Abstract
Toll-like receptors (TLRs) 3, 7, and 9 recognize microbial nucleic acids in endolysosomes and initiate innate and adaptive immune responses. TLR7/9 in dendritic cells (DCs) also respond to self-derived RNA/DNA, respectively, and drive autoantibody production. Remarkably, TLR7 and 9 appear to have mutually opposing, pathogenic or protective, impacts on lupus nephritis in MRL/lpr mice. Little is known, however, about the contrasting relationship between TLR7 and 9. We show that TLR7 and 9 are inversely linked by Unc93B1, a multiple membrane-spanning endoplasmic reticulum (ER) protein. Complementation cloning with a TLR7-unresponsive but TLR9-responsive cell line revealed that amino acid D34 in Unc93B1 repressed TLR7-mediated responses. D34A mutation rendered Unc93B1-deficient DCs hyperresponsive to TLR7 ligand but hyporesponsive to TLR9 ligand, with TLR3 responses unaltered. Unc93B1 associates with and delivers TLR7/9 from the ER to endolysosomes for ligand recognition. The D34A mutation up-regulates Unc93B1 association with endogenous TLR7 in DCs, whereas Unc93B1 association with TLR9 was down-regulated by the D34A mutation. Consistently, the D34A mutation up-regulated ligand-induced trafficking of TLR7 but down-regulated that of TLR9. Collectively, TLR response to nucleic acids in DCs is biased toward DNA-sensing by Unc93B1.
Toll-like receptors (TLRs) can sense a variety of microbial products such as microbial membrane lipids or nucleic acids. Cell-surface TLR dimers including TLR4/MD-2, TLR1/2, and TLR6/2 recognize microbial membrane lipids, whereas TLR3, 7, 8, and 9 reside in intracellular organelles and recognize microbial nucleic acids (1, 2). Although membrane lipids like lipopolysaccharides or lipopeptides are structurally unique to microbes, microbial nucleic acids share the basic structure with host-derived nucleic acids. Indeed, TLR9 is able to respond to mammalian DNA if expressed on the cell surface (3). TLR7 also responds to host-derived, single-stranded RNA (4). Given the potential autoreactivity, nucleic acid–sensing TLR7/9 have to be tightly controlled to avoid autoimmune reaction. Nucleic acid sensing in endolysosomes rather than on the cell surface is thought to be a safety mechanism avoiding response to self–nucleic acid, because self–nucleic acids are rapidly degraded before reaching endolysosomes (3). Viral nucleic acid is, on the other hand, protected by capsid proteins and able to reach endolysosomes.
Self-derived nucleic acids, however, may reach endolysosomes in an inflammatory or autoimmune situation, where a variety of nucleic acid–binding proteins like autoantibodies, antimicrobial peptides, and HMGB-1 (5–7) are complexed with host nucleic acids. These complexes are resistant to degradation, reach endolysosomes, and stimulate TLR7 and 9. To avoid unnecessary stimulation of TLR7 and 9, another safety mechanism is working that TLR7 and 9 reside in the ER and traffic to endolysosomes only upon activation. The TLR7/9 trafficking is controlled by Unc93B1, a multitransmembrane ER-resident protein. In mice, a missense mutation in the Unc93b1 gene (H412R mutation) abrogates signaling via TLR3, 7, and 9 without compromising other TLRs (8). These mutant mice (3d mice) show increased susceptibility to infection by a variety of pathogens. Functional null mutations of Unc93B1 are responsible for herpes simplex encephalitis in children (9). As in 3d mice, cells from Unc93B1-deficient patients are defective in signaling via the nucleic acid–sensing TLRs 3, 7, 8, and 9. Unc93B1 was shown to bind to TLR3/7/9/13 via the transmembrane domains of the TLRs (10). The point mutation H412R of Unc93B1 abolishes these interactions. Unc93B1 was further reported to deliver TLR7/9 from the ER to endolysosomes, where TLR7/9 recognize their ligands (11).
Despite the fail-safe mechanisms limiting the access of both ligands and TLR7/9 to endolysosomes, TLR7 and 9 predispose to autoimmune diseases. Overexpression of TLR7 in the Yaa or transgenic mice predisposes to lupus nephritis (12, 13), whereas the lack of the TLR7 gene ameliorates disease progression in lupus-prone mice (14, 15). TLR9 is more complicated than TLR7 in its link with autoimmune disease. Despite the reports showing a pathogenic role for TLR9 in psoriasis, lupus nephritis, adjuvant-induced arthritis, or a mouse model of multiple sclerosis (16–20), opposing results were also described that TLR9 deficiency in autoimmune-prone MRL/lpr mice exacerbates clinical diseases, including more severe glomerulonephritis, a significantly shortened lifespan, and in some models, elevated titers of autoantibodies reactive with RNA-associated autoantigens (14, 21). The disease severity and RNA-skewed autoantibody repertoire are reminiscent of the phenotype of Yaa autoimmune-prone mice in which TLR7 is overexpressed. TLR9 signaling might protect disease progression at least in part by antagonizing TLR7. Indeed, overexpression of TLR9 inhibits TLR7 (22), and TLR9 ligand strongly inhibited the development of inflammatory arthritis (23).
TLR7 and 9 are both expressed in B cells and DCs, and share downstream signaling pathways (24, 25). If TLR7 and 9 work differently, TLR7 and 9 need to be coordinated in a single cell. The present study revealed that TLR7 and 9 are inversely linked by Unc93B1. Moreover, Unc93B1 biases DC responses toward TLR9 and against TLR7. Given the pathogenic and protective roles for TLR7 and 9 in lupus nephritis, respectively, the present study may have revealed another safety mechanism avoiding autoimmune diseases by balancing TLR7 and 9 in DCs.
RESULTS
Unc93B1 is required for TLR7-unresponsive and TLR9-responsive cell lines
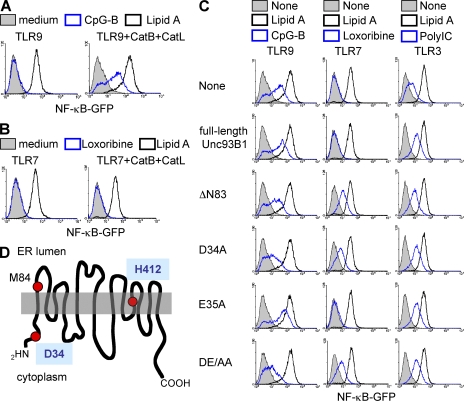
We previously established the pro–B cell line Ba/F3, in which TLR-dependent NF-κB activation was easily detected by GFP induction (26). Lipid A induced GFP when TLR4/MD-2 was expressed (Fig. 1 A, black histogram). GFP was, however, not induced by the TLR9 ligand CpG-B despite expression of TLR9 (Fig. 1 A, left, blue histogram). Functional cloning identified cathepsin B/L as molecules able to confer TLR9 responsiveness on Ba/F3 cells (Fig. 1 A, right) (26). We next studied TLR7 responsiveness in Ba/F3 cells. Overexpressed TLR7 failed to induce NF-κB–dependent GFP expression upon stimulation with a TLR7 ligand loxoribine in Ba/F3 cells (Fig. 1 B, left, blue histogram). Expression of cathepsin B/L was unable to confer TLR7 responses (Fig. 1 B, right). Therefore, we hypothesized that a molecule is missing in Ba/F3 cells that is required for TLR7 but not TLR9. Functional cloning was conducted again, and a cDNA encoding Unc93B1 conferred TLR7 responsiveness on Ba/F3 cells (Fig. 1 C, middle, ΔN83). The Unc93B1 cDNA obtained by complementation cloning lacked the 5′ region up to 102 bases from the initiating codon, suggesting that N-terminally truncated Unc93B1 is expressed from the second methionine M84 (Fig. 1 D). Expression of the truncated Unc93B1 (referred to as ΔN83) was examined by flow cytometry and biochemical analyses of the truncated cDNA tagged with GFP or hemagglutinin (HA), respectively, to the C terminus (Fig. S1, A and B). Expression of ΔN83 protein was confirmed by both flow cytometry and immunoprecipitation but seemed to be lower than that of full-length Unc93B1, probably because of proteolytic degradation (Fig. S1 B). RT-PCR analyses revealed that Unc93B1 mRNA is expressed in Ba/F3 cells as well as in DCs and RAW264.7 cells (Fig. S1 C), suggesting that full-length Unc93B1 endogenously expressed in Ba/F3 cells did not enable TLR7 response in Ba/F3 cells. Indeed, further expression of full-length Unc93B1 did not rescue TLR7 response (Fig. 1 C, middle). These results suggest that N-terminally truncation of Unc93B1 is critical for conferring TLR7 responsiveness on Ba/F3 cells.
Figure 1.
