Abstract
Immunoglobulin class switch recombination (CSR) is initiated by DNA breaks triggered by activation-induced cytidine deaminase (AID). These breaks activate DNA damage response proteins to promote appropriate repair and long-range recombination. Aberrant processing of these breaks, however, results in decreased CSR and/or increased frequency of illegitimate recombination between the immunoglobulin heavy chain locus and oncogenes like c-myc. Here, we have examined the contribution of the DNA damage sensors Parp1 and Parp2 in the resolution of AID-induced DNA breaks during CSR. We find that although Parp enzymatic activity is induced in an AID-dependent manner during CSR, neither Parp1 nor Parp2 are required for CSR. We find however, that Parp1 favors repair of switch regions through a microhomology-mediated pathway and that Parp2 actively suppresses IgH/c-myc translocations. Thus, we define Parp1 as facilitating alternative end-joining and Parp2 as a novel translocation suppressor during CSR.
The B cell repertoire is diversified during immune responses through somatic hypermutation (SHM) and class switch recombination (CSR) to generate highly specific and adapted humoral responses. SHM introduces point mutations in the variable region of Ig genes, thereby increasing antibody affinity for antigen (1). CSR modulates antibody effector functions by replacing the antibody isotype expressed (from IgM to IgG, IgE, or IgA), while retaining the antigen-binding specificity of the receptor (2). SHM and CSR require the expression of activation-induced cytidine deaminase (AID) (3, 4), an enzyme that deaminates cytidines in DNA and that generates U:G mismatches in Ig genes (5, 6). Lesions induced by AID are processed by base excision repair and/or mismatch repair enzymes (including uracyl DNA glycosylase [UNG], APE1, APE2, MSH2, and MSH6) to generate mutations or double-stranded DNA breaks (DSBs) in Ig genes (1, 2). CSR is a region-specific recombination reaction that involves the joining of repetitive, but nonhomologous, switch region DNA sequences that can be separated by up to 200 kb and that requires DSBs as intermediates (2, 7). These DNA breaks activate DNA damage response proteins, including the PI3-like protein kinase ataxia-telangiectasia mutated (ATM), the histone variant H2AX, the MRN complex (Nbs1, Mre11, and Rad50), MDC1, and 53BP1 to promote appropriate repair and efficient long-range recombination (7). Consistent with this, deficiency in any of these genes results in defective CSR (2, 7). The joining step of the reaction was believed to be primarily mediated by the nonhomologous end-joining pathway (NHEJ) (2, 7). However, recent evidence indicates that an alternative pathway that is independent of XRCC4 and DNA ligase IV, and which is biased toward microhomology usage, has a significant contribution in the resolution of AID-induced DNA breaks during CSR (8–10).
Despite the numerous pathways and proteins involved in sensing and mediating the repair of DNA damage, AID-induced DNA breaks can be aberrantly resolved in cis to produce internal deletions within the Ig heavy chain (IgH) locus (11–14), and in trans to produce chromosomal translocations (15–19) that have the potential to promote cellular transformation (20). Indeed, translocations involving the IgH locus are frequently found in almost all cancer-associated chromosomal translocations in mature B cell lymphomas and in multiple myeloma (21). Strong evidence supporting the hypothesis that reciprocal translocations involving the IgH and oncogenes like c-myc are byproducts of the SHM and CSR reactions has been recently provided (15–19). The generation of IgH/c-myc translocations is dependent on AID expression, AID's catalytic activity, on the processing of AID-induced U:G mismatches in DNA by UNG, and on the transcriptional status of the c-myc locus (15–19). In addition, suppression of IgH/c-myc translocations requires the establishment of p53-mediated checkpoints through the activation of Nbs1, ATM, and/or the tumor suppressor p19Arf (17). Furthermore, DSB resolution into chromosomal translocations seems to be independent of the NHEJ components Ku80, XRCC4, and DNA ligase IV, indicating that an alternative NHEJ is involved in mediating aberrant interchromosomal joining (9, 17).
Poly(ADP)-ribose polymerases catalyze the covalent attachment of poly(ADP)-ribose units on amino acid residues of acceptor proteins using β-NAD+ as a substrate (22). The resulting poly(ADP)ribosylation is an immediate and transient posttranslational modification of target proteins that has been involved in modulating many essential cellular processes, including transcription, replication, and DNA repair (22). Among the 17 members of the Parp family of proteins described to date, the best studied and characterized are the founding member Parp1 and its close homologue Parp2 (23). These two enzymes are unique in that they are the only members of the family that recognize and are activated by DNA breaks and are believed to be DNA damage sensors (23–25). Consistent with this, their inactivation in mice leads to increased sensitivity to ionizing radiation and alkylating agents and to genomic instability (24, 26). Furthermore, genetic interactions between Parp1, Parp2, and ATM have been described and ATM−/−Parp1−/−, ATM−/−Parp2−/−, and Parp1−/−Parp2−/− mice display embryonic lethality (26–28). Here, we have examined the role of Parp1 and Parp2 in responding to AID-induced DNA breaks generated during CSR. We show that Parp1 and Parp2 have nonredundant roles and that Parp1 facilitates repair through the alternative NHEJ, whereas Parp2 actively suppresses the generation of IgH/c-myc translocations induced by AID during Ig CSR.
RESULTS
AID-dependent Parp activity is induced in B cells undergoing CSR
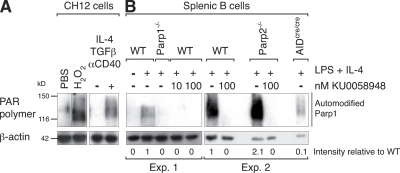
Induction of DNA damage results in the activation of Parp1 and Parp2, which are enzymes that catalyze the poly(ADP) ribosylation of multiple target proteins to promote DNA repair (23). This activity can be readily visualized by Western blotting using an antibody specific for the polymer of ADP-ribose (PAR). As one of the main targets of Parp activity is Parp1 itself, the antibody primarily reveals the automodified form of Parp1 and a smear above corresponding to Parp1 containing polymers of different length (29). To determine whether Parp activity is induced during CSR we stimulated CH12 cells with TGF-β, Il-4 and anti-CD40 antibody. Under these conditions, the IgM+ CH12 lymphoma cell line undergoes very efficient CSR to IgA (30). After 72 h in culture we assayed for Parp activity by Western blotting (Fig. 1 A). We found that Parp activity is induced in CH12 cells at levels comparable to those observed in cells that were exposed to H2O2 (Fig. 1 A), treatment known to induce a strong Parp activity (31). To determine whether Parp activity is also induced in primary splenic B cells, we cultured purified resting B cells from wild-type mice in the presence of LPS and IL-4 for up to 72 h. Western blot analysis using the anti-PAR antibody revealed that CSR induction resulted in robust Parp activity (Fig. 1 B). This activity was completely abolished by the addition of 10 or 100 nM KU0058948, a potent Parp inhibitor that has been described to inhibit both Parp1 and Parp2 at 100 nM (Fig. 1 B) (32). As expected, the automodified form of Parp1 was not observed in stimulated Parp1−/− B cells (Fig. 1 B). In addition, we found that Parp1 automodification was increased on average by twofold (as determined by Western blot quantification) in Parp2-deficient B cells (Fig. 1 B). Thus, Parp1 activity is increased in Parp2−/− B cells and most likely Parp1 compensates for the lack of Parp2. This finding is consistent with previous reports of increased Parp1 activity in Parp2-deficient B cells (33) and with our own observations in other Parp2-deficient cell systems (unpublished data). To determine whether the induction of Parp activity is dependent on AID expression, we cultured AID-deficient B cells (obtained from mice bearing a targeted insertion of the CRE recombinase cDNA into AID's exon 1 [AIDcre/cre]; reference 19) and which display the same phenotype as AID−/− mice (3) in the presence of LPS and IL-4. We found that Parp activity was reduced by ∼90% in AID-deficient B cells when compared with wild-type B cells (Fig. 1 B). The remaining Parp activity may correspond to break-independent Parp activation, to other modes of physiological Parp activation, or to activation of other Parp family members. We conclude that Parp activity is induced in B cells undergoing CSR, that this activity is increased in Parp2−/− B cells, and that it is dependent on AID expression.
