Abstract
DNA cytosine methylation is crucial for retrotransposon silencing and mammalian development. In a computational search for enzymes that could modify 5-methylcytosine (5mC), we identified TET proteins as mammalian homologs of the trypanosome proteins JBP1 and JBP2, which have been proposed to oxidize the 5-methyl group of thymine. We show here that TET1, a fusion partner of the MLL gene in acute myeloid leukemia, is a 2-oxoglutarate (2OG)- and Fe(II)-dependent enzyme that catalyzes conversion of 5mC to 5-hydroxymethylcytosine (hmC) in cultured cells and in vitro. hmC is present in the genome of mouse embryonic stem cells, and hmC levels decrease upon RNA interference–mediated depletion of TET1. Thus, TET proteins have potential roles in epigenetic regulation through modification of 5mC to hmC.
5-methylcytosine (5mC) is a minor base in mammalian DNA: It constitutes ∼1% of all DNA bases and is found almost exclusively as symmetrical methylation of the dinucleotide CpG (1). The majority of methylated CpG is found in repetitive DNA elements, suggesting that cytosine methylation evolved as a defense against transposons and other parasitic elements (2). Methylation patterns change dynamically in early embryogenesis, when CpG methylation is essential for X-inactivation and asymmetric expression of imprinted genes (3). In somatic cells, promoter methylation often shows a correlation with gene expression: CpG methylation may directly interfere with the binding of certain transcriptional regulators to their cognate DNA sequences or may enable recruitment of methyl-CpG binding proteins that create a repressed chromatin environment (4). DNA methylation patterns are highly dysregulated in cancer: Changes in methylation status have been postulated to inactivate tumor suppressors and activate oncogenes, thus contributing to tumorigenesis (5).
Trypanosomes contain base J (β-d-glucosyl hydroxymethyluracil), a modified thymine produced by sequential hydroxylation and glucosylation of the methyl group of thymine (fig. S1A) (6). J biosynthesis requires JBP1 and JBP2, enzymes of the 2OG- and Fe(II)-dependent oxygenase superfamily predicted to catalyze the first step of J biosynthesis (7, 8). Like 5-methylcytosine, base J has an association with gene silencing: It is present in silenced copies of the genes encoding the variable surface glycoprotein (VSG) responsible for antigenic variation in the host but is absent from the single expressed copy (6). We performed a computational search for homologs of JBP1 and JBP2 in the hope of identifying mammalian enzymes that modified 5mC.
Iterative sequence profile searches using the predicted oxygenase domains of JBP1 and JBP2 recovered homologous regions in three paralogous human proteins TET1, TET2, and TET3 and their orthologs found throughout metazoa (e < 10−5), as well as homologous domains in fungi and algae (fig. S2 and SOM Text). Secondary structure predictions suggested the existence of an N-terminal α helix followed by a continuous series of β strands, typical of the double-stranded β helix (DSBH) fold of the 2OG-Fe(II) oxygenases (fig. S3) (9). A multiple sequence alignment showed that the new TET/JBP family displayed all of the typical features of 2OG-Fe(II) oxygenases, including conservation of residues predicted to be important for coordination of the cofactors Fe(II) and 2OG (fig. S3 and SOM Text). The metazoan TET proteins contain a unique conserved cysteine-rich region, contiguous with the N terminus of the DSBH region (Fig. 1A and SOM Text). Vertebrate TET1 and TET3, and their orthologs from all other animals, also possess a CXXC domain, a binuclear Zn-chelating domain, found in several chromatin-associated proteins, that in certain cases has been shown to discriminate between methylated and unmethylated DNA (10).
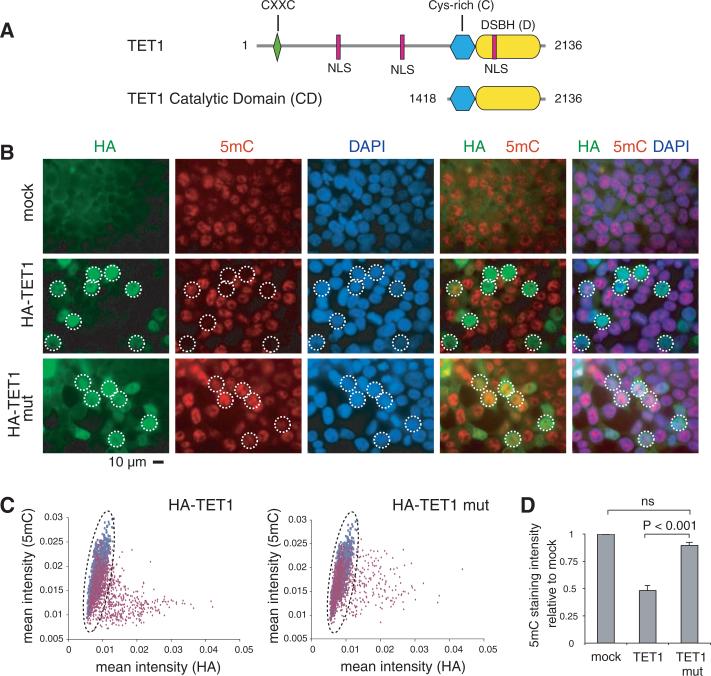
Fig. 1.
Expression of TET1 in HEK293 cells results in decreased 5mC staining. (A) Predicted domain architecture of human TET1 showing the CXXC-type zinc-binding domain (amino acid 584 to 624); the cysteine-rich region (Cys-rich) (amino acid 1418 to 1610); the DSBH domain (amino acid 1611 to 2074); and three bipartite nuclear localization sequences (NLS). (B) HEK293 cells overexpressing wild-type or mutant HA-TET1 were costained with antibodies specific for the HA epitope (green) and 5mC (red). To orient the reader, some HA-expressing cells are circled. Scale bar, 10 μm. (C) Staining intensities of HA and 5mC were measured in individual nuclei. Data from HA-TET1–expressing cell populations are presented as dot plots (red), superimposed on dot plots from mock-transfected cells (blue), with each dot representing an individual cell. (D) Quantification of 5mC staining intensity of HA-positive cells compared with that of mock-transfected cells (set to 1). Data shown are mean ± SEM and are representative of three experiments.
As a first step in determining whether TET proteins operate on 5mC, we transfected human embryonic kidney (HEK) 293 cells with full-length hemagglutinin (HA)–tagged TET1, then stained the transfected cells for 5mC and the HA epitope. Mock-transfected cells showed substantial variation in 5mC staining intensity (fig. S4; Fig. 1B, top panel; quantified in Fig. 1C), either because 5mC levels vary from cell to cell or because the accessibility of 5mC to the antibody differs among cells because of technical considerations (e.g., incomplete denaturation of DNA). Cells transfected with wild-type TET1 showed a strong correlation of HA positivity with decreased staining for 5mC, both visually (Fig. 1B, middle panel) and by quantification (Fig. 1C, left panel, and Fig. 1D). Untransfected HA-low cells showed a spread of 5mC staining intensity similar to that of mock-transfected cells (Fig. 1C, left panel; note overlapping red and blue dots at low HA intensities), whereas productively transfected HA-high cells showed uniformly low 5mC staining intensity (Fig. 1C, left panel, red HA-high dots). Cells transfected with mutant TET1 bearing H1671Y, D1673A substitutions predicted to impair Fe(II) binding did not show decreased staining for 5mC (Fig. 1B, bottom panel; Fig. 1C, right panel; and Fig. 1D).
To determine quantitatively whether TET1 overexpression affects intracellular 5mC levels, we measured the ratio of 5mC to C at a subset of genomic CpG sites in cells expressing either full-length TET1 (TET1-FL) or the predicted catalytic domain of TET1 [TET1-CD, comprising the Cys-rich (C) and DSBH (D) regions; Fig. 1A]. HEK293 cells were transiently transfected with plasmids in which TET1 expression was coupled to expression of human CD25 from an internal ribosome entry site (IRES). Genomic DNA from CD25-expressing cells was digested with MspI, which cleaves DNA at the sequence C^CGG regardless of whether the second C is methylated. The resulting fragments, whose 5′ ends derive from the dinucleotide CpG and contain either C or 5mC, were end-labeled and digested to yield 5′ phosphorylated dNMPs that were resolved using thin-layer chromatography (TLC) (Fig. 2A) (11).
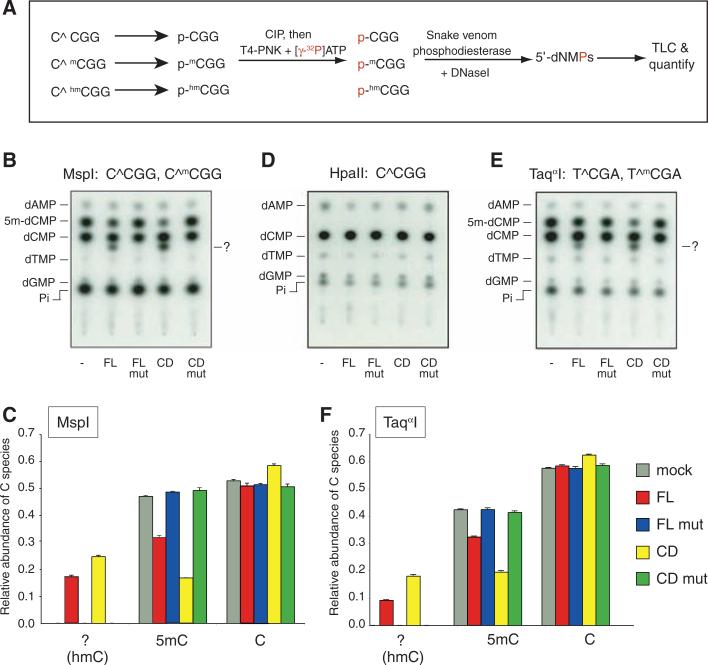
Fig. 2.
Genomic DNA of TET1-overexpressing cells contains a modified nucleotide within the dinucleotide CG. (A) Description of experimental design. (B to F) Genomic DNA was purified from HEK293 cells overexpressing TET1 and cleaved with [(B) and (C)] MspI, (D) HpaII, or [(E) and (F)] TaqαI. The fragments were end-labeled, digested to 5′ dNMPs, and resolved by TLC. A modified nucleotide (subsequently identified as hm-dCMP) is indicated by ?. Neither 5m-dCMP nor the modified nucleotide are observed when the DNA is digested with HpaII. [(C) and (F)] Quantification of the relative abundance of dCMP, 5m-dCMP, and the modified nucleotide. Data shown are mean ± SD of three independent transfections and are representative of at least three experiments.
MspI-digested DNA from cells transfected with the control vector yielded predominantly dCMP and 5m-dCMP as expected (Fig. 2B, lane 1), whereas DNA from cells expressing wild-type TET1-FL or TET1-CD yielded an additional unidentified labeled species migrating more slowly than dCMP (“?,” Fig. 2B, lanes 2 and 4). This new species was not detected when the DNA was digested with HpaII, the methylation-sensitive isoschizomer of MspI (Fig. 2D) but was clearly observed when the DNA was digested with TaqαI, a methylation-insensitive enzyme that cuts at a different sequence, T^CGA (Fig. 2E). The new species was not observed in MspI- or TaqαI-digested DNA from cells transfected with mutant TET1-FL or TET1-CD (Fig. 2, B and E, lanes 3 and 5), and its appearance was associated with decreased abundance of 5m-dCMP (Fig. 2, B and E, lanes 2 and 4; quantified in Fig. 2, C and F). In all experiments, expression of wild-type TET1-CD correlated with a small but significant increase in the abundance of dCMP (Fig. 2, C and F). Together these results suggest that the unidentified species is derived through modification of 5m-dCMP and may be an intermediate in the passive or active conversion of 5mC to C.
We used high-resolution mass spectrometry (MS) to identify the novel nucleotide. Genomic DNA was prepared from HEK293 cells overexpressing wild-type or mutant TET1-CD. A singly charged species with an observed mass/charge ratio (m/z) of 336.0582, consistent with a molecular formula of C10H15NO8P–, was the only species migrating at the expected position on TLC that exhibited a large (by a factor of about 19) difference in abundance between the wild-type and mutant samples (Fig. 3, A and B). Based on this result, our computational analyses, and the data of Fig. 2, we hypothesized that the unidentified species was 5-hydroxymethylcytosine (hmC), produced by TET1 through hydroxylation of the methyl group of 5mC (fig. S1). As a standard, we prepared authentic hm-dCMP from unglucosylated DNA of T4 phage grown in Escherichia coli ER1656, a strain deficient in the glucose donor molecule UDP-glucose (abbreviated T4* DNA) (12). TLC assays showed that the novel nucleotide generated in unsorted cells expressing TET1-CD migrated similarly to hm-dCMP from T4* DNA (Fig. 3A). Tandem mass spectrometry (MS-MS) fragmentation experiments at several collision energies (15 and 25 V) in both positive and negative ion modes confirmed that the fragmentation pattern of the 336.0582 dalton ion was identical to that of hm-dCMP (Fig. 3C and fig. S5).
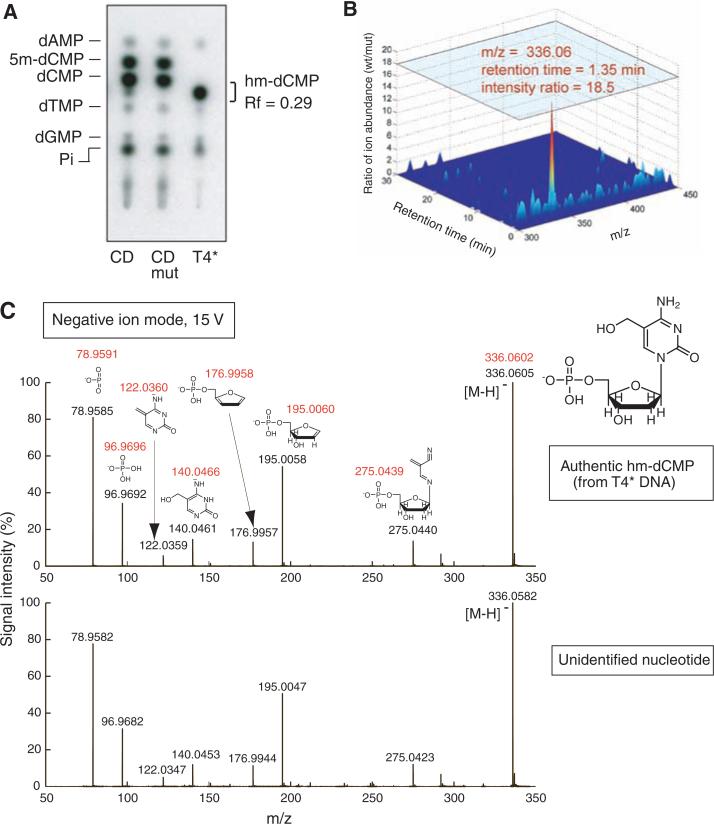
Fig. 3.
