Abstract
Proliferation and survival of Hodgkin and Reed/Sternberg (HRS) cells, the malignant cells of classical Hodgkin lymphoma (cHL), are dependent on constitutive activation of nuclear factor κB (NF-κB). NF-κB activation through various stimuli is negatively regulated by the zinc finger protein A20. To determine whether A20 contributes to the pathogenesis of cHL, we sequenced TNFAIP3, encoding A20, in HL cell lines and laser-microdissected HRS cells from cHL biopsies. We detected somatic mutations in 16 out of 36 cHLs (44%), including missense mutations in 2 out of 16 Epstein-Barr virus–positive (EBV+) cHLs and a missense mutation, nonsense mutations, and frameshift-causing insertions or deletions in 14 out of 20 EBV− cHLs. In most mutated cases, both TNFAIP3 alleles were inactivated, including frequent chromosomal deletions of TNFAIP3. Reconstitution of wild-type TNFAIP3 in A20-deficient cHL cell lines revealed a significant decrease in transcripts of selected NF-κB target genes and caused cytotoxicity. Extending the mutation analysis to primary mediastinal B cell lymphoma (PMBL), another lymphoma with constitutive NF-κB activity, revealed destructive mutations in 5 out of 14 PMBLs (36%). This report identifies TNFAIP3 (A20), a key regulator of NF-κB activity, as a novel tumor suppressor gene in cHL and PMBL. The significantly higher frequency of TNFAIP3 mutations in EBV− than EBV+ cHL suggests complementing functions of TNFAIP3 inactivation and EBV infection in cHL pathogenesis.
Classical Hodgkin lymphoma (cHL) is one of the most common malignant lymphomas. It is characterized by the presence of rare Hodgkin and Reed/Sternberg (HRS) cells embedded in an extensive inflammatory infiltrate. Constitutive activation of NF-κB in HRS cells that transcriptionally regulates expression of multiple antiapoptotic factors and proinflammatory cytokines plays a central role in the pathogenesis of cHL (1, 2). In a nonstimulated condition, NF-κB proteins are rendered inactive by binding to inhibitors of NF-κB (IκBs), which sequester them in the cytoplasm. Stimulation of multiple receptors activates the IκB kinase (IKK) complex that phosphorylates IκB at two specific serine residues, followed by its ubiquitination and proteasomal degradation, thereby releasing NF-κB proteins and allowing their nuclear translocation (3). Recently, two studies provided further insights into the molecular mechanisms of IKK activation upon TNF stimulation (4, 5). Activation of the IKK complex and subsequent NF-κB activation requires Lys63 polyubiquitination of RIP1, a kinase that is recruited to the receptor upon TNF stimulation. IKK-γ (NF-κB essential modulator), the regulatory subunit of the IKK complex, specifically recognizes these Lys63-linked polyubiquitins attached to RIP1 and thereby activates IKK and NF-κB (4, 5).
A20 is a ubiquitin-modifying enzyme that inhibits NF-κB activation in succession of TNF receptor– and Toll-like receptor–induced signals (6–8). This enzyme removes Lys63-linked ubiquitin chains from RIP1 and adds Lys48 polyubiquitins to RIP1, thereby targeting this factor for proteasomal degradation, thus explaining the molecular mechanism of NF-κB inhibition by A20 (6). A20 also likely inhibits NF-κB activity by additional means, including interaction with TRAF1 and TRAF2 (9).
The TNFAIP3 gene, encoding A20, is located in chromosome band 6q23, a region that is frequently deleted in B cell lymphomas (10, 11). Recently, studies applying high-resolution, genome-wide cytogenetic techniques such as array-based comparative genomic hybridization (aCGH) or single nucleotide polymorphism (SNP) chip analysis on non-Hodgkin lymphoma and cHL reported a region of minimal common loss at 6q23, including TNFAIP3 (12–15). However, mutations in this gene have not been reported in these studies (12–15).
To test whether mutational inactivation of A20 contributes to the pathogenesis of cHL and primary mediastinal B cell lymphoma (PMBL), another lymphoma with constitutive NF-κB activity (16), we sequenced TNFAIP3 in these lymphomas, and performed functional studies with cHL cell lines.
RESULTS AND DISCUSSION
Lack of A20 in consequence of mutations in TNFAIP3 in cHL cell lines
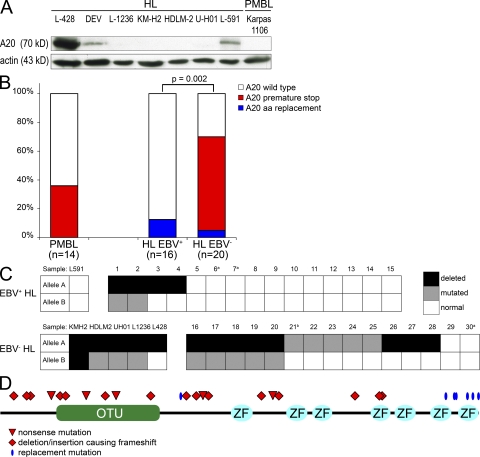
Because the underlying mechanisms of constitutive NF-κB activity in HL and PMBL are only partly understood (17), we analyzed the A20 protein by Western blotting in HL and PMBL cell lines (Fig. 1 A). Although TNFAIP3, a direct NF-κB target gene, was reported to be strongly expressed on a transcriptional level in virtually all HL cases (18), no A20 protein was detected in four out of seven HL cell lines analyzed, as well as the PMBL cell line Karpas-1106P. DNA sequence analysis of the entire coding region of TNFAIP3 revealed a nonsense mutation, a duplication, and deletions in the A20 protein–negative HL cell lines (Table I). Only the mutated alleles were identified, explaining the absence of detectable protein in the respective cell lines. In accordance with these findings, an SNP chip analysis in L-1236, HDLM-2, and U-HO1 showed loss of heterozygosity (LOH) in 6q23, including the TNFAIP3 locus (Fig. S1). A homozygous deletion in the coding sequence of TNFAIP3 in cell line KM-H2 was previously reported (19). Because the DEV cell line originates from nodular lymphocyte–predominant HL, it was excluded from further analysis.