Figure 1.
AID-dependent Parp activity is induced in B cells undergoing CSR. Western blot analysis of poly(ADP)ribosylation using antibodies specific for the polymer of ADP-ribose (PAR; top) and β-actin (bottom) in whole-cell extracts from CH12 lymphoma and splenic B lymphocytes. (A) CH12 cells extracts were prepared before or after treatment with PBS or 1 mM H2O2 for 10 min as controls, and before or after stimulation for 3 d with IL-4, TGF-β, and anti-CD40 antibody. (B) Splenic B cell extracts from wild-type (WT), Parp1−/− (24), Parp2−/− (26), and AID-deficient (AIDcre/cre; reference 19) B lymphocytes were prepared before or after 2–3 d of stimulation with LPS and IL-4 in the presence or absence of 10 and 100 nM KU0058948 Parp inhibitor. Automodified Parp1 is indicated by a vertical line. Numbers below the panel reflect the intensity of bands representing automodified Parp1 relative to wild type and after normalization to β-actin. Data from two independent experiments are shown (samples were processed at 72 h [experiment 1] and 48 h [experiment 2]). Molecular weight sizes in kilodaltons (kD) are indicated.
Inhibition of Parp activity results in increased CSR in transformed B cell lines
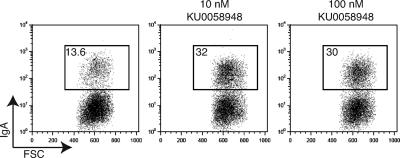
To determine whether pharmacological inhibition of Parp activity affects the efficiency of CSR, we stimulated CH12 cells in the presence or absence of the Parp inhibitor KU0058948. Treatment of CH12 cells with this inhibitor, at concentrations that inhibit Parp activity in B cells (Fig. 1) and other systems (32), had no effect on cell survival or proliferation (not depicted). We found, however, that the addition of KU0058948 resulted in a two- to threefold increase in CSR efficiency as determined by flow cytometry (Fig. 2). This observation is consistent with the finding that Parp inhibition in the I.29μ transformed B cell line also results in enhanced CSR (34).
Figure 2.
Pharmacological inhibition of Parp activity results in increased CSR in CH12 cells. CH12 cells were stimulated with IL-4, TGF-β, and anti-CD40 antibody, in the presence or absence of 10 and 100 nM KU0058948 Parp inhibitor. After 3 d in culture, cells were analyzed by flow cytometry for cell surface expression of IgA in three independent experiments. Representative flow cytometry profiles are shown. The percentage of switched cells is indicated in each plot.
Parp1 and Parp2 are not required for CSR in splenic B cells
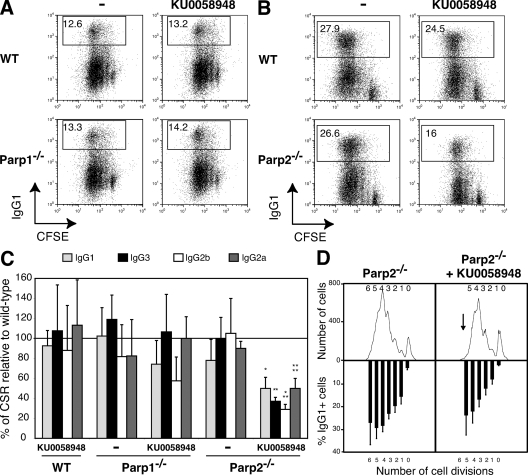
To directly assess the contribution of Parp1 and Parp2 in responding to AID-mediated DNA breaks during CSR, we tested the intrinsic ability of Parp1−/− and Parp2−/− B lymphocytes to undergo CSR in vitro in the presence or absence of KU0058948 Parp inhibitor (Fig. 3). We purified mature resting B cells from the spleen, labeled them with CFSE to track proliferation, and stimulated them to undergo CSR to various isotypes by using in vitro culture conditions known to induce switching to precise isotypes. After 72 h in culture, cells were analyzed by flow cytometry for proliferation (CFSE dye dilution) and Ig surface expression. We found that deficiency in Parp1 or Parp2 had no effect on proliferation in response to LPS, LPS + IL-4, or LPS + IFN-γ stimulation (Fig. 3, A and B, and not depicted). Surprisingly, in contrast to CH12 cells, we found that Parp1−/− and Parp2−/− B cells underwent CSR to IgG1, IgG2b, IgG3, and IgG2a at wild-type frequencies (Fig. 3, A–C). Although in our hands CSR induction to IgA with LPS + IL-5 + TGF-β is not very efficient (<2%), we did not observe an increase in CSR in Parp1−/− or wild-type B cells treated with the Parp inhibitor (not depicted). To determine whether Parp2 compensates for Parp1 deficiency and vice versa, we tested the ability of Parp1−/− and Parp2−/− B cells to undergo CSR in the presence of KU0058948. Inhibition of Parp activity in a Parp1-deficient background had no effect in either proliferation or in CSR efficiency (Fig. 3, A and C). A significant reduction in the frequency of IgG+ cells was observed in Parp2−/− B cells in the presence of KU0058948 (Fig. 3, B and C). As CSR is tightly correlated with cell division (35), this effect could be explained by reduced proliferation of Parp2−/− B cells in the presence of the Parp inhibitor (Fig. 3 D). Consistent with this, differences in CSR were not significant when cells were analyzed for Ig surface expression by gating on cell populations having undergone a defined number of cell divisions (Fig. 3 D). Furthermore, we found that AID expression (as determined by RT-PCR and Western blot) was unaffected by deficiency in Parp1 or Parp2, or by the pharmacological inhibition of Parp activity (not depicted). The level of switch region sterile transcripts and circle transcripts, generated only after successful CSR (36), were detected at similar levels in all experimental groups and were not different from those observed in wild-type B cells (not depicted). We conclude that Parp1 and Parp2 are not required for CSR in freshly isolated splenic B cells.