Unc93B1 is selectively required for TLR7 response. (A) Ba/F3 cells expressing TLR9, CD14/TLR4/MD-2, and NF-κB–GFP (left) were further transfected with vectors encoding cathepsin B+L (right). These cells were left unstimulated or were stimulated for 24 h with 100 nM CpG-B or 1 µg/ml lipid A, as indicated. GFP induction was determined by flow cytometry. (B) Experiments were conducted as in A, except that TLR7 was expressed instead of TLR9, and cells were stimulated with 100 µM loxoribine or lipid A, as indicated in the figure. (C) Ba/F3 cells expressing CD14/TLR4/MD-2, NF-κB–GFP, and cathepsin B+L were further transfected with cDNA encoding TLR9 (left), 7 (middle), or 3 (right). These cells were then transfected with full-length Unc93B1, or its mutants lacking the N-terminal 83 aa (ΔN83) or containing an alanine substitution at D34, E35, or both (D34A, E35A, or DE/AA). GFP induction is shown in response to 1 µg/ml lipid A (black lines) or TLR3/7/9 ligand (100 nM CpG-B, 100 µM loxoribine, or 25 µg/ml poly(I:C); blue lines). These experiments were repeated three times, and the represented data are shown. (D) Schematic representation of Unc93B1. D34 and H412 (mutated in 3d mice) are indicated. The second M at 84 is also shown.
D34 in Unc93B1 is required for repression of TLR7-mediated responses
A missense mutation in the Unc93B1 gene in mice abrogates signaling via TLR3/7/9 without compromising other TLRs (8). Despite the requirement for Unc93B1 in both TLR7 and 9, functional cloning suggested that the N-terminal 83 aa negatively regulates TLR7 but not TLR9 in Ba/F3 cells. To narrow down the N-terminal region critical for repressing TLR7-mediated responses, we examined Unc93B1 deletion mutants. The mutants lacking up to N-terminal 33 aa (ΔN18, ΔN23, ΔN30, and ΔN33) had no effect on TLR7 response, whereas those lacking beyond 36 aa (ΔN36, ΔN37, ΔN55, and ΔN83) conferred TLR7 response (Fig. S2 A). The mutant lacking N-terminal 155 aa was unable to confer TLR7 responsiveness. These mutants suggested that DEL (34-36) is critical for negatively regulating TLR7 responses. We tested the Unc93B1 mutants containing an alanine substitution at D34, E35, or both (D34A, E35A, or DE/AA). D34A and DE/AA but not E35A mutants conferred TLR7-dependent GFP induction (Fig. 1 C, middle). D34 is conserved in human Unc93B1 (Fig. S2 B) and predicted to locate in the cytoplasm, whereas H412 (the position of 3d mutation) is in the transmembrane region of the C-terminal half (Fig. 1 D). The Unc93B1 mutants were also expressed in Ba/F3 cells expressing TLR3 or 9 as well as cathepsin B/L. All of the mutants able to confer TLR7 responsiveness (ΔN83, D34A, and DE/AA) down-regulated CpG-B–dependent GFP induction, likely through their dominant-negative effect on endogenously expressed Unc93B1 (Fig. 1 C, left). TLR3-dependent responses were up-regulated by overexpression of all of the Unc93B1 mutants as well as full-length Unc93B1 (Fig. 1 C, right). N-terminal mutations had no effect on TLR3 responsiveness in Ba/F3 cells.
D34A mutation biases DC response toward TLR7 and against TLR9
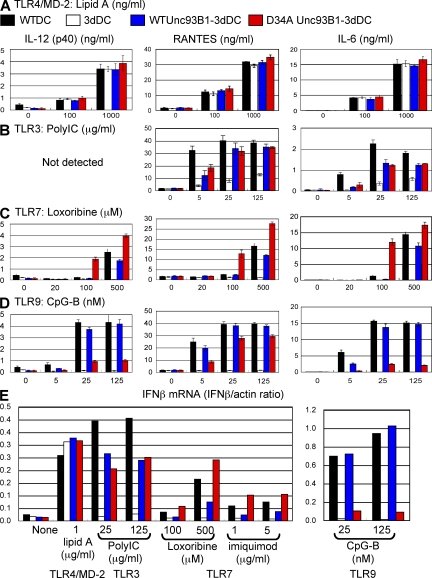
WT or D34A Unc93B1 was next expressed in BM-derived DCs (BM-DCs) from 3d mice harboring the H412R missense mutation (WT or D34A Unc93B1-3dDC, respectively). Given that H412R mutation abolishes Unc93B1 association with TLR3/7/9 (10), H412R Unc93B1 endogenously expressed in 3dDCs unlikely affects interaction between TLR3/7/9 and overexpressed Unc93B1. Retrovirus vector encoding WT or D34A Unc93B1-GFP was transduced into 3d BM cells that were then induced to differentiate into DCs by GM-CSF. All of the BM-DCs expressed the equivalent amount of Unc93B1-GFP, which was easily detected with flow cytometry (Fig. S3). DCs were stimulated with a variety of TLR ligands and cytokine production was determined by ELISA. Lipid A–induced production of IL-12, RANTES, and IL-6 was equivalently induced by WTDC, 3dDC, WT Unc93B1-3dDC, and D34A Unc93B1-3dDC (Fig. 2 A). Poly(I:C)-induced production of RANTES and IL-6 was not completely abolished in 3dDCs, which can be explained by the cytoplasmic RNA sensors RIG-I or MDA-5 (Fig. 2 B, white bars) (27). TLR3-dependent poly(I:C) responses were equally rescued by WT and D34A Unc93B1 (Fig. 2 B, blue and red bars). By sharp contrast, TLR7 ligand induced much more pronounced production of IL-12, RANTES, and IL-6 in D34A Unc93B1-3dDCs than in WTDCs or in WT Unc93B1-3dDCs (Fig. 2 C). WTDCs were unable to respond at 100 µM loxoribine, as were 3dDCs, but D34A-3dDC was able to produce cytokines (Fig. 2 C). On the contrary, D34A Unc93B1 failed to rescue TLR9-dependent production of IL-12 (p40) and IL-6 in 3dDCs (Fig. 2 D). For an unknown reason, RANTES production in 3dDCs was appreciably rescued by D34A Unc93B1. Induction of IFN-β mRNA was next studied by semiquantitative real-time RT-PCR (Fig. 2 E). No alteration was seen between WT and D34A Unc93B1 in rescuing IFN-β mRNA induction upon stimulation with poly(I:C) in 3dDC. WT Unc93B1 only partially rescued TLR7-dependent IFN-β mRNA up to ∼50% of the WTDC response, whereas D34A Unc93B1 conferred on 3dDC a higher induction of IFN-β mRNA than WTDCs (Fig. 2 E). TLR9-dependent induction of IFN-β mRNA in 3dDCs was rescued by WT Unc93B1 but not by D34A Unc93B1 (Fig. 2 E). It is possible that D34A Unc93B1 influences TLR7/9 responses by modulating their mRNA expression. We examined expression of mRNA encoding TLR3/7/8/9/13 in WTDC, 3dDC, WT Unc93B1-3dDC, or D34A Unc93B1-3dDC. No apparent change was caused by transduction of WT or D34A Unc93B1 (Fig. S4 A). These results clearly demonstrate that 34D in Unc93B1 leads to DC polarization toward TLR9 over TLR7 without influencing TLR3.
Figure 2.
Unc93B1 D34A mutant reveals a mutually exclusive link between TLR7 and 9 in DCs. (A–D) DCs from WT mice were transduced with a retrovirus vector encoding GFP (WTDC; black bars). DCs from 3d mice (3dDC) were also transduced by retroviral vector encoding GFP (white bars), WT Unc93B1-GFP (blue bars), or D34A Unc93B1-GFP (red bars). These DCs were stimulated with (A) lipid A, (B) poly(I:C), (C) loxoribine, or (D) CpG-B at the indicated concentrations. Production of IL-12 (left), RANTES (middle), and IL-6 (right) was determined by ELISA. (E) To determine induction of IFN-β mRNA, semiquantitative real-time PCR was conducted. The results were represented by mean values (A–E) with SD (A–D) from triplicate wells. These experiments were repeated twice and the represented data are shown.
