3′, probe 5′-CAGCACGACTTCTTC-3′.
Immunoblotting
One hundred oocytes, eggs, or embryos were transferred to 0.5-ml microcentrifuge tubes containing 10 μl of 2X Laemmli sample buffer [24] (Sigma-Aldrich). The extracts were subjected to SDS-PAGE (13% acrylamide gel) and transferred to a PVDF membrane using a semi-dry transfer apparatus. The membrane was blocked for 1 h at room temperature with ECL Advance Blocking Agent (2% w/v; Amersham Biosciences, Piscataway, NY) dissolved in Tris-buffered saline (TBS; pH 7.5) containing 0.1% Tween-20 (TBST). The membrane was then probed with the UBE2I antibody at a 1:1000 dilution overnight at 4°C, followed by horseradish peroxidase (HRP)-conjugated secondary antibody (donkey anti-goat IgG; Jackson ImmunoResearch, West Grove, PA) at a 1:20 000 dilution for 1 h at room temperature. The membrane was washed several times in TBST after each incubation. The membrane was re-probed with the TUBB antibody at a 1:50 000 dilution for 1 h at room temperature, followed by HRP-conjugated secondary antibody (donkey anti-mouse IgG; Jackson ImmunoResearch) at a 1:100 000 dilution for 1 h at room temperature. The signal was detected using an ECL Advance Western Blotting System (Amersham Biosciences).
Immunocytochemistry
Immunocytochemistry was performed as described previously [25] with minor modifications. Oocytes, eggs, preimplantation embryos, and HeLa S3 cells grown on coverslips were fixed for 20 min in PBS containing 2.5% paraformaldehyde at room temperature, followed by permeabilization with PBS containing 0.1% Triton X-100 for 15 min. After blocking in PBS containing 1% BSA and 0.01% Tween-20 (blocking solution), the cells were sequentially incubated with each primary antibody and secondary antibody for 60 min at room temperature in blocking solution. After washing with PBS, DNA was stained by an additional 30-min incubation at room temperature with 1 μM of SYTOX Green (Molecular Probes, Eugene, OR) or 4′,6′-diamidino-2-phenylindole (DAPI). Fluorescence was detected on a Leica TCS SP laser-scanning confocal microscope.
Quantification of UBE2I-Containing Bodies
The number of UBE2I-containing bodies in meiotically incompetent and competent oocytes was quantified by counting these structures from a z-stack series of confocal images. The diameters of the large-sized bodies was then measured in arbitrary units.
BrUTP Incorporation Transcription Assay
The BrUTP run-on incorporation assay was performed as described [26] with minor modifications. Briefly, oocytes were washed in PBS, and the plasma membrane was permeabilized for 2 min with 0.05% Triton X-100 in physiological buffer (PB) that consisted of 100 mM potassium acetate, 30 mM KCl, 1 mM MgCl2, 10 mM Na2HPO4, 1 mM ATP supplemented with 1 mM dithiothreitol, 0.2 mM PMSF, and 80 units/ml of RNasin (Promega, Madison, WI). Following this treatment, the oocytes were briefly washed with PB and then transferred to PB buffer containing 2 mM ATP, 0.4 mM each of GTP, CTP, and BrUTP, and 2 mM MgCl2. After a 10-min incubation at 37°C, the oocytes were washed briefly with PB, and the nuclear membrane was fixed with 2.5% paraformaldehyde in PBS for 20 min and permeabilized by a 5-min treatment in PBS containing 0.2% Triton X-100. The incorporated BrUTP was detected by immunostaining with anti-BrdU antibody.
Statistical Analysis
Quantitative Image analysis of BrUTP incorporation was performed using ImageJ v1.40 software ( http://rsb.info.nih.gov/ij ). Three high-power fields of each nucleus were selected and averaged for analysis (with the average of background subtracted). The data were analyzed with Statview software (Statview 5.0; Abacus Concepts, Berkeley, CA). Comparisons of the intensity were performed with one-way ANOVA. Differences were considered significant at P < 0.05.
RESULTS
Temporal and Spatial Patterns of Expression of UBE2I During Oocye Growth and Preimplantation Development
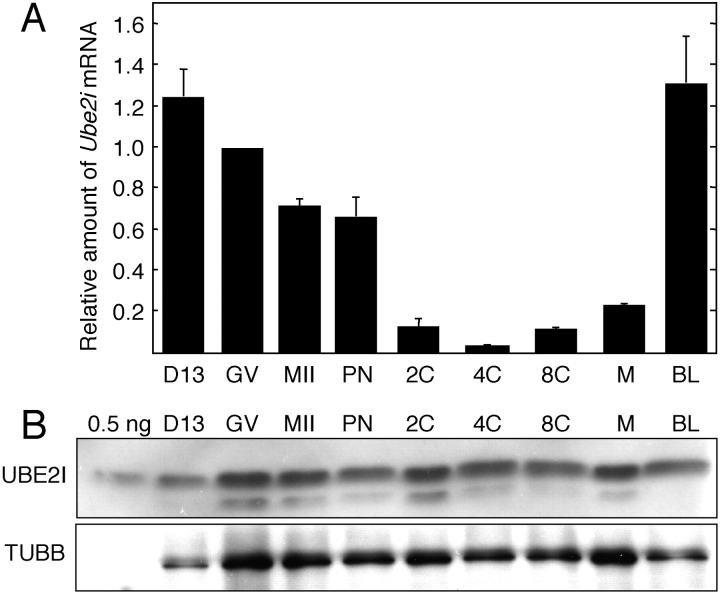
We first assessed the temporal pattern of Ube2i mRNA expression during oocyte and preimplantation development using quantitative RT-PCR (Fig. 1A). The decrease in relative abundance of Ube2i mRNA that occurs during oocyte growth is actually more pronounced because the volume of a fully grown oocyte is ∼3–3.5 times greater than that of oocytes obtained from mice 13 days of age. As with many mRNAs, Ube2i mRNA degradation initiated with the onset of oocyte maturation and continued during early preimplantation development. The increase in relative abundance of Ube2i mRNA between the 4-cell and 8-cell stages likely represented zygotic transcription. This transcription profile is similar to many maternal mRNAs (e.g., actin) that are degraded following initiation of maturation and then replaced with zygotic transcripts around the 2- to 4/8-cell stage.