Figure 1.
Inactivation of A20 in cHL and PMBL. (A) Mutations in TNFAIP3 correlate with the absence of detectable A20 protein (∼70 kD) in lymphoma cell lines. Immunoblotting using anti-A20 antibody was performed with each 100 µg of whole-cell extracts of PMBL and HL cell lines. β-Actin was used as loading control. No truncated proteins were detected. (B) Frequency and pattern of TNFAIP3 mutations in PMBL and cHL arranged by EBV status (A20 wild-type: cases carrying exclusively wild-type TNFAIP3 sequence; A20 premature stop: cases with destructive mutations [nonsense mutations and deletions or insertions causing premature stop codons]; A20 missense: cases with missense mutations). Numbers in brackets indicate the numbers of cases analyzed. p-values were calculated using Fisher’s exact test. Case 19 harbors both missense and premature stop, and is depicted among the group with premature stop. (C) Graphic representation of allelic distribution of TNFAIP3 mutations of cHL grouped according to EBV status. The columns represent the cases, and the rows represent the two alleles. a, the loss of one allele cannot be excluded as cases were not evaluable by FISH and lacked heterozygous sequence polymorphisms; b, the case harbors two mutations, but their allelic distribution could not be determined. (D) Schematic representation of location of TNFAIP3 mutations translated to A20 protein in cHL and PMBL, as described in Tables I and II. OTU, ovarian tumor domain; ZF, A20 zinc finger domains.
Table I.
Sequence and gene copy number analysis of TNFAIP3 from cHL cell lines and primary HRS cells
| Sample | EBV | Subtype | Nucleotide changea | Amino acid changeb | LOH/Delc | Both alleles mutated |
| Cell lines | ||||||
| L-591 | + | NS | − | − | − | − |
| L-428 | − | NS | − | − | +d | − |
| HDLM-2 | − | NS (T cell origin) | Dupl. 586–614 | Frameshift aa 174 | +d | + |
| KM-H2 | − | NS | Δ intron 2–exon 6 | Frameshift aa 99 | +e | + |
| L-1236 | − | MC | G491A | W142STOP | +d | + |
| U-H01 | − | NS | Δ 193–200 | Frameshift aa 43 | +d | + |
| HL biopsies | ||||||
| 1 | + | NS | C2275A | Q737K | + | + |
| 2 | + | UN | G2317A | E751K | + | + |
| 3 | + | UN | − | − | + | − |
| 4 | + | LR | − | − | + | − |
| 5 | + | MC | G2317Rf | M788I | − | − |
| 6 | + | MC | − | − | n.e. | − |
| 7 | + | NS | − | − | n.e. | − |
| 8 | + | NS | − | − | − | − |
| 9 | + | MC | − | − | − | − |
| 10 | + | MC | − | − | − | − |
| 11 | + | NS | − | − | − | − |
| 12 | + | MC | − | − | − | − |
| 13 | + | NS | − | − | − | − |
| 14 | + | MC | − | − | − | − |
| 15 | + | UN | − | − | − | − |
| 16 | − | NS | G320A | W85STOP | + | + |
| 17 | − | NS | Δ 1,824–1,875 | Frameshift aa 586 | + | + |
| 18 | − | NS | G1420T | E452STOP | + | + |
| 19 | − | UN | Δ GG 132–133/T971C | Frameshift aa 22/L302P | + | + |
| 20 | − | NS | G2323A | A753T | +g | + |
| 21 | − | NS | Δ TGTTCAG 215–221/Δ C 1436 | Frameshifts aa 50, 457 | − | n.e.h |
| 22 | − | NS | Δ GTTCTCG 811–817 | Frameshift aa 249 | − | − |
| 23 | − | NS | Ins T 992–993 | Frameshift aa 309 | − | − |
| 24 | − | NS | Δ TG 1,945–1,946 | Frameshift aa 627 | − | − |
| 25 | − | NS | Δ GC 1,361–1,362 | Frameshift aa 432 | − | − |
| 26 | − | NS | − | − | + | − |
| 27 | − | NS | − | − | + | − |
| 28 | − | NS | − | − | + | − |
| 29 | − | MC | − | − | − | − |
| 30 | − | NS | − | − | n.e. | − |
CD30+ HRS cells were analyzed in groups of 10–20 cells. In cases of mutation, TNFAIP3 was additionally sequenced from single HRS and nonneoplastic cells. cHLs were further tested for allelic losses using interphase cytogenetics. EBV status was determined by LMP1 immunohistochemical staining and/or EBV-encoded RNA in situ hybridization. Del, deletion; n.e., not evaluable; Δ, deletion; Dupl., duplication; Ins, insertion; LR, lymphocyte rich; MC, mixed cellularity; NS, nodular sclerosis; UN, unclassifiable.
Corresponding to GenBank/EMBL/DDBJ accession no. NM_006290.2.
Corresponding to PDB accession no. NP_006281.
As indicated by interphase cytogenetics, SNP chip analysis, and/or sequence analysis. Because of the frequent hyperploidy of the HRS cells, it cannot be excluded that cases with diploid signal patterns in the FISH analysis also carry (subclonal) losses of the TNFAIP3 locus (from a hyperploid clone; Table S1, cases 21, 22, and 24). A high intratumoral variability of the signal patterns is typical for cHL. In four mutated primary cHL (cases 1, 2, 17, and 18), allelic losses were identified by the detection of only the mutated alleles. The latter three cases were not evaluable by interphase cytogenetics, but case 1 may harbor an uniparental disomy. In case 23, the presence of two alleles was evident from the concurrent detection of mutated and unmutated sequences. In cases 5, 15, and 29, presence of two alleles was evident by heterozygous constellation of polymorphisms (Fig. S2).
LOH shown by GeneChip SNP chip analysis.
Homozygous deletion (reference 19).
Sequence variation was also found in nontumor cells and, hence, does not represent a somatic mutation but, presumably, a polymorphism.
Signal constellation of FISH analysis indicated biallelic loss of TNFAIP3 in a fraction of HRS cells (Table S1).
Allelic distribution could not be determined because of the long distance of mutations on genomic DNA.