Figure 3.
Parp1 and Parp2 are not required for CSR in splenic B cells. Wild-type (WT), Parp1−/−, and Parp2−/− resting splenic B cells were stained with CFSE and cultured for 3 d with LPS (CSR to IgG3 and IgG2b); LPS + IL-4 (CSR to IgG1); LPS + IFN-γ (CSR to IgG2a); and in the presence or absence of 100 nM KU0058948 Parp inhibitor. Ig surface expression was determined by flow cytometry within the live cell population (gated using ToPro-3 exclusion) and using isotype-specific antibodies in at least five independent experiments. Cell division was measured by CFSE dye dilution. (A) IgG1 cell surface expression of WT and Parp1−/− B cells stimulated with LPS + IL-4 in the presence or absence of 100 nM KU0058948. The percentage of switched cells is indicated in each plot. (B) IgG1 cell surface expression of WT and Parp2−/− B cells stimulated with LPS + IL-4 in the presence or absence of 100 nM KU0058948. The percentage of switched cells is indicated in each plot. (C) The mean percentages (± standard deviation) of CSR relative to WT for the different isotypes tested are shown (IgG1, light gray bars; IgG3, black bars; IgG2b, white bars; IgG2a, dark gray bars). CSR in WT B cells was set to 100% (horizontal line). Statistical significance as determined by an unpaired Student's t test is indicated as follows: *, P = 0.0073; **, P = 0.0008; ***, P < 0.0001; ****, P = 0.0004. (D) Flow cytometric analysis of IgG1 expression on CFSE-labeled Parp2−/− B cells stimulated with LPS plus IL-4 for 3 d in the presence or absence of 100 nM KU0058948 Parp inhibitor. Cell division as measured by CFSE dye dilution is shown at top. The mean percentages (± standard deviation) of cells expressing IgG1 after a specific number of cell divisions is indicated on the bottom.
Reduced microhomology at switch recombination junctions in Parp1−/− B cells
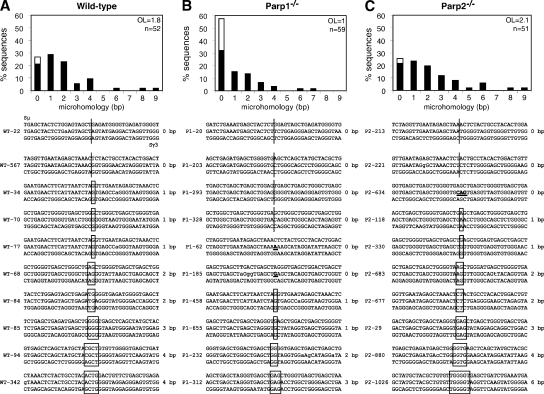
Switch regions are joined by classical and alternative end-joining mechanisms and are characterized by small insertions and short regions of microhomology at the junction. In the absence of the NHEJ proteins XRCC4 or DNA ligase IV, switch junctions display a significant increase in microhomology and a complete absence of sequences with no microhomology (9). A similar increase in homology was also observed in patients with hypomorphic mutations in DNA ligase IV (10). To determine the pathway that catalyzes CSR in the absence of Parp1 or Parp2, we cloned and sequenced Sμ/Sγ3 switch junctions from LPS-stimulated wild-type, Parp1−/−, and Parp2−/− B cells (Fig. 4 and Figs. S1–S3). Comparison of CSR junctions from Parp1-deficient B cells to those of controls revealed a significant difference. Wild-type sequences contained direct joints (21.2%) and small (1–4 nt) insertions (5.8%), with most of the remainder sequences bearing short homologies (Fig. 4 A). The mean length of overlap at the junction was 1.8 bp. Interestingly, Parp1-deficient sequences displayed a clear shift toward no microhomology (Fig. 4 B), and 57.6% of the sequences had no homology and were either direct joints (32.2%) or contained small insertions (25.4%). Furthermore, the mean length of overlap at the junction was reduced to 1.0 bp (Fig. 4 B; P = 0.003 vs. wild type). In contrast to Parp1−/−, no increase in the proportion of sequences with no microhomology was observed in Parp2−/− sequences (Fig. 4 C), which were more similar to wild-type in their distribution (Fig. 4 C). The minor increase in the mean length of overlap at the junction in Parp2−/− sequences (2.1 bp) was not statistically significant when compared with wild type (Fig. 4 C; P = 0.22 vs. wild type). We conclude that switch recombination junctions in Parp1−/− B cells are biased toward no microhomology. Thus, in the absence of Parp1, DNA ends generated during CSR are primarily processed through an end-joining mechanism not relying on microhomology.
Figure 4.
Reduced microhomology at switch recombination junctions in Parp1−/− B cells. Histograms showing the percentage of Sμ/Sγ3 switch junction sequences with indicated nucleotide overlap and obtained from wild-type (A; n = 52), Parp1−/− (B; n = 59), and Parp2−/− (C; n = 51) LPS-stimulated B cells in at least 5 independent experiments. Mean length of overlap in base pairs (OL) and the number of sequences analyzed (n) is indicated. White portion of bars indicates the percentage of sequences with small (1–4 nt) insertions and was as follows: wild type, 5.8%; Parp1−/−, 25.4%; and Parp2−/−, 3.9%. Switch junction alignments are shown below each panel (additional sequences in Figs. S1–S3). Overlap was determined by identifying the longest region at the switch junction of perfect uninterrupted donor/acceptor identity. Sequences with insertions at the junction were scored as having no microhomology. Sμ/Sγ3 sequences are shown in the middle. The Sμ and Sγ3 germline sequences (chromosome 12 genomic sequence: NT-114985) are shown above and below each junction sequence, respectively. Homology at the junctions is boxed, and the length of overlap is indicated on the right. Lowercase letters indicate mutations. Insertions are bolded and underlined. Statistical significance was determined by a Mann-Whitney test: wild-type versus Parp1−/−, P = 0.003; Parp1−/− versus Parp2−/−, P = 0.0006; wild-type versus Parp2−/−, P = 0.22.