The modified nucleotide is identified as 5-hydroxymethylcytosine. (A) Genomic DNA from T4 phage grown in UDP-glucose–deficient E. coli ER1656 (T4*) and HEK293 cells transfected with wild-type or mutant TET1-CD were digested with TaqαI. The fragments were end-labeled, digested to mononucleotides, and separated by TLC. The modified nucleotide present in TET1-CD–expressing cells migrates similarly to authentic hm-dCMP. (B) Comparison of liquid chromatography–electrospray ionization MS ions present at an Rf = 0.29 in genomic DNA from cells expressing wild-type (wt) or mutant (mut) TET1-CD. A species with an observed m/z = 336.06 was more abundant by a factor of 18.5 in the wild-type sample compared with the mutant sample. (C) Mass spectrometry fragmentation (MS/MS) analysis of authentic hm-dCMP (top), and the m/z = 336.06 species isolated from cells expressing TET1-CD (bottom). Expected m/z values are shown in red; observed m/z values are shown in black (anticipated mass accuracy is within 0.002 Da).
To establish that TET1 was directly responsible for hmC production, we expressed Flag-HA-tagged wild-type and mutant TET1-CD in Sf9 insect cells, purified the recombinant proteins to near homogeneity (fig. S6A), and assayed their catalytic activity on fully methylated double-stranded DNA oligonucleotides. Wild-type, but not mutant, TET1-CD catalyzed robust conversion of 5mC to hmC and displayed an absolute requirement for both Fe(II) and 2OG (Fig. 4A; quantified in Fig. 4B). Omission of ascorbate did not result in a significant decrease in catalytic activity, most likely because we included dithiothreitol in the reaction to counteract the strong tendency of TET1-CD to oxidize (fig. S6, A to D) (13-15). Recombinant TET1-CD was specific for 5mC: We did not detect conversion of thymine to hydromethyluracil (hmU) (fig. S7).
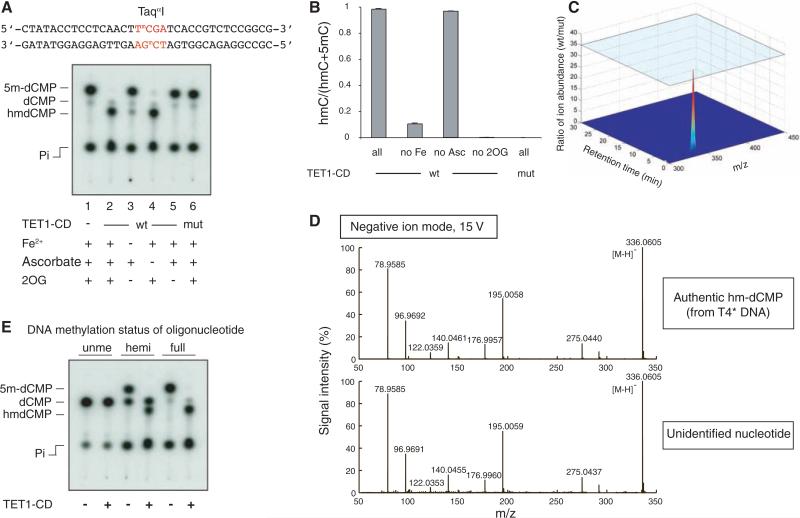
Fig. 4.
Recombinant Flag-HA-TET1-CD purified from Sf9 cells converts 5mC to hmC in methylated DNA oligonucleotides in vitro. (A) Double-stranded DNA oligonucleotides containing a fully methylated TaqαI site were incubated with wt or mut Flag-HA-TET1-CD (1:10 enzyme to substrate ratio). Recovered oligonucleotides were digested with TaqαI and analyzed by TLC. The faint dCMP spot in each lane is derived from end-labeling of the C at the 5′ end of each strand of the substrate. (B) The extent of conversion of 5mC to hmC is shown as the mean ratio [hmC/(hmC+5mC)] ± SD. (C) Comparison of species at an Rf of 0.29 in products resulting from incubation with wt or mut Flag-HA-TET1-CD. (D) MS fragmentation analysis of authentic hm-dCMP (top) and the nucleotide generated by Flag-HA-TET1-CD (bottom). Observed masses are shown in black (mass accuracy was within 0.002 Da). (E) Recombinant Flag-HA-TET1-CD is able to hydroxylate 5mC in fully methylated (full, lanes 5 and 6) and hemimethylated (hemi, lanes 3 and 4) substrates. Unme, unmethylated DNA oligonucleotide (lanes 1 and 2).
We used high-resolution MS to demonstrate, as before, that a singly charged species at m/z of 336.0582 was the only species migrating at the expected position that differed significantly (by a factor of 35) in abundance when comparing substrates incubated with wild-type and mutant proteins (Fig. 4C). MS-MS experiments confirmed that the fragmentation pattern of the species produced by recombinant TET1-CD was identical to that of authentic hm-dCMP in unglucosylated T4 DNA (Fig. 4D). TET1-CD was also able to oxidize 5mC to hmC in hemimethylated double-stranded DNA (Fig. 4E).
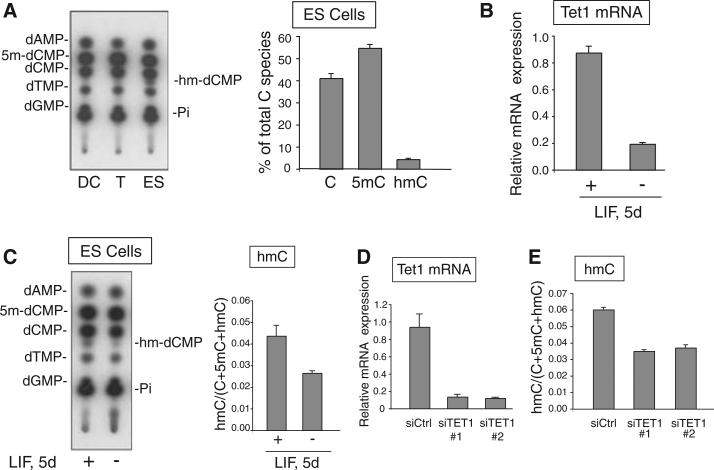
We asked whether hmC was a physiological constituent of mammalian DNA. Using the TLC assay, we observed a clear spot corresponding to labeled hmC in mouse embryonic stem (ES) cells but not in previously activated human T cells or mouse dendritic cells (Fig. 5A). Quantification of multiple experiments indicated that hmC and 5mC constituted 4 to 6% and 55 to 60%, respectively, of all cytosine species in MspI cleavage sites (C^CGG) in ES cells (Fig. 5A). Tet1 mRNA levels declined by 80% in response to leukemia inhibitory factor (LIF) withdrawal for 5 days, compared with the levels observed in undifferentiated ES cells (Fig. 5B); in parallel, hmC levels diminished from 4.4 to 2.6% of total C species, a decline of ∼40% from control levels (Fig. 5C). The difference might be due to the compensatory activity of other Tet-family proteins. Similarly, RNA interference (RNAi)–mediated depletion of endogenous Tet1 resulted in an 87% decrease in Tet1 mRNA levels and a parallel ∼40% decrease in hmC levels (Fig. 5, D and E). Again, the difference is likely due to the presence of Tet2 and Tet3, which are both expressed in ES cells.
Fig. 5.
hmC is present in ES cell DNA, and its abundance decreases upon differentiation or Tet1 depletion. (A) (Left) TLC showing that hmC is detected in the genome of undifferentiated ES cells but not dendritic cells or T cells. (Right) Quantification of the relative abundance by PhosphorImager of 5mC, C, and hmC in the genomic DNA of undifferentiated ES cells. (B) Tet1 mRNA levels decline by ∼80% in ES cells induced to differentiate by withdrawal of LIF for 5 days. (C) The same differentiated ES cells show ∼40% decrease in hmC levels. (D) Transfection of ES cells using two different RNAi duplexes directed against Tet1 decreases Tet1 mRNA levels by ∼75%. (E) The same Tet1-depleted ES cells show ∼40% decline in hmC levels. Data shown are mean ± SD and are representative of 2 to 3 experiments.
Together these data strongly support the hypothesis that Tet1, and potentially other Tet family members, are responsible for hmC generation in ES cells under physiological conditions (fig. S8A). CpG dinucleotides are ∼0.8% of all dinucleotides in the mouse genome (16); thus, hmC (which constitutes ∼4% of all cytosine species in CpG dinucleotides located in MspI cleavage sites) is ∼0.032% of all bases (∼1 in every 3000 nucleotides, or ∼2 × 106 bases per haploid genome). For comparison, 5mC is 55 to 60% of all cytosines in CpG dinucleotides in MspI cleavage sites (Fig. 5A), about 14 times as high as hmC (hmC may not be confined to CpG) (SOM Text). An important question is whether hmC and TET proteins are localized to specific regions of ES cell DNA—for instance, genes that are involved in maintaining pluripotency or that are poised to be expressed upon differentiation. A full appreciation of the biological importance of hmC will require the development of tools that allow hmC, 5mC, and C to be distinguished unequivocally (SOM Text).
As a potentially stable base (SOM Text), hmC may influence chromatin structure and local transcriptional activity by recruiting selective hmC-binding proteins or excluding methyl-CpG–binding proteins (MBPs) that normally recognize 5mC, thus displacing chromatin-modifying complexes recruited by MBPs (fig. S8B, center). Indeed, it has already been demonstrated that the methyl-binding protein MeCP2 does not recognize hmC (17). Alternatively, conversion of 5mC to hmC may facilitate passive DNA demethylation by excluding the maintenance DNA methyltransferase DNMT1, which recognizes hmC poorly (fig. S8B, left) (18). Even a minor reduction in the fidelity of maintenance methylation would be expected to result in an exponential decrease in CpG methylation over the course of many cell cycles. Finally, hmC may be an intermediate in a pathway of active DNA demethylation (fig. S8B, right). hmC has been shown to yield cytosine through loss of formaldehyde in photooxidation experiments (19) and at high pH (20, 21), leaving open the possibility that hmC could convert to cytosine under certain conditions in cells. A related possibility is that specific DNA repair mechanisms replace hmC or its derivatives with C (22, 23). In support of this hypothesis, a glycosylase activity specific for hmC was reported in bovine thymus extracts (24). Moreover, several DNA glycosylases, including TDG and MBD4, have been implicated in DNA demethylation, although none of them has shown convincing activity on 5mC in in vitro enzymatic assays (25-27). Cytosine deamination has also been implicated in demethylation of DNA (26-28); in this context, deamination of hmC yields hmU, and high levels of hmU:G glycosylase activity have been reported in fibroblast extracts (29).
These studies alter our perception of how cytosine methylation may be regulated in mammalian cells. Notably, disruptions of the TET1 and TET2 genetic loci have been reported in association with hematologic malignancies (SOM Text). A fusion of TET1 with the histone methyltransferase MLL has been identified in several cases of acute myeloid leukemia (AML) associated with t(10;11)(q22;q23) translocation (30, 31). Homozygous null mutations and chromosomal deletions involving the TET2 locus have been found in myeloproliferative disorders, suggesting a tumor suppressor function for TET2 (32, 33). It will be important to test the involvement of TET proteins and hmC in oncogenic transformation and malignant progression.
Supplementary Material
Supporting Online Material for
Conversion of 5-Methylcytosine to 5-Hydroxymethylcytosine in Mammalian DNA by MLL Partner TET1
Mamta Tahiliani, Kian Peng Koh, Yinghua Shen, William A. Pastor, Hozefa Bandukwala, Yevgeny Brudno, Suneet Agarwal, Lakshminarayan M. Iyer, David R. Liu,* L. Aravind,* Anjana Rao*
*To whom correspondence should be addressed. E-mail: arao@idi.harvard.edu (A.R.); aravind@ncbi.nlm.nih.gov (L.A.); drliu@fas.harvard.edu (D.L.)
Published 16 April 2009 on Science Express DOI: 10.1126/science.1170116
This PDF file includes:
Materials and Methods
SOM Text
Figs. S1 to S8
References
Supplementary Text
Identification of a novel family of 2OG-Fe(II) oxygenases with predicted 5-methylpyrimidine oxidase activity. To identify candidate enzymes that catalyze the oxidative modification of 5-methylcytosine, we created a position-specific score matrix (profile) of known 2OG-Fe(II) oxygenases that included the predicted oxygenase domains of JBP1 and JBP2 (Fig. S1). We used this profile to conduct a systematic search of the non-redundant database using the PSI-BLAST program (1). This search recovered homologous domains in the gp2 proteins from the mycobacteriophages Cooper and Nigel and a related prophage from the Frankia alni genome (e<10−4). We then generated a new profile which included the gp2 protein sequences, and performed a second search of the protein sequence database of microbes from environmental samples. This search detected numerous homologous proteins potentially derived from uncultured marine phages and prophages. A further search of the non-redundant database, with the newly-detected proteins from the environmental sequences added to the profile, recovered homologous regions in the three paralogous human oncogenes TET1 (CXXC6), TET2 and TET3 and their orthologs found throughout metazoa (e<10−5). Homologous domains were also found in fungi and algae. In PSI-BLAST searches, these groups of homologous domains consistently recovered each other prior to recovering any other member of the 2OG-Fe(II) oxygenase superfamily, suggesting that they constitute a distinct family (LM Iyer, L Aravind, manuscript in preparation).