Inactivating mutation in TNFAIP3 in primary HRS cells of EBV− cHL
Extending the study to primary biopsies of 30 cHLs, we individually laser-microdissected CD30+ HRS cells, pooled 10–20 cells, and sequenced DNA of the entire coding sequence of TNFAIP3 after two rounds of seminested amplification. In cases harboring mutations, single HRS and nonneoplastic cells were additionally analyzed to confirm clonality and somatic origin of the mutations identified. Chromosomal deletions of TNFAIP3 were investigated by interphase cytogenetics (i.e., fluorescence in situ hybridization [FISH] or the combined fluorescence immunophenotyping and interphase cytogenetics [FICTION] technique; Table I, Fig. 2, and Table S1). We ascertained somatic, clonal mutations in 12 out of 30 cHL cases analyzed. Additionally, 1 out of the 30 cases showed a sequence variation that was also present in the respective nontumor cells and, hence, presumably represents a polymorphism (Table I and Fig. S2). Including the cell lines, 11 out of the 16 mutated cases showed deletion of the other allele of TNFAIP3. Mutation frequency and mutation patterns differed significantly (P = 0.002) according to the EBV status of the cHL: among 16 EBV+ cHLs, only 2 cases (12.5%) each harbored a single somatic missense mutation in the last exon of TNFAIP3, whereas 14 out of 20 EBV− cHLs showed clonal mutations (70%; Fig. 1 B). In 13 out of these 14 mutated EBV− cHLs, we detected nonsense mutations and insertions or deletions causing frameshifts. In 9 out of the 14 cases carrying mutations generating premature stop codons, both gene copies of TNFAIP3 were genetically inactivated as the second alleles of the gene were deleted (Table I, Fig. 1 C, and Fig. S2). We analyzed 21 additional cHLs by interphase cytogenetics for losses in 6q23 and detected signal constellations indicating deletions of TNFAIP3 in 9 cases, confirming the high incidence of deletions of the chromosomal locus of TNFAIP3 in cHL (Table S1). This is also supported by a recent aCGH study of HRS cell–rich cHL cases revealing monoallelic losses of the TNFAIP3 locus in 6 out of 10 cases (15).
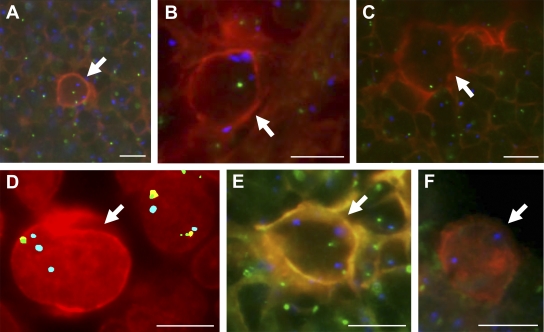
Figure 2.
Chromosomal deletion of TNFAIP3 in cHL detected by interphase cytogenetics. FICTION analyses of representative cHL cases combining CD30 expression (red) and FISH probes for TNFAIP3 and chromosome 6 centromere (blue [b]). In the double-color assays in A–C and E and F, the TNFAIP3 probe is labeled in green (g); in the triple-color assay applied in D, the TNFAIP3 probe gives a g/orange colocalized (co) signal. Two different strategies are used to display double- and triple-color FISH assays in combination with CD30 immunofluorescence; i.e., double-color assays (A–C and E and F) are shown using a triple-color display, whereas a false multicolor display as obtained by Isis software is applied for the triple-color assay (D) to simultaneously show four colors (i.e., CD30 [r], TNFAIP3 [g] and orange, and chromosome 6 centromere [b]). In contrast to the triple-color assay, the quadruple-color assay is based on the overlay of two displays generated by Isis software. To this end, the different channels needed to be individually enhanced so that the final integrated image shows multicolor signals. Fluorescent signals are shown in a false-color display. Arrows point to CD30+ HRS cells. For each case, between 6 and 26 evaluable HRS cells (mean = 12) were considered. (A) HRS cell (case 41) with normal diploid signal pattern (2g + 2b). (B) HRS cell (case 36) exhibiting one copy of TNFAIP3 (1g) and three copies of chromosome 6 centromere (3b), indicating the presence of a chromosomal deletion of TNFAIP3. (C) HRS cell (case 16) exhibiting one copy of TNFAIP3 (1g) and two copies of chromosome 6 centromere (2b), indicating the presence of a chromosomal deletion of TNFAIP3. (D) HRS cell (case 19) exhibiting one copy of TNFAIP3 (1co) and three copies of chromosome 6 centromere (3b), indicating the presence of a chromosomal deletion of TNFAIP3. (E) HRS cell (case 35) exhibiting two copies of TNFAIP3 (2g) and three copies of chromosome 6 centromere (3b), indicating the presence of a chromosomal deletion of TNFAIP3. (F) HRS cell (case 33) lacking signals for TNFAIP3 (0g) but showing two copies of chromosome 6 centromere (2b), indicating the presence of a chromosomal (homozygous?) deletion of TNFAIP3. Neighboring bystander CD30− cells mostly show normal signal patterns (2g or 2co + 2b), although truncation artifacts do occur (some signals are out of the focus plane). Bars, 10 µm.
Inactivating mutation in TNFAIP3 in PMBL
Global gene expression profiling revealed remarkable similarities between the transcriptional profiles of PMBL and cHL (16). Furthermore, survival and proliferation of tumor cells of PMBL also depend on constitutive NF-κB activity (16, 20). Hence, we expanded the sequence analysis on primary biopsies of PMBL and the PMBL cell line Karpas-1106P, shown to be A20 protein negative (Fig. 1 A). TNFAIP3 was sequenced after one round of amplification from whole-tissue DNA. We identified mutations in 5 out of 14 cases (36%), with each mutated case carrying at least one mutation leading to A20 inactivation (Fig. 1 B). In three primary cases in which nontumor cells could be isolated by microdissection, the somatic origin of the mutations was confirmed. In cases with more than one mutation (PMBLs 2 and 3), TNFAIP3 amplicons were further cloned to identify allelic distribution of mutations, revealing bialleic distribution of the mutations in at least one of the two cases (Table II and Fig. S2). Remarkably, deletions in 6q23.3–6q24.1 have recently been identified by aCGH in ∼30% of PMBLs (21, 22).