FIG. 1.
Temporal pattern of Ube2i mRNA and UBE2I protein expression during oocyte growth and preimplantation development. A) Ube2i transcript abundance was determined by quantitative RT-PCR and expressed relative to fully grown GV-intact oocytes. The results shown are the mean ± SEM from three independent experiments. B) Immunoblot analysis of UBE2I in oocyte and preimplantation embryos. Immunoblots were performed using 0.5 ng of UBE2I recombinant protein or extracts derived from 100 oocytes or embryos. The blots were probed with either anti-UBE2I antibody (upper panel) or anti-TUBB antibody (lower panel). The results shown are representative of three independent experiments. 0.5 ng, 0.5 ng of recombinant UBE2I protein; D13, Day 13 incompetent oocyte; GV, fully grown oocyte; MII, metaphase II; PN, pronuclear embryo; 2C, 4C, 8C, 2-cell, 4-cell, 8-cell stage embryo, respectively; M, morula; BL, blastocyst.
Although pronounced changes in the relative abundance of Ube2i mRNA occur during oocyte growth and preimplantation development, the relative amount of UBE2I protein remained essentially constant (Fig. 1B); the faster migrating species could be a degradation product. When accounting for the difference in volume of growing and fully grown oocytes, the concentration of UBE2I in oocytes didn't markedly change. These results suggest that UBE2I is very stable. Consistent with this conclusion is that oocytes injected with Ube2i double-strand RNA (dsRNA) and then cultured in milrinone-containing medium to inhibit resumption of meiosis for 48 h resulted in a 97% decrease in Ube2i mRNA, but no detectable decrease in the amount of UBE2I protein (data not shown). In addition, although the amount of Ube2i mRNA was reduced by >60% in fully grown oocytes obtained from transgenic Ube2i that use the oocyte-specific Zp3 promoter to drive the expression of a long hairpin Ube2i dsRNA [27], no significant differences in the amount of UBE2I protein were noted when compared to nontransgenic oocytes (data not shown).
UBE2I Is Mainly Localized in the Nucleus
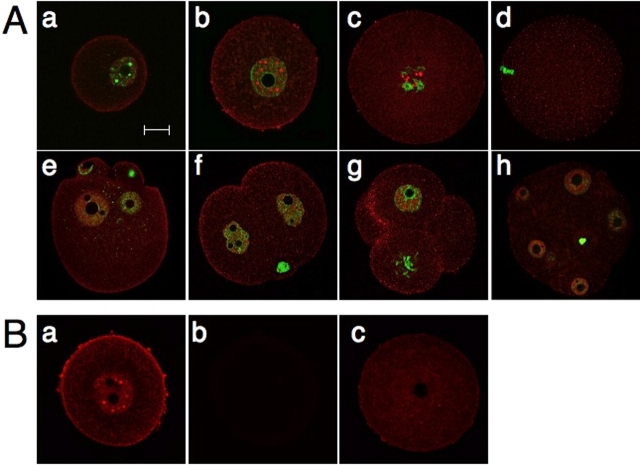
Immunocytochemical analysis revealed that endogenous UBE2I localized mainly in the nucleoplasm, with a punctate staining pattern in meiotically incompetent and fully grown oocytes (Fig. 2A, a and b). The staining observed in the region of the plasma membrane could reflect an interaction with plasma membrane-associated protein, e.g., the TGFB receptor [28]. Oocyte growth was associated with an increase in both the number and size of the UBE2I-containing structures. The specificity of the anti-UBE2I antibody was confirmed by demonstrating that any signal was barely detected when the primary antibody was omitted and that the signal intensity was substantially reduced when the antibody was incubated in the presence of recombinant UBE2I protein (Fig. 2B). UBE2I was also localized in the nucleus of preimplantation embryos, but the staining did not exhibit the pronounced punctate staining observed in oocytes (Fig. 2A, e–h).
FIG. 2.