D34A mutation enhances TLR7 trafficking and down-regulates TLR9 trafficking
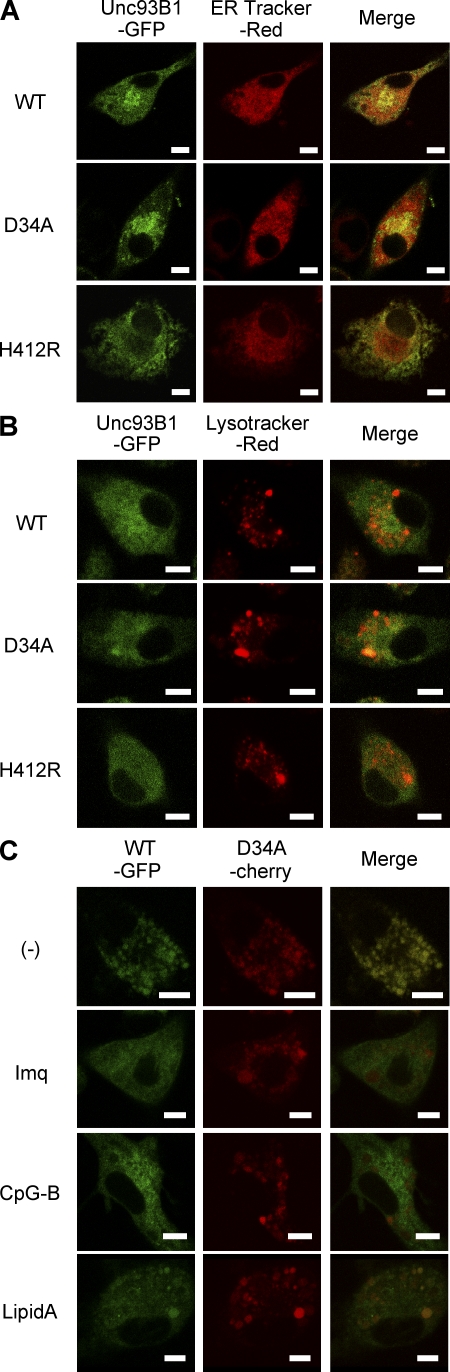
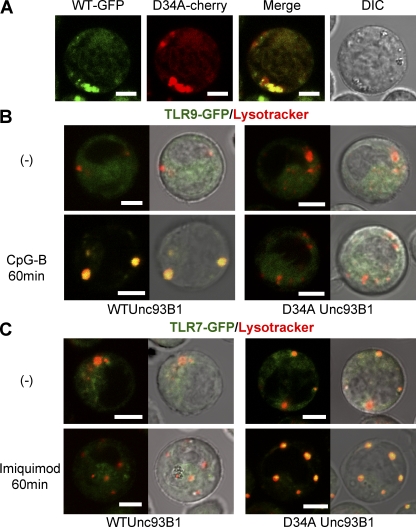
To gain insight into a mechanism differentially regulating TLR7 and 9 by Unc93B1, we compared the subcellular distribution of WT, D34A, or H412R (3d) Unc93B1-GFP in DCs (Fig. 3, A and B). These three types of Unc93B1-GFP all showed colocalization with an ER marker but not with a lysosomal marker. Unc93B1 was reported to associate with TLR7/9 in the ER and deliver TLR7/9 to endolysosome (11). The subcellular distribution of WT and D34A Unc93B1 before and after stimulation with TLR ligands was next compared by analyzing BM-DCs expressing WT Unc93B1-GFP and D34A Unc93B1-cherry (Fig. 3 C). Stimulation with three TLR ligands all induced partial accumulation of D34A Unc93B1 in vesicular structure, probably endolysosomes (Fig. 3 C, middle). WT Unc93B1 also accumulated in vesicular structure (Fig. 3 C, left). Further, the subcellular distribution of WT and D34A Unc93B1 was also indistinguishable in Ba/F3 cells (Fig. 4 A). These results suggest that D34A mutation does not alter the subcellular distribution of Unc93B1.
Figure 3.
D34A mutation does not influence the subcellular distribution of Unc93B1. (A and B) WT, D34A, or H412R Unc93B1-GFP was expressed in DCs from 3d mice. Their subcellular distribution was determined by confocal microscopy (left). Cells were also stained with a marker locating (A) ER or (B) lysosome (center). (right) Merged images. (C) WT Unc93B1-GFP (left) and D34A Unc93B1-cherry (center) were coexpressed in DCs from 3d mice. These cells were left unstimulated or stimulated with 5 µg/ml imiquimod, 1 µM CpG-B, or 1 µg/ml lipid A for 3 h, as indicated in the figure. Green and red images were merged (right). These experiments were repeated three times and the represented images are shown. Bars, 5 µm.
Figure 4.
Differential regulation of ligand-induced TLR7/9 trafficking by Unc93B1 (A) WT Unc93B1-GFP and D34A Unc93B1-cherry were coexpressed in Ba/F3 cells, and live cells were imaged. The merged and differential interference contrast (DIC) images are shown. (B) Ba/F3 cells expressing cathepsin B+L and TLR9-GFP were further transfected with WT (left) or D34A (right) Unc93B1. These cells were left unstimulated (top) or stimulated with 100nM CpG-B for 60 min (bottom). Cells were stained with a lysosome marker. Green and red images were merged (left), or merged images were further imposed on DIC images (right). (C) TLR9-GFP in B was replaced with TLR7-GFP and 5 µg/ml imiquimod was used as the stimulant. These experiments were repeated four times and represented images are shown. Bars, 5 µm.
Unc93B1-dependent TLR7/9 trafficking was next studied. TLR9- or TLR7-GFP was expressed with WT or D34A Unc93B1 in Ba/F3 cells. CpG-B–induced TLR9 accumulation to endolysosomes was appreciably detected in WT Unc93B1–expressing cells, but little, if any, was detected in D34A Unc93B1–expressing cells (Fig. 4 B). In contrast, only a small fraction of TLR7 showed ligand-induced trafficking into endolysosomes in WT Unc93B1–expressing cells, whereas more TLR7 relocated into endolysosomes in D34A Unc93B1–expressing cells (Fig. 4 C). These results suggest that D34A Unc93B1 enhances TLR7 trafficking, whereas it down-regulates TLR9 trafficking.
D34A mutation up-regulates Unc93B1 association with TLR7/8/13
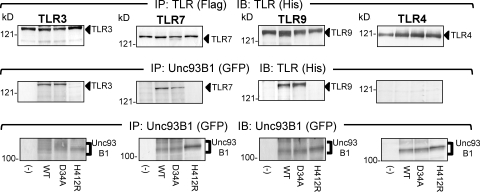
Considering that Unc93B1 controls TLR7/9 trafficking through physical association (10, 11), D34A mutation may influence Unc93B1 association with TLR7/9. Physical association between TLR3/4/7/9 and Unc93B1 was therefore examined. TLR3/4/7/9-flag and Unc93B1-GFP were expressed in Ba/F3 cells. Unc93B1 was immunoprecipitated, and coprecipitation of TLR3/4/7/9 was detected. As reported previously (10), TLR3/7/9, but not TLR4, was coprecipitated with WT Unc93B1. The 3d mutation (H412R) completely abolished Unc93B1 association with TLR3/7/9 (Fig. 5, middle). In contrast, D34A Unc93B1 was indistinguishable from WT Unc93B1 in association with TLR3/7/9 (Fig. 5).
Figure 5.
D34A Unc93B1 mutant is not impaired in association with TLR3, 7, and 9. WT, D34A, or H412R Unc93B1 was expressed in Ba/F3 cells expressing TLR3, 7, 9, or 4, as indicated in the figure. TLRs were tagged with flag and his, whereas Unc93B1 was tagged with GFP. These cells were subjected to immunoprecipitation and immunoprobing with anti-flag (for immunoprecipitation of TLR), anti-his (for immunoprobing TLR), or anti-GFP antibody (for Unc93B1), as indicated in the figure. These experiments were repeated three times and the presented data are shown.