Parp2 suppresses IgH/c-myc translocations during CSR
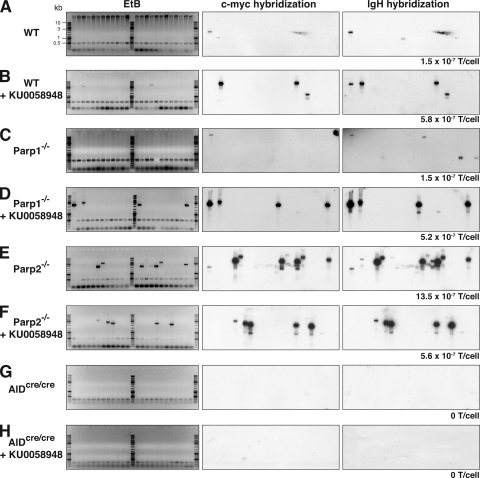
DNA DSBs induced by AID are believed to be obligatory intermediates in CSR. Aberrant processing of these breaks results in defective CSR, increased frequency of short-range recombination events (11–14), and generation of AID-dependent chromosomal translocations involving oncogenes such as c-myc (15–19). To determine the involvement of Parp1 and Parp2 in suppressing IgH/c-myc translocations during CSR, we cultured B cells obtained from wild-type, Parp1−/−, and Parp2−/− mice with LPS + IL-4 in the presence or absence of 100 nM KU0058948 Parp inhibitor. After 72 h in culture, we determined the frequency of IgH/c-myc translocations by long-range PCR (15) and Southern blotting (Fig. 5) and analyzed the translocations by sequencing (Figs. S4 and S5). Consistent with previous reports (17), we found that IgH/c-myc translocations were induced at low frequency in wild-type B cells (1.5 × 10−7 translocations/cell; Fig. 5 A). Interestingly, inhibition of Parp activity in wild-type B cells resulted in a 3.8-fold increase in the frequency of IgH/c-myc translocations (5.8 × 10−7 translocations/cell; P = 0.0193; Fig. 5 B), suggesting a role for the catalytic activity of Parp1 and/or Parp2 in suppressing these translocations. In Parp1−/− B cells, IgH/c-myc translocations accumulated at a frequency that was similar to that observed in wild-type B cells (1.5 × 10−7 translocations/cell; P = 0.6574; Fig. 5 C), indicating that Parp1 does not play an active role in suppressing this type of translocation, or alternatively, that Parp2 enzymatic activity may compensate for the lack of Parp1. To discriminate between these two possibilities, we assayed for translocations in Parp1−/− B cells in the presence of KU0058948 Parp inhibitor. Pharmacological inhibition of Parp activity in a Parp1-deficient background resulted in a 3.4-fold increase in the translocation frequency (5.2 × 10−7 translocations/cell; Fig. 5 D) when compared with wild-type B cells (P = 0.0318), indicating that Parp1 and Parp2 have nonredundant roles and that the catalytic activity of Parp2 is required to suppress IgH/c-myc translocations during CSR. Consistent with this, we found that Parp2 deficiency resulted in a ninefold increase in the translocation frequency (13.5 × 10−7 translocations/cell; Fig. 5 E) when compared with wild-type B cells (P < 0.0001). Thus, Parp2 is required to suppress IgH/c-myc translocations. To determine whether Parp1 activity can modulate the Parp2-dependent suppression, we cultured Parp2−/− B cells in the presence of the inhibitor. Under these conditions, the translocation frequency (5.6 × 10−7 translocations/cell; Fig. 5 F) was 3.7-fold higher than in wild-type B cells (P = 0.0139), similar to wild-type + KU0058948 (P = 0.5472) or Parp1−/− + KU0058948 (P = 0.4837), but reduced when compared with Parp2−/− B cells (P = 0.0006). Thus, increased Parp1 activity in Parp2−/− B cells correlates with increased translocation frequency and suggests that Parp1 modulates the suppressor function of Parp2 by favoring repair through alternative NHEJ. To determine whether the increase in translocation frequency triggered by Parp inhibition is dependent on AID expression, we cultured AIDcre/cre B cells in the presence or absence of KU0058948. Consistent with previous reports (15–19), IgH/c-myc translocations were not observed in AID-deficient B cells (0 translocations/cell; Fig. 5 G). Furthermore, the increase in translocation frequency triggered by the inhibition of Parp activity was AID dependent (Fig. 5 H), as we did not detect any translocations in AIDcre/cre B cells that were cultured in the presence of KU0058948 (0 translocations/cell; P = 0.0005 vs. wild type + KU0058948; P = 0.0008 vs. Parp1−/− + KU0058948; P = 0.0003 vs. Parp2−/− + KU0058948). We conclude that the translocation suppressor function of Parp2 is AID dependent. Translocation sequence analysis revealed no significant differences between groups in either breakpoint distribution (in Sμ and in c-myc) or in the amount of donor/acceptor homology at the junction (Figs. S4 and S5). We conclude that Parp1 and Parp2 have nonredundant activities and that although Parp1 has no protective role, Parp2 suppresses AID-mediated IgH/c-myc translocations during CSR.
Figure 5.
Parp2 suppresses IgH/c-myc translocations. Wild-type (WT), Parp1−/−, Parp2−/− B cells, and AIDcre/cre B cells were cultured for 3 d with LPS plus IL-4, in the presence or absence of 100 nM KU0058948 Parp inhibitor, and assayed for IgH/c-myc translocations by long-range PCR in three to five independent experiments. Representative ethidium bromide stained agarose gels (EtB; left) and corresponding Southern blots using c-myc (middle) and IgH (right) probes are shown. The number of translocations found (T) and individual PCR assays done (with template DNA corresponding to 105 cells) was as follows: WT, T=3, n = 200; WT + KU0058948, T=12, n = 208; Parp1−/−, T=3, n = 200; Parp1−/− + KU0058948, T=12, n = 232; Parp2−/−, T=40, n = 296; Parp2−/− + KU0058948, T=18, n = 319; AIDcre/cre, T=0, n = 188; AIDcre/cre + KU0058948, T=0, n = 188. The corresponding frequency of translocations per cell (T/cell) is indicated underneath each panel and was as follows: WT, 1.5 × 10−7; WT + KU0058948, 5.8 × 10−7 (P = 0.0193 vs. WT); Parp1−/−, 1.5 × 10−7 (P = 0.6574 vs. WT); Parp1−/− + KU0058948, 5.2 × 10−7 (P = 0.0318 vs. WT; P = 0.0318 vs. Parp1−/−); Parp2−/−, 13.5 × 10−7 (P < 0.0001 vs. WT; P < 0.0001 vs. Parp1−/−; P = 0.0009 vs. Parp1−/− + KU0058948); Parp2−/− + KU0058948, 5.6 × 10−7 (P = 0.0139 vs. WT; P = 0.5472 vs. WT + KU0058948; P = 0.4837 vs. Parp1−/− + KU0058948; P = 0.0006 vs. Parp2−/−); AIDcre/cre, 0 (P = 0.1360 vs. WT; P = 0.1360 vs. Parp1−/−; P < 0.0001 vs. Parp2−/−); AIDcre/cre + KU0058948, 0 (P = 0.1360 vs. WT; P = 0.0005 vs. WT + KU0058948; P = 0.0008 vs. Parp1−/− + KU0058948; P = 0.0003 vs. Parp2−/− + KU0058948). Statistical significance was determined using one-tailed exact Fisher's test. Molecular weight sizes in kilobasepairs (kb) are indicated in A and are the same for all panels.