Subcellular localization of UBE2I in oocytes and preimplantation embryos. A) UBE2I was stained with the anti-UBE2I antibody (red). The nuclei were stained with SYTOX green (green). a–h) Day 13 incompetent oocyte (a), GV oocyte (b), GVBD oocyte (c), MII (d), pronuclear embryo (e), 2-cell embryo (f), 4-cell embryo (g), and morula (h). The results shown are representative of three independent experiments in which at least 30 oocytes or embryos were examined. B) Antibody specificity. Oocytes were stained with (a) or without (b) the anti-UBE2I antibody or prior to immunostaining (c); the anti-UBE2I antibody was initially incubated with recombinant UBE2I. Bar = μm.
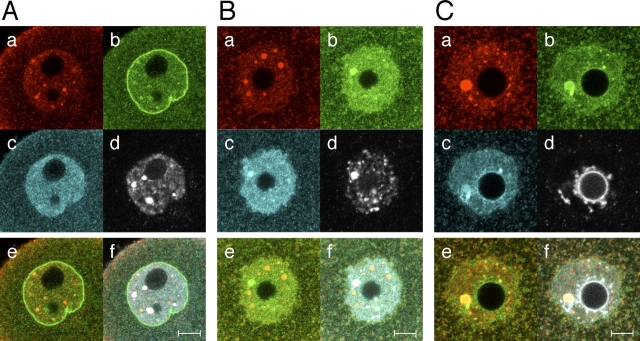
To analyze further the localization of UBE2I during oocyte development, we compared the localization of UBE2I in Day13 meiotically incompetent, transcriptionally active oocytes to that in fully grown oocytes that displayed the nonsurrounded nucleolus (NSN) DNA configuration, which are associated with reduced developmental competence and are still transcriptionally active, or those that displayed surrounded nucleolus (SN) DNA configuration, which are transcriptionally quiescent [29] (Fig. 3). In addition, because UBE2I is the principal E2-conjugating enzyme, we also examined the localization of SUMO1, and SUMO2 and SUMO3.
FIG. 3.
Subcellular localization of UBE2I, SUMO1, and SUMO2 and SUMO3 in the nucleus of oocytes at different stages of development. A) Day 13 meiotically incompetent oocyte. B) Fully grown NSN oocyte. C) Fully grown SN oocyte. In each panel, oocytes were stained with anti-UBE2I antibody (a), anti-SUMO1 antibody (b), and anti-SUMO2 and -SUMO3 antibody (c), sequentially. The nuclei were stained with DAPI (d). e) Merge of UBE2I and SUMO1 image. f) Merge of UBE2I, SUMO1, SUMO2 and SUMO3, and DAPI staining. The experiment was conducted at least four times, and shown are representative images; at least eight images were taken for each type of oocyte. Bars = 10 μm.
In incompetent oocytes, small UBE2I-containing structures were mainly detected in nucleus (Figs. 2A and 3A). Meiotically incompetent oocytes contained 45 ± 4 (mean ± SEM, n = 10) UBE2I-containing structures. The number was greater in meiotically competent oocytes that displayed either the NSN or SN configuration, 102 ± 33 (n = 11) and 115 ± 13 (n = 11), respectively. In addition, the mean diameter (in arbitrary units) of the larger sized UBE2I-containing structures was similar in meiotically incompetent oocytes and NSN competent oocytes—2.0 ± 0.1 (n = 18) and 2.0 ± 0.1 (n = 32), respectively—but was larger in SN competent oocytes, 2.7 ± 0.2 (n = 31; Fig. 3, A–C). This difference in diameter corresponds to a ∼2.5-fold increase in the volume of the larger sized UBE2I-containing structures.
Several punctate UBE2I-containing structures colocalized with SUMO1, but SUMO1 was principally localized on nuclear membrane (Fig. 3A). In contrast, SUMO2 and SUMO3 were diffusely localized in nucleoplasm and were not enriched on the nuclear membrane. The situation differed for fully grown oocytes. Interestingly, the anti-SUMO1 antibody stained the nucleoplasm, but no staining was observed on the nuclear membrane in NSN oocytes (Fig. 3B). In NSN oocytes, the SUMO1-containing structures that stained the most intensely colocalized with chromatin, whereas those that displayed a weaker intensity colocalized with UBE2I-containing bodies. On the other hand, SUMO2 and SUMO3 localized in nucleoplasm and with chromatin, but were not colocalized with UBE2I-containing bodies (Fig. 3B).
In SN oocytes, the SUMO1 frequently colocalized with UBE2I-containing structures and was only weakly detected with chromatin around the nucleolus and on the nuclear membrane (Fig. 3C). SUMO2 and SUMO3 colocalized with the large UBE2I- and SUMO1-containing bodies and localized throughout the nucleoplasm and on chromatin (Fig. 3C). In summary, UBE2I, SUMO1, and SUMO2 and SUMO3 displayed dynamic temporal changes in their distribution during the course of oocyte development.
SUMO1 staining of the nuclear envelope in meiotically incompetent oocytes could reflect sumoylation of RANGAP1, which localizes to nuclear pore complexes [30]. Thus, the loss of SUMO1 signal at the nuclear envelope could suggest that RANGAP1 sumoylation and localization are developmentally regulated. RANGAP1, however, was localized to the nuclear membrane in meiotically incompetent, as well as NSN and SN, oocytes (Supplemental Fig. S1, available online at www.biolreprod.org ). Because RANGAP1 likely remains sumoylated in NSN and SN oocytes—sumoylation is required for RANGAP1 localization to nuclear pores [30, 31]—lack of intense SUMO1 staining at the nuclear envelope may reflect a developmental change in accessibility of the anti-SUMO1 antibody and perhaps changes in nuclear import.