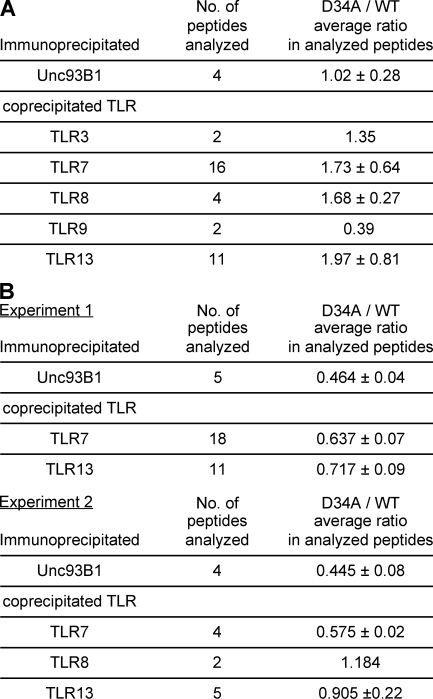
TLR7 and 9 reside in the ER (11, 28), whereas TLR3 resides outside the ER (29). TLR7/9 trafficking from the ER to endolysosomes is not ligand specific, because TLR7/9 traffics to endolysosomes upon LPS stimulation (11). In infection or inflammation, TLR7 may compete with TLR9 but not with TLR3 for association with Unc93B1 in the ER. We hypothesized that a role for D34 would be revealed only in a situation where TLR7 and 9 compete for Unc93B1 association, but not in a situation where each TLR and Unc93B1 were overexpressed, as in Fig. 5. To address this possibility, it was important to use primary DCs but not a cell line like RAW264.7 cells, because expression of nucleic acid–sensing TLRs is apparently different between RAW264.7 cells and BM-DCs (Fig. S4 B). WT or D34A Unc93B1-GFP was expressed in 3dDCs, which endogenously express TLR3, 7, 8, 9, and 13 (Fig. S4). Unc93B1 was immunoprecipitated, and whole immunoprecipitates were analyzed on liquid chromatography tandem mass spectrometry (LC-MS/MS) for a label-free semiquantitative analysis. TLR3, 7, 8, 9, and 13 were all coprecipitated with D34A or WT Unc93B1 (Fig. 6 A). All of the identified peptide sequences, with their scores and the extracted ion current (XIC) values, are shown in Table S1. A previous report described Unc93B1 association with TLR3, 7, 9, and 13, but not TLR8, by using RAW264.7 cells (10), probably because TLR8 is not expressed in RAW cells (Fig. S4 B). The present study was able to show for the first time that TLR8 associates with Unc93B1 in DCs because TLR8 mRNA was detected in DCs (Fig. S4 B).
Figure 6.
D34A mutation up-regulates Unc93B1 association with TLR7/8/13. (A and B) D34A or WT Unc93B1-GFP was expressed in BM-DCs from 3d mice. Unc93B1-GFP was immunoprecipitated by anti-GFP mAb, and precipitates were analyzed on LC-MS/MS analyses. (A) For label-free semiquantitative analysis, the XIC values of each peptide from Unc93B1 and TLR3, 7, 8, 9, and 13 were calculated directly by the MSQuant program. Immunoprecipitated peptides from TLR3, 7, 8, 9, and 13 associated with D34A or WT Unc93B1 in BM-DCs were compared by dividing the value from D34A by that from WT in each peptide. The number of analyzed peptides and the mean ratio are shown. Detailed results are shown in Table S1. The experiments were repeated twice and similar results were obtained. (B) For SILAC experiments, BM-DCs expressing WT or D34A Unc93B1-GFP were cultured in the presence of normal lysine and arginine or the heavy isotopic forms of these aa (experiment 1), or of arginine or the heavy isotopic form of arginine (experiment 2), respectively. The XIC values of each peptide from Unc93B1 and TLR7 and 9 were calculated using the MSQuant program. The ratios represent the relative abundance of the heavy to the light peptide, indicating the binding ratio (D34/WT). The number of analyzed peptides, the mean ratio, and the SD are shown. Detailed information for each peptides is shown in Table S2. These experiments were repeated twice and all of the data are shown.
Semiquantitative analyses with the MSQuant program revealed a difference between WT and D34A Unc93B1 (Fig. 6 A). The amount of immunoprecipitated Unc93B1 was equivalent between WT and D34A Unc93B1 (1.02 in the D34A/WT ratio). Despite that only the two peptides were identified from TLR9, both peptides suggested that TLR9 was coprecipitated much less with D34A Unc93B1 than WT Unc93B1 (0.39). TLR7, 8, and 13 were coprecipitated more with D34A Unc93B1 than WT Unc93B1 (1.73, 1.68, and 1.97, respectively). mRNA expression of TLR9 was equivalent to TLR7 or 8 in BM-DCs and was not influenced by expression of WT or D34A Unc93B1-GFP (Fig. S4). These results suggested that D34 differentially regulates Unc93B1 association with TLR9 or with TLR7/8/13. The D34A/WT ratio of TLR3 was between TLR9 and TLR7/8/13 (Fig. 6 A).
To confirm the results with semiquantitative analysis using XIC-based quantification, highly accurate quantitative analysis by stable isotope labeling with aa (SILAC) was conducted twice. In experiment 1, both arginine and lysine were labeled with isotopes, and only a small number of BM-DCs were obtained, probably because of low concentrations of these two aa in culture medium. To improve cell recovery, only arginine was labeled in experiment 2. Although a larger number of BM-DCs were recovered, the number of peptides analyzed was reduced, probably because of inefficient labeling with only one isotope. Despite these results, SILAC confirmed that larger amounts of TLR7/8/13 are coprecipiated with D34A Unc93B1 than WT Unc93B1 (Fig. 6 B and Table S2). In contrast, SILAC failed to detect a peptide from TLR9 or 3.
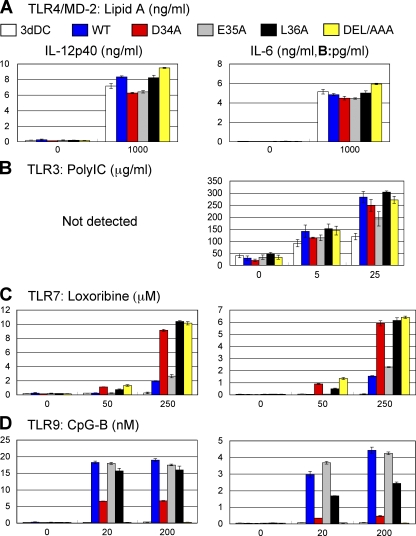
TLR7/9 responses are inversely regulated by the DEL sequence in the N-terminal region of Unc93B1
To gain further insight into a mechanism of how the N-terminal region of Unc93B1 influences Unc93B1 association with TLR7/8/13, we further studied the additional requirement for aa adjacent to D34. Given that the negative regulation was abolished by the lack of the N-terminal 36 aa or further (Fig. S2), we focused on L36 and used two additional Unc93B1 mutants, L36A and DEL/AAA, in which L36 or DEL (34-36) was replaced with alanine, respectively. These mutants together with WT, D34A, and E35A Unc93B1 were expressed in BM-DCs from 3d mice. BM-DCs were stimulated with the indicated TLR ligands, and production of IL-12 and IL-6 was determined by ELISA. L36A and DEL/AAA mutations did not influence BM-DC responses to lipid A or poly(I:C) (Fig. 7, A and B). L36A and DEL/AAA were both as effective as D34A in enhancing TLR7 responses (Fig. 7 C). L36A was less effective than D34A in down-regulating TLR9 responses (Fig. 7 D). DEL/AAA completely abolished TLR9 responses (Fig. 7 D). As expected from the results with Ba/F3 cells (Fig. 1 C), E35A had no effect on TLR7/9 responses. These results revealed that the DEL sequence in the N-terminal region is important in differentially regulating TLR7 and 9 responses.
Figure 7.
D34 and L36 are required for differential regulation of TLR7 and 9 in BM-DCs. (A–D) DCs from 3d mice were transduced by retroviral vector encoding GFP (3dDC; white bars), WT Unc93B1-GFP (WT; blue bars), D34A Unc93B1-GFP (D34A; red bars), E35A Unc93B1-GFP (E35A; gray bars), L36A Unc93B1-GFP (L36A; black bars), or DEL/AAA Unc93B1-GFP (DEL/AAA; yellow bars). These DCs were stimulated with (A) lipid A, (B) poly(I:C), (C) loxoribine, or (D) CpG-B at the indicated concentrations. Production of IL-12p40 (left) and IL-6 (right) was determined by ELISA. The results are represented by mean values with SD from triplicate wells. These experiments were repeated twice and the represented data are shown.
The N-terminal region of Unc93B1 is expressed in TLR7-responsive cells
The results that the N-terminal region of Unc93B1 represses TLR7 responses raise a question of how the N-terminal portion of Unc93B1 is controlled in TLR7-responsive cells like RAW264.7 cells. Alternative splicing or alternative transcription start may generate two mRNA isoforms encoding full-length or N-terminally truncated Unc93B1. To detect the mRNA encoding the N-terminally truncated Unc93B1, we conducted RT-PCR amplifying the 5′ end of Unc93B1 mRNA by using oligo-capped cDNA (30). Only a single isoform of Unc93B1 mRNA was detected, and no difference was seen between TLR7-responsive RAW264.7 cells and TLR7-unresponsive Ba/F3 cells (Fig. S5). Even if mRNA encodes only the full-length Unc93B1, Unc93B1 protein may be proteolytically cleaved, generating the N-terminally truncated Unc93B1. TLR7-unresponsive Ba/F3 cells may be different from TLR7-responsive RAW264.7 cells in the proteolytic cleavage of Unc93B1. This possibility was, however, unlikely, because no difference was seen between Unc93B1-GFP expressed in Ba/F3 or RAW264.7 cells (Fig. S1 B). The N-terminal region of Unc93B1 is likely to be intact in TLR7-responsive cells. The truncated Unc93B1 cDNA obtained in this case was probably derived from incomplete cDNA synthesis. It has to be noted that the cDNA library used in this study also contained the full-length Unc93B1 cDNA.