Table II.
Sequence analysis of TNFAIP3 from primary PMBLs and cell line Karpas-1106P
| Sample | Nucleotide changea | Amino acid changeb | Both alleles mutated |
| Cell line | |||
| Karpas-1106P | Δ TCATC 1,037–1,041c | Frameshift aa 324 | + |
| Biopsies | |||
| 1 | Δ AG 1,294–1,295c | Frameshift aa 410 | + |
| 2 | C635M/Δ CAGAGAAAACAAA 1,959–1,971 + 8 bp (splice donor site)d | S190STOP/frameshift aa 631 | + |
| 3 | Ins A389/T2382K/A2409W/T2437Y/A2439R | Frameshift aa 108/N772K/E781D/STOP>Q/STOP>Q | n.e.e |
| 4 | T1070Y | L335STOP | − |
| 5 | − | − | − |
| 6 | − | − | − |
| 7 | − | − | − |
| 8 | − | − | − |
| 9 | − | − | − |
| 10 | − | − | − |
| 11 | − | − | − |
| 12 | − | − | − |
| 13 | − | − | − |
Analysis was performed on whole-tissue section DNA. Δ, deletion; Ins, insertion.
Corresponding to GenBank/EMBL/DDBJ accession no. NM_006290.2.
Corresponding to PDB accession no. NP_006281.
LOH as indicated by sequence analysis; in Karpas-1106P and one mutated primary PMBL (case 1), allelic losses were identified by the detection of only the mutated alleles (Fig. S2 C).
Mutations on separate alleles as indicated by cloning of PCR product spanning both sites of mutations.
Four missense mutations (at positions 2,382–2,438) located on one allele, as indicated by cloning. Because of the long distance between the insertion (position 389) and missense mutations (positions 2,382–2,438) on genomic DNA, allelic distribution of insertion and missense mutations could not be determined.
Collectively, we identified 16 out of 36 cHLs and 5 out of 14 PMBLs harboring somatic mutations in TNFAIP3. The majority of mutations represent nonsense mutations and deletions or insertions leading to frameshifts (Fig. 1 D). No mutation hotspot was identified (Tables I and II; and Fig. 1 D). However, most missense mutations are located in the last exon of TNFAIP3 coding for most C-terminal A20 zinc fingers. The question of the oncogenic potential of these amino acid replacements remains open.
Loss of A20 contributes to constitutive NF-κB activity in cHL cell lines
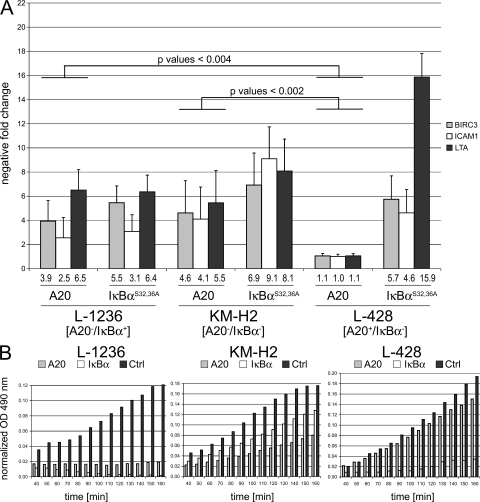
Based on this genetic evidence, TNFAIP3 appears to be a tumor suppressor gene in cHL and PMBL. To functionally test this hypothesis, we expressed A20 by lentiviral gene transfer in cHL cell lines harboring inactivating TNFAIP3 mutations (L-1236 and KM-H2) and as a control in a cHL cell line expressing wild-type A20 (L-428; Fig. 1 A). To this end, we generated lentiviral expression constructs allowing coexpression of GFP and either A20, IκBαS32,36A superrepressor or luciferase by inserting the Thosea asigna insect virus T2A sequence between the coding sequences of the genes. Ribosome skipping occurs during translation of the T2A sequence, leading to generation of two separate proteins from one transcript (23). In the IκBαS32,36A superrepressor, which served in our experiments as a control for NF-κB repression, two serine residues of positions 32 and 36 are replaced by alanines, thus preventing the phosphorylation required for proteasomal degradation of this NF-κB inhibitor (24). To assess A20 expression levels mediated by lentiviruses and efficient ribosome skipping, we analyzed A20 and GFP protein expression of isolated GFP-expressing L-1236 cells by Western blotting. This revealed the efficiency of ribosome skipping mediated by the T2A sequence by the absence of detectable fusion protein (Fig. S3). The analysis also indicated that lentivirally transduced cells express an approximately fourfold level of A20 compared with L-428 cells, expressing endogenous wild-type A20. To investigate the consequences of reconstitution of A20 in cHL cell lines, we FACS sorted 2,500 cells transduced with the respective lentiviruses according to GFP expression, and determined by quantitative PCR (qPCR) mRNA levels of selected NF-κB target genes (BIRC3, ICAM1, and LTA), which are negatively regulated upon inhibition of NF-κB in cHL (2). qPCR values of NF-κB target genes were normalized to GAPDH, and the resulting data from luciferase-expressing cells, which served as negative control, were subtracted from A20- and IκBαS32,36A-transduced cells (ΔΔCt method). Upon reconstitution of A20 in cell lines carrying inactivated TNFAIP3 (KM-H2 and L-1236), we observed a significant decrease of mRNA levels of the selected NF-κB target genes, indicating a globally attenuated transcriptional activity of NF-κB (Fig. 3 A). Mean transcriptional reduction of these genes by A20 ranged between 4.1- and 5.5-fold in KM-H2 and 2.5- and 6.5-fold in L-1236 as compared with cells transduced with luciferase-expressing viruses, respectively. A similar decrease of transcription of NF-κB target genes was seen in KM-H2 and L-1236 cells expressing IκBαS32,36A (mean reduction values = 6.9–9.1 for KM-H2 and 3.1–6.4 for L-1236). Remarkably, besides inactive TNFAIP3, KM-H2 cells also harbor inactivating mutations in NFKBIA (IκBα) (25). In contrast, L-428 cells, strongly expressing endogenous wild-type A20, did not show down-regulation of NF-κB transcriptional activity by A20 gene transfer (mean = 1–1.1), indicating that this increased A20 expression does not impair NF-κB activity in these cells. Notably, L-428 cells carry inactivating mutations in the NF-κB inhibitors NFKBIA and NFKBIE (IκBϵ) (25, 26). In accordance with the known effect of A20 on RIP protein stability, we observed strongly reduced levels of RIP protein in A20-reconstituted L-1236 cells as compared with nonreconstituted L-1236 cells (Fig. S4).