To confirm the relationship of the newly-identified proteins (hereafter referred to as the TET/JBP family; see legend to Fig. 1) with classical 2OG-Fe(II) oxygenases, we prepared a multiple alignment of their shared conserved domains and used this to generate a hidden markov model (HMM). A profile-profile comparison of this HMM against a library of HMM's generated for all structurally-characterized domains from the Protein Database (PDB) with the HHpred program (2) resulted in recovery of prolyl hydroxylase (e<10−12), a canonical member of the 2OG-Fe(II) oxygenase superfamily, as the best hit. Secondary structure predictions suggested the existence of an N-terminal α-helix followed by a continuous series of β-strands, typical of the double-stranded β-helix (DSBH) fold of the 2OG-Fe(II) oxygenases (Fig. 1A, bottom) (3). A multiple sequence alignment (Fig. 1A) further showed that the new TET/JBP family displayed all of the typical features of 2OG-Fe(II) oxygenases including: (i) the HxD signature, just downstream of an N terminal β-strand which chelates Fe(II) (x is any amino acid); (ii) a small residue, usually glycine, at the beginning of the strand immediately downstream of the HxD motif, which helps in positioning the active site arginine; (iii) the Hxs motif (where s is a small residue) in the C-terminal part, in which the H chelates the Fe(II) and the small residue helps in binding the 2-oxo acid; (iv) the Rx5a signature (where a is an aromatic residue) downstream of the above motif – the R in this motif forms a salt bridge with the 2-oxo acid and the aromatic residue helps position the first metal-chelating histidine (the key residues are marked with asterisks in Fig 1A). These observations strongly suggested that members of the TET/JBP family, including TET1, 2 and 3, are catalytically-active 2OG-Fe(II) oxygenases.
Additionally, the metazoan TET proteins contain a unique conserved cysteine-rich region, contiguous with the N-terminus of the DSBH region, which possesses at least eight conserved cysteines and one histidine that are likely to comprise a binuclear metal cluster (Fig. 1A, top). Vertebrate TET1 and TET3, and their orthologs from all other animals, also possess a CXXC domain, a binuclear Zn-chelating domain with eight conserved cysteines and one histidine, located N-terminal to the 2OG-Fe(II) oxygenase domain (shown schematically for TET1 in Fig. 1B). Analysis of the architectures of CXXC domain proteins suggests that the CXXC domain is an accessory DNA-binding domain, that tends to be combined in the same polypeptide with a variety of domains that possess diverse chromatin-modifying and modification recognition activities. The CXXC domain is found in several chromatin-associated proteins, including the methyl-DNA-binding protein MBD1, the histone methyltransferase MLL, the DNA methyltransferase DNMT1 (4) and the lysine demethylases KDM2A (JHDM1A/ FBXL11) and KDM2B (JHDM1B/FBLX10, which also contains a ubiquitin E3 ligase domain) (5), and in certain cases has been shown to discriminate between methylated and unmethylated DNA (6).
Taken together, the contextual information gleaned from these domain architectures and from gene neighborhoods in the bacteriophage members (Supplementary Information) supported a conserved DNA modification function for the entire TET/ JBP family, namely oxidation of 5-methyl-pyrimidines. In this study we tested the specific hypothesis that TET proteins might operate on 5mC to catalyze oxidation or oxidative removal of the methyl group, focusing on TET1 as a mammalian example of the TET/JBP family.
hmC may not be confined to CpGs. Nearest-neighbour analyses have shown that 15−20% of total 5mC in ES cells is present in CpT, CpA and (to a minor extent) CpC sequences; in contrast, somatic tissues have negligible non-CpG methylation (7).
Previous studies on hmC. With the exception of two papers that reported high levels of hmC in genomic DNA (8), most previous studies have described hmC as a rare base that is a probable oxidation product of 5mC (9, 10).
Methods of distinguishing hmC, 5mC and C. We show here that two of the three most commonly used techniques do not adequately distinguish between C, 5mC and hmC. A widely-used mouse monoclonal antibody to 5mC apparently does not recognize hmC by immunocytochemistry (Fig. 2A), thus it will be important to reevaluate previous reports of DNA demethylation based solely on the use of this antibody. Similarly, the methylation-sensitive restriction enzyme, HpaII, fails to cut hmC (Fig. 3D) as previously reported (11), raising the possibility that in some instances hmC-modified DNA was incorrectly judged to be methylated. Another methylation-sensitive restriction enzyme, McrBC, is already known to cleave 5mC- and hmC-containing DNA equivalently (12), and therefore also does not allow these two nucleotides to be distinguished.
It is yet to be determined how bisulfite modification analysis interprets the presence of hmC in DNA. Treatment of DNA with sodium bisulfite promotes the spontaneous deamination of cytosine to uracil, while leaving 5mC unaffected; amplification of the sequence of interest followed by sequencing allows the precise methylation patterns at a given sequence to be determined (13). It is known that bisulfite reacts rapidly with hmC at the C5 to form a stable cytosine 5-methylenesulfonate adduct, which is not readily deaminated (14). This substituted species, which is expected to form base pairs similar to those formed by cytosine, could be read by polymerases as C during the amplification steps, resulting in the sequence being interpreted as containing 5mC. Alternatively, polymerases may not copy cytosine 5-methylenesulfonate efficiently, in which case the DNA containing this adduct would not be amplified effectively and the sequence containing the original hmC modification would be underrepresented in the amplified DNA.
Stability of hmC versus nitrogen-linked hydroxymethyl groups. The stability of hmC in the genome of T-even phages (15) contrasts with the well-documented instability of N-linked hydroxymethyl adducts generated by the DNA repair enzymes AlkB and the JmjC domain-containing histone demethylases, which are also 2OG and Fe(II)-dependent oxygenases. These nitrogen-linked adducts spontaneously resolve, yielding the unmethylated amino group and formaldehyde (16); in contrast, oxidation of carbon-linked methyl groups by JBP1/2 and thymine hydroxylase does not result in removal of the methyl adduct via oxidation (17, 18). This difference, which is due to the fact that carbon is a much poorer leaving group than nitrogen, explains our ability to detect hmC in genomic DNA.
Mechanisms of DNA demethylation. The mammalian DNA methyltransferases DNMT1, DNMT3A and DNMT3B, establish and maintain DNA methylation patterns (19, 20). Preventing maintenance methylation through successive replication cycles progressively dilutes the methyl mark and results in “passive” DNA demethylation (21). “Active” (replication-independent) DNA demethylation has been convincingly demonstrated only for the paternal genome shortly after fertilization (22−24); potential mechanisms could involve thermodynamically unfavorable cleavage of the carbon-carbon bond linking the methyl group to the pyrimidine, resulting in release of the methyl moiety, or a repair-like process in which the methylated base or nucleotide is excised and the lesion is then repaired to replace the original 5mC with an unmethylated C (reviewed in (25, 26)). This latter mechanism occurs in plants, where DEMETER, a bifunctional glycosylase restricted to flowering plants, cleaves the glycosidic bond of 5mC and stimulates insertion of an unmethylated C through base-excision repair (27). A variety of candidate mammalian demethylase activities have been described, but unfortunately, none of these reports have been confirmed by other laboratories (reviewed in (19, 28−30)).
Roles of TET proteins in cancer. The human LCX (leukemia-associated protein with a CXXC domain) / Tet1 (ten-eleven translocation-1) gene was originally defined as a novel MLL fusion partner in cases of pediatric and adult AML with t(10;11)(q22;23) (4, 31). In a recent study, the MLL/Tet1 fusion accounted for 3/759 (0.4%) of MLL-associated leukemia (32), with two cases of ALL, together showing occurrence of the MLL/Tet1 fusion in both pediatric and adult ALL and AML. Possible pathogenic mechanisms of the MLL/Tet1 fusion include altered genomic targeting of Tet1 catalytic activity and/or disrupted recruitment of cofactors to sites of Tet-1 mediated hmC generation. Homozygous Tet2 deletions / loss of function mutations have recently been reported in approximately 14% of patients with JAK2V617F-positive and negative myeloproliferative neoplasms (including polycythemia vera (PV), essential thrombocythemia (ET), primary myelofibrosis (MF), post-PV MF, post-ET MF, and blast phase PV/ET/MF) (33, 34); approximately 29% of patients with systemic mastocytosis (35); and other myeloid malignancies including chronic myelomonocytic leukemia, myelodysplastic syndrome, and acute myeloid leukemia (36, 37). It will be interesting to investigate the association of Tet2 loss of function, epigenetic alterations such as promoter hypermethylation that characterize myeloproliferative neoplasms, and response to epigenetic therapy (38).
Fig. S1. Modification of 5-methylpyrimidines. (A) Trypanosomes contain a modified version of thymine, called base J (β-D-glucosyl hydroxymethyluracil), in their genome. It has been proposed that base J is synthesized by sequential hydroxylation and glucosylation of the methyl group of thymine (17). (B) TET1, and presumably the other TET family members, oxidize the methyl group of 5-methylcytosine (5mC) generating 5-hydroxymethylcytosine (hmC).
Fig. S2. Sequences used to generate the position-specific score matrix (PSSM). The figure shows the sequences used to create the position specific score matrix that retrieved the TET proteins in searches with the PSI-BLAST program. Each sequence is identified by the gi number allowing its recovery from the GenBank database and the numbers flanking the alignment provide the limits of the aligned region in the sequence. The JBP proteins from kinetoplastids are marked in red. The searches were performed using the checkpoint start option –B, with–h set to 0.01 and –F was set to F and the all JBP sequences were cycled through the –i options for individual searches.
Fig. S3. Multiple sequence alignment of the TET/JBP family and representative 2OG-Fe(II) dependent dioxygenase domains. The cysteine-rich region (C) and the core catalytic domain (D) are aligned separately. Proteins are labeled by their gene name and species, separated by underscores. The 95% consensus was calculated from a larger alignment of the TET/JBP proteins. The consensus for the TET-specific C-terminal strands was calculated separately from a larger alignment specifically of the metazoan TET homologs. In addition to the TET/JBP family, the alignment also contains the structurally characterized representatives of the dioxygenase superfamily, the Chlamydomonas prolyl hydroxylase (P4H) and the E. coli AlkB (2FD8) along with their PDB codes.
Fig. S4. Image analysis using CellProfiler. Nuclei were outlined based on DAPI fluorescence using the IdentifyPrimAutomatic module and denoted as Outlined Nuclei. Objects within a pre-set diameter range of 10−35 pixel units are outlined in green and included in analysis. Objects outside the range (including cell clusters) as well as those touching the borders are outlined in red and excluded from analysis. To account for HA staining at nuclear boundaries, an IdentifySecondary module was added to expand the nuclear outlines by 2 pixels, denoted as ExpandedNuclei Outlines. The MeasureObjectIntensity module was then used to apply the ExpandedNuclei Outlines on the corresponding HA image, and the Outlined Nuclei on the corresponding 5mC image (not shown), to measure pixel intensities of HA and 5mC staining respectively. Pixel numbers are indicated at the axes of images, which are taken at a magnification of 20x.
Fig. S5. Mass spectrometry fragmentation analyis (MS/MS) of authentic hm-dCMP from T4* phage (top) and the unidentified nucleotide species present in genomic DNA from HEK293 cells overexpressing TET1-CD (bottom). (A) MS/MS analysis was performed in negative ion mode with a collision energy of 25 V. (B) MS/MS analysis was performed in positive ion mode with a collision energy of 25 V. Observed masses are shown (anticipated mass accuracy was within 0.003 Da).
Fig. S6. Purification of recombinant Flag-HA-TET1-CD from Sf9 cells and characterization of TET1-CD fragment. (A) Coomassie-stained SDS-PAGE gel of wild-type and mutant Flag-HA-TET1-CD purified from Sf9 cells to near homogeneity by affinity chromatography with anti-Flag antibody conjugated beads. Known amounts of BSA were loaded on the same gel for comparison. Wild-type and mutant TET1-CD (79 KDa, indicated by the arrow) both demonstrate a strong tendency to oxidize and form disulfide-linked dimers (158 KDa) and higher-order multimers, which are resistant to DTT. Asterisks denote degradation products that are not detected by immunoblotting (see (B)), probably due to loss of the N-terminal Flag-HA epitope tag. (B) The bands of apparent higher molecular weight were identified as TET1 oxidation products by immunoblotting with an anti-HA antibody. (C) Immunoblot with anti-Flag antibody showing that the multimeric forms TET1-CD increase with increased processing time of lysates and concomitant exposure to oxidizing conditions. A twenty minute lysis in 10 mM DTT-containing RIPA buffer results in the detectable presence of a TET1-CD dimer (lane 1). This dimer is not detected when cell pellets from Flag-HA-TET1-CD overexpressing cells are lysed directly in denaturing Laemlli SDS sample buffer containing 700 mM β-ME (lane 3). The tendency of TET1-CD to oxidize appears to be due at least in part to disulfide bond formation by the Cys-Rich region (C) that is N-terminal to the DSBH (D) region (lanes 2, 4). Removal of the N-terminal Cys-Rich region resulted in expression of a protein (Flag-HA-TET1-D) that runs at its expected molecular weight (57.6 KDa) after extraction in either RIPA buffer or SDS sample buffer. Together the data shown in (A-C) suggest that intermolecular disulfide bonds that are resistant to reducing agent are formed during extraction. (D) Overexpression of Flag-HA-TET1-CD in HEK293 cells, but not Flag-HA-TET1-D, results in decreased staining by an anti-5mC antibody.
Fig. S7. Recombinant Flag-HA-TET1-CD purified from Sf9 cells does not convert thymine (another 5-methylpyrimidine) to hmU in vitro. (A) Synthetic double-stranded DNA oligonucleotides containing a fully-methylated TaqαI (T^mCGA) or a SalI (G^TCGAC) site were incubated with purified Flag-HA-TET1-CD or mutant Flag-HA-TET1-CD for 5 hours at 37 C (1:10 enzyme to substrate ratio). (B) Recovered oligonucleotides were digested with TaqαI or SalI, end-labeled, hydrolyzed to dNMP's and resolved using TLC. TET1-CD is able to hydroxylate 5mC, but is not able to act on thymine in the context of a SalI site in a double-stranded oligonucleotide substrate in vitro. A double-stranded DNA oligonucleotide terminating in hmU was end-labelled and hydrolyzed to generate a standard for hm-dUMP migration (lane 5). SalI has been demonstrated to cleave G(hmU)CGAC equivalently to GTCGAC (39).