DISCUSSION
The present study showed that the N-terminal, putatively cytoplasmic portion of Unc93B1 represses TLR7 responses and enhances TLR9 responses in BM-DCs. This conclusion is based on the following results. First, Unc93B1 N-terminally truncated or D34A mutant enables TLR7 responses in Ba/F3 cells (Fig. 1). Second, Unc93B1 D34A mutant rendered Unc93B1-deficient BM-DCs hyperresponsive to TLR7 ligand but hyporesponsive to TLR9 ligand (Fig. 2). LC-MS/MS analyses revealed the difference between TLR9 and 7. D34A mutation enhanced Unc93B1 association with TLR7, whereas that with TLR9 was suggested to be down-regulated by D34A mutation (Fig. 6). Further, D34A mutation enhanced TLR7 trafficking but suppressed TLR9 trafficking to endolysosomes (Fig. 4). The present study demonstrated that Unc93B1 actively biases TLR-dependent nucleic acid sensing against RNA-sensing. Polarization of DC responses against TLR7-mediated responses might be understood as a mechanism avoiding hazardous autoimmune reaction, because previous reports showed that TLR7 is pathogenic in a variety of autoimmune diseases (Fig. S6) (12–15).
LC-MS/MS analyses clearly revealed that Unc93B1 association with TLR7/8/13 is up-regulated by D34A mutation (Fig. 6, Table S1, and Table S2). In contrast, Unc93B1 association with TLR3 or 9 was hard to detect, particularly in SILAC (Fig. 6 B and Table S2). Brinkmann et al. clearly demonstrated Unc93B1 association with endogenous TLR3/7/9/13 in a macrophage cell line, RAW264.7, by using a large number of cells (4 × 109) (10). The present study, on the other hand, focused on BM-DCs and studied Unc93B1 interactions with endogenous TLRs in BM-DCs. In this regard, differences in TLR mRNA expression between RAW264.7 cells and BM-DCs are noteworthy in that RAW264.7 cells express TLR3/7 much higher than BM-DCs without expressing TLR8 (Fig. S4 B). Along this line, Brinkmann et al. detected Unc93B1 association only with TLR7/9 in a B cell line, A20 (10). Unc93B1 association with TLRs is likely to change with cell types, and it was important to study Unc93B1 association with TLRs in BM-DCs in the present study. Unfortunately, we were unable to obtain as many BM-DCs as RAW264.7 cells used by the previous study because of the lack of an in vitro DC line. However, semiquantitative analyses with LC-MS/MS suggested that D34A mutation weakens Unc93B1 association with TLR9 (Fig. 6 A and Table S1). This finding is consistent with the results that D34A mutation down-regulated TLR9 trafficking and TLR9 responses in BM-DCs (Figs. 2 and 4). Down-regulation of TLR9 responses and trafficking is not explained by up-regulation of Unc93B1 association with TLR7/8/13, suggesting that strengthened Unc93B1 association with TLR7/8/13, in turn, down-regulates that with TLR9. Although a ligand for TLR13 has not been identified, the present study suggests the possibility that TLR13 responds to microbial RNA. RNA-sensing TLRs might compete with DNA-sensing TLR9 for association with Unc93B1 in the ER (Fig. S6).
The present study looked for a difference between TLR7-responsive RAW264.7 and TLR7-unresponsive Ba/F3 cells, and found that the N-terminal portion of Unc93B1 represses TLR7 responses. These results raise a question of how negative regulation by the N-terminal region of Unc93B1 is controlled in TLR7-responsive cells like RAW264.7 cells. It is possible that Unc93B1 mRNA in TLR7-responsive cells is under the control of alternative splicing or alternative transcription start, leading to the lack of the 5′ portion encoding the N-terminal region. This is, however, unlikely, because PCR amplification of the 5′ region of Unc93B1 mRNA failed to detect such alternative Unc93B1 mRNA isoforms in RAW264.7 and Ba/F3 cells (Fig. S5). Another possibility is that the N-terminal region of Unc93B1 may be proteolytically cleaved off in TLR7-responsive cells, as was TLR9 (26, 31, 32). Unc93B1-GFP expressed in RAW264.7 cells, however, did not appear to be truncated when compared with that in Ba/F3 cells (Fig. S1 B, compare left and right lanes).
The N-terminal region of Unc93B1 is left uncleaved in TLR7-responsive cells, and is likely to be inactivated by an as yet unknown mechanism. In this regard, it is noteworthy that the DEL (34-36) sequence required for repressing TLR7 response agrees with the motif (D/E-X-Φ) that is known to interact with the class III PDZ (PDS95/dlg/ZO-1) domain (33). Requirement for 34D and 36L but not 35E is consistent with the lack of any restriction (X) in the second aa in the D/E-X-Φ motif (Fig. 1 C; Fig. 7; and Fig. S2). The class III PDZ domain is not found in TLRs, suggesting that DEL (34-36) is required for interaction with a non-TLR, class III PDZ domain–containing molecule, which may have a role in controlling the activity of the N-terminal region of Unc93B1.
Although not experimentally proven yet, the lack of an N-terminal hydrophobic signal sequence suggests that the N-terminal region of Unc93B1 faces the cytosol. The N terminus of Unc93B1 may have a role not only in differential association of Unc93B1 with TLR7 and 9 but also in modulating signaling pathways downstream of TLR7 or 9. In this regard, it is of note that TLR9-dependent RANTES production was more resistant to D34A mutation than production of IL-12 or IL-6 (Fig. 2 D). It will be interesting to see, in future studies, the effect of D34A mutation in signaling pathways downstream of TLR7 and 9.
Immune cells such as DCs or macrophages express multiple TLRs, which are concomitantly activated in response to pathogens, because single microbes or viruses express a variety of TLR ligands. Given that multiple TLRs simultaneously respond to pathogens, their distribution and activation need to be orchestrated for optimal immune responses. Indeed, a synergistic relationship between TLR4/MD-2 and TLR7/9 has been recently reported in the triggering of IL-12 and other Th1-promoting cytokines by DCs (34, 35). TLR3 and TLR7/9 also show synergistic responses (36–38). Dual recognition of Mycobacterium tuberculosis by TLR2 and 9 is required for efficient responses (39). HSVs were reported to be recognized by both TLR2 and 9 (40). We have shown in this study that multiple TLRs in DCs are connected not only by these additive or synergistic links but also by an inversely regulated link.
Simultaneous triggering of TLR7 and 9 may be thought to be a less likely combination in viral infection, as viruses are classified into either DNA or RNA viruses. The DNA genomes from viruses like HSV-1, HSV-2, and mouse CMV are rich in CpG motifs, which activates TLR9 (40), whereas TLR7 recognizes single-stranded RNA from RNA viruses like influenza and vesicular stomatitis virus (41). Concurrent infection with a DNA virus and an RNA virus is a rare event. However, a DNA virus like herpesvirus has been recently suggested to stimulate both TLR7 and 9 (42). DNA and RNA derived from bacterial pathogens are reported to stimulate TLR3, 7, and 9 (43), as well as cell-surface TLR2 and TLR4/MD-2. Finally, self-derived DNA and RNA derived from dead cells are also able to simultaneously stimulate TLR7 and 9 expressed in B cells or DCs during inflammation. Concurrent activation of TLR7 and 9 is likely to occur in infectious and noninfectious diseases.
Type I IFNs are tightly regulated cytokines, and overexpression of type I IFN can be detrimental to the host. Highly elevated levels of type I IFN have been implicated as etiologic for systemic lupus erythematosus (44). Increased serum type I IFN has been shown to correlate directly with disease severely in human systemic lupus erythematosus (45). TLR7 and 9 are known to be strong promoters of type I IFN secretion from DCs. If TLR7 and 9 are both activated without any limit, type I IFN is likely to be excessively produced, predisposing to autoimmune diseases. DCs may inversely link TLR7 with TLR9 to keep the production of type I IFN under the control and to avoid excessive production of type I IFN.
Previous reports have revealed pathogenic roles for TLR7 in lupus nephritis. Overexpression of TLR7 in the Yaa or transgenic mice predisposes to lupus nephritis (12, 13), whereas the lack of the TLR7 gene ameliorates disease progression in lupus-prone mice (14, 15). DCs need to have a mechanism limiting TLR7 activation. Overexpression of TLR9 inhibits TLR7 responses (22). Another mechanism has been revealed in this study that TLR7 responsiveness was down-regulated by the N-terminal cytoplasmic portion of Unc93B1. Given the reciprocal link between TLR7 and 9, Unc93B1 balances TLR7 and 9 to warrant sufficient TLR7 responsiveness without detrimental autoimmune responses (Fig. S6).