Figure 3.
A20 reconstitution in cHL cell lines harboring inactivating TNFAIP3 mutations results in reduction of transcriptional NF-κB activity and reduction of cellular metabolism. cHL cell lines harboring inactive TNFAIP3 (L-1236 and KM-H2) or wild-type TNFAIP3 (L-428) were lentivirally transduced with expression constructs encoding the reporter GFP and either A20, IκBαS32,36A superrepressor or luciferase as control (Ctrl). (A) GFP+ cells were analyzed by qPCR for expression of representative NF-κB target genes (BIRC3, ICAM1, and LTA). Reduction of NF-κB target gene expression upon TNFAIP3 or IκBαS32,36A gene transfer was determined by subtraction of ΔCts of luciferase-expressing cells, respectively (ΔΔCt method). Bars represent mean values of down-regulation resulting from at least three independent infections that were each analyzed in several independent replicates. p-values were calculated per gene using the Wilcoxon rank-sum test to determine the level of significance between negative regulation of NF-κB target genes in L-1236 and KM-H2 cells in comparison to L-428 cells in response to A20 expression. (B) GFP+ cells were FACS sorted, cultured for 48 h, and analyzed by MTS assay for metabolic activity. Mean OD values, reflecting conversion of MTS into a formazan product, are normalized to the empty medium control. The quantity of formazan product as measured by the amount of 490-nm absorbance is directly proportional to the number of living cells in culture. The experiments were performed in duplicates (Fig. S5). Each value is based on four measurements.
A20 reconstitution confers cytotoxicity to A20-deficient cHL cell lines
To test the consequence of A20 reconstitution on proliferation and/or survival in A20-negative cell lines, we performed an MTS assay with sorted GFP+ cells expressing either A20, IκBαS32,36A, or luciferase. This revealed a strong cytotoxic effect of A20 reexpression in L-1236 cells and a more moderate one in KM-H2 cells compared with the luciferase-expressing negative control cells (Fig. 3 B). In contrast, overexpression of A20 in L-428 cells had little or no effect on these cells (also indicating that the experimental overexpression of A20 per se is not cytotoxic), whereas IκBαS32,36A expression showed strong cytotoxicity (Fig. 3 B and Fig. S5).
Concluding remarks
We identify TNFAIP3 as a novel tumor suppressor gene in cHL and PMBL by showing frequent somatic and clonal biallelic inactivation of the gene, and presenting evidence that loss of A20 function contributes to the constitutive activity of the transcription factor NF-κB and the survival and/or proliferation of the cells. The detection of destructive somatic mutations within the gene and frequent complete inactivation, together with the functional data, are indeed critical findings to verify this hypothesis. The chromosomal region containing TNFAIP3 is recurrently affected by monoallelic deletions in different lymphomas and the fact that, in follicular lymphoma for example, which is not characterized by strong NF-κB activity, monoallelic deletions including TNFAIP3 were found, but no somatic mutations within the gene in 50 cases analyzed (14), indicated that another gene is the target of the deletions in follicular lymphoma.
The striking clustering of unequivocally destructive mutations with EBV− cases of cHL defines the first example of a genetic lesion that distinguishes EBV+ from EBV− cHL cases. This clustering suggests complementing functions of A20 inactivation and EBV transformation, and thereby supports important pathogenetic roles of both events. On the other hand, there is also indication from the cHL cell lines that multiple transforming events in the NF-κB pathway can cooperate, such as concurrent TNFAIP3 and NFKBIA mutations in KM-H2. This represents one of the few examples in which multiple genetic lesions occur in the same pathway in one tumor clone. Whether concurrent mutations in TNFAIP3 and NFKBIA, which is mutated in ∼10–20% of cHL cases (25, 27, 28), also occur in primary cases of cHL remains to be identified. In PMBL, NFKBIA mutations have not been found (29). The important role of A20 as a key regulator of NF-κB activity in multiple immune functions was recently impressively demonstrated (8, 30, 31), and its role as a tumor suppressor gene may well go beyond cHL and PMBL, as shown by the recent detection of TNFAIP3 mutations in marginal zone B cell lymphomas (32).
MATERIALS AND METHODS
Patient samples and cell lines.
Lymph node samples from 54 patients with cHL were collected from the Department of Pathology, Hematopathology Section and Lymph Node Registry at the University of Kiel and the Senkenberg Institute of Pathology at the University of Frankfurt. According to morphological and immunohistochemical criteria, 26 cases were classified as nodular sclerosis cHL, 20 cases were classified as mixed cellularity, 1 case was classified as lymphocyte depleted, 1 case was classified as lymphocyte rich, and 6 cases were unclassified. In cases for sequence analysis, EBV status was determined by LMP1 immunohistochemical staining and/or EBV-encoded RNA in situ hybridization. PMBLs were obtained from the Institute of Pathology at the University of Heidelberg and the Department of Pathology at the University of Kiel. PMBL cell line Karpas-1106P and cHL cell lines L-428, L-591, L-1236, HDLM-2, KM-H2, and the recently established line U-HO1 (33) were analyzed. Approval by the Institutional Review Boards in Kiel and Frankfurt was obtained for these studies.
Laser microdissection and pressure catapulting of HRS and nontumor cells.
5-µm frozen lymph node sections of primary cHL biopsies were mounted on membrane-covered slides (PALM) and stained with anti-CD30 antibody (BerH2; Dako). Single CD30+ HRS cells were microdissected using laser microdissection and pressure catapulting (PALM) into PCR buffer, and pooled into groups of 10–20 cells. Nonneoplastic cells were microdissected in groups of 20–50 cells.