Fig. S8. Model speculating on the biological role of hmC and its potential as an intermediate in DNA demethylation. (A) Integration of hmC into the known pathways of DNA methylation and passive replication-dependent demethylation. Known pathways are shown in black and the new findings in this study in red. Dashed lines and question marks are used to indicate pathways that may exist but have not been experimentally established. (B) Potential biological mechanisms involving hmC. hmC may recruit specialized binding proteins; and conversion of 5mC to hmC may displace methyl-binding proteins from DNA (center). hmC may also be an intermediate that facilitates passive DNA demethylation: conversion of 5mC to hmC in hemi-methylated DNA may interfere with recognition by DNMT1 and associated SRA-domain proteins (left). Finally, hmC may convert to cytosine through a spontaneous or enzymatic process or it may be recognized by specialized DNA repair proteins in specific cell types, and so function as an intermediate in active DNA demethylation (right).
Methods
Computational and bioinformatic analyses. The non-redundant (NR) database of protein sequences (National Center for Biotechnology Information, NIH, Bethesda) was searched using the PSI-BLAST programs (1). Profile searches using the PSI-BLAST program were conducted either with a single sequence or a sequence with a PSSM used as the query, with a profile inclusion expectation (E) value threshold of 0.01, and were iterated until convergence (1). For all compositionally biased queries the correction using composition-based statistics was used in the PSI-BLAST searches (40). Multiple alignments were constructed using the Kalign program (41), followed by manual correction based on the PSI-BLAST results. The multiple alignment was used to create a HMM using the Hmmbuild program of the HMMER package (42). It was then optimized with Hmmcaliberate and the resulting profile was used to search a database of completely sequenced genomes using the Hmmsearch program of the HMMER package (42). Profile-profile searches were performed using the HHpred program (2, 43). The JPRED program (44) and the COILS program were used to predict secondary structure. Globular domains were predicted using the SEG program with the following parameters: window size 40, trigger complexity=3.4; extension complexity=3.75 (45).
The Swiss-PDB viewer (46) and Pymol programs were used to carry out manipulations of PDB files. Reconstruction of exon-intron boundaries was done using the NCBI Splign program with the tblastn searches against chromosomes as a guide. Gene neighborhoods were determined using a custom script that uses completely sequenced genomes or whole genome shot gun sequences to derive a table of gene neighbors centered on a query gene. Then the BLASTCLUST program is used to cluster the products in the neighborhood and establish conserved co-occurring genes. These conserved gene neighborhood are then sorted as per a ranking scheme based on occurrence in at least one other phylogenetically distinct lineage (“phylum” in NCBI Taxonomy database), complete conservation in a particular lineage (“phylum”) and physical closeness on the chromosome indicating sharing of regulatory −10 and −35 elements. Phylogenetic trees were constructed with the MEGA4 package.
TET1 expression plasmids TET1 ORF was amplified from SY5Y cDNA and the human clone and inserted into XhoI and NotI sites of an Flag-HA tagged pOZ-N. Mutant TET1 (H1671D, Y1673A) was generated using the QuikChange Mutagenesis Kit (Stratagene). The sequences of all clones were confirmed by conventional DNA sequencing. Wild-type and mutant Flag-HA-TET1-CD was amplified and cloned into Acc651 and XbaI sites of pEF1 (Invitrogen). The IRES-CD25 of pOZ-N was amplified and cloned into the the XbaI and BstB1 sites of Flag-HA-TET1-CD-pEF1. Wild-type and mutant Flag-HA-TET1-CD was amplified from Flag-HA-TET1-CD-pOZ and inserted into SalI and NotI sites of pFastBac (Invitrogen).
Immunocytochemistry Cells were plated on sterile coverglass in 24-well plates at 1.5 × 105 cells/well and grown overnight before transient transfection with pEF1-TET1 expression constructs or empty vector (mock) using TransIT™-293 transfection reagent (Mirus, Madison, WI) according to manufacturer's instructions. At 42−44 hr post-transfection, cells were fixed for 15 min in 4% paraformaldehyde in PBS and permeabilized with 0.2% Triton X-100 in PBS for 15 min at room temperature. For detection of 5-methylcytosine, cells were treated with 2N HCl at room temperature for 30 min and subsequently neutralized for 10 min with 100 mM Tris-HCl buffer, pH 8. After extensive washes in PBS, cells were blocked for 1 hour at room temperature in 1%BSA, 0.05% Tween 20 in PBS. Rabbit anti-HA polyclonal antibody (diluted at 1:400; Santa Cruz Biotechnology, Santa Cruz, CA ) and mouse anti-5 methylcytosine clone 162 33 D3 antibody (diluted at 1:2500−3000; Calbiochem, San Diego, CA) were added in blocking buffer for 2−3 hours at room temperature and detected concurrently by secondary antibodies coupled with Cy2 or Cy3 respectively. DNA was stained with 250 ng/ml of4’,6-diamidino-2-phenylindole (DAPI) and mounted in SlowFade® Gold antifade reagent (Molecular Probes, Eugene, OR). Images were recorded digitally on a Zeiss Axiovert 200 inverted microscope equipped with a CCD camera by using OpenLab imaging software (Improvision, Coventry, UK).
CellProfiler™ cell image analysis. Three fields, each containing 200−400 cells, were imaged from each well of transfected cells using an 20x objective and pooled for analysis in each experiment. Greyscale images (tiffs) were uploaded on CellProfiler as three individual files for every field captured under the three excitation wavelengths respectively for DAPI, GFP (detection of HA) and Cy3 (detection of 5mC) (47). Nuclear outlines were profiled based on DAPI staining, with a secondary module included to expand the nuclear outlines by another 2 pixels (denoted as “expanded nuclei”) to account for HA staining at nuclear boundaries. Clustered cells and cells at the edge of fields were excluded. Staining intensities of HA and 5mC within individual cells profiled were measured as mean pixel intensities of GFP and Cy3 signals, respectively, within the “expanded nuclei” and original nuclei profiles, respectively. The raw data were plotted as dot plots of 5mC mean pixel intensity versus HA mean pixel intensity for each transfection sample. The same set of mock-transfected cells is shown in the two panels in Fig. 1C. The population average of 5mC staining intensity of HA-expressing cells (arbitrarily categorized as cells with mean HA pixel intensity above the highest intensity observed in mock-transfected cells) is compared to mock transfected cells. Values are background-subtracted before normalization and are mean ± SEM from 3 experiments; statistical comparisons are based on ANOVA and Bonferroni's post-hoc test.
Transfection and Sorting of HEK293T cells expressing hCD25 HEK293T cells were transfected with TET1-CD-IRES-CD25-pEF1 vectors using TransIT transfection reagent (Mirius). Control cells (mock) were transfected with a corresponding empty vector that drove expression of CD25 alone. After 48 hours, 30 × 106 cells were stained with anti-hCD25-PE antibody (1:200) (Becton Dickinson) in 1X FACS Buffer (1XPBS, 2% FBS, 1 mM EDTA, 0. 1% NaAzide) for 30 min at 4 C. Cells were washed twice with 1X FACS Buffer. Cells were then stained with anti-PE microbeads (Miltenyi) for 25 min, 4 C and then washed two times with 1X FACS Buffer and then once with 1X MACS buffer (1X PBS, 0.5% BSA, 0.09% Na Azide and 2 mM EDTA). Cells were resuspended in 2 ml of ice-cold 1X MACS Buffer and CD25-expressing cells were sorted using an AutoMACS cells sorter (Possel). Input, flow-through and collected samples were analyzed by FACS to confirm enrichment for CD25-positive cells in collected sample.
Analysis of 5mC levels using thin-layer chromatography Nuclei were prepared from CD25-positive cells by resuspension in 1 ml NPB (240 mM sucrose, 7.5 mM Tris, pH 7.5, 3.75 mM MgCl2, 0,75% Triton-X-100, with 100 μg RNAseA/ml (Qiagen)) and placing on ice for 20 minutes. Cells were spun at 1300 g for 15 min, 4 C and then washed once in NPB. Nuclei were lysed in 650 μl of 1X LB ((10 mM Tris, pH 8.0, 300 mM NaAcetate, pH 7.2, 0.5% SDS, 5 mM EDTA, 100 μg RNAseA/ml and 300 μg/ml Proteinase K (Roche)) and incubated overnight at 55 C. An extra 300 μg/ml Proteinase K was added in the morning and the samples were left at 55 C for 5 hours. Samples were extracted with equal volumes of phenol, phenol: chloroform: isoamyl alcohol (25:24:1) and chloroform: isoamyl alcohol (24:1) and then precipitated with 2 volumes of ethanol. Genomic DNA was washed twice with 1 ml of 70% EtOH, dried and resuspended in 10 mM Tris, 0.1 mM EDTA, pH 8.0 and allowed to resuspend overnight at 32 C.
2 μg of genomic DNA was digested with 100 units of MspI, HpaII or Taqα1 and 100 μg of RNaseA (Qiagen) overnight. An extra 100 units of restriction enzyme was added in the morning and incubations were continued for 6 hours. 10 units of calf intestinal phosphatase (CIP) (NEB) was added and incubated for 1 hour at 37 C. DNA was purified using Qiaquick Nucleotide Removal Kit (Qiagen) as per the manufacturer's instructions. 400 ng of eluted DNA fragments were end-labeled with T4 Polynucleotide Kinase (T4 PNK) (NEB) and 10 μCi of [γ32-P]-ATP for 1 hour at 37 C. Labeled fragments were precipitated by the addition of 30 μg of linear polyacrylamide, 1/10 volume of 3 M NaAcetate, pH 7.2 and 2.5 volumes of ethanol at left at −80 C for 1 hour. Samples were spun at 14,000 rpm, for 20 minutes at 4 C and washed twice with 70% EtOH at 25 C. Pellets were resuspended in 30 mM Tris, pH 8.9, 15 mM MgCl2, 2 mM CaCl, with 10 μg of DNaseI (Worthington) and 10 μg SVPD (Worthington) and incubated for 3 hours at 37 C. 3 μl was spotted on cellulose TLC plates (20 cm × 20 cm, Merck) and developed in isobutyric acid: H20: NH3 (66:20:1). Plates were analyzed by phosphorimager scanning using Phosphorimager Storm 860 scanner software. The low-level labeling of other nucleotides reflects DNA shearing or contaminating endonucleolytic activity.
Preparation of unglucosylated T4 phage DNA for preparation of hm-dCMP standard T4 phage stock was titred by spotting 10 μl of serial 10X dilutions on an LB plate on which 100 μl of an overnight culture of E.coli CR63 in 3 ml of T4 top agar was poured and allowed to solidify. The plate was incubated overnight at 37 C. 10 ml of E.coli CR63 OD600 of 0.5 was infected with a single plaque of T4 phage and incubated with shaking at 37°C until the culture cleared (about 2.5 hours). The culture was incubated on ice for 10 minutes and then lysed was completed by the addition of several drops of chloroform and gentle mixing. The lysate was titred as described above.
E.coli ER1656 was grown in LB to OD600 of 0.5 and then infected with 0.2 phage per bacterium and incubated at 37°C with shaking until the culture cleared (about 8 hours).. The culture was chilled on ice for 10 minutes and then lysis was completed by the addition of 1 ml of chloroform. DNase I was added to 1 mg/ml and the culture was incubated for 2 hours at 4°C. The lysate was centrifuged at 12,000g for 10 min at 4 C to pellet debris. The supernatant was collected and phage were pelleted by centrifugation at 23,500g for 1.5 hours at 4 C. The phage pellet was left covered in TE overnight to resuspend. Phage DNA was extracted using an equal volume of phenol, phenol: chloroform: isoamyl alcohol (25:24:1) and chloroform: isoamyl alcohol (24:1). The extracted phage DNA was dialyzed into TE overnight with 2 changes of buffer.
Mass spectrometry experiments. Genomic DNA from HEK293 cells transfected with TET1 wild-type or mutant CD or T4 phage grown in E.coli 13656 were hydrolyzed to dNMP's with SVPD and DNaseI and resolved using TLC. Spots corresponding to paricular dNMP's were scraped, extracted with water, lyophilized, and re-suspended in water for liquid chromatography/mass spectrometry (LC/MS) analysis using an Acquity UPLC/Q-TOF Premier electrospray LC/ESI-MS system (Waters Corp., Milford, MA). Liquid chromatography (LC) was performed with a Waters HSS C18 column (1.0mm i.d. × 50mm, 1.8-um particles) using a linear gradient of 0% to 50% methanol in 0.1% aqueous ammonium formate, pH 6.0. The flow rate was 0.05 mL per min and the eluant was directly injected into the mass spectrometer. Mass spectra were recorded in continuum mode and converted to centroid mode to generate accurate mass spectra. Data was analyzed with Masslynx 4.1 software (Waters).
Recombinant Protein Expression and Purification Bacmid DNA was generated using DH10Bac™ E. coli E.coli (Invitrogen) as directed by the manufacturer. Transposition into the correct site was confirmed using PCR. Baculovirus was amplified for three generations using suspension adapted Sf9 cells. Sf9 cells were then infected with baculovirus for 4 days. The resulting cell pellet was kept on ice for 30 minutes in 40 mM Tris, pH 7.4, 300 mM NaCl, 0.2% NP40, 0.4% Triton, 5 mM DTT, 1X protease inhibitors without EDTA (Roche) and then at 12,000 rpm (SLA-TC600), 30 min, 4 C. The supernantant was then incubated with anti-Flag antibody-conjugated beads (Invitrogen) for 5 hours at 4 C. The beads were washed 4 times in 40 mM Tris, pH 7.4, 300 mM NaCl, 0.2% NP40, 8% Glycerol, 1X PI, 5 mM DTT and then eluted in 195 mM Tris, pH 7.4, 110 mM NaCL, 0.14% NP40, 5.8% Glycerol, 0.37X PI, 3.7mM DTT, 365 μg/ml Flag peptide. The homogeneity of the eluted protein was determined using SDS-PAGE followed by Coomassie blue staining and immunoblotting using an anti-Flag antibody (Sigma).