Distribution of UBE2I Is Associated with Transcriptional Activity
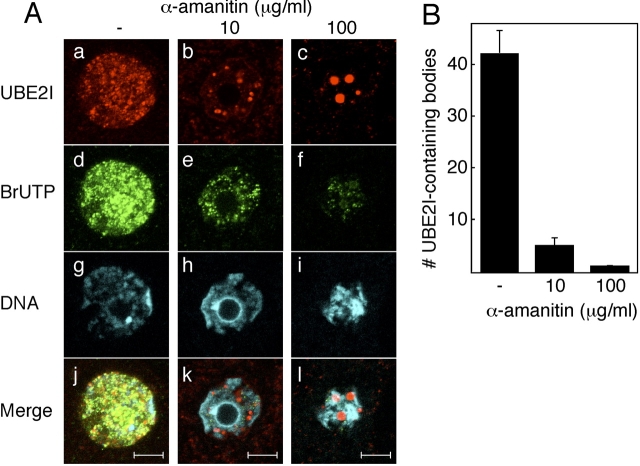
Sumoylation of transcription factors and cofactors often leads to transcription repression. We noted that associated with oocyte growth and the associated global inhibition of transcription were that the number and size of the UBE2I-containing structures increased. To explore this correlation further, we treated meiotically incompetent, transcriptionally active oocytes with α-amanitin to inhibit transcription and then assessed the effect on the UBE2I-containing structures. In the absence of α-amanitin the majority of UBE2I-containing structures colocalized with sites of nascent transcription, i.e., sites of BrUTP incorporation (Fig. 4A). Whether this reflects true colocalization or is merely the consequence of overlap due to the widespread BrUTP stained cannot be resolved at this time (Supplemental Fig. S2, available online at www.biolreprod.org ). When incompetent oocytes were incubated in medium containing α-amanitin (10 μg/ml and 100 μg/ml) for 12 h or 24 h, the size of the UBE2I-containing bodies dramatically increased, and the number of UBE2I-containing bodies decreased in a concentration-dependent manner (Fig. 4, A and B), but the UBE2I-containing bodies did not colocalize with the condensed chromatin (Supplemental Fig. S1). These changes are consistent with the UBE2I-containing structures coalescing in response to inhibiting transcription. Results of these experiments suggest there may be a relationship between transcription and the UBE2I-containing bodies and therefore a role for sumoylation.
FIG. 4.
Reorganization of UBE2I-containing bodies in response to α-amanitin. A) Localization of UBE2I-containing bodies and sites of transcription. Day 13 incompetent oocytes were incubated for 12 h without (a, d, g, and j) or with 10 μg/ml (b, e, h, and k) or 100 μg/ml (c, f, i, and l) of α-amanitin. a–c) UBE2I was stained with anti-UBE2I antibody. d–f) BrUTP was stained with anti-BrUTP antibody. g–i) Chromatin was stained with DAPI. j–l) Merge of UBE2I, BrUTP, and DAPI staining images. The experiment was conducted three times, and shown are representative images; at least five images were taken for each type of oocyte. Bar = 10 μm. B) Quantification of the effect of α-amanitin on the number of UBE2I-containing bodies. The number of UBE2I-containing dots in the nucleus of each ooocyte was counted after reconstruction of 3D images. Three oocytes were examined in each of three independent experiments. The data are presented as mean ± SEM.
Transcription Is Stimulated by Overexpression of UBE2I, Independent of Its Catalytic Activity
To explore further a role for UBE2I in transcription, we generated nontagged wild-type UBE2I (UBE2I WT) and a catalytically inactive and potentially dominant-negative version of UBE2I (UBE2I C93S), in which C93 was changed to a serine reside; C93 is essential for forming the thioester linkage critical for its E2 ligase activity [32]. We used an untagged version of UBC because the localization of tagged UBE2I, such as GFP-, HA-, and T7-UBE2I differed from that of endogenous UBE2I (Supplemental Fig. S3, available online at www.biolreprod.org , and data not shown). The inappropriate localization of the fusion or epitope-tagged UBE2I proteins may inhibit UBE2I interactions with binding partners or substrates. In fact, C-terminal epitopes (Myc and Flag) suppressed the activity of yeast Ube2i in vivo, possibly through altered interactions with other components of the SUMO conjugation pathway [33].