DISCUSSION
We have found that Parp activity is induced in an AID-dependent manner in B cells undergoing CSR, and therefore induced in response to DNA breaks triggered by AID. Despite the induction of Parp activity, we found that disruption of Parp1 or Parp2, or the pharmacological inhibition of their catalytic activities, had no significant impact on the efficiency of the CSR reaction in cultured splenic B cells. Surprisingly, this is in contrast to the increased CSR detected in the transformed cell lines CH12 (this study) and 1.29μ (34) treated with a Parp inhibitor. Although we are unable to provide a molecular explanation for this observation, we speculate that the differential effect of Parp inhibition observed in primary B cells compared with CH12 cells might be related to the transformed state of the cell line, to unsuspected chromosomal aberrations (i.e., CH12 cells have three copies of the IgH locus; unpublished data), to differences in the frequency of AID-induced DNA breaks, or, alternatively, to the relative sequence homology between the Sμ and Sα switch regions.
The intrinsic ability of splenic B cells to undergo CSR to all isotypes tested is not compromised in Parp1−/− or Parp2−/− B cells. Consistent with this, we found that AID expression, which was robust and detectable as early as day 2 after stimulation, was not affected by deficiency in either Parp1 or Parp2 or by the inhibition of Parp activity. Furthermore, we found no differences in the levels of switch region sterile transcripts, and circle transcripts, which are generated only after successful CSR. Our results differ from a recent report showing that Parp1−/− mice display abnormal levels of circulating serum Igs (elevated IgG2a and IgA and reduced IgG2b). This study also showed that in vitro–stimulated Parp1−/− B cells secrete higher levels of IgG2a and IgA, but wild-type levels of IgG2b antibodies, despite decreased levels of serum IgG2b (33). Furthermore, Parp1−/− B cells secreted wild-type levels of IgG3 and IgG2b in response to LPS stimulation, despite undetectable levels of AID mRNA at day 3 and a prominent delay in AID mRNA accumulation by day 7 (33). A possible explanation for the differences observed between both studies may be related to the assays used to analyze the CSR reaction. In this work, we assessed the intrinsic ability of purified resting B cells to undergo CSR in response to different stimuli in vitro by analyzing the cell surface expression of Igs by flow cytometry and not their capacity to secrete antibodies into the culture medium. Furthermore, alterations in the level of circulating serum Igs in vivo may be influenced by other factors such as the frequency of plasma cells, survival of switched B cells, or abnormal B–T cell interactions. Indeed, both Parp1−/− and Parp2−/− mice display alterations within the T cell compartment (33, 37).
AID-induced DNA breaks generated during CSR are ultimately joined by pathways that require the Ku70 and Ku80 subunits of DNA-PK (14, 38, 39). Given this requirement, it was believed that DNA breaks occurring within switch regions during CSR were ultimately repaired through NHEJ (2, 7). This view however, was recently challenged by experiments showing that CSR is only moderately diminished (25–50% reduction) in the absence of the NHEJ core components XRCC4 or DNA ligase IV (8, 9), indicating that an alternative pathway, independent of XRCC4 and DNA ligase IV, can efficiently join AID-induced DNA breaks (8, 9). Analysis of switch region junctions in XRCC4- or DNA ligase IV–deficient B cells revealed that this alternative pathway relies on microhomology (9, 10). Furthermore, a substantial fraction of XRCC4-deficient B cells displayed chromosomal aberrations, the majority of which involved translocations, supporting the hypothesis that this alternative end-joining pathway significantly contributes to repairing DNA breaks during CSR and that it frequently produces translocations in response to AID-induced DNA breaks (9).
DNA breaks generated during CSR are complex and heterogeneous in nature, and some DNA ends may be rapidly joined through the classical NHEJ, whereas other breaks may require extensive end processing, a search for homology, and the cleavage of nonhomologous nucleotides before ligation. These types of DNA ends may be repaired at a slower rate and may be better substrates for the alternative NHEJ (9). Thus, it has been proposed that in B cells undergoing CSR, the classical and alternative NHEJ are in direct competition for DNA ends generated in switch region sequences (9). Consistent with this, we have found that Parp1 deficiency results in a qualitative effect on the CSR reaction, as indicated by the nature of the switch junction sequences. Switch junctions obtained from Parp1-deficient B cells displayed a clear bias toward no homology, indicating that the alternative NHEJ was suppressed in the absence of Parp1, and that Parp1 favors the usage of a microhomology-based end-joining mechanism. This is consistent with the proposed role for Parp1 in competing with Ku80 for accessibility to DNA ends (40) and with the proposed role for Parp1, XRCC1, and DNA ligase III in mediating alternative end-joining (41–44). Our results support the hypothesis that Parp1 is part of the alternative NHEJ pathway in mammalian cells and implicate the base excision repair proteins XRCC1 and DNA ligase III in the processing of AID-induced DNA breaks during CSR.
We have found that Parp1 and Parp2 have specific and nonredundant roles, and that although Parp1 has no protective role, Parp2 suppresses AID-mediated IgH/c-myc translocations during CSR. The translocation suppressor function of Parp2 appeared, however, to be down-modulated by the addition of a Parp inhibitor, suggesting that the catalytic activity of Parp1 may regulate the suppressor function of Parp2. Given the role of Parp1 in facilitating repair through a microhomology-based end-joining mechanism that frequently produces translocations (9) and the increased Parp1 activity that we observed in Parp2−/− B cells (Fig. 1 B), it is possible that the translocation frequency observed in Parp2−/− B cells results from the synergistic effect of a bias toward alternative NHEJ driven by increased Parp1 activity and by the lack of suppression by Parp2. Thus, inhibiting Parp activity in wild-type, Parp1−/−, and Parp2−/− B cells reveals the unique contribution of Parp2 in suppressing IgH/c-myc translocations.
Despite the established shared functions of Parp1 and Parp2 in the cellular response to DNA damage and in the maintenance of heterochromatin integrity, recent advances reveal unique functions that have only begun to be clarified (23). Parp1 and Parp2 have different substrates and targets both in DNA and in proteins (23), and they display substantial structural differences that could account for their differential requirement in facilitating alternative end-joining and in suppressing IgH/c-myc translocations. For example, Parp1 and Parp2 differ in the structure of their DNA-binding domains (23, 25, 45), and as such may not recognize the same structures or be activated by the same type of DNA lesion. Indeed, it has been suggested previously that Parp2 displays higher affinity than Parp1 for gaps or flaps (25, 46), structures that we speculate may correspond to DNA ends that require substantial processing before joining and that may represent translocation intermediates. Parp1 and Parp2 also differ in their protein–protein interaction domains, and this may result in differential responses caused by specific partner recruitment. In addition, Parp2 contains a 3-aa insertion in the vicinity of the catalytic pocket, which may contribute to fine tuning substrate selection and downstream effector specificity (47). Notably, Parp1 accumulates at sites of DNA damage with faster kinetics than Parp2 (48), suggesting that Parp1 may play a more prominent role in sensing DNA damage and that Parp2 may be required at later steps during DNA repair. Furthermore, XRCC1 recruitment to DNA breaks is dependent on Parp1 and not Parp2 (48). Thus, it is possible that faster recruitment of Parp1 to DNA breaks followed by XRCC1 accumulation contributes to its specific role in facilitating alternative end-joining. In addition to a direct role in sensing DNA breaks, with the potential of being channeled into an interchromosomal translocation, it is possible that Parp2 exercises its suppressor function by enforcing the p53-dependent signal for oncogenic stress (17). This could be achieved by modulating the activity of the tumor suppressor p19Arf via the physical association of Parp2 with B23 (also known as nucleophosmin 1; reference 31), a nucleolar protein (49) that regulates the activity of p19Arf (50–55) and that has been previously implicated in CSR (56). In addition, it is possible that Parp2 suppresses IgH/c-myc translocations through p53-independent pathways that would involve the posttranslational modification of effector proteins that have not yet been identified.