Preparation of double-stranded oligonucleotide substrates Synthetic oligonucleotides were purchased from IDT. All oligonucleotides were 35 nucleotides in length with the modifications shown below.
F: 5’-CTATACCTCCTCAACTTCGATCACCGTCTCCGGCG-3’
FMe: 5’-CTATACCTCCTCAACTT(mC)GATCACCGTCTCCGGCG-3’
R: 5’-Biotin-CGCCGGAGACGGTGATCGAAGTTGAGGAGGTATAG-3’
RMe: 5’-Biotin-CGCCGGAGACGGTGAT(mC)GAAGTTGAGGAGGTAT AG-3’
Oligonucleotides were annealed to the appropriate complementary oligonucleotide in 100 mM KAc, 30 mM HEPES, pH 7.5. The mixture was boiled for 5 minutes then slowly cooled to room temperature overnight. Double-stranded oligonucleotides were purified by polyacrylamide gel electrophoresis.
In vitro Enzymatic Assays 7.5 μl of recombinant protein (about 3 μg) was incubated with 2 μg of oligonucleotide substrates in 50 mM HEPES, pH 8, 50 mM NaCl, 2 mM Ascorbic Acid, 1mM 2OG, 100 μM FAS (Fe2+), and 1 mM DTT for 3 hours at 37 C. Oligonucleotide substrates were purified using Qiaquick Nucleotide Removal Kit (Qiagen) and then digested with Taqa1 overnight, treated with CIP for 1 hour and purified once more with Qiaquick Nucleotide Removal Kit (Qiagen). Purified DNA oligonucleotides were end-labeled with T4 Polynucleotide Kinase (NEB) and 10 μCi of γ32P-ATP for 1 hour at 37 C. Labeled fragments were precipitated by the addition of 30 μg of linear polyacrylamide, 1/10 volume of 3 M Sodium Acetate, pH 7.2 and 2.5 volumes of ethanol followed by incubation at −80 C for 1 hour. Samples were spun at 14,000 rpm, for 20 minutes at 4 C in a refrigerated microcentrifuge. Unincorporated radionucleotide was removed by washing two times with 70% EtOH and spinning at room temperature for 10 minutes. Pellets were resuspended in 10 μl 30 mM Tris, 15 mM MgCl2, 2 mM CaCl, pH 8.9 with 10 μg of DNaseI (Worthington) and 10 μg SVPD (Worthington) and incubated for 3 hours at 37 C. 3 μl was spotted on cellulose TLC plates (Merck) and developed in isobutyric acid: water: ammonia (66:20:1). Plates were analyzed by phosphorimager scanning using Phosphorimager Storm 860 scanner software. The faint dCMP spot in each lane is derived from end-labelling of the C at the 5’ end of each strand of the substrate. T4 PNK is not able to phophorylate blunt ends as efficiently as the 5’ overhangs generated by restriction enzyme cleavage.
ES cell culture V6.5 mouse ES cells were maintained on mitomycin C-inactivated primary mouse embryonic fibroblasts in ES medium containing DMEM (Invitrogen, Carlsbad, CA), 15% ES FBS (Omega Scientific, Tarzana, CA) , 0.1 mM each of nonessential amino acids (Invitrogen), 2 mM L-glutamine (Invitrogen), 0.1 mM β-mercaptoethanol (Invitrogen), 50 units/ml penicillin/streptomycin (Invitrogen) and 1000 U/ml ESGRO® (LIF; Chemicon). For all experiments described, cells were trypsinized and plated for 30 min on standard tissue culture dishes to remove feeder cells before floating ES cells were collected and re-plated on gelatin-coated dishes or wells. For LIF withdrawal assays, cells were plated at a density of 2−3 × 105 cells per 10-cm dish and LIF was removed the day after (day 0). RNA interference (RNAi) experiments were performed as previously described (48) using Dharmacon siGENOME siRNA duplexes (Thermo Fisher Scientific Inc, Boulder, CO) against mouse Tet1 (Cat. # D-062861−01/02). The Dharmacon siGENOME non-targeting siRNA#2 (Cat. # D-001210−02) was used as a negative control. Mouse ES cells were seeded in gelatin-coated 12-well at a density of 1 × 105 cells per well and transfected the day after (day 0) with 50 nM siRNA using Lipofectamine RNAiMAX reagent (Invitrogen) according to the manufacturer's instructions. Retransfections were performed on pre-adherant cells at day 2 at a split of 1:4 and finally at day 4 at a split of 1:2 in 6-well plates. Cells were harvested at Day 5 for RNA and thin-layer chromatography analyses.
RNA Isolation, cDNA synthesis and Quantitative Real-Time PCR Total RNA was isolated with an RNeasy kit (Qiagen, Chatsworth, CA) with on-column DNase treatment. cDNA was synthesized with 0.5 μg total RNA using SuperScript III reverse transcriptase (Invitrogen). Quantitative PCR was performed using FastStart Universal SYBR Green Master mix (Roche, Mannheim, Germany) on a StepOnePlus real-time PCR system (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions. The levels of gene expression were normalized to Gapdh. Primer sequences are: Tet1 forward 5’-GAGCCTGTTCCTCGATGTGG-3’, Tet1 reverse 5’-CAAACCCACCTGAGGC TGTT-3’; Gapdh forward 5’-GTGTTCCTACCCCCAATG TGT-3’, Gapdh reverse 5’-ATTGTCATACCAGGAAATGAGCTT-3’.
References
1. S. F. Altschul et al., Nucleic Acids Res 25, 3389 (Sep 1, 1997).
2. J. Soding, A. Biegert, A. N. Lupas, Nucleic Acids Res 33, W244 (Jul 1, 2005).
3. L. Aravind, E. V. Koonin, Genome Biol 2, RESEARCH0007 (2001).
4. R. Ono et al., Cancer Res 62, 4075 (Jul 15, 2002).
5. Y. Tsukada et al., Nature 439, 811 (Feb 16, 2006).
6. M. D. Allen et al., Embo J 25, 4503 (Oct 4, 2006).
7. B. H. Ramsahoye et al., Proc Natl Acad Sci U S A 97, 5237 (May 9, 2000).
8. N. W. Penn, R. Suwalski, C. O'Riley, K. Bojanowski, R. Yura, Biochem J 126, 781 (Feb, 1972).
9. J. R. Pratt et al., Transplant Proc 38, 3344 (Dec, 2006).
10. V. Valinluck, L. C. Sowers, Cancer Res 67, 946 (Feb 1, 2007).
11. S. Tardy-Planechaud, J. Fujimoto, S. S. Lin, L. C. Sowers, Nucleic Acids Res 25, 553 (Feb 1, 1997).
12. E. Sutherland, L. Coe, E. A. Raleigh, J Mol Biol 225, 327 (May 20, 1992).
13. T. Rein, M. L. DePamphilis, H. Zorbas, Nucleic Acids Res 26, 2255 (May 15, 1998).
14. H. Hayatsu, M. Shiragami, Biochemistry 18, 632 (Feb 20, 1979).
15. G. R. Wyatt, S. S. Cohen, Biochem J 55, 774 (Dec, 1953).
16. C. Loenarz, C. J. Schofield, Nat Chem Biol 4, 152 (Mar, 2008).
17. Z. Yu et al., Nucleic Acids Res 35, 2107 (2007).
18. J. A. Smiley, M. Kundracik, D. A. Landfried, V. R. Barnes, Sr., A. A. Axhemi, Biochim Biophys Acta 1723, 256 (May 25, 2005).
19. A. Bird, Genes Dev 16, 6 (Jan 1, 2002).
20. M. G. Goll, T. H. Bestor, Annu Rev Biochem 74, 481 (2005).
21. D. U. Lee, S. Agarwal, A. Rao, Immunity 16, 649 (May, 2002).
22. W. Mayer, A. Niveleau, J. Walter, R. Fundele, T. Haaf, Nature 403, 501 (Feb 3, 2000).
23. J. Oswald et al., Curr Biol 10, 475 (Apr 20, 2000).
24. F. Santos, B. Hendrich, W. Reik, W. Dean, Dev Biol 241, 172 (Jan 1, 2002).
25. H. Cedar, G. L. Verdine, Nature 397, 568 (Feb 18, 1999).
26. H. D. Morgan, F. Santos, K. Green, W. Dean, W. Reik, Hum Mol Genet 14 Spec No 1, R47 (Apr 15, 2005).
27. M. Gehring et al., Cell 124, 495 (Feb 10, 2006).
28. A. P. Wolffe, P. L. Jones, P. A. Wade, Proc Natl Acad Sci U S A 96, 5894 (May 25, 1999).
29. S. K. Ooi, T. H. Bestor, Cell 133, 1145 (Jun 27, 2008).
30. J. Jiricny, M. Menigatti, Cell 135, 1167 (Dec 26, 2008).
31. R. B. Lorsbach et al., Leukemia 17, 637 (Mar, 2003).
32. C. Meyer et al., Leukemia (Mar 5, 2009).
33. F. Delhommeau et al., paper presented at the ASH Annual Meeting and Exposition, San Francisco, CA, December 9, 2008 2008.
34. A. Tefferi et al., Leukemia (Mar 5, 2009).
35. A. Tefferi et al., Leukemia (Mar 5, 2009).
36. F. Viguie et al., Leukemia 19, 1411 (Aug, 2005).
37. A. Tefferi et al., Leukemia (Mar 19, 2009).
38. A. Tefferi, Leuk Lymphoma 49, 2231 (Dec, 2008).
39. M. Hori et al., Nucleic Acids Res 31, 1191 (Feb 15, 2003).
40. A. A. Schaffer et al., Nucleic Acids Res 29, 2994 (Jul 15, 2001).
41. T. Lassmann, E. L. Sonnhammer, Nucleic Acids Res 34, W596 (Jul 1, 2006).
42. S. R. Eddy, Bioinformatics 14, 755 (1998).
43. J. Soding, Bioinformatics 21, 951 (Apr 1, 2005).
44. J. A. Cuff, G. J. Barton, Proteins 40, 502 (Aug 15, 2000).
45. J. C. Wootton, Comput Chem 18, 269 (Sep, 1994).
46. N. Guex, M. C. Peitsch, Electrophoresis 18, 2714 (Dec, 1997).
47. A. E. Carpenter et al., Genome Biol 7, R100 (2006).
48. L. S. Lim et al., Mol Biol Cell 18, 1348 (Apr, 2007).
References and Notes
- 1.Ehrlich M, Wang RY. Science. 1981;212:1350. doi: 10.1126/science.6262918. [DOI] [PubMed] [Google Scholar]
- 2.Goll MG, Bestor TH. Annu. Rev. Biochem. 2005;74:481. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 3.Reik W. Nature. 2007;447:425. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 4.Bird A. Genes Dev. 2002;16:6. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 5.Gal-Yam EN, Saito Y, Egger G, Jones PA. Annu. Rev. Med. 2008;59:267. doi: 10.1146/annurev.med.59.061606.095816. [DOI] [PubMed] [Google Scholar]
- 6.Borst P, Sabatini R. Annu. Rev. Microbiol. 2008;62:235. doi: 10.1146/annurev.micro.62.081307.162750. [DOI] [PubMed] [Google Scholar]
- 7.Yu Z, et al. Nucleic Acids Res. 2007;35:2107. doi: 10.1093/nar/gkm049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cliffe LJ, et al. Nucleic Acids Res. 2009;37:1452. doi: 10.1093/nar/gkn1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aravind L, Koonin EV. Genome Biol. 2001;2:RESEARCH0007. doi: 10.1186/gb-2001-2-3-research0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allen MD, et al. EMBO J. 2006;25:4503. doi: 10.1038/sj.emboj.7601340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cedar H, Solage A, Glaser G, Razin A. Nucleic Acids Res. 1979;6:2125. doi: 10.1093/nar/6.6.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wyatt GR, Cohen SS. Biochem. J. 1953;55:774. doi: 10.1042/bj0550774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Que L, Jr., Ho RY. Chem. Rev. 1996;96:2607. doi: 10.1021/cr960039f. [DOI] [PubMed] [Google Scholar]
- 14.Loenarz C, Schofield CJ. Nat. Chem. Biol. 2008;4:152. doi: 10.1038/nchembio0308-152. [DOI] [PubMed] [Google Scholar]
- 15.Netto LE, Stadtman ER. Arch. Biochem. Biophys. 1996;333:233. doi: 10.1006/abbi.1996.0386. [DOI] [PubMed] [Google Scholar]
- 16.Simmen MW. Genomics. 2008;92:33. doi: 10.1016/j.ygeno.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 17.Valinluck V, et al. Nucleic Acids Res. 2004;32:4100. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valinluck V, Sowers LC. Cancer Res. 2007;67:946. doi: 10.1158/0008-5472.CAN-06-3123. [DOI] [PubMed] [Google Scholar]
- 19.Privat E, Sowers LC. Chem. Res. Toxicol. 1996;9:745. doi: 10.1021/tx950182o. [DOI] [PubMed] [Google Scholar]
- 20.Flaks JG, Cohen SS. J. Biol. Chem. 1959;234:1501. [PubMed] [Google Scholar]
- 21.Alegria AH. Biochim. Biophys. Acta. 1967;149:317. doi: 10.1016/0005-2787(67)90159-1. [DOI] [PubMed] [Google Scholar]
- 22.Ooi SK, Bestor TH. Cell. 2008;133:1145. doi: 10.1016/j.cell.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 23.Jiricny J, Menigatti M. Cell. 2008;135:1167. doi: 10.1016/j.cell.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 24.Cannon SV, Cummings A, Teebor GW. Biochem. Biophys. Res. Commun. 1988;151:1173. doi: 10.1016/s0006-291x(88)80489-3. [DOI] [PubMed] [Google Scholar]
- 25.Zhu B, et al. Proc. Natl. Acad. Sci. U.S.A. 2000;97:5135. [Google Scholar]
- 26.Metivier R, et al. Nature. 2008;452:45. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- 27.Kangaspeska S, et al. Nature. 2008;452:112. doi: 10.1038/nature06640. [DOI] [PubMed] [Google Scholar]
- 28.Rai K, et al. Cell. 2008;135:1201. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rusmintratip V, Sowers LC. Proc. Natl. Acad. Sci. U.S.A. 2000;97:14183. doi: 10.1073/pnas.97.26.14183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ono R, et al. Cancer Res. 2002;62:4075. [PubMed] [Google Scholar]
- 31.Lorsbach RB, et al. Leukemia. 2003;17:637. doi: 10.1038/sj.leu.2402834. [DOI] [PubMed] [Google Scholar]
- 32.Viguie F, et al. Leukemia. 2005;19:1411. doi: 10.1038/sj.leu.2403818. [DOI] [PubMed] [Google Scholar]
- 33.Delhommeau F, et al. paper presented at the American Society of Hematology Annual Meeting and Exposition; San Francisco, CA. December 9, 2008. [Google Scholar]
- 34.We thank K. Kreuzer for the gift of the T4 phage and E. coli ER1656 and CR63; Charles Richardson, Udi Qimron, and Ben Beauchamp for advice and assistance in culturing T4 phage; Melissa Call for advice on production of recombinant proteins in insect cells; and Patrick Hogan for many helpful discussions. This work was supported by NIH grant AI44432, a Scholar Award from the Juvenile Diabetes Research Foundation, and a Seed grant from the Harvard Stem Cell Institute (to A.R.); Howard Hughes Medical Institute and NIH/NIGMS (R01GM065865) funding (to D.R.L.); an American Heart Association postdoctoral fellowship (to K.P.K.); intramural funds of the National Library of Medicine, NIH (to L.A. and L.I.); a Lady Tata Memorial postdoctoral fellowship (to H.B.); NIH award K08 HL089150 (to S.A.); NSF Graduate Fellowships (to W.A.P. and Y.B.); and a Department of Defense graduate fellowship (to W.A.P.).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Online Material for
Conversion of 5-Methylcytosine to 5-Hydroxymethylcytosine in Mammalian DNA by MLL Partner TET1
Mamta Tahiliani, Kian Peng Koh, Yinghua Shen, William A. Pastor, Hozefa Bandukwala, Yevgeny Brudno, Suneet Agarwal, Lakshminarayan M. Iyer, David R. Liu,* L. Aravind,* Anjana Rao*
*To whom correspondence should be addressed. E-mail: arao@idi.harvard.edu (A.R.); aravind@ncbi.nlm.nih.gov (L.A.); drliu@fas.harvard.edu (D.L.)