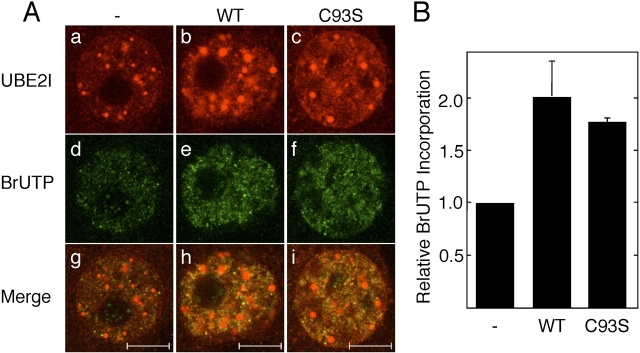
When meiotically incompetent oocytes were injected with nontagged Ube2i mRNA encoding either UBE2I WT or UBE2I C93S, the size of UBE2I-containing bodies in the nucleus and the intensity of the nucleus and cytoplasm increased relative to controls (Fig. 5A), demonstrating overexpression of UBE2I. Compared to water-injected oocytes, BrUTP incorporation was increased to a similar extent whether the WT or catalytically inactive versions were expressed (Fig. 5, A and B). Injection of nontagged UBE2I WT or UBE2I C93S recombinant protein also stimulated BrUTP incorporation (Supplemental Fig. S4, available online at www.biolreprod.org ). These results suggest that the relationship between the increase in size of UBE2I-containing bodies and the global decrease in transcription that occurs during oocyte growth (see above) is not causal. Moreover, the results strongly imply that the observed increase in transcription observed in response to UBE2I is independent of its catalytic activity.
FIG. 5.
UBE2I stimulates BrUTP incorporation independent of catalytic activity. A) Effect of UBE2I on BrUTP incorporation. Day 13 meiotically incompetent oocytes were injected with water (a, d, and g), wild-type Ube2i mRNA (b, e, and h), or Ube2i C93S mRNA (c, f, and i). The oocytes were then cultured for 18 h and then immunostained for UBE2I (a–c) and an anti-BrUTP antibody (d–f). g–i) Merged images of UBE2I and BrUTP staining. The experiment was performed three times, and shown are representative images. At least 50 oocytes were examined. The bars represent 10 μm. B) Quantification of BrUTP incorporation. For each experiment, the control was set to a value of 1, and the data (mean ± SEM) are expressed relative to the control, water-injected oocytes.
UBE2I Colocalizes with SFRS2
SFRS2 is a major component of nuclear speckles, which are nuclear organelles that control pre-mRNA splicing and mRNA export [34], and SFRS2-containing structures have been observed in mouse oocytes [35]. Interestingly, inhibiting transcription in oocytes results in a dramatic decrease in the number of SFRS2 foci as well as an increase in their size [36], a finding similar to that we observed for UBE2I-containing structures following α-amanitin treatment. Accordingly, we ascertained whether UBE2I colocalizes with nuclear speckles in oocytes (both incompetent and competent) and somatic cells (Fig. 6).
FIG. 6.
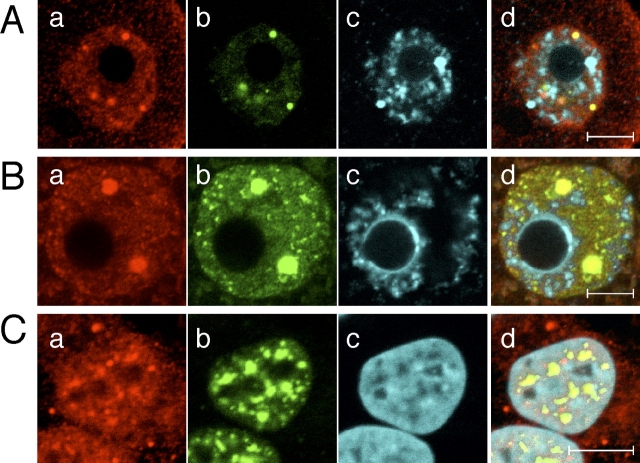
Colocalization of UBE2I with SFRS2. Subcellular localization of UBE2I and SFRS2 in the nucleus of a Day 13 incompetent oocyte (A), a fully grown oocyte (B), and Hela S3 cells (C). a–d) UBE2I (a), SFRS2 (b), DNA (DAPI) (c), and merged image (d) of UBE2I, SFRS2, and DAPI staining. The experiment was performed three times, and shown are representative images. At least 30 oocytes were examined. Bars = 10 μm.
In meiotically incompetent oocytes, several UBE2I-containing bodies did not overlap with nuclear speckles, but all UBE2I-containing bodies overlapped with nuclear speckles in fully grown oocytes (Fig. 6, A and B). Although the staining pattern of UBE2I in HeLa cells differed from that in incompetent and fully grown oocytes, the bulk of UBE2I colocalized with SFRS2-containing bodies (Fig. 6C). Similar results were observed with HEK293T cells (data not shown). These data indicate that UBE2I is a component of nuclear speckles and therefore is implicated in transcription via RNA processing.
DISCUSSION
Many nuclear proteins implicated in gene expression, e.g., transcription regulators and promyelocytic leukemia nuclear bodies (PML-NBs)-associated proteins, can be sumoylated [1–3]. Because oocyte growth is associated with a global decrease in transcription, we explored whether UBE2I is involved in this change. Results of our studies provide tantalizing evidence of a novel role for UBE2I in transcription in particular, because it colocalizes in nuclear speckles.
Our immunocytochemical data also suggest that SUMO1 and SUMO2 and SUMO3 play different roles for oocyte development. In somatic cells SUMO1 is diffusely distributed in nucleoplasm, is associated with the nuclear membrane, nucleoli, and chromatin, and is concentrated within PML-NBs [3]. In contrast, SUMO2 and SUMO3 do not accumulate on the nuclear membrane [37, 38]. Although SUMO2 and SUMO3 are diffusely distributed in nucleoplasm and associate with chromatin during oocyte growth, changes in localization of SUMO1 are readily observed between incompetent oocytes and fully grown oocytes displaying either the NSN or SN DNA configuration. SUMO1 localizes to the nuclear membrane in incompetent oocytes, colocalizes with heterochromatic regions but minimally with UBE2I-containing bodies in NSN oocytes, and colocalizes with UBE2I-containing bodies in SN oocytes. These changes in localization of SUMO proteins suggest that different proteins are sumoylated during oocyte growth and may contribute to changes in the global patterns of gene expression that likely underlie acquisition of meiotic competence.