Our results establish that Parp1 and Parp2 respond to programmed DNA damage in B cells undergoing CSR, define a role for Parp1 in favoring DNA repair through the alternative NHEJ, and uncover Parp2 as a novel translocation suppressor during CSR.
MATERIALS AND METHODS
Mice.
Parp1−/− (24), Parp2−/− (26), and AIDcre/cre (19) mice were bred and maintained under specific pathogen–free conditions. Age-matched littermates (8–12 wk old) obtained from heterozygous crosses were used in all experiments. All animal work was performed under protocols approved by the Direction des Services Vétérinaires du Bas-Rhin, France (authorization no. 67–343).
Lymphocyte cultures and flow cytometry.
Resting B lymphocytes were isolated from the spleen using CD43 Microbeads (Miltenyi Biotec) labeled with 5 µM CFSE (Invitrogen) and cultured with 50 µg/ml LPS (Sigma-Aldrich), 5 ng/ml IL-4 (Sigma-Aldrich), or 100 ng/ml IFN-γ (Peprotech). CH12 cells were cultured with 5 ng/ml IL-4, 3 ng/ml hTGF-β (R&D Systems), and 100 ng/ml monoclonal anti-CD40 antibody (eBioscience). The KU0058948 Parp inhibitor (AstraZeneca/KuDOS) (32) was diluted in PBS and used at a final concentration of 10 or 100 nM. To induce Parp activity, cells were treated for 10 min with 1 mM H2O2. For flow cytometry, cells were stained with biotin anti-IgG1 (BD), biotin anti-IgG3 (BD), biotin anti-IgG2b (BioLegend), biotin anti-IgG2a (BD), PE anti-IgA (SouthernBiotech), and PE-Streptavidin (Beckman Coulter). Dead cells were excluded from the analysis by staining with 50 nM ToPro-3 (Invitrogen). Data were collected on a FACSCalibur (BD) and analyzed with the FlowJo software (Tree Star, Inc.).
Translocation PCR.
PCR for detecting IgH/c-myc translocations was performed using previously described primers and PCR conditions (15, 17). In brief, genomic DNA corresponding to 105 cells was submitted to 2 rounds of 25 cycles of nested PCR using the Expand Long template PCR system (Roche). As positive control for amplification of all DNA samples, AID-specific primers (forward, 5′-GGACCCAACCCAGGAGGCAGATGT-3′; reverse, 5′-CCTCTAAGGCTTCGCTGTTATTACCAC-3′) were included in the first round of amplification. For PCR on AIDcre/cre genomic DNA, Cre specific primer (5′-CACTCGTTGCATCGACCGGTAATG-3′) was used in combination with AID forward primer. PCR products were analyzed by Southern blot using IgH and c-myc specific probes as previously described (15, 17). Genomic DNA obtained from at least three independent experiments and corresponding to ∼20-30 × 106 cells was assayed for each genotype.
Switch junction analysis.
Sμ-Sγ3 switch junctions were amplified using previously described primers (57) and conditions (14). PCR products were cloned using TOPO-TA cloning kit (Invitrogen) and sequenced using M13 universal primers. Sequence analysis was performed using Sequence Manager II software (DNASTAR).
Western blotting.
The antibodies used were: rabbit anti-PAR antibody (BioMol; 29), mouse anti-Parp1 (EGT69; 31), mouse anti–β-actin (clone AC-74; Sigma-Aldrich). Whole-cell extracts were prepared using standard procedures. Proteins were fractionated by SDS-PAGE and transferred to Immobilon-P membranes (Millipore) before Western blotting.
Online supplemental material.
Sμ/Sγ3 switch junction sequences obtained from wild-type, Parp1−/−, and Parp2−/− are shown in Figs. S1–S3, respectively. Fig. S4 contains the analysis of microhomology at the junction and breakpoint distribution of IgH/c-myc translocations. IgH/c-myc translocation junction sequence alignments are shown in Fig. S5. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20082468/DC1.
Acknowledgments
We thank Gilbert de Murcia, Jean-Christophe Amé, Valérie Schreiber and members of the Reina-San-Martin laboratory for discussions; Jean-Pierre de Villartay and Anne Durandy for comments on the manuscript; Delphine Schwartz for assistance with RT-PCR experiments; Rosy El Ramy for animal care; and Bondo Monga and Doulaye Dembele for advice on statistical analysis. We also dedicate this manuscript to Josiane Ménissier-de Murcia for her tremendous impact in the PARP field.
This work was supported by grants to B. Reina-San-Martin from the Agence Nationale pour la Recherche and the Institut National de la Santé et de la Recherche Médicale (AVENIR-INSERM). B. Reina-San-Martin was a fellow of the Fondation Recherche Médicale and is an AVENIR-INSERM young investigator.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used: AID, activation-induced cytidine deaminase; ATM, ataxia-telangiectasia mutated; CSR, class switch recombination; DSB, double-stranded DNA break; IgH, Ig heavy chain; NHEJ, nonhomologous end-joining pathway; SHM, somatic hypermutation; UNG, uracyl DNA glycosylase.