Published 16 April 2009 on Science Express DOI: 10.1126/science.1170116
This PDF file includes:
Materials and Methods
SOM Text
Figs. S1 to S8
References
Supplementary Text
Identification of a novel family of 2OG-Fe(II) oxygenases with predicted 5-methylpyrimidine oxidase activity. To identify candidate enzymes that catalyze the oxidative modification of 5-methylcytosine, we created a position-specific score matrix (profile) of known 2OG-Fe(II) oxygenases that included the predicted oxygenase domains of JBP1 and JBP2 (Fig. S1). We used this profile to conduct a systematic search of the non-redundant database using the PSI-BLAST program (1). This search recovered homologous domains in the gp2 proteins from the mycobacteriophages Cooper and Nigel and a related prophage from the Frankia alni genome (e<10−4). We then generated a new profile which included the gp2 protein sequences, and performed a second search of the protein sequence database of microbes from environmental samples. This search detected numerous homologous proteins potentially derived from uncultured marine phages and prophages. A further search of the non-redundant database, with the newly-detected proteins from the environmental sequences added to the profile, recovered homologous regions in the three paralogous human oncogenes TET1 (CXXC6), TET2 and TET3 and their orthologs found throughout metazoa (e<10−5). Homologous domains were also found in fungi and algae. In PSI-BLAST searches, these groups of homologous domains consistently recovered each other prior to recovering any other member of the 2OG-Fe(II) oxygenase superfamily, suggesting that they constitute a distinct family (LM Iyer, L Aravind, manuscript in preparation).
To confirm the relationship of the newly-identified proteins (hereafter referred to as the TET/JBP family; see legend to Fig. 1) with classical 2OG-Fe(II) oxygenases, we prepared a multiple alignment of their shared conserved domains and used this to generate a hidden markov model (HMM). A profile-profile comparison of this HMM against a library of HMM's generated for all structurally-characterized domains from the Protein Database (PDB) with the HHpred program (2) resulted in recovery of prolyl hydroxylase (e<10−12), a canonical member of the 2OG-Fe(II) oxygenase superfamily, as the best hit. Secondary structure predictions suggested the existence of an N-terminal α-helix followed by a continuous series of β-strands, typical of the double-stranded β-helix (DSBH) fold of the 2OG-Fe(II) oxygenases (Fig. 1A, bottom) (3). A multiple sequence alignment (Fig. 1A) further showed that the new TET/JBP family displayed all of the typical features of 2OG-Fe(II) oxygenases including: (i) the HxD signature, just downstream of an N terminal β-strand which chelates Fe(II) (x is any amino acid); (ii) a small residue, usually glycine, at the beginning of the strand immediately downstream of the HxD motif, which helps in positioning the active site arginine; (iii) the Hxs motif (where s is a small residue) in the C-terminal part, in which the H chelates the Fe(II) and the small residue helps in binding the 2-oxo acid; (iv) the Rx5a signature (where a is an aromatic residue) downstream of the above motif – the R in this motif forms a salt bridge with the 2-oxo acid and the aromatic residue helps position the first metal-chelating histidine (the key residues are marked with asterisks in Fig 1A). These observations strongly suggested that members of the TET/JBP family, including TET1, 2 and 3, are catalytically-active 2OG-Fe(II) oxygenases.
Additionally, the metazoan TET proteins contain a unique conserved cysteine-rich region, contiguous with the N-terminus of the DSBH region, which possesses at least eight conserved cysteines and one histidine that are likely to comprise a binuclear metal cluster (Fig. 1A, top). Vertebrate TET1 and TET3, and their orthologs from all other animals, also possess a CXXC domain, a binuclear Zn-chelating domain with eight conserved cysteines and one histidine, located N-terminal to the 2OG-Fe(II) oxygenase domain (shown schematically for TET1 in Fig. 1B). Analysis of the architectures of CXXC domain proteins suggests that the CXXC domain is an accessory DNA-binding domain, that tends to be combined in the same polypeptide with a variety of domains that possess diverse chromatin-modifying and modification recognition activities. The CXXC domain is found in several chromatin-associated proteins, including the methyl-DNA-binding protein MBD1, the histone methyltransferase MLL, the DNA methyltransferase DNMT1 (4) and the lysine demethylases KDM2A (JHDM1A/ FBXL11) and KDM2B (JHDM1B/FBLX10, which also contains a ubiquitin E3 ligase domain) (5), and in certain cases has been shown to discriminate between methylated and unmethylated DNA (6).
Taken together, the contextual information gleaned from these domain architectures and from gene neighborhoods in the bacteriophage members (Supplementary Information) supported a conserved DNA modification function for the entire TET/ JBP family, namely oxidation of 5-methyl-pyrimidines. In this study we tested the specific hypothesis that TET proteins might operate on 5mC to catalyze oxidation or oxidative removal of the methyl group, focusing on TET1 as a mammalian example of the TET/JBP family.
hmC may not be confined to CpGs. Nearest-neighbour analyses have shown that 15−20% of total 5mC in ES cells is present in CpT, CpA and (to a minor extent) CpC sequences; in contrast, somatic tissues have negligible non-CpG methylation (7).
Previous studies on hmC. With the exception of two papers that reported high levels of hmC in genomic DNA (8), most previous studies have described hmC as a rare base that is a probable oxidation product of 5mC (9, 10).
Methods of distinguishing hmC, 5mC and C. We show here that two of the three most commonly used techniques do not adequately distinguish between C, 5mC and hmC. A widely-used mouse monoclonal antibody to 5mC apparently does not recognize hmC by immunocytochemistry (Fig. 2A), thus it will be important to reevaluate previous reports of DNA demethylation based solely on the use of this antibody. Similarly, the methylation-sensitive restriction enzyme, HpaII, fails to cut hmC (Fig. 3D) as previously reported (11), raising the possibility that in some instances hmC-modified DNA was incorrectly judged to be methylated. Another methylation-sensitive restriction enzyme, McrBC, is already known to cleave 5mC- and hmC-containing DNA equivalently (12), and therefore also does not allow these two nucleotides to be distinguished.
It is yet to be determined how bisulfite modification analysis interprets the presence of hmC in DNA. Treatment of DNA with sodium bisulfite promotes the spontaneous deamination of cytosine to uracil, while leaving 5mC unaffected; amplification of the sequence of interest followed by sequencing allows the precise methylation patterns at a given sequence to be determined (13). It is known that bisulfite reacts rapidly with hmC at the C5 to form a stable cytosine 5-methylenesulfonate adduct, which is not readily deaminated (14). This substituted species, which is expected to form base pairs similar to those formed by cytosine, could be read by polymerases as C during the amplification steps, resulting in the sequence being interpreted as containing 5mC. Alternatively, polymerases may not copy cytosine 5-methylenesulfonate efficiently, in which case the DNA containing this adduct would not be amplified effectively and the sequence containing the original hmC modification would be underrepresented in the amplified DNA.
Stability of hmC versus nitrogen-linked hydroxymethyl groups. The stability of hmC in the genome of T-even phages (15) contrasts with the well-documented instability of N-linked hydroxymethyl adducts generated by the DNA repair enzymes AlkB and the JmjC domain-containing histone demethylases, which are also 2OG and Fe(II)-dependent oxygenases. These nitrogen-linked adducts spontaneously resolve, yielding the unmethylated amino group and formaldehyde (16); in contrast, oxidation of carbon-linked methyl groups by JBP1/2 and thymine hydroxylase does not result in removal of the methyl adduct via oxidation (17, 18). This difference, which is due to the fact that carbon is a much poorer leaving group than nitrogen, explains our ability to detect hmC in genomic DNA.
Mechanisms of DNA demethylation. The mammalian DNA methyltransferases DNMT1, DNMT3A and DNMT3B, establish and maintain DNA methylation patterns (19, 20). Preventing maintenance methylation through successive replication cycles progressively dilutes the methyl mark and results in “passive” DNA demethylation (21). “Active” (replication-independent) DNA demethylation has been convincingly demonstrated only for the paternal genome shortly after fertilization (22−24); potential mechanisms could involve thermodynamically unfavorable cleavage of the carbon-carbon bond linking the methyl group to the pyrimidine, resulting in release of the methyl moiety, or a repair-like process in which the methylated base or nucleotide is excised and the lesion is then repaired to replace the original 5mC with an unmethylated C (reviewed in (25, 26)). This latter mechanism occurs in plants, where DEMETER, a bifunctional glycosylase restricted to flowering plants, cleaves the glycosidic bond of 5mC and stimulates insertion of an unmethylated C through base-excision repair (27). A variety of candidate mammalian demethylase activities have been described, but unfortunately, none of these reports have been confirmed by other laboratories (reviewed in (19, 28−30)).
Roles of TET proteins in cancer. The human LCX (leukemia-associated protein with a CXXC domain) / Tet1 (ten-eleven translocation-1) gene was originally defined as a novel MLL fusion partner in cases of pediatric and adult AML with t(10;11)(q22;23) (4, 31). In a recent study, the MLL/Tet1 fusion accounted for 3/759 (0.4%) of MLL-associated leukemia (32), with two cases of ALL, together showing occurrence of the MLL/Tet1 fusion in both pediatric and adult ALL and AML. Possible pathogenic mechanisms of the MLL/Tet1 fusion include altered genomic targeting of Tet1 catalytic activity and/or disrupted recruitment of cofactors to sites of Tet-1 mediated hmC generation. Homozygous Tet2 deletions / loss of function mutations have recently been reported in approximately 14% of patients with JAK2V617F-positive and negative myeloproliferative neoplasms (including polycythemia vera (PV), essential thrombocythemia (ET), primary myelofibrosis (MF), post-PV MF, post-ET MF, and blast phase PV/ET/MF) (33, 34); approximately 29% of patients with systemic mastocytosis (35); and other myeloid malignancies including chronic myelomonocytic leukemia, myelodysplastic syndrome, and acute myeloid leukemia (36, 37). It will be interesting to investigate the association of Tet2 loss of function, epigenetic alterations such as promoter hypermethylation that characterize myeloproliferative neoplasms, and response to epigenetic therapy (38).
Fig. S1. Modification of 5-methylpyrimidines. (A) Trypanosomes contain a modified version of thymine, called base J (β-D-glucosyl hydroxymethyluracil), in their genome. It has been proposed that base J is synthesized by sequential hydroxylation and glucosylation of the methyl group of thymine (17). (B) TET1, and presumably the other TET family members, oxidize the methyl group of 5-methylcytosine (5mC) generating 5-hydroxymethylcytosine (hmC).
Fig. S2. Sequences used to generate the position-specific score matrix (PSSM). The figure shows the sequences used to create the position specific score matrix that retrieved the TET proteins in searches with the PSI-BLAST program. Each sequence is identified by the gi number allowing its recovery from the GenBank database and the numbers flanking the alignment provide the limits of the aligned region in the sequence. The JBP proteins from kinetoplastids are marked in red. The searches were performed using the checkpoint start option –B, with–h set to 0.01 and –F was set to F and the all JBP sequences were cycled through the –i options for individual searches.
Fig. S3. Multiple sequence alignment of the TET/JBP family and representative 2OG-Fe(II) dependent dioxygenase domains. The cysteine-rich region (C) and the core catalytic domain (D) are aligned separately. Proteins are labeled by their gene name and species, separated by underscores. The 95% consensus was calculated from a larger alignment of the TET/JBP proteins. The consensus for the TET-specific C-terminal strands was calculated separately from a larger alignment specifically of the metazoan TET homologs. In addition to the TET/JBP family, the alignment also contains the structurally characterized representatives of the dioxygenase superfamily, the Chlamydomonas prolyl hydroxylase (P4H) and the E. coli AlkB (2FD8) along with their PDB codes.