Since UBE2I was identified [39], UBE2I has been reported to localize at nuclear envelope and nucleoplasm [40–42]. We find that in oocytes, UBE2I displays a highly punctate localization within the nucleoplasm. Of interest is that transcriptionally quiescent SN oocytes have larger UBE2I-containing bodies than transcriptionally active, meiotically incompetent growing oocytes or NSN oocytes, and that genome activation during the 2-cell stage is associated with the appearance of smaller UBE2I-containing bodies. This finding suggests that this change may be linked to the global decrease in transcription associated with oocyte growth, a linkage consistent with the dramatic increase in size of the UBE2I-containing bodies in response to inhibiting transcription in meiotically incompetent oocytes. Such an interpretation, however, is clearly simplistic, because overexpressing UBE2I, which results in an increase in the size of the UBE2I-containing bodies, also stimulates transcription. Strikingly, this stimulation of transcription is independent of UBE2I's catalytic activity and the potential for this catalytically inactive form to function as a dominant-negative. The molecular basis for UBE2I's ability to stimulate transcription independent of its ligase activity is not understood. Noteworthy is that PIAS (protein inhibitor of activated STAT) proteins, which are SUMO E3 ligases and enhance the efficiency of sumoylation [43], can modulate subnuclear targeting or assembly of transcription complexes independent of PIAS ligase activity [43]. Like PIAS proteins, UBE2I can bind to many proteins [43], which may account for its ability to stimulate transcription independent of catalytic activity.
The nucleus contains many well-studied subnuclear organelles, e.g., nucleoli, PML-NBs, Cajal bodies, and nuclear speckles [34]. UBE2I colocalizes with SFRS2, a main component of nuclear speckles. Nuclear speckles are subnuclear structures that control pre-mRNA splicing and mRNA export [17]. Although UBE2I is necessary for formation of PML-NBs [44], to date, there are no reports of UBE2I colocalizing with nuclear speckles or any other subnuclear structures involved in mRNA processing and nuclear export.
Nuclear speckles contain not only mRNA processing factors, but also many different factors involved in transcription and production of functional mRNAs, such as transcription factors, RNA polymerase II subunits, cleavage and polyadenylation factors, and RNA export proteins [45]. The finding that overexpressing UBE2I results in an increase in nuclear speckle size and stimulates transcription independent of catalytic activity suggests that UBE2I may serve as a scaffold protein for nuclear speckle assembly. The increase in nuclear speckle size presumably reflects recruitment of additional factors involved in mRNA maturation that in turn could result in an increase in overall transcription. In addition, UBE2I localization to nuclear speckles may indicate that these structures also serve as “sumoylation factories” in which proteins are sumoylated (or become associated with UBE2I independent of catalytic activity) as they transiently pass through, also accounting for the low signal intensity of SUMO in the nuclear speckles. In fact, nuclear speckles are thought to coordinate splicing and gene expression [17, 46, 47]. Recently, several proteins involved in mRNA processing, such as HNRNP family proteins, SART1, CPSF73, symplekinand, and poly(A) polymerase, have been reported to be SUMO-binding proteins or sumoylated proteins [48–52], and indeed, poly(A) polymerase interacts strongly with UBE2I [52]. PIAS1 was identified as a spliceosome-associated factor in a proteomic analysis of the human spliceosome [53]. Thus, sumoylation appears associated with the RNA processing machinery, and some substrates may be sumoylated during their transit through speckle bodies. Taken together, our results suggest a novel function for UBE2I and therefore sumoylation in regulation of gene expression by coordinating transcription and RNA processing.
Supplementary Material
Acknowledgments
We are grateful to Drs. G. Dreyfuss (University of Pennsylvania) and A. Kikuchi and S. Tashiro (Hiroshima University) for helpful discussions.
Footnotes
1This work was supported by a grant from the National Institutes of Health (HD22681) to R.M.S. and the Naito Foundation to M.I.