TLR7/9 agonists and antagonists are being developed for therapeutic intervention in infectious diseases, cancer, allergic diseases, and autoimmune diseases (46–48). The present study showed that the effect of these agonists/antagonists has to be carefully evaluated, because specific agonists/antagonists for TLR7 or 9 may have an indirect effect on TLR9 or 7, respectively. Such indirect effects may cause unexpected action on diseases. Given that TLR7 hyperresponsiveness and TLR9 hyporesponsiveness both predispose to autoimmune diseases (12–14), the N-terminal portion of Unc93B1 may be a novel target for a therapeutic intervention in autoimmune diseases that modulates the balance between TLR7 and 9 in DCs.
MATERIALS AND METHODS
Reagents and antibodies.
CpG-B (5′-TCCATGACGTTCCTGATGCT-3′) was synthesized by Hokkaido System Science. Lipid A was provided by K. Fukase (Osaka University, Osaka, Japan). Loxoribine, imiquimod, and poly(I:C) were purchased from InvivoGen. Recombinant mouse stem cell factor, mouse IL-3, and mouse IL-6 were purchased from PeproTech. Recombinant mouse GM-CSF was purchased from R&D Systems. Texas red–conjugated ER-tracker and Texas red–conjugated Lysotracker were purchased from Invitrogen. PE-conjugated anti-CD11c antibody was purchased from eBioscience. Rabbit anti-GFP antibody was purchased from MBL International. The affinity-purified rat mAbs against 6xHis (clone 333) and GFP (clone FM264) were generated in our laboratory. Anti-flag antibody–conjugated beads, G418, and puromycin were purchased from Sigma-Aldrich. Anti-HA antibody–conjugated beads and anti-HA antibody (clone 3F10) were purchased from Roche.
Mice and cell lines.
WT C57BL/6 mice were purchased from Japan SLC, and 3d mice were obtained from K. Tabeta (Niigata University, Niigata, Japan). Mice were kept under specific pathogen-free conditions in the animal center at the University of Tokyo. The experiments were performed according to institutional ethical guidelines for animal experiments at the University of Tokyo. All animal experiments were approved by the Animal Care and Use Committee of the University of Tokyo. Ba/F3 and RAW 264.7 cells were maintained as previously described (49).
Plasmid constructs.
The epitopes (flag-6xHis, GFP, mCherry, or HA) were tagged to the C termini of mouse TLR3, 7, and 9, and Unc93B1, as indicated in the figures. They were generated by PCR and cloned into retroviral pMX, pMXpuro, or pMXneo vectors. Mouse TLR4, MD-2, CD14, NF-κB–hrGFP, cathepsin B, and cathepsin L were cloned into expression vectors as previously described (19). Truncated Unc93B1 and aa-mutated Unc93B1 were generated by the QuickChange Site-Directed Mutagenesis Kit (Agilent Technologies) and cloned into C-terminally tagged pMX or pMXpuro vectors. The primers for aa mutant Unc93B1 were constructed to change their DNA as follows: H412R, CAC (His) to CGC (Arg); D34A, GAC (Asp) to GCC (Ala); E35A, GAA (Glu) to GCA (Ala); and L36A, CTC (Leu) to GCC (Ala). The DEAA mutant was generated by sequential mutation of D34A and E35A. The DEL/AAA mutant was generated by combining the three mutations.
Retroviral transduction.
pMX, pMXpuro, and pMXneo vectors were transfected into Plat-E packaging cells with FuGene6 (Roche). After 2 d of incubation, supernatants were obtained as virus suspensions. Cells were infected by the mixture of virus suspension and DOTAP (Roche).
ELISA.
Cytokines in culture supernatants were measured with ELISA kits (R&D Systems).
Quantitative real-time PCR.
Total RNA were prepared by ISOGEN (NIPPON GENE) or RNAeasy (QIAGEN) and reverse transcribed with ReverTra Ace (TOYOBO) or the Super Script II RT kit (Invitrogen). Quantitative real-time PCR analyses were performed using 7300 Fast Real-Time PCR System (Applied Biosystems), as previously described (50).
Immunofluoresence staining and confocal microscopy.
DCs were incubated on a poly–l-lysine–coated glass-bottom dish overnight to allow them to adhere. Cells were stimulated by 1 µM CpG-B or 5 µg/ml imiquimod for 60 min. After stimulation, cells were incubated with Texas red–conjugated ER-tracker or Lysotracker for staining ERs or lysosomes, respectively. Nonstimulated cells were stained the same. Stained cells were washed two times with HBSS. ER-tracker–stained cells were fixed with 3.7% formaldehyde in PBS for 10 min at room temperature. Lysotracker-stained cells were not fixed and suspended in 10% FCS containing PBS. Confocal microscopy was performed on a confocal microscope (LSM510; Carl Zeiss, Inc.).
Immunoprecipitation.
Cells were collected and washed with HBSS two times. Washed cells were lysed with 1% digitonin lysis buffer in Fig. 4 (150 mM NaCl, 50 mM Tris/HCl [pH 7.4], 5 mM EDTA [pH 8], 10 µg/ml leupepsin, 10 µg/ml aprotinin, and 1 mM PMSF) or 0.5% Triton lysis-washing buffer in Fig. S2 (137 mM NaCl, 20 mM Tris/HCl [pH 7.4], 1 mM EDTA [pH 8], 10 µg/ml leupepsin, 10 µg/ml aprotinin, 1 mM PMSF). Lysates were rotated with antibody-conjugated beads overnight. Beads were washed with 0.1% digitonin washing buffer (150 mM NaCl, 50 mM Tris/Hcl [pH 7.4], 5 mM EDTA [pH 8]) three times. Immunoprecipitates were subjected to SDS-PAGE after boiling in SDS-PAGE sample buffer (125 mM Tris/HCl [pH 6.8], 20% glycerol, 4% SDS, 10% 2-ME, 0.005% bromophenol blue). After electrophoresis, samples were transferred to PVDF membrane and subjected to immunoblotting.
Complementation cloning.
cDNA was synthesized from RAW264.7 cells and cloned into a retrovirus vector. About 2 million independent colonies were obtained in each library. These cDNA libraries were packaged and transduced into Ba/F3 cells expressing CD14/TLR4/MD-2, TLR7-flag, and pNF-κB–hrGFP. These cells were stimulated with 100 µM loxoribine for 24 h, and GFP-positive cells were selected by sorting with flow cytometry. Recovered cells were allowed to grow and subjected to a second sorting. After the third sorting, cDNAs derived from the library were recovered from genomic DNA by PCR and retransduced to confirm the complementation activity.
Transduction into BM-DCs.
BM cells were isolated from WT or 3d mutant mice that had been injected intraperitoneally with 5 mg 5-fluorouracil. After 48 h of culture, cells were transduced with retroviral vector containing the puromycin resistance gene on two successive days. After the second transduction, cells were washed and cultured in the presence of 10 ng/ml GM-CSF. During BM-DC induction, 2 µg/ml puromycin was included to enrich BM-DCs expressing Unc93B1. For double selection, 1 µg/ml puromycin and 200 µg/ml neomycin were included.
LC-MS/MS analyses.
BM-DCs expressing WT or D34A Unc93B1-GFP were subjected to lysis with digitonin and immunoprecipitation with anti-GFP mAb. Bound molecules released by acid elution were digested with trypsin directly in the solution, desalted, and concentrated to a volume of ∼20 µl. The samples were then injected into a direct nanoflow LC system (Dina; KYA Technologies), and sprayed into a quadrupole time-of-flight tandem mass spectrometer (QSTAR Elite; Applied Biosystems).
Label-free semiquantitative analysis using the XIC-based peptide quantification.
Database searches were performed with Mascot (http://www.matrixscience.com). The score shown in the tables is the Mascot ion score representing the quality of the match of the identified peptide. The XIC is a measure that is proportional to the peptide’s abundance. The XIC values of each peptide from Unc93B1 and TLR3, 7, 8, 9, and 13 were calculated directly without isotope labeling by using the MSQuant program (http://msquant.sourceforge.net/). The XIC values were retrieved directly for the corresponding peptides’ m/z signals between the two states. The values of peptides derived from BM-DCs expressing D34A Unc93B1-GFP were divided with the values from WT Unc93B1 to compare the binding ratio. The mean and SD were calculated for each protein (Tables S1 and S2).
SILAC.
BM-DCs expressing WT or D34A Unc93B1-GFP were grown in the presence of normal lysine and arginine (unlabeled) or heavy isotopic forms of these aa (labeled) in experiment 1 (Fig. 6). In experiment 2, only arginine was labeled. BM-DCs were cultured for 11 d (51) and subjected to lysis with digitonin, and lysates from labeled and unlabeled cells were combined in a 1:1 ratio according to their protein concentrations estimated by BCA assay (Thermo Fisher Scientific). The mixed lysate was immunoprecipitated with anti-GFP mAb. SILAC methods were used (52).