PCR amplification and sequencing of TNFAIP3.
All coding exons of TNFAIP3 were amplified using primer sequences displayed in Table S2. DNA of cell lines and of whole-tissue sections of PMBLs was amplified in one round of PCR using first-round primers and 35 cycles. Nested amplification of the individual exons was performed using identical conditions with internal primers and 40 cycles in the second round of PCR. Sequencing was performed on an ABI 3130 sequencing apparatus (Applied Biosystems).
Immunoblotting.
Western blot analysis was conducted using antibody clone 59A426 (NatuTec) for A20, antibody clone I-19 (Santa Cruz Biotechnology, Inc.) for actin, antibody AB513 for TurboGFP (Evrogen), and antibody clone 38/RIP for RIP (BD), applying standard techniques.
Interphase cytogenetics.
FISH and FICTION analyses were performed as previously described (34) using differently labeled bacterial artificial chromosome clones RP11-783B20 (spanning TNFAIP3 and extending in the centromeric direction, labeled with SpectrumGreen) and (in some cases) RP11-703G8 (spanning TNFAIP3 and extending in the telomeric direction, labeled with SpectrumOrange), as well as a probe for centromere 6 (CEP6, labeled with SpectrumAqua; all from Abbott/Vysis) as an internal control. For FICTION, immunofluorescence with monoclonal anti-CD30 antibody (Dako) detected with an Alexa Fluor 594–conjugated secondary antibody (Invitrogen) was applied in combination with the TNFAIP3-CEP6 probe. Slides were analyzed using a fluorescence microscope (Axio Imager.A1; Carl Zeiss, Inc.) equipped with the appropriate filter sets (AHF) and were documented using an Isis imaging system (MetaSystems). Nuclei from HRS cells were identified by virtue of their larger size, frequent hyperploid genomic status, and CD30 expression (the latter only by FICTION). Cases showing lower copy numbers for TNFAIP3 than for CEP6 were classified as deleted.
SNP microarray analysis.
The genome-wide human SNP Array 6.0 (Affymetrix) was used according to the protocol provided by the manufacturer. Microarrays were washed and stained with the Fluidics Station 450 (Affymetrix) and scanned with the GeneChip Scanner 3000 (Affymetrix) using the Genotyping Console software (version 3.0; Affymetrix). The Birdseed v2 algorithm was used to genotype tumor samples. Copy number analysis, LOH analysis, and segmentation were calculated using Genotyping Console software. Segments with aberrant copy number were considered as copy number aberration only if they consisted of at least 20 consecutive SNPs and comprised a minimal size of 100 kb. Custom 100K GeneChip Mapping SNP Array analyses were performed at Affymetrix. The complete GeneChip datasets have been deposited in the Gene Expression Omnibus under accession no. GSE15264.
Construction of lentiviral vectors.
Lentiviral vectors were constructed by PCR and cloning based on pGIPZshRNAmir (Open Biosystems). A T2A sequence for ribosome skipping was cloned in frame between coding sequences of TNFAIP3, IκBαS32,36A (provided by J. Feuillard, Centre National de la Recherche Scientifique, CHU Dupuytren, Limoges, France) or luciferase (provided by B. Jungnickel, Helmholtz Center Munich, Munich, Germany) and GFP. Expression was driven by a CMV promoter. Virus production was performed by cotransfection of 293T cells using GeneJuice (EMD) with the plasmids pGIPZ-NheI (Fig. S3), psPAX2 (plasmid 12260; Addgene), and pMD2.G (plasmid 12259; Addgene; the latter two were constructed by D. Trono, Ecole Polytechnique Fédérale de Lausanne, Lausanne, Switzerland). 3 d after transfection, supernatants were collected and concentrated by ultracentrifugation for 1.5 h at 100,000 g (Beckman Coulter). Titration of lentiviruses was performed using 293T cells.
Transduction of cHL cell lines, FACS sorting, and qPCR of NF-κB target genes.
cHL cell line cells were infected using a multiplicity of infection of 10. GFP+ cells were FACS sorted 80 h after infection, excluding propidium iodide–positive cells. For qPCR analysis, 2,500 GFP-expressing cells were sorted into RLT lysis buffer (QIAGEN). RNA was extracted using the RNeasy Micro Kit (QIAGEN) according to the manufacturer’s instructions. qPCR analysis was conducted using gene expression assays (TaqMan; Applied Biosystems) and analyzed on an ABI Prism 7900HT Fast Real-Time PCR System (Applied Biosystems). For each cell line, transfections and sorting of cells were done at least in triplicates, of which mostly three or four independent qPCR measurements were performed.
MTS assay.
cHL cell line cells were FACS sorted 80 h after infection in aliquots of 10,000 GFP-expressing cells in PBS/0.5% BSA. After centrifugation, the aliquots were resuspended in 100 µl of conditioned culture medium each in a 96-well plate. After 48 h, CellTiter 96 AQueous One Solution reagent (Promega) was added to each well and analyzed at 490 nm. The conversion of MTS into a formazan product was accomplished by dehydrogenase enzymes found in metabolically active cells. The quantity of formazan product as measured by the amount of 490-nm absorbance is directly proportional to the number of living cells in culture.
Online supplemental material.
Genome-wide human SNP Array 6.0 analyses of cHL cell lines revealing deletions and LOH at the TNFAIP3 locus are shown in Fig. S1. Fig. S2 exemplifies TNFAIP3 sequences of primary HRS cells, PMBLs, and corresponding nontumor cells. Expression and function of the A20-T2A-GFP lentiviral construct is shown in Fig. S3. Fig. S4 shows reduction of RIP protein levels upon A20 reexpression in L-1236 cells. Additional data on A20 reconstitution in cHL cell lines harboring inactivating TNFAIP3 mutations are depicted in Fig. S5. Table S1 provides detailed results of the interphase cytogenetic analyses. Table S2 shows primer sequences used for seminested two-round genomic DNA amplification of TNAIP3. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20090528/DC1.