REFERENCES
- Johnson E.Protein modification by SUMO. Annu Rev Biochem 2004; 73: 355–382.. [DOI] [PubMed] [Google Scholar]
- Zhao J.Sumoylation regulates diverse biological processes. Cell Mol Life Sci 2007; 64: 3017–3033.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss-Friedlander R, Melchior F.Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol 2007; 8: 947–956.. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Seki M, Maeda D, Wang W, Kawabe Y, Seki T, Saitoh H, Fukagawa T, Yagi H, Enomoto T.Ubc9 is essential for viability of higher eukaryotic cells. Exp Cell Res 2002; 280: 212–221.. [DOI] [PubMed] [Google Scholar]
- Nowak M, Hammerschmidt M.Ubc9 regulates mitosis and cell survival during zebrafish development. Mol Biol Cell 2006; 17: 5324–5336.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacerddine K, Lehembre F, Bhaumik M, Artus J, Cohen-Tannoudji M, Babinet C, Pandolfi P, Dejean A.The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev Cell 2005; 9: 769–779.. [DOI] [PubMed] [Google Scholar]
- Schultz RM.Oogenesis and the control of meiotic maturation. Pedersen R, Rossant J.Experimental Approaches to Mammalian Embryonic Development New York: Cambridge University Press;1986: 195–237.. [Google Scholar]
- Sorensen R, Wassarman P.Relationship between growth and meiotic maturation of the mouse oocyte. Dev Biol 1976; 50: 531–536.. [DOI] [PubMed] [Google Scholar]
- Wickramasinghe D, Albertini D.Centrosome phosphorylation and the developmental expression of meiotic competence in mouse oocytes. Dev Biol 1992; 152: 62–74.. [DOI] [PubMed] [Google Scholar]
- Wickramasinghe D, Ebert K, Albertini D.Meiotic competence acquisition is associated with the appearance of M-phase characteristics in growing mouse oocytes. Dev Biol 1991; 144: 162–172.. [DOI] [PubMed] [Google Scholar]
- Eppig J, Schroeder A.Capacity of mouse oocytes from preantral follicles to undergo embryogenesis and development to live young after growth, maturation, and fertilization in vitro. Biol Reprod 1989; 41: 268–276.. [DOI] [PubMed] [Google Scholar]
- Moore G.The RNA polymerase activity of the preimplantation mouse embryo. J Embryol Exp Morphol 1975; 275: 447–458.. [PubMed] [Google Scholar]
- Moore G, Lintern-Moore S.Transcription of the mouse oocyte genome. Biol Reprod 1978; 18: 865–870.. [DOI] [PubMed] [Google Scholar]
- De La Fuente R, Viveiros M, Burns K, Adashi E, Matzuk M, Eppig J.Major chromatin remodeling in the germinal vesicle (GV) of mammalian oocytes is dispensable for global transcriptional silencing but required for centromeric heterochromatin function. Dev Biol 2004; 275: 447–458.. [DOI] [PubMed] [Google Scholar]
- Girdwood D, Tatham M, Hay R.SUMO and transcriptional regulation. Semin Cell Dev Biol 2004; 15: 201–210.. [DOI] [PubMed] [Google Scholar]
- Hay R.SUMO: a history of modification. Mol Cell 2005; 18: 1–12.. [DOI] [PubMed] [Google Scholar]
- Hall L, Smith K, Byron M, Lawrence J.Molecular anatomy of a speckle. Anat Rec A Discov Mol Cell Evol Biol 2006; 288: 664–675.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda P, Stein P, Hayashi H, Schultz R.Selective reduction of dormant maternal mRNAs in mouse oocytes by RNA interference. Development 2000; 127: 4147–4156.. [DOI] [PubMed] [Google Scholar]
- Svoboda P, Stein P, Schultz R.RNAi in mouse oocytes and preimplantation embryos: effectiveness of hairpin dsRNA. Biochem Biophys Res Commun 2001; 287: 1099–1104.. [DOI] [PubMed] [Google Scholar]
- Chatot CL, Ziomek CA, Bavister BD, Lewis JL, Torres I.An improved culture medium supports development of random-bred 1-cell mouse embryos in vitro. J Reprod Fertil 1989; 86: 679–688.. [DOI] [PubMed] [Google Scholar]
- Erbach GT, Lawitts JA, Papaioannou VE, Biggers JD.Differential growth of the mouse preimplantation embryo in chemically defined media. Biol Reprod 1994; 50: 1027–1033.. [DOI] [PubMed] [Google Scholar]
- Whitten WK.Nutrient requirements for the culture of preimplantation mouse embryo in vitro. Adv Biosci 1971; 6: 129–139.. [Google Scholar]
- Kurasawa S, Schultz R, Kopf G.Egg-induced modifications of the zona pellucida of mouse eggs: effects of microinjected inositol 1,4,5-trisphosphate. Dev Biol 1989; 133: 295–304.. [DOI] [PubMed] [Google Scholar]
- Laemmli UK.Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970; 227: 680–685.. [DOI] [PubMed] [Google Scholar]
- Anger M, Stein P, Schultz R.CDC6 requirement for spindle formation during maturation of mouse oocytes. Biol Reprod 2005; 72: 188–194.. [DOI] [PubMed] [Google Scholar]
- Aoki F, Worrad D, Schultz R.Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Dev Biol 1997; 182: 296–307.. [DOI] [PubMed] [Google Scholar]
- Stein P, Svoboda P, Schultz R.Transgenic RNAi in mouse oocytes: a simple and fast approach to study gene function. Dev Biol 2003; 127: 4147–4156.. [DOI] [PubMed] [Google Scholar]