Detection of full-length mRNA.
Total RNA from Ba/F3 or RAW264.7 cells were prepared with RNAeasy kit according to the manufacturer’s instruction. Total RNA was subjected to oligo capping, as previously described (30). Oligo-capped full-length mRNA were reverse transcribed with the Super Script II RT kit with an oligo-dT primer. Synthesized cDNA were used as the PCR template. The PCR reaction was performed by a forward primer on oligo capping (5′-TCGAGTCGGCCTTGTTGGCCTACTG-3′) and a reverse primer on Unc93B1 (5′-TGGCGTAAGCGAAAGTCACGCACGTG-3′; 1,063–1,038). The expected size is ∼1,100 bp.
Online supplemental material.
Fig. S1 shows the expression of full-length and truncated Unc93B1. Fig. S2 depicts the effect of N-terminal deletion of Unc93B1 on TLR7 responses. Fig. S3 shows expression of Unc93B1-GFP and CD11c in BM-DCs. Fig. S4 depicts TLR mRNA expression in BM-DCs, Ba/F3 cells, and RAW264.7 cells. Fig. S5 shows no difference in Unc93B1 mRNA transcription start between Raw264.7 and Ba/F3 cells. Fig. S6 depicts RNA- versus DNA-sensing in DCs controlled by a Unc93B1-dependent inverse link between TLR7 and 9. Table S1 shows the XIC-based peptide quantification for Unc93B1 and coprecipitated TLRs. Table S2 provides the calculated XIC values, ions scores, and SILAC ratio (D34/WT) of each peptide from Unc93B1 and TLR7 and 9. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20082316/DC1.
Acknowledgments
We thank Drs. R. Jennings and T. Kaisho for critically reviewing the manuscript. We thank Drs. K. Abe and S. Sugano for technical advice and support in preparation of oligo-capped full-length mRNA. We also thank T. Adachi for processing LC-MS/MS data.
This work was supported in part by a strategic Japanese–Korean Cooperative Program on Basic Medical Research; a contract research fund from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) for a Program of Founding Research Centers for Emerging and Reemerging Infectious Diseases; strategic cooperation to control emerging and reemerging infections funded by the Special Coordination Funds for Promoting Science and Technology of MEXT; and Grants-in-Aid for Scientific Research on Priority Areas, for Scientific Research (B), and for Exploratory Research from MEXT.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used: BM-DC, BM-derived DC; HA, hemagglutinin; LC-MS/MS, liquid chromatography tandem mass spectrometry; SILAC, stable isotope labeling with aa; TLR, Toll-like receptor; XIC, extracted ion current.
References
- Beutler B., Jiang Z., Georgel P., Crozat K., Croker B., Rutschmann S., Du X., Hoebe K. 2006. Genetic analysis of host resistance: Toll-like receptor signaling and immunity at large.Annu. Rev. Immunol. 24:353–389 [DOI] [PubMed] [Google Scholar]
- Kaisho T., Akira S. 2006. Toll-like receptor function and signaling.J. Allergy Clin. Immunol. 117:979–987 [DOI] [PubMed] [Google Scholar]
- Barton G.M., Kagan J.C., Medzhitov R. 2006. Intracellular localization of Toll-like receptor 9 prevents recognition of self DNA but facilitates access to viral DNA.Nat. Immunol. 7:49–56 [DOI] [PubMed] [Google Scholar]
- Diebold S.S., Massacrier C., Akira S., Paturel C., Morel Y., Reis e Sousa C. 2006. Nucleic acid agonists for Toll-like receptor 7 are defined by the presence of uridine ribonucleotides.Eur. J. Immunol. 36:3256–3267 [DOI] [PubMed] [Google Scholar]
- Tian J., Avalos A.M., Mao S.Y., Chen B., Senthil K., Wu H., Parroche P., Drabic S., Golenbock D., Sirois C., et al. 2007. Toll-like receptor 9-dependent activation by DNA-containing immune complexes is mediated by HMGB1 and RAGE.Nat. Immunol. 8:487–496 [DOI] [PubMed] [Google Scholar]
- Marshak-Rothstein A., Rifkin I.R. 2007. Immunologically active autoantigens: the role of toll-like receptors in the development of chronic inflammatory disease.Annu. Rev. Immunol. 25:419–441 [DOI] [PubMed] [Google Scholar]
- Lande R., Gregorio J., Facchinetti V., Chatterjee B., Wang Y.H., Homey B., Cao W., Wang Y.H., Su B., Nestle F.O., et al. 2007. Plasmacytoid dendritic cells sense self-DNA coupled with antimicrobial peptide.Nature. 449:564–569 [DOI] [PubMed] [Google Scholar]
- Tabeta K., Hoebe K., Janssen E.M., Du X., Georgel P., Crozat K., Mudd S., Mann N., Sovath S., Goode J., et al. 2006. The Unc93b1 mutation 3d disrupts exogenous antigen presentation and signaling via Toll-like receptors 3, 7 and 9.Nat. Immunol. 7:156–164 [DOI] [PubMed] [Google Scholar]
- Casrouge A., Zhang S.Y., Eidenschenk C., Jouanguy E., Puel A., Yang K., Alcais A., Picard C., Mahfoufi N., Nicolas N., et al. 2006. Herpes simplex virus encephalitis in human UNC-93B deficiency.Science. 314:308–312 [DOI] [PubMed] [Google Scholar]
- Brinkmann M.M., Spooner E., Hoebe K., Beutler B., Ploegh H.L., Kim Y.M. 2007. The interaction between the ER membrane protein UNC93B and TLR3, 7, and 9 is crucial for TLR signaling.J. Cell Biol. 177:265–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.M., Brinkmann M.M., Paquet M.E., Ploegh H.L. 2008. UNC93B1 delivers nucleotide-sensing toll-like receptors to endolysosomes.Nature. 452:234–238 [DOI] [PubMed] [Google Scholar]
- Pisitkun P., Deane J.A., Difilippantonio M.J., Tarasenko T., Satterthwaite A.B., Bolland S. 2006. Autoreactive B cell responses to RNA-related antigens due to TLR7 gene duplication.Science. 312:1669–1672 [DOI] [PubMed] [Google Scholar]
- Deane J.A., Pisitkun P., Barrett R.S., Feigenbaum L., Town T., Ward J.M., Flavell R.A., Bolland S. 2007. Control of toll-like receptor 7 expression is essential to restrict autoimmunity and dendritic cell proliferation.Immunity. 27:801–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen S.R., Shupe J., Nickerson K., Kashgarian M., Flavell R.A., Shlomchik M.J. 2006. Toll-like receptor 7 and TLR9 dictate autoantibody specificity and have opposing inflammatory and regulatory roles in a murine model of lupus.Immunity. 25:417–428 [DOI] [PubMed] [Google Scholar]
- Berland R., Fernandez L., Kari E., Han J.H., Lomakin I., Akira S., Wortis H.H., Kearney J.F., Ucci A.A., Imanishi-Kari T. 2006. Toll-like receptor 7-dependent loss of B cell tolerance in pathogenic autoantibody knockin mice.Immunity. 25:429–440 [DOI] [PubMed] [Google Scholar]
- Deng G.M., Nilsson I.M., Verdrengh M., Collins L.V., Tarkowski A. 1999. Intra-articularly localized bacterial DNA containing CpG motifs induces arthritis.Nat. Med. 5:702–705 [DOI] [PubMed] [Google Scholar]
- Ronaghy A., Prakken B.J., Takabayashi K., Firestein G.S., Boyle D., Zvailfler N.J., Roord S.T., Albani S., Carson D.A., Raz E. 2002. Immunostimulatory DNA sequences influence the course of adjuvant arthritis.J. Immunol. 168:51–56 [DOI] [PubMed] [Google Scholar]
- Ehlers M., Fukuyama H., McGaha T.L., Aderem A., Ravetch J.V. 2006. TLR9/MyD88 signaling is required for class switching to pathogenic IgG2a and 2b autoantibodies in SLE.J. Exp. Med. 203:553–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asagiri M., Hirai T., Kunigami T., Kamano S., Gober H.J., Okamoto K., Nishikawa K., Latz E., Golenbock D.T., Aoki K., et al. 2008. Cathepsin K-dependent toll-like receptor 9 signaling revealed in experimental arthritis.Science. 319:624–627 [DOI] [PubMed] [Google Scholar]
- Prinz M., Garbe F., Schmidt H., Mildner A., Gutcher I., Wolter K., Piesche M., Schroers R., Weiss E., Kirschning C.J., et al. 2006. Innate immunity mediated by TLR9 modulates pathogenicity in an animal model of multiple sclerosis.J. Clin. Invest. 116:456–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P., Wellmann U., Kunder S., Quintanilla-Martinez L., Jennen L., Dear N., Amann K., Bauer S., Winkler T.H., Wagner H. 2006. Toll-like receptor 9-independent aggravation of glomerulonephritis in a novel model of SLE.Int. Immunol. 18:1211–1219 [DOI] [PubMed] [Google Scholar]