References
- Di Noia J.M., Neuberger M.S. 2007. Molecular mechanisms of antibody somatic hypermutation.Annu Rev Biochem. 76:1–22 [DOI] [PubMed] [Google Scholar]
- Stavnezer J., Guikema J.E., Schrader C.E. 2008. Mechanism and Regulation of Class Switch Recombination.Annu. Rev. Immunol. 26:261–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu M., Kinoshita K., Fagarasan S., Yamada S., Shinkai Y., Honjo T. 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme.Cell. 102:553–563 [DOI] [PubMed] [Google Scholar]
- Revy P., Muto T., Levy Y., Geissmann F., Plebani A., Sanal O., Catalan N., Forveille M., Dufourcq-Labelouse R., Gennery A., et al. 2000. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2).Cell. 102:565–575 [DOI] [PubMed] [Google Scholar]
- Petersen-Mahrt S.K., Harris R.S., Neuberger M.S. 2002. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification.Nature. 418:99–103 [DOI] [PubMed] [Google Scholar]
- Rada C., Williams G.T., Nilsen H., Barnes D.E., Lindahl T., Neuberger M.S. 2002. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice.Curr. Biol. 12:1748–1755 [DOI] [PubMed] [Google Scholar]
- Ramiro A., San-Martin B.R., McBride K., Jankovic M., Barreto V., Nussenzweig A., Nussenzweig M.C. 2007. The role of activation-induced deaminase in antibody diversification and chromosome translocations.Adv. Immunol. 94:75–107 [DOI] [PubMed] [Google Scholar]
- Soulas-Sprauel P., Le Guyader G., Rivera-Munoz P., Abramowski V., Olivier-Martin C., Goujet-Zalc C., Charneau P., de Villartay J.P. 2007. Role for DNA repair factor XRCC4 in immunoglobulin class switch recombination.J. Exp. Med. 204:1717–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C.T., Boboila C., Souza E.K., Franco S., Hickernell T.R., Murphy M., Gumaste S., Geyer M., Zarrin A.A., Manis J.P., et al. 2007. IgH class switching and translocations use a robust non-classical end-joining pathway.Nature. 449:478–482 [DOI] [PubMed] [Google Scholar]
- Pan-Hammarstrom Q., Jones A.M., Lahdesmaki A., Zhou W., Gatti R.A., Hammarstrom L., Gennery A.R., Ehrenstein M.R. 2005. Impact of DNA ligase IV on nonhomologous end joining pathways during class switch recombination in human cells.J. Exp. Med. 201:189–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley D.D., Manis J.P., Zarrin A.A., Kaylor L., Tian M., Alt F.W. 2002. Internal IgH class switch region deletions are position-independent and enhanced by AID expression.Proc. Natl. Acad. Sci. USA. 99:9984–9989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reina-San-Martin B., Chen H.T., Nussenzweig A., Nussenzweig M.C. 2004. ATM is required for recombination between immunoglobulin switch regions.J. Exp. Med. 200:1103–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reina-San-Martin B., Chen J., Nussenzweig A., Nussenzweig M.C. 2007. Enhanced intra-switch region recombination during immunoglobulin class switch recombination in 53BP1−/− B cells.Eur. J. Immunol. 37:235–239 [DOI] [PubMed] [Google Scholar]
- Reina-San-Martin B., Difilippantonio S., Hanitsch L., Masilamani R.F., Nussenzweig A., Nussenzweig M.C. 2003. H2AX is required for recombination between immunoglobulin switch regions but not for intra-switch region recombination or somatic hypermutation.J. Exp. Med. 197:1767–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramiro A.R., Jankovic M., Eisenreich T., Difilippantonio S., Chen-Kiang S., Muramatsu M., Honjo T., Nussenzweig A., Nussenzweig M.C. 2004. AID is required for c-myc/IgH chromosome translocations in Vivo.Cell. 118:431–438 [DOI] [PubMed] [Google Scholar]
- Unniraman S., Zhou S., Schatz D.G. 2004. Identification of an AID-independent pathway for chromosomal translocations between the Igh switch region and Myc.Nat. Immunol. 5:1117–1123 [DOI] [PubMed] [Google Scholar]
- Ramiro A.R., Jankovic M., Callen E., Difilippantonio S., Chen H.T., McBride K.M., Eisenreich T.R., Chen J., Dickins R.A., Lowe S.W., et al. 2006. Role of genomic instability and p53 in AID-induced c-myc-Igh translocations.Nature. 440:105–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett Y., Robbiani D.F., Jankovic M., Reina-San-Martin B., Eisenreich T.R., Nussenzweig M.C. 2007. A role for AID in chromosome translocations between c-myc and the IgH variable region.J. Exp. Med. 204:2225–2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani D.F., Bothmer A., Callen E., Reina-San-Martin B., Dorsett Y., Difilipantonio S., Bolland D.J., Chen H.T., Corcoran A.E., Nussenzweig A., Nussenzweig M.C. 2008. AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations.Cell. 135:1028–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Yebenes V.G., Ramiro A.R. 2006. Activation-induced deaminase: light and dark sides.Trends Mol. Med. 12:432–439 [DOI] [PubMed] [Google Scholar]
- Kuppers R. 2005. Mechanisms of B-cell lymphoma pathogenesis.Nat. Rev. Cancer. 5:251–262 [DOI] [PubMed] [Google Scholar]
- Schreiber V., Dantzer F., Ame J.C., de Murcia G. 2006. Poly(ADP-ribose): novel functions for an old molecule.Nat. Rev. Mol. Cell Biol. 7:517–528 [DOI] [PubMed] [Google Scholar]
- Yelamos J., Schreiber V., Dantzer F. 2008. Toward specific functions of poly(ADP-ribose) polymerase-2.Trends Mol. Med. 14:169–178 [DOI] [PubMed] [Google Scholar]
- de Murcia J.M., Niedergang C., Trucco C., Ricoul M., Dutrillaux B., Mark M., Oliver F.J., Masson M., Dierich A., LeMeur M., et al. 1997. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells.Proc. Natl. Acad. Sci. USA. 94:7303–7307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ame J.C., Rolli V., Schreiber V., Niedergang C., Apiou F., Decker P., Muller S., Hoger T., Menissier-de Murcia J., de Murcia G. 1999. PARP-2, A novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase.J. Biol. Chem. 274:17860–17868 [DOI] [PubMed] [Google Scholar]
- Menissier de Murcia J., Ricoul M., Tartier L., Niedergang C., Huber A., Dantzer F., Schreiber V., Ame J.C., Dierich A., LeMeur M., et al. 2003. Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse.EMBO J. 22:2255–2263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menisser-de Murcia J., Mark M., Wendling O., Wynshaw-Boris A., de Murcia G. 2001. Early embryonic lethality in PARP-1 Atm double-mutant mice suggests a functional synergy in cell proliferation during development.Mol. Cell. Biol. 21:1828–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber A., Bai P., de Murcia J.M., de Murcia G. 2004. PARP-1, PARP-2 and ATM in the DNA damage response: functional synergy in mouse development.DNA Repair (Amst.). 3:1103–1108 [DOI] [PubMed] [Google Scholar]