- Kang JS, Saunier EF, Akhurst RJ, Derynck R.The type I TGF-beta receptor is covalently modified and regulated by sumoylation. Nat Cell Biol 2008; 10: 654–664.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Fuente R.Chromatin modifications in the germinal vesicle (GV) of mammalian oocytes. Dev Biol 2006; 292: 1–12.. [DOI] [PubMed] [Google Scholar]
- Wilson VG, Rangasamy D.Intracellular targeting of proteins by sumoylation. Exp Cell Res 2001; 271: 57–65.. [DOI] [PubMed] [Google Scholar]
- Saitoh N, Uchimura Y, Tachibana T, Sugahara S, Saitoh H, Nakao M.In situ SUMOylation analysis reveals a modulatory role of RanBP2 in the nuclear rim and PML bodies. Exp Cell Res 2006; 312: 1418–1430.. [DOI] [PubMed] [Google Scholar]
- Schwarz SE, Matuschewski K, Liakopoulos D, Scheffner M, Jentsch S.The ubiquitin-like proteins SMT3 and SUMO-1 are conjugated by the UBC9 E2 enzyme. Proc Natl Acad Sci U S A 1998; 95: 560–564.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Waardenburg R, Duda D, Lancaster C, Schulman B, Bjornsti M.Distinct functional domains of Ubc9 dictate cell survival and resistance to genotoxic stress. Mol Cell Biol 2006; 26: 4958–4969.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handwerger K, Gall J.Subnuclear organelles: new insights into form and function. Trends Cell Biol 2006; 16: 19–26.. [DOI] [PubMed] [Google Scholar]
- Parfenov VN, Pochukalina GN, Davis DS, Reinbold R, Scholer HR, Murti KG.Nuclear distribution of Oct-4 transcription factor in transcriptionally active and inactive mouse oocytes and its relation to RNA polymerase II and splicing factors. J Cell Biochem 2003; 89: 720–732.. [DOI] [PubMed] [Google Scholar]
- Borsuk E, Vautier D, Szöllösi M, Besombes D, Debey P.Development-dependent localization of nuclear antigens in growing mouse oocytes. Mol Reprod Dev 1996; 43: 376–386.. [DOI] [PubMed] [Google Scholar]
- Ayaydin F, Dasso M.Distinct in vivo dynamics of vertebrate SUMO paralogues. Mol Biol Cell 2004; 15: 5208–5218.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh H, Hinchey J.Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J Biol Chem 2000; 275: 6252–6258.. [DOI] [PubMed] [Google Scholar]
- Seufert W, Futcher B, Jentsch S.Role of a ubiquitin-conjugating enzyme in degradation of S- and M-phase cyclins. Nature 1995; 373: 78–81.. [DOI] [PubMed] [Google Scholar]
- Lee G, Melchior F, Matunis M, Mahajan R, Tian Q, Anderson P.Modification of Ran GTPase-activating protein by the small ubiquitin-related modifier SUMO-1 requires Ubc9, an E2-type ubiquitin-conjugating enzyme homologue. J Biol Chem 1998; 273: 6503–6507.. [DOI] [PubMed] [Google Scholar]
- Saitoh H, Pizzi M, Wang J.Perturbation of SUMOlation enzyme Ubc9 by distinct domain within nucleoporin RanBP2/Nup358. J Biol Chem 2002; 277: 4755–4763.. [DOI] [PubMed] [Google Scholar]
- Zhang H, Saitoh H, Matunis M.Enzymes of the SUMO modification pathway localize to filaments of the nuclear pore complex. Mol Cell Biol 2002; 22: 6498–6508.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palvimo JJ.PIAS proteins as regulators of small ubiquitin-related modifier (SUMO) modifications and transcription. Biochem Soc Trans 2007; 35: 1405–1408.. [DOI] [PubMed] [Google Scholar]
- Bernardi R, Pandolfi P.Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol 2007; 8: 1006–1016.. [DOI] [PubMed] [Google Scholar]
- Saitoh N, Spahr C, Patterson S, Bubulya P, Neuwald A, Spector D.Proteomic analysis of interchromatin granule clusters. Mol Biol Cell 2004; 15: 3876–3890.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley D.Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol 2005; 17: 251–256.. [DOI] [PubMed] [Google Scholar]
- Komili S, Silver P.Coupling and coordination in gene expression processes: a systems biology view. Nat Rev Genet 2008; 9: 38–48.. [DOI] [PubMed] [Google Scholar]
- Li T, Evdokimov E, Shen R, Chao C, Tekle E, Wang T, Stadtman E, Yang D, Chock P.Sumoylation of heterogeneous nuclear ribonucleoproteins, zinc finger proteins, and nuclear pore complex proteins: a proteomic analysis. Proc Natl Acad Sci U S A 2004; 101: 8551–8556.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassileva M, Matunis M.SUMO modification of heterogeneous nuclear ribonucleoproteins. Mol Cell Biol 2004; 24: 3623–3632.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vertegaal A, Ogg S, Jaffray E, Rodriguez M, Hay R, Andersen J, Mann M, Lamond A.A proteomic study of SUMO-2 target proteins. J Biol Chem 2004; 279: 33791–33798.. [DOI] [PubMed] [Google Scholar]
- Vethantham V, Rao N, Manley J.Sumoylation modulates the assembly and activity of the pre-mRNA 3' processing complex. Mol Cell Biol 2007; 27: 8848–8858.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vethantham V, Rao N, Manley J.Sumoylation regulates multiple aspects of mammalian poly(A) polymerase function. Genes Dev 2008; 22: 499–511.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J, Ryder U, Lamond A, Mann M.Large-scale proteomic analysis of the human spliceosome. Genome Res 2002; 12: 1231–1245.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.