Fig. S4. Image analysis using CellProfiler. Nuclei were outlined based on DAPI fluorescence using the IdentifyPrimAutomatic module and denoted as Outlined Nuclei. Objects within a pre-set diameter range of 10−35 pixel units are outlined in green and included in analysis. Objects outside the range (including cell clusters) as well as those touching the borders are outlined in red and excluded from analysis. To account for HA staining at nuclear boundaries, an IdentifySecondary module was added to expand the nuclear outlines by 2 pixels, denoted as ExpandedNuclei Outlines. The MeasureObjectIntensity module was then used to apply the ExpandedNuclei Outlines on the corresponding HA image, and the Outlined Nuclei on the corresponding 5mC image (not shown), to measure pixel intensities of HA and 5mC staining respectively. Pixel numbers are indicated at the axes of images, which are taken at a magnification of 20x.
Fig. S5. Mass spectrometry fragmentation analyis (MS/MS) of authentic hm-dCMP from T4* phage (top) and the unidentified nucleotide species present in genomic DNA from HEK293 cells overexpressing TET1-CD (bottom). (A) MS/MS analysis was performed in negative ion mode with a collision energy of 25 V. (B) MS/MS analysis was performed in positive ion mode with a collision energy of 25 V. Observed masses are shown (anticipated mass accuracy was within 0.003 Da).
Fig. S6. Purification of recombinant Flag-HA-TET1-CD from Sf9 cells and characterization of TET1-CD fragment. (A) Coomassie-stained SDS-PAGE gel of wild-type and mutant Flag-HA-TET1-CD purified from Sf9 cells to near homogeneity by affinity chromatography with anti-Flag antibody conjugated beads. Known amounts of BSA were loaded on the same gel for comparison. Wild-type and mutant TET1-CD (79 KDa, indicated by the arrow) both demonstrate a strong tendency to oxidize and form disulfide-linked dimers (158 KDa) and higher-order multimers, which are resistant to DTT. Asterisks denote degradation products that are not detected by immunoblotting (see (B)), probably due to loss of the N-terminal Flag-HA epitope tag. (B) The bands of apparent higher molecular weight were identified as TET1 oxidation products by immunoblotting with an anti-HA antibody. (C) Immunoblot with anti-Flag antibody showing that the multimeric forms TET1-CD increase with increased processing time of lysates and concomitant exposure to oxidizing conditions. A twenty minute lysis in 10 mM DTT-containing RIPA buffer results in the detectable presence of a TET1-CD dimer (lane 1). This dimer is not detected when cell pellets from Flag-HA-TET1-CD overexpressing cells are lysed directly in denaturing Laemlli SDS sample buffer containing 700 mM β-ME (lane 3). The tendency of TET1-CD to oxidize appears to be due at least in part to disulfide bond formation by the Cys-Rich region (C) that is N-terminal to the DSBH (D) region (lanes 2, 4). Removal of the N-terminal Cys-Rich region resulted in expression of a protein (Flag-HA-TET1-D) that runs at its expected molecular weight (57.6 KDa) after extraction in either RIPA buffer or SDS sample buffer. Together the data shown in (A-C) suggest that intermolecular disulfide bonds that are resistant to reducing agent are formed during extraction. (D) Overexpression of Flag-HA-TET1-CD in HEK293 cells, but not Flag-HA-TET1-D, results in decreased staining by an anti-5mC antibody.
Fig. S7. Recombinant Flag-HA-TET1-CD purified from Sf9 cells does not convert thymine (another 5-methylpyrimidine) to hmU in vitro. (A) Synthetic double-stranded DNA oligonucleotides containing a fully-methylated TaqαI (T^mCGA) or a SalI (G^TCGAC) site were incubated with purified Flag-HA-TET1-CD or mutant Flag-HA-TET1-CD for 5 hours at 37 C (1:10 enzyme to substrate ratio). (B) Recovered oligonucleotides were digested with TaqαI or SalI, end-labeled, hydrolyzed to dNMP's and resolved using TLC. TET1-CD is able to hydroxylate 5mC, but is not able to act on thymine in the context of a SalI site in a double-stranded oligonucleotide substrate in vitro. A double-stranded DNA oligonucleotide terminating in hmU was end-labelled and hydrolyzed to generate a standard for hm-dUMP migration (lane 5). SalI has been demonstrated to cleave G(hmU)CGAC equivalently to GTCGAC (39).
Fig. S8. Model speculating on the biological role of hmC and its potential as an intermediate in DNA demethylation. (A) Integration of hmC into the known pathways of DNA methylation and passive replication-dependent demethylation. Known pathways are shown in black and the new findings in this study in red. Dashed lines and question marks are used to indicate pathways that may exist but have not been experimentally established. (B) Potential biological mechanisms involving hmC. hmC may recruit specialized binding proteins; and conversion of 5mC to hmC may displace methyl-binding proteins from DNA (center). hmC may also be an intermediate that facilitates passive DNA demethylation: conversion of 5mC to hmC in hemi-methylated DNA may interfere with recognition by DNMT1 and associated SRA-domain proteins (left). Finally, hmC may convert to cytosine through a spontaneous or enzymatic process or it may be recognized by specialized DNA repair proteins in specific cell types, and so function as an intermediate in active DNA demethylation (right).
Methods
Computational and bioinformatic analyses. The non-redundant (NR) database of protein sequences (National Center for Biotechnology Information, NIH, Bethesda) was searched using the PSI-BLAST programs (1). Profile searches using the PSI-BLAST program were conducted either with a single sequence or a sequence with a PSSM used as the query, with a profile inclusion expectation (E) value threshold of 0.01, and were iterated until convergence (1). For all compositionally biased queries the correction using composition-based statistics was used in the PSI-BLAST searches (40). Multiple alignments were constructed using the Kalign program (41), followed by manual correction based on the PSI-BLAST results. The multiple alignment was used to create a HMM using the Hmmbuild program of the HMMER package (42). It was then optimized with Hmmcaliberate and the resulting profile was used to search a database of completely sequenced genomes using the Hmmsearch program of the HMMER package (42). Profile-profile searches were performed using the HHpred program (2, 43). The JPRED program (44) and the COILS program were used to predict secondary structure. Globular domains were predicted using the SEG program with the following parameters: window size 40, trigger complexity=3.4; extension complexity=3.75 (45).
The Swiss-PDB viewer (46) and Pymol programs were used to carry out manipulations of PDB files. Reconstruction of exon-intron boundaries was done using the NCBI Splign program with the tblastn searches against chromosomes as a guide. Gene neighborhoods were determined using a custom script that uses completely sequenced genomes or whole genome shot gun sequences to derive a table of gene neighbors centered on a query gene. Then the BLASTCLUST program is used to cluster the products in the neighborhood and establish conserved co-occurring genes. These conserved gene neighborhood are then sorted as per a ranking scheme based on occurrence in at least one other phylogenetically distinct lineage (“phylum” in NCBI Taxonomy database), complete conservation in a particular lineage (“phylum”) and physical closeness on the chromosome indicating sharing of regulatory −10 and −35 elements. Phylogenetic trees were constructed with the MEGA4 package.
TET1 expression plasmids TET1 ORF was amplified from SY5Y cDNA and the human clone and inserted into XhoI and NotI sites of an Flag-HA tagged pOZ-N. Mutant TET1 (H1671D, Y1673A) was generated using the QuikChange Mutagenesis Kit (Stratagene). The sequences of all clones were confirmed by conventional DNA sequencing. Wild-type and mutant Flag-HA-TET1-CD was amplified and cloned into Acc651 and XbaI sites of pEF1 (Invitrogen). The IRES-CD25 of pOZ-N was amplified and cloned into the the XbaI and BstB1 sites of Flag-HA-TET1-CD-pEF1. Wild-type and mutant Flag-HA-TET1-CD was amplified from Flag-HA-TET1-CD-pOZ and inserted into SalI and NotI sites of pFastBac (Invitrogen).
Immunocytochemistry Cells were plated on sterile coverglass in 24-well plates at 1.5 × 105 cells/well and grown overnight before transient transfection with pEF1-TET1 expression constructs or empty vector (mock) using TransIT™-293 transfection reagent (Mirus, Madison, WI) according to manufacturer's instructions. At 42−44 hr post-transfection, cells were fixed for 15 min in 4% paraformaldehyde in PBS and permeabilized with 0.2% Triton X-100 in PBS for 15 min at room temperature. For detection of 5-methylcytosine, cells were treated with 2N HCl at room temperature for 30 min and subsequently neutralized for 10 min with 100 mM Tris-HCl buffer, pH 8. After extensive washes in PBS, cells were blocked for 1 hour at room temperature in 1%BSA, 0.05% Tween 20 in PBS. Rabbit anti-HA polyclonal antibody (diluted at 1:400; Santa Cruz Biotechnology, Santa Cruz, CA ) and mouse anti-5 methylcytosine clone 162 33 D3 antibody (diluted at 1:2500−3000; Calbiochem, San Diego, CA) were added in blocking buffer for 2−3 hours at room temperature and detected concurrently by secondary antibodies coupled with Cy2 or Cy3 respectively. DNA was stained with 250 ng/ml of4’,6-diamidino-2-phenylindole (DAPI) and mounted in SlowFade® Gold antifade reagent (Molecular Probes, Eugene, OR). Images were recorded digitally on a Zeiss Axiovert 200 inverted microscope equipped with a CCD camera by using OpenLab imaging software (Improvision, Coventry, UK).
CellProfiler™ cell image analysis. Three fields, each containing 200−400 cells, were imaged from each well of transfected cells using an 20x objective and pooled for analysis in each experiment. Greyscale images (tiffs) were uploaded on CellProfiler as three individual files for every field captured under the three excitation wavelengths respectively for DAPI, GFP (detection of HA) and Cy3 (detection of 5mC) (47). Nuclear outlines were profiled based on DAPI staining, with a secondary module included to expand the nuclear outlines by another 2 pixels (denoted as “expanded nuclei”) to account for HA staining at nuclear boundaries. Clustered cells and cells at the edge of fields were excluded. Staining intensities of HA and 5mC within individual cells profiled were measured as mean pixel intensities of GFP and Cy3 signals, respectively, within the “expanded nuclei” and original nuclei profiles, respectively. The raw data were plotted as dot plots of 5mC mean pixel intensity versus HA mean pixel intensity for each transfection sample. The same set of mock-transfected cells is shown in the two panels in Fig. 1C. The population average of 5mC staining intensity of HA-expressing cells (arbitrarily categorized as cells with mean HA pixel intensity above the highest intensity observed in mock-transfected cells) is compared to mock transfected cells. Values are background-subtracted before normalization and are mean ± SEM from 3 experiments; statistical comparisons are based on ANOVA and Bonferroni's post-hoc test.
Transfection and Sorting of HEK293T cells expressing hCD25 HEK293T cells were transfected with TET1-CD-IRES-CD25-pEF1 vectors using TransIT transfection reagent (Mirius). Control cells (mock) were transfected with a corresponding empty vector that drove expression of CD25 alone. After 48 hours, 30 × 106 cells were stained with anti-hCD25-PE antibody (1:200) (Becton Dickinson) in 1X FACS Buffer (1XPBS, 2% FBS, 1 mM EDTA, 0. 1% NaAzide) for 30 min at 4 C. Cells were washed twice with 1X FACS Buffer. Cells were then stained with anti-PE microbeads (Miltenyi) for 25 min, 4 C and then washed two times with 1X FACS Buffer and then once with 1X MACS buffer (1X PBS, 0.5% BSA, 0.09% Na Azide and 2 mM EDTA). Cells were resuspended in 2 ml of ice-cold 1X MACS Buffer and CD25-expressing cells were sorted using an AutoMACS cells sorter (Possel). Input, flow-through and collected samples were analyzed by FACS to confirm enrichment for CD25-positive cells in collected sample.
Analysis of 5mC levels using thin-layer chromatography Nuclei were prepared from CD25-positive cells by resuspension in 1 ml NPB (240 mM sucrose, 7.5 mM Tris, pH 7.5, 3.75 mM MgCl2, 0,75% Triton-X-100, with 100 μg RNAseA/ml (Qiagen)) and placing on ice for 20 minutes. Cells were spun at 1300 g for 15 min, 4 C and then washed once in NPB. Nuclei were lysed in 650 μl of 1X LB ((10 mM Tris, pH 8.0, 300 mM NaAcetate, pH 7.2, 0.5% SDS, 5 mM EDTA, 100 μg RNAseA/ml and 300 μg/ml Proteinase K (Roche)) and incubated overnight at 55 C. An extra 300 μg/ml Proteinase K was added in the morning and the samples were left at 55 C for 5 hours. Samples were extracted with equal volumes of phenol, phenol: chloroform: isoamyl alcohol (25:24:1) and chloroform: isoamyl alcohol (24:1) and then precipitated with 2 volumes of ethanol. Genomic DNA was washed twice with 1 ml of 70% EtOH, dried and resuspended in 10 mM Tris, 0.1 mM EDTA, pH 8.0 and allowed to resuspend overnight at 32 C.
2 μg of genomic DNA was digested with 100 units of MspI, HpaII or Taqα1 and 100 μg of RNaseA (Qiagen) overnight. An extra 100 units of restriction enzyme was added in the morning and incubations were continued for 6 hours. 10 units of calf intestinal phosphatase (CIP) (NEB) was added and incubated for 1 hour at 37 C. DNA was purified using Qiaquick Nucleotide Removal Kit (Qiagen) as per the manufacturer's instructions. 400 ng of eluted DNA fragments were end-labeled with T4 Polynucleotide Kinase (T4 PNK) (NEB) and 10 μCi of [γ32-P]-ATP for 1 hour at 37 C. Labeled fragments were precipitated by the addition of 30 μg of linear polyacrylamide, 1/10 volume of 3 M NaAcetate, pH 7.2 and 2.5 volumes of ethanol at left at −80 C for 1 hour. Samples were spun at 14,000 rpm, for 20 minutes at 4 C and washed twice with 70% EtOH at 25 C. Pellets were resuspended in 30 mM Tris, pH 8.9, 15 mM MgCl2, 2 mM CaCl, with 10 μg of DNaseI (Worthington) and 10 μg SVPD (Worthington) and incubated for 3 hours at 37 C. 3 μl was spotted on cellulose TLC plates (20 cm × 20 cm, Merck) and developed in isobutyric acid: H20: NH3 (66:20:1). Plates were analyzed by phosphorimager scanning using Phosphorimager Storm 860 scanner software. The low-level labeling of other nucleotides reflects DNA shearing or contaminating endonucleolytic activity.