- Wang J., Shao Y., Bennett T.A., Shankar R.A., Wightman P.D., Reddy L.G. 2006. The functional effects of physical interactions among Toll-like receptors 7, 8, and 9.J. Biol. Chem. 281:37427–37434 [DOI] [PubMed] [Google Scholar]
- Wu H.J., Sawaya H., Binstadt B., Brickelmaier M., Blasius A., Gorelik L., Mahmood U., Weissleder R., Carulli J., Benoist C., Mathis D. 2007. Inflammatory arthritis can be reined in by CpG-induced DC-NK cell cross talk.J. Exp. Med. 204:1911–1922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki N., Ho S., Antonenko S., Malefyt R.W., Kastelein R.A., Bazan F., Liu Y.J. 2001. Subsets of human dendritic cell precursors express different toll-like receptors and respond to different microbial antigens.J. Exp. Med. 194:863–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernasconi N.L., Onai N., Lanzavecchia A. 2003. A role for Toll-like receptors in acquired immunity: up-regulation of TLR9 by BCR triggering in naive B cells and constitutive expression in memory B cells.Blood. 101:4500–4504 [DOI] [PubMed] [Google Scholar]
- Matsumoto F., Saitoh S., Fukui R., Kobayashi T., Tanimura N., Konno K., Kusumoto Y., Akashi-Takamura S., Miyake K. 2008. Cathepsins are required for Toll-like receptor 9 responses.Biochem. Biophys. Res. Commun. 367:693–699 [DOI] [PubMed] [Google Scholar]
- Yoneyama M., Onomoto K., Fujita T. 2008. Cytoplasmic recognition of RNA.Adv. Drug Deliv. Rev. 60:841–846 [DOI] [PubMed] [Google Scholar]
- Latz E., Schoenemeyer A., Visintin A., Fitzgerald K.A., Monks B.G., Knetter C.F., Lien E., Nilsen N.J., Espevik T., Golenbock D.T. 2004. TLR9 signals after translocating from the ER to CpG DNA in the lysosome.Nat. Immunol. 5:190–198 [DOI] [PubMed] [Google Scholar]
- Matsumoto K., Watanabe N., Akikusa B., Kurasawa K., Matsumura R., Saito Y., Iwamoto I., Saito T. 2003. Fc receptor-independent development of autoimmune glomerulonephritis in lupus-prone MRL/lpr mice.Arthritis Rheum. 48:486–494 [DOI] [PubMed] [Google Scholar]
- Maruyama K., Sugano S. 1994. Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides.Gene. 138:171–174 [DOI] [PubMed] [Google Scholar]
- Park B., Brinkmann M.M., Spooner E., Lee C.C., Kim Y.M., Ploegh H.L. 2008. Proteolytic cleavage in an endolysosomal compartment is required for activation of Toll-like receptor 9.Nat. Immunol. 9:1407–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald S.E., Lee B.L., Lau L., Wickliffe K.E., Shi G.P., Chapman H.A., Barton G.M. 2008. The ectodomain of Toll-like receptor 9 is cleaved to generate a functional receptor.Nature. 456:658–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemth P., Gianni S. 2007. PDZ domains: folding and binding.Biochemistry. 46:8701–8708 [DOI] [PubMed] [Google Scholar]
- Gautier G., Humbert M., Deauvieau F., Scuiller M., Hiscott J., Bates E.E., Trinchieri G., Caux C., Garrone P. 2005. A type I interferon autocrine-paracrine loop is involved in Toll-like receptor-induced interleukin-12p70 secretion by dendritic cells.J. Exp. Med. 201:1435–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napolitani G., Rinaldi A., Bertoni F., Sallusto F., Lanzavecchia A. 2005. Selected Toll-like receptor agonist combinations synergistically trigger a T helper type 1-polarizing program in dendritic cells.Nat. Immunol. 6:769–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmore M.M., DeVeer M.J., Edling A., Oates R.K., Simons B., Lindner D., Williams B.R. 2004. Synergistic activation of innate immunity by double-stranded RNA and CpG DNA promotes enhanced antitumor activity.Cancer Res. 64:5850–5860 [DOI] [PubMed] [Google Scholar]
- Warger T., Osterloh P., Rechtsteiner G., Fassbender M., Heib V., Schmid B., Schmitt E., Schild H., Radsak M.P. 2006. Synergistic activation of dendritic cells by combined Toll-like receptor ligation induces superior CTL responses in vivo.Blood. 108:544–550 [DOI] [PubMed] [Google Scholar]
- Roelofs M.F., Joosten L.A., Abdollahi-Roodsaz S., van Lieshout A.W., Sprong T., van den Hoogen F.H., van den Berg W.B., Radstake T.R. 2005. The expression of toll-like receptors 3 and 7 in rheumatoid arthritis synovium is increased and costimulation of toll-like receptors 3, 4, and 7/8 results in synergistic cytokine production by dendritic cells.Arthritis Rheum. 52:2313–2322 [DOI] [PubMed] [Google Scholar]
- Bafica A., Scanga C.A., Feng C.G., Leifer C., Cheever A., Sher A. 2005. TLR9 regulates Th1 responses and cooperates with TLR2 in mediating optimal resistance to Mycobacterium tuberculosis.J. Exp. Med. 202:1715–1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato A., Linehan M.M., Iwasaki A. 2006. Dual recognition of herpes simplex viruses by TLR2 and TLR9 in dendritic cells.Proc. Natl. Acad. Sci. USA. 103:17343–17348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry A.K., Chen G., Zheng D., Tang H., Cheng G. 2005. The host type I interferon response to viral and bacterial infections.Cell Res. 15:407–422 [DOI] [PubMed] [Google Scholar]
- Zucchini N., Bessou G., Traub S., Robbins S.H., Uematsu S., Akira S., Alexopoulou L., Dalod M. 2008. Cutting edge: Overlapping functions of TLR7 and TLR9 for innate defense against a herpesvirus infection.J. Immunol. 180:5799–5803 [DOI] [PubMed] [Google Scholar]
- Kariko K., Buckstein M., Ni H., Weissman D. 2005. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA.Immunity. 23:165–175 [DOI] [PubMed] [Google Scholar]
- Banchereau J., Pascual V. 2006. Type I interferon in systemic lupus erythematosus and other autoimmune diseases.Immunity. 25:383–392 [DOI] [PubMed] [Google Scholar]
- Ronnblom L., Alm G.V. 2001. An etiopathogenic role for the type I IFN system in SLE.Trends Immunol. 22:427–431 [DOI] [PubMed] [Google Scholar]
- Krieg A.M. 2007. Antiinfective applications of toll-like receptor 9 agonists.Proc. Am. Thorac. Soc. 4:289–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg A.M. 2007. Development of TLR9 agonists for cancer therapy.J. Clin. Invest. 117:1184–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrat F.J., Coffman R.L. 2008. Development of TLR inhibitors for the treatment of autoimmune diseases.Immunol. Rev. 223:271–283 [DOI] [PubMed] [Google Scholar]
- Kobayashi M., Saitoh S., Tanimura N., Takahashi K., Kawasaki K., Nishijima M., Fujimoto Y., Fukase K., Akashi-Takamura S., Miyake K. 2006. Regulatory roles for MD-2 and TLR4 in ligand-induced receptor clustering.J. Immunol. 176:6211–6218 [DOI] [PubMed] [Google Scholar]
- Takahashi K., Shibata T., Akashi-Takamura S., Kiyokawa T., Wakabayashi Y., Tanimura N., Kobayashi T., Matsumoto F., Fukui R., Kouro T., et al. 2007. A protein associated with Toll-like receptor (TLR) 4 (PRAT4A) is required for TLR-dependent immune responses.J. Exp. Med. 204:2963–2976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama M., Kozuka-Hata H., Tasaki S., Semba K., Hattori S., Sugano S., Inoue J., Yamamoto T. 2009. Temporal perturbation of tyrosine phosphoproteome dynamics reveals the system-wide regulatory networks.Mol. Cell. Proteomics. 8:226–231 [DOI] [PubMed] [Google Scholar]
- Harsha H.C., Molina H., Pandey A. 2008. Quantitative proteomics using stable isotope labeling with amino acids in cell culture.Nat. Protoc. 3:505–516 [DOI] [PubMed] [Google Scholar]