- Affar E.B., Duriez P.J., Shah R.G., Winstall E., Germain M., Boucher C., Bourassa S., Kirkland J.B., Poirier G.G. 1999. Immunological determination and size characterization of poly(ADP-ribose) synthesized in vitro and in vivo.Biochim. Biophys. Acta. 1428:137–146 [DOI] [PubMed] [Google Scholar]
- Nakamura M., Kondo S., Sugai M., Nazarea M., Imamura S., Honjo T. 1996. High frequency class switching of an IgM+ B lymphoma clone CH12F3 to IgA+ cells.Int. Immunol. 8:193–201 [DOI] [PubMed] [Google Scholar]
- Meder V.S., Boeglin M., de Murcia G., Schreiber V. 2005. PARP-1 and PARP-2 interact with nucleophosmin/B23 and accumulate in transcriptionally active nucleoli.J. Cell Sci. 118:211–222 [DOI] [PubMed] [Google Scholar]
- Farmer H., McCabe N., Lord C.J., Tutt A.N., Johnson D.A., Richardson T.B., Santarosa M., Dillon K.J., Hickson I., Knights C., et al. 2005. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy.Nature. 434:917–921 [DOI] [PubMed] [Google Scholar]
- Ambrose H.E., Willimott S., Beswick R.W., Dantzer F., de Murcia J.M., Yelamos J., Wagner S.D. 2008. Poly(ADP-ribose) polymerase-1 (Parp-1)-deficient mice demonstrate abnormal antibody responses.Immunology. doi:10.1111/j.1365-2567.2008.02921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shockett P., Stavnezer J. 1993. Inhibitors of poly(ADP-ribose) polymerase increase antibody class switching.J. Immunol. 151:6962–6976 [PubMed] [Google Scholar]
- Hodgkin P.D., Lee J.H., Lyons A.B. 1996. B cell differentiation and isotype switching is related to division cycle number.J. Exp. Med. 184:277–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita K., Harigai M., Fagarasan S., Muramatsu M., Honjo T. 2001. A hallmark of active class switch recombination: transcripts directed by I promoters on looped-out circular DNAs.Proc. Natl. Acad. Sci. USA. 98:12620–12623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelamos J., Monreal Y., Saenz L., Aguado E., Schreiber V., Mota R., Fuente T., Minguela A., Parrilla P., de Murcia G., et al. 2006. PARP-2 deficiency affects the survival of CD4+CD8+ double-positive thymocytes.EMBO J. 25:4350–4360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casellas R., Nussenzweig A., Wuerffel R., Pelanda R., Reichlin A., Suh H., Qin X.F., Besmer E., Kenter A., Rajewsky K., Nussenzweig M.C. 1998. Ku80 is required for immunoglobulin isotype switching.EMBO J. 17:2404–2411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manis J.P., Gu Y., Lansford R., Sonoda E., Ferrini R., Davidson L., Rajewsky K., Alt F.W. 1998. Ku70 is required for late B cell development and immunoglobulin heavy chain class switching.J. Exp. Med. 187:2081–2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Wu W., Wu W., Rosidi B., Zhang L., Wang H., Iliakis G. 2006. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways.Nucleic Acids Res. 34:6170–6182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosidi B., Wang M., Wu W., Sharma A., Wang H., Iliakis G. 2008. Histone H1 functions as a stimulatory factor in backup pathways of NHEJ.Nucleic Acids Res. 36:1610–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audebert M., Salles B., Weinfeld M., Calsou P. 2006. Involvement of polynucleotide kinase in a poly(ADP-ribose) polymerase-1-dependent DNA double-strand breaks rejoining pathway.J. Mol. Biol. 356:257–265 [DOI] [PubMed] [Google Scholar]
- Audebert M., Salles B., Calsou P. 2008. Effect of double-strand break DNA sequence on the PARP-1 NHEJ pathway.Biochem. Biophys. Res. Commun. 369:982–988 [DOI] [PubMed] [Google Scholar]
- Audebert M., Salles B., Calsou P. 2004. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining.J. Biol. Chem. 279:55117–55126 [DOI] [PubMed] [Google Scholar]
- Pion E., Ullmann G.M., Ame J.C., Gerard D., de Murcia G., Bombarda E. 2005. DNA-induced dimerization of poly(ADP-ribose) polymerase-1 triggers its activation.Biochemistry. 44:14670–14681 [DOI] [PubMed] [Google Scholar]
- Schreiber V., Ricoul M., Amé J.C., Dantzer F., Meder V.S., Spenlehauer C., Stiegler P., Niedergang C., Sabatier L. 2004. PARP-2, structure-function relationship. In Poly(ADP-ribosyl)ation. Burkle A., editor Landes Bioscience, Georgetown: 2004. 13–31 [Google Scholar]
- Oliver A.W., Ame J.C., Roe S.M., Good V., de Murcia G., Pearl L.H. Crystal structure of the catalytic fragment of murine poly(ADP-ribose) polymerase-2.Nucleic Acids Res. 32:456–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortusewicz O., Ame J.C., Schreiber V., Leonhardt H. 2007. Feedback-regulated poly(ADP-ribosyl)ation by PARP-1 is required for rapid response to DNA damage in living cells.Nucleic Acids Res. 35:7665–7675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuwaki M. 2008. The structure and functions of NPM1/Nucleophsmin/B23, a multifunctional nucleolar acidic protein.J. Biochem. (Tokyo). 143:441–448 [DOI] [PubMed] [Google Scholar]
- Yogev O., Saadon K., Anzi S., Inoue K., Shaulian E. 2008. DNA damage-dependent translocation of B23 and p19 ARF is regulated by the Jun N-terminal kinase pathway.Cancer Res. 68:1398–1406 [DOI] [PubMed] [Google Scholar]
- Rizos H., McKenzie H.A., Ayub A.L., Woodruff S., Becker T.M., Scurr L.L., Stahl J., Kefford R.F. 2006. Physical and functional interaction of the p14ARF tumor suppressor with ribosomes.J. Biol. Chem. 281:38080–38088 [DOI] [PubMed] [Google Scholar]
- Lee C., Smith B.A., Bandyopadhyay K., Gjerset R.A. 2005. DNA damage disrupts the p14ARF-B23(nucleophosmin) interaction and triggers a transient subnuclear redistribution of p14ARF.Cancer Res. 65:9834–9842 [DOI] [PubMed] [Google Scholar]
- Korgaonkar C., Hagen J., Tompkins V., Frazier A.A., Allamargot C., Quelle F.W., Quelle D.E. 2005. Nucleophosmin (B23) targets ARF to nucleoli and inhibits its function.Mol. Cell. Biol. 25:1258–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertwistle D., Sugimoto M., Sherr C.J. 2004. Physical and functional interactions of the Arf tumor suppressor protein with nucleophosmin/B23.Mol. Cell. Biol. 24:985–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto T., Lindstrom M.S., Jin A., Ke H., Zhang Y. 2006. Essential role of the B23/NPM core domain in regulating ARF binding and B23 stability.J. Biol. Chem. 281:18463–18472 [DOI] [PubMed] [Google Scholar]
- Borggrefe T., Wabl M., Akhmedov A.T., Jessberger R. 1998. A B-cell-specific DNA recombination complex.J. Biol. Chem. 273:17025–17035 [DOI] [PubMed] [Google Scholar]
- Guikema J.E., Linehan E.K., Tsuchimoto D., Nakabeppu Y., Strauss P.R., Stavnezer J., Schrader C.E. 2007. APE1- and APE2-dependent DNA breaks in immunoglobulin class switch recombination.J. Exp. Med. 204:3017–3026 [DOI] [PMC free article] [PubMed] [Google Scholar]