Acknowledgments
We thank E. Tiacci, E. Maggio, B. Jungnickel, I. Pfeil, M. Grez, and I. Vogler for helpful discussions; J. Feuillard, B. Jungnickel, and D. Trono for reagent supply; and G. Lorenz, K. Lennartz, K. Waldhelm, R. Zühlke-Jenisch, and R. Lieberz for excellent technical assistance. We thank F. Chen for inspiring this work with his excellent review.
This work was supported by grants from the Wilhelm Sander-Stiftung (2005.168.2), the Deutsche Krebshilfe, the Mildred Scheel-Stiftung (107736 and 107748), the Deutsche Forschungsgemeinschaft, and the Kinder-Krebs-Initiative Buchholz/Holm Seppensen.
The authors have no conflicting financial interests.
References
- Bargou R.C., Emmerich F., Krappmann D., Bommert K., Mapara M.Y., Arnold W., Royer H.D., Grinstein E., Greiner A., Scheidereit C., Dorken B. 1997. Constitutive nuclear factor-kappaB-RelA activation is required for proliferation and survival of Hodgkin’s disease tumor cells.J. Clin. Invest. 100:2961–2969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz M., Lemke P., Anagnostopoulos I., Hacker C., Krappmann D., Mathas S., Dorken B., Zenke M., Stein H., Scheidereit C. 2002. Nuclear factor κB–dependent gene expression profiling of Hodgkin’s disease tumor cells, pathogenetic significance, and link to constitutive signal transducer and activator of transcription 5a activity.J. Exp. Med. 196:605–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden M.S., Ghosh S. 2008. Shared principles in NF-kappaB signaling.Cell. 132:344–362 [DOI] [PubMed] [Google Scholar]
- Wu C.J., Conze D.B., Li T., Srinivasula S.M., Ashwell J.D. 2006. Sensing of Lys 63-linked polyubiquitination by NEMO is a key event in NF-kappaB activation.Nat. Cell Biol. 8:398–406 [DOI] [PubMed] [Google Scholar]
- Ea C.K., Deng L., Xia Z.P., Pineda G., Chen Z.J. 2006. Activation of IKK by TNFalpha requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO.Mol. Cell. 22:245–257 [DOI] [PubMed] [Google Scholar]
- Wertz I.E., O’Rourke K.M., Zhou H., Eby M., Aravind L., Seshagiri S., Wu P., Wiesmann C., Baker R., Boone D.L., et al. 2004. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling.Nature. 430:694–699 [DOI] [PubMed] [Google Scholar]
- Lee E.G., Boone D.L., Chai S., Libby S.L., Chien M., Lodolce J.P., Ma A. 2000. Failure to regulate TNF-induced NF-kappaB and cell death responses in A20-deficient mice.Science. 289:2350–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone D.L., Turer E.E., Lee E.G., Ahmad R.C., Wheeler M.T., Tsui C., Hurley P., Chien M., Chai S., Hitotsumatsu O., et al. 2004. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses.Nat. Immunol. 5:1052–1060 [DOI] [PubMed] [Google Scholar]
- Song H.Y., Rothe M., Goeddel D.V. 1996. The tumor necrosis factor-inducible zinc finger protein A20 interacts with TRAF1/TRAF2 and inhibits NF-kappaB activation.Proc. Natl. Acad. Sci. USA. 93:6721–6725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Weber-Matthiesen K., Siebert R., Matthiesen P., Schlegelberger B. 1997. Frequent deletions of 6q23-24 in B-cell non-Hodgkin’s lymphomas detected by fluorescence in situ hybridization.Genes Chromosomes Cancer. 18:310–313 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Matthiesen P., Harder S., Siebert R., Castoldi G., Calasanz M.J., Wong K.F., Rosenwald A., Ott G., Atkin N.B., Schlegelberger B. 2000. A 3-cM commonly deleted region in 6q21 in leukemias and lymphomas delineated by fluorescence in situ hybridization.Genes Chromosomes Cancer. 27:52–58 [DOI] [PubMed] [Google Scholar]
- Chanudet E., Ye H., Ferry J., Bacon C., Adam P., Muller-Hermelink H., Radford J., Pileri S., Ichimura K., Collins V., et al. 2009. A20 deletion is associated with copy number gain at the TNFA/B/C locus and occurs preferentially in translocation-negative MALT lymphoma of the ocular adnexa and salivary glands.J. Pathol. 217:420–430 [DOI] [PubMed] [Google Scholar]
- Honma K., Tsuzuki S., Nakagawa M., Karnan S., Aizawa Y., Kim W.S., Kim Y.D., Ko Y.H., Seto M. 2008. TNFAIP3 is the target gene of chromosome band 6q23.3-q24.1 loss in ocular adnexal marginal zone B cell lymphoma.Genes Chromosomes Cancer. 47:1–7 [DOI] [PubMed] [Google Scholar]
- Ross C.W., Ouillette P.D., Saddler C.M., Shedden K.A., Malek S.N. 2007. Comprehensive analysis of copy number and allele status identifies multiple chromosome defects underlying follicular lymphoma pathogenesis.Clin. Cancer Res. 13:4777–4785 [DOI] [PubMed] [Google Scholar]
- Hartmann S., Martin-Subero I., Gesk S., Hüsken S., Giefing M., Nagel I., Riemke J., Chott A., Klapper W., Parrens M., et al. 2008. Detection of genomic imbalances in microdissected Hodgkin- and Reed-Sternberg cells of classical Hodgkin lymphoma by array based comparative genomic hybridization.Haematologica. 93:1318–1326 [DOI] [PubMed] [Google Scholar]
- Savage K.J., Monti S., Kutok J.L., Cattoretti G., Neuberg D., De Leval L., Kurtin P., Dal Cin P., Ladd C., Feuerhake F., et al. 2003. The molecular signature of mediastinal large B-cell lymphoma differs from that of other diffuse large B-cell lymphomas and shares features with classical Hodgkin lymphoma.Blood. 102:3871–3879 [DOI] [PubMed] [Google Scholar]