Preparation of unglucosylated T4 phage DNA for preparation of hm-dCMP standard T4 phage stock was titred by spotting 10 μl of serial 10X dilutions on an LB plate on which 100 μl of an overnight culture of E.coli CR63 in 3 ml of T4 top agar was poured and allowed to solidify. The plate was incubated overnight at 37 C. 10 ml of E.coli CR63 OD600 of 0.5 was infected with a single plaque of T4 phage and incubated with shaking at 37°C until the culture cleared (about 2.5 hours). The culture was incubated on ice for 10 minutes and then lysed was completed by the addition of several drops of chloroform and gentle mixing. The lysate was titred as described above.
E.coli ER1656 was grown in LB to OD600 of 0.5 and then infected with 0.2 phage per bacterium and incubated at 37°C with shaking until the culture cleared (about 8 hours).. The culture was chilled on ice for 10 minutes and then lysis was completed by the addition of 1 ml of chloroform. DNase I was added to 1 mg/ml and the culture was incubated for 2 hours at 4°C. The lysate was centrifuged at 12,000g for 10 min at 4 C to pellet debris. The supernatant was collected and phage were pelleted by centrifugation at 23,500g for 1.5 hours at 4 C. The phage pellet was left covered in TE overnight to resuspend. Phage DNA was extracted using an equal volume of phenol, phenol: chloroform: isoamyl alcohol (25:24:1) and chloroform: isoamyl alcohol (24:1). The extracted phage DNA was dialyzed into TE overnight with 2 changes of buffer.
Mass spectrometry experiments. Genomic DNA from HEK293 cells transfected with TET1 wild-type or mutant CD or T4 phage grown in E.coli 13656 were hydrolyzed to dNMP's with SVPD and DNaseI and resolved using TLC. Spots corresponding to paricular dNMP's were scraped, extracted with water, lyophilized, and re-suspended in water for liquid chromatography/mass spectrometry (LC/MS) analysis using an Acquity UPLC/Q-TOF Premier electrospray LC/ESI-MS system (Waters Corp., Milford, MA). Liquid chromatography (LC) was performed with a Waters HSS C18 column (1.0mm i.d. × 50mm, 1.8-um particles) using a linear gradient of 0% to 50% methanol in 0.1% aqueous ammonium formate, pH 6.0. The flow rate was 0.05 mL per min and the eluant was directly injected into the mass spectrometer. Mass spectra were recorded in continuum mode and converted to centroid mode to generate accurate mass spectra. Data was analyzed with Masslynx 4.1 software (Waters).
Recombinant Protein Expression and Purification Bacmid DNA was generated using DH10Bac™ E. coli E.coli (Invitrogen) as directed by the manufacturer. Transposition into the correct site was confirmed using PCR. Baculovirus was amplified for three generations using suspension adapted Sf9 cells. Sf9 cells were then infected with baculovirus for 4 days. The resulting cell pellet was kept on ice for 30 minutes in 40 mM Tris, pH 7.4, 300 mM NaCl, 0.2% NP40, 0.4% Triton, 5 mM DTT, 1X protease inhibitors without EDTA (Roche) and then at 12,000 rpm (SLA-TC600), 30 min, 4 C. The supernantant was then incubated with anti-Flag antibody-conjugated beads (Invitrogen) for 5 hours at 4 C. The beads were washed 4 times in 40 mM Tris, pH 7.4, 300 mM NaCl, 0.2% NP40, 8% Glycerol, 1X PI, 5 mM DTT and then eluted in 195 mM Tris, pH 7.4, 110 mM NaCL, 0.14% NP40, 5.8% Glycerol, 0.37X PI, 3.7mM DTT, 365 μg/ml Flag peptide. The homogeneity of the eluted protein was determined using SDS-PAGE followed by Coomassie blue staining and immunoblotting using an anti-Flag antibody (Sigma).
Preparation of double-stranded oligonucleotide substrates Synthetic oligonucleotides were purchased from IDT. All oligonucleotides were 35 nucleotides in length with the modifications shown below.
F: 5’-CTATACCTCCTCAACTTCGATCACCGTCTCCGGCG-3’
FMe: 5’-CTATACCTCCTCAACTT(mC)GATCACCGTCTCCGGCG-3’
R: 5’-Biotin-CGCCGGAGACGGTGATCGAAGTTGAGGAGGTATAG-3’
RMe: 5’-Biotin-CGCCGGAGACGGTGAT(mC)GAAGTTGAGGAGGTAT AG-3’
Oligonucleotides were annealed to the appropriate complementary oligonucleotide in 100 mM KAc, 30 mM HEPES, pH 7.5. The mixture was boiled for 5 minutes then slowly cooled to room temperature overnight. Double-stranded oligonucleotides were purified by polyacrylamide gel electrophoresis.
In vitro Enzymatic Assays 7.5 μl of recombinant protein (about 3 μg) was incubated with 2 μg of oligonucleotide substrates in 50 mM HEPES, pH 8, 50 mM NaCl, 2 mM Ascorbic Acid, 1mM 2OG, 100 μM FAS (Fe2+), and 1 mM DTT for 3 hours at 37 C. Oligonucleotide substrates were purified using Qiaquick Nucleotide Removal Kit (Qiagen) and then digested with Taqa1 overnight, treated with CIP for 1 hour and purified once more with Qiaquick Nucleotide Removal Kit (Qiagen). Purified DNA oligonucleotides were end-labeled with T4 Polynucleotide Kinase (NEB) and 10 μCi of γ32P-ATP for 1 hour at 37 C. Labeled fragments were precipitated by the addition of 30 μg of linear polyacrylamide, 1/10 volume of 3 M Sodium Acetate, pH 7.2 and 2.5 volumes of ethanol followed by incubation at −80 C for 1 hour. Samples were spun at 14,000 rpm, for 20 minutes at 4 C in a refrigerated microcentrifuge. Unincorporated radionucleotide was removed by washing two times with 70% EtOH and spinning at room temperature for 10 minutes. Pellets were resuspended in 10 μl 30 mM Tris, 15 mM MgCl2, 2 mM CaCl, pH 8.9 with 10 μg of DNaseI (Worthington) and 10 μg SVPD (Worthington) and incubated for 3 hours at 37 C. 3 μl was spotted on cellulose TLC plates (Merck) and developed in isobutyric acid: water: ammonia (66:20:1). Plates were analyzed by phosphorimager scanning using Phosphorimager Storm 860 scanner software. The faint dCMP spot in each lane is derived from end-labelling of the C at the 5’ end of each strand of the substrate. T4 PNK is not able to phophorylate blunt ends as efficiently as the 5’ overhangs generated by restriction enzyme cleavage.
ES cell culture V6.5 mouse ES cells were maintained on mitomycin C-inactivated primary mouse embryonic fibroblasts in ES medium containing DMEM (Invitrogen, Carlsbad, CA), 15% ES FBS (Omega Scientific, Tarzana, CA) , 0.1 mM each of nonessential amino acids (Invitrogen), 2 mM L-glutamine (Invitrogen), 0.1 mM β-mercaptoethanol (Invitrogen), 50 units/ml penicillin/streptomycin (Invitrogen) and 1000 U/ml ESGRO® (LIF; Chemicon). For all experiments described, cells were trypsinized and plated for 30 min on standard tissue culture dishes to remove feeder cells before floating ES cells were collected and re-plated on gelatin-coated dishes or wells. For LIF withdrawal assays, cells were plated at a density of 2−3 × 105 cells per 10-cm dish and LIF was removed the day after (day 0). RNA interference (RNAi) experiments were performed as previously described (48) using Dharmacon siGENOME siRNA duplexes (Thermo Fisher Scientific Inc, Boulder, CO) against mouse Tet1 (Cat. # D-062861−01/02). The Dharmacon siGENOME non-targeting siRNA#2 (Cat. # D-001210−02) was used as a negative control. Mouse ES cells were seeded in gelatin-coated 12-well at a density of 1 × 105 cells per well and transfected the day after (day 0) with 50 nM siRNA using Lipofectamine RNAiMAX reagent (Invitrogen) according to the manufacturer's instructions. Retransfections were performed on pre-adherant cells at day 2 at a split of 1:4 and finally at day 4 at a split of 1:2 in 6-well plates. Cells were harvested at Day 5 for RNA and thin-layer chromatography analyses.
RNA Isolation, cDNA synthesis and Quantitative Real-Time PCR Total RNA was isolated with an RNeasy kit (Qiagen, Chatsworth, CA) with on-column DNase treatment. cDNA was synthesized with 0.5 μg total RNA using SuperScript III reverse transcriptase (Invitrogen). Quantitative PCR was performed using FastStart Universal SYBR Green Master mix (Roche, Mannheim, Germany) on a StepOnePlus real-time PCR system (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions. The levels of gene expression were normalized to Gapdh. Primer sequences are: Tet1 forward 5’-GAGCCTGTTCCTCGATGTGG-3’, Tet1 reverse 5’-CAAACCCACCTGAGGC TGTT-3’; Gapdh forward 5’-GTGTTCCTACCCCCAATG TGT-3’, Gapdh reverse 5’-ATTGTCATACCAGGAAATGAGCTT-3’.
References
1. S. F. Altschul et al., Nucleic Acids Res 25, 3389 (Sep 1, 1997).
2. J. Soding, A. Biegert, A. N. Lupas, Nucleic Acids Res 33, W244 (Jul 1, 2005).
3. L. Aravind, E. V. Koonin, Genome Biol 2, RESEARCH0007 (2001).
4. R. Ono et al., Cancer Res 62, 4075 (Jul 15, 2002).
5. Y. Tsukada et al., Nature 439, 811 (Feb 16, 2006).
6. M. D. Allen et al., Embo J 25, 4503 (Oct 4, 2006).
7. B. H. Ramsahoye et al., Proc Natl Acad Sci U S A 97, 5237 (May 9, 2000).
8. N. W. Penn, R. Suwalski, C. O'Riley, K. Bojanowski, R. Yura, Biochem J 126, 781 (Feb, 1972).
9. J. R. Pratt et al., Transplant Proc 38, 3344 (Dec, 2006).
10. V. Valinluck, L. C. Sowers, Cancer Res 67, 946 (Feb 1, 2007).
11. S. Tardy-Planechaud, J. Fujimoto, S. S. Lin, L. C. Sowers, Nucleic Acids Res 25, 553 (Feb 1, 1997).
12. E. Sutherland, L. Coe, E. A. Raleigh, J Mol Biol 225, 327 (May 20, 1992).
13. T. Rein, M. L. DePamphilis, H. Zorbas, Nucleic Acids Res 26, 2255 (May 15, 1998).
14. H. Hayatsu, M. Shiragami, Biochemistry 18, 632 (Feb 20, 1979).
15. G. R. Wyatt, S. S. Cohen, Biochem J 55, 774 (Dec, 1953).
16. C. Loenarz, C. J. Schofield, Nat Chem Biol 4, 152 (Mar, 2008).
17. Z. Yu et al., Nucleic Acids Res 35, 2107 (2007).
18. J. A. Smiley, M. Kundracik, D. A. Landfried, V. R. Barnes, Sr., A. A. Axhemi, Biochim Biophys Acta 1723, 256 (May 25, 2005).
19. A. Bird, Genes Dev 16, 6 (Jan 1, 2002).
20. M. G. Goll, T. H. Bestor, Annu Rev Biochem 74, 481 (2005).
21. D. U. Lee, S. Agarwal, A. Rao, Immunity 16, 649 (May, 2002).
22. W. Mayer, A. Niveleau, J. Walter, R. Fundele, T. Haaf, Nature 403, 501 (Feb 3, 2000).
23. J. Oswald et al., Curr Biol 10, 475 (Apr 20, 2000).
24. F. Santos, B. Hendrich, W. Reik, W. Dean, Dev Biol 241, 172 (Jan 1, 2002).
25. H. Cedar, G. L. Verdine, Nature 397, 568 (Feb 18, 1999).
26. H. D. Morgan, F. Santos, K. Green, W. Dean, W. Reik, Hum Mol Genet 14 Spec No 1, R47 (Apr 15, 2005).
27. M. Gehring et al., Cell 124, 495 (Feb 10, 2006).
28. A. P. Wolffe, P. L. Jones, P. A. Wade, Proc Natl Acad Sci U S A 96, 5894 (May 25, 1999).
29. S. K. Ooi, T. H. Bestor, Cell 133, 1145 (Jun 27, 2008).
30. J. Jiricny, M. Menigatti, Cell 135, 1167 (Dec 26, 2008).
31. R. B. Lorsbach et al., Leukemia 17, 637 (Mar, 2003).
32. C. Meyer et al., Leukemia (Mar 5, 2009).
33. F. Delhommeau et al., paper presented at the ASH Annual Meeting and Exposition, San Francisco, CA, December 9, 2008 2008.
34. A. Tefferi et al., Leukemia (Mar 5, 2009).
35. A. Tefferi et al., Leukemia (Mar 5, 2009).
36. F. Viguie et al., Leukemia 19, 1411 (Aug, 2005).
37. A. Tefferi et al., Leukemia (Mar 19, 2009).
38. A. Tefferi, Leuk Lymphoma 49, 2231 (Dec, 2008).
39. M. Hori et al., Nucleic Acids Res 31, 1191 (Feb 15, 2003).
40. A. A. Schaffer et al., Nucleic Acids Res 29, 2994 (Jul 15, 2001).
41. T. Lassmann, E. L. Sonnhammer, Nucleic Acids Res 34, W596 (Jul 1, 2006).
42. S. R. Eddy, Bioinformatics 14, 755 (1998).
43. J. Soding, Bioinformatics 21, 951 (Apr 1, 2005).
44. J. A. Cuff, G. J. Barton, Proteins 40, 502 (Aug 15, 2000).
45. J. C. Wootton, Comput Chem 18, 269 (Sep, 1994).
46. N. Guex, M. C. Peitsch, Electrophoresis 18, 2714 (Dec, 1997).
47. A. E. Carpenter et al., Genome Biol 7, R100 (2006).
48. L. S. Lim et al., Mol Biol Cell 18, 1348 (Apr, 2007).