- Courtois G., Gilmore T.D. 2006. Mutations in the NF-kappaB signaling pathway: implications for human disease.Oncogene. 25:6831–6843 [DOI] [PubMed] [Google Scholar]
- Durkop H., Hirsch B., Hahn C., Foss H.D., Stein H. 2003. Differential expression and function of A20 and TRAF1 in Hodgkin lymphoma and anaplastic large cell lymphoma and their induction by CD30 stimulation.J. Pathol. 200:229–239 [DOI] [PubMed] [Google Scholar]
- Giefing M., Arnemann J., Martin-Subero J.I., Nieländer I., Bug S., Hartmann S., Arnold N., Tiacci E., Frank M., Hansmann M.-L., et al. 2008. Identification of candidate tumor suppressor gene loci for Hodgkin and Reed-Sternberg cells by characterization of homozygous deletions in classical Hodgkin lymphoma cell lines.Br. J. Haematol. 142:916–924 [DOI] [PubMed] [Google Scholar]
- Feuerhake F., Kutok J.L., Monti S., Chen W., LaCasce A.S., Cattoretti G., Kurtin P., Pinkus G.S., de Leval L., Harris N.L., et al. 2005. NFkappaB activity, function, and target-gene signatures in primary mediastinal large B-cell lymphoma and diffuse large B-cell lymphoma subtypes.Blood. 106:1392–1399 [DOI] [PubMed] [Google Scholar]
- Wessendorf S., Barth T.F., Viardot A., Mueller A., Kestler H.A., Kohlhammer H., Lichter P., Bentz M., Dohner H., Moller P., Schwaenen C. 2007. Further delineation of chromosomal consensus regions in primary mediastinal B-cell lymphomas: an analysis of 37 tumor samples using high-resolution genomic profiling (array-CGH).Leukemia. 21:2463–2469 [DOI] [PubMed] [Google Scholar]
- Kimm L.R., deLeeuw R.J., Savage K.J., Rosenwald A., Campo E., Delabie J., Ott G., Muller-Hermelink H.K., Jaffe E.S., Rimsza L.M., et al. 2007. Frequent occurrence of deletions in primary mediastinal B-cell lymphoma.Genes Chromosomes Cancer. 46:1090–1097 [DOI] [PubMed] [Google Scholar]
- Szymczak A.L., Workman C.J., Wang Y., Vignali K.M., Dilioglou S., Vanin E.F., Vignali D.A. 2004. Correction of multi-gene deficiency in vivo using a single ‘self-cleaving’ 2A peptide-based retroviral vector.Nat. Biotechnol. 22:589–594 [DOI] [PubMed] [Google Scholar]
- Traenckner E.B., Pahl H.L., Henkel T., Schmidt K.N., Wilk S., Baeuerle P.A. 1995. Phosphorylation of human I kappa B-alpha on serines 32 and 36 controls I kappa B-alpha proteolysis and NF-kappa B activation in response to diverse stimuli.EMBO J. 14:2876–2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabannes E., Khan G., Aillet F., Jarrett R.F., Hay R.T. 1999. Mutations in the IkBa gene in Hodgkin’s disease suggest a tumour suppressor role for IkappaBalpha.Oncogene. 18:3063–3070 [DOI] [PubMed] [Google Scholar]
- Emmerich F., Theurich S., Hummel M., Haeffker A., Vry M.S., Dohner K., Bommert K., Stein H., Dorken B. 2003. Inactivating I kappa B epsilon mutations in Hodgkin/Reed-Sternberg cells.J. Pathol. 201:413–420 [DOI] [PubMed] [Google Scholar]
- Emmerich F., Meiser M., Hummel M., Demel G., Foss H.D., Jundt F., Mathas S., Krappmann D., Scheidereit C., Stein H., Dorken B. 1999. Overexpression of I kappa B alpha without inhibition of NF-kappaB activity and mutations in the I kappa B alpha gene in Reed-Sternberg cells.Blood. 94:3129–3134 [PubMed] [Google Scholar]
- Jungnickel B., Staratschek-Jox A., Brauninger A., Spieker T., Wolf J., Diehl V., Hansmann M.L., Rajewsky K., Küppers R. 2000. Clonal deleterious mutations in the IκBα gene in the malignant cells in Hodgkin’s lymphoma.J. Exp. Med. 191:395–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H., Feuerhake F., Monti S., Kutok J.L., Aster J.C., Shipp M.A. 2006. Lack of IKBA coding region mutations in primary mediastinal large B-cell lymphoma and the host response subtype of diffuse large B-cell lymphoma.Blood. 107:844–845 [DOI] [PubMed] [Google Scholar]
- Coornaert B., Baens M., Heyninck K., Bekaert T., Haegman M., Staal J., Sun L., Chen Z.J., Marynen P., Beyaert R. 2008. T cell antigen receptor stimulation induces MALT1 paracaspase-mediated cleavage of the NF-kappaB inhibitor A20.Nat. Immunol. 9:263–271 [DOI] [PubMed] [Google Scholar]
- Hitotsumatsu O., Ahmad R.C., Tavares R., Wang M., Philpott D., Turer E.E., Lee B.L., Shiffin N., Advincula R., Malynn B.A., et al. 2008. The ubiquitin-editing enzyme A20 restricts nucleotide-binding oligomerization domain containing 2-triggered signals.Immunity. 28:381–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak U., Rinaldo A., Kwee I., Nandula S.V., Rancoita P.M.V., Compagno M., Cerri M., Rossi D., Murty V.V., Zucca E., et al. 2009. The NK-kappaB negative regulator TNFAIP3 (A20) is inactivated by somatic mutations and genomic deletions in marginal zone B-cell lymphomas.Blood. doi:10.1182/blood-2008-08-174110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mader A., Brüderlein S., Wegener S., Melzner I., Popov S., Müller-Hermelink H.K., Barth T.F., Viardot A., Möller P. 2007. U-HO1, a new cell line derived from a primary refractory classical Hodgkin lymphoma.Cytogenet. Genome Res. 119:204–210 [DOI] [PubMed] [Google Scholar]
- Martin-Subero J.I., Chudoba I., Harder L., Gesk S., Grote W., Novo F.J., Calasanz M.J., Siebert R. 2002. Multicolor-FICTION: expanding the possibilities of combined morphologic, immunophenotypic, and genetic single cell analyses.Am. J. Pathol. 161:413–420 [DOI] [PMC free article] [PubMed] [Google Scholar]