Abstract
Repetitive DNA sequences in the immunoglobulin switch μ region form RNA-containing secondary structures and undergo hypermutation by activation-induced deaminase (AID). To examine how DNA structure affects transcription and hypermutation, we mapped the position of RNA polymerase II molecules and mutations across a 5-kb region spanning the intronic enhancer to the constant μ gene. For RNA polymerase II, the distribution was determined by nuclear run-on and chromatin immunoprecipitation assays in B cells from uracil-DNA glycosylase (UNG)–deficient mice stimulated ex vivo. RNA polymerases were found at a high density in DNA flanking both sides of a 1-kb repetitive sequence that forms the core of the switch region. The pileup of polymerases was similar in unstimulated and stimulated cells from Ung−/− and Aid−/−Ung−/− mice but was absent in cells from mice with a deletion of the switch region. For mutations, DNA was sequenced from Ung−/− B cells stimulated in vivo. Surprisingly, mutations of A nucleotides, which are incorporated by DNA polymerase η, decreased 10-fold before the repetitive sequence, suggesting that the polymerase was less active in this region. We propose that altered DNA structure in the switch region pauses RNA polymerase II and limits access of DNA polymerase η during hypermutation.
Class switch recombination is initiated by activation-induced deaminase (AID) in switch (S) regions preceding each CH gene (1). AID acts by deaminating cytosine to uracil in DNA, and the rogue uracil causes mutations and DNA strand breaks by error-prone processing. Mutations of C:G bp could be produced by replication past uracil or an abasic site caused by removal of uracil by uracil-DNA glycosylase (UNG). Mutations can also arise from the combined actions of MSH2-MSH6 mismatch repair proteins and the low fidelity DNA polymerase (pol) η to produce mutations of A:T bp (2). Strand breaks produced by cleavage of abasic sites on both strands by an abasic endonuclease (3) would produce substrates for nonhomologous end joining to different S regions. In the S region preceding the Cμ gene, mutations start downstream of an intronic (Iμ) promoter located in the enhancer (Eμ) region (4), accumulate for 2 kb, and then decrease before the Cμ gene (5). A similar pattern of mutations occurs downstream of intronic promoters in the S regions of other CH genes as well (5). This distribution supports the prevailing hypothesis that AID either travels with the transcription complex or is brought to the single-strand regions formed during transcription (6, 7).
The Sμ DNA sequence is unique in two aspects: it contains an abundance of hot spot motifs, and it has a stable secondary structure that exposes single-strand DNA for AID to act on. The substructure of this region contains a 1-kb highly repetitive sequence, which is comprised of tandem WGCW (W is A or T) motifs and clusters of three to four Gs on the nontranscribed strand. WGCW is a hot spot for AID activity, and the G clusters have been proposed to form secondary structures during transcription, such as R-loops and G-loops containing G-rich RNA and C-rich transcribed DNA (8, 9). Although the nontranscribed strand is mostly single stranded in these structures, the spectrum of mutations in Ung−/− Msh2−/− mice indicates that both strands are targeted for mutation by AID (5). The mechanism by which AID deaminates the transcribed strand containing an RNA-DNA hybrid is unknown, although three theories have been proposed: (a) the DNA upstream of an elongating RNA pol II may be supercoiled and unwound, which would allow AID access to both strands (10); (b) the DNA in R-loops may be collapsed by endogenous RNase H digestion, which would expose single-strand regions on the transcribed strand (11); and (c) single-strand DNA on the transcribed strand could be generated during antisense transcription. Antisense transcripts have been detected at low levels in S regions, but it is not known if they participate in hypermutation (12).
The DNA region upstream of the repetitive sequence has fewer WGCW motifs and G clusters, but it can also form secondary structure and undergo hypermutation (5, 13, 14). Huang et al. (11) have recently mapped the 5′ and 3′ boundaries of R-loops in the Sμ region and found that they begin 600 bp upstream and end 600 bp downstream of the 1-kb repetitive sequence. The existence of R-loops and associated single-strand DNA in the upstream region may also limit the participation of MSH2-MSH6 and DNA pol η during hypermutation, which would reduce the frequency of mutations of A:T bp.
RESULTS AND DISCUSSION
Sμ DNA structure affects the location of RNA pol II molecules
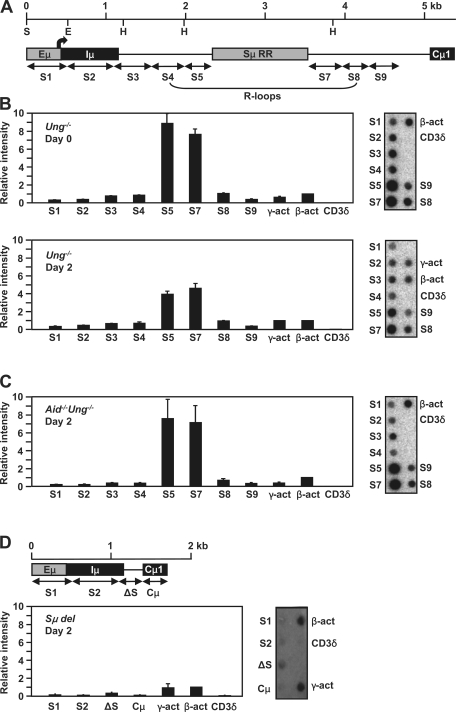
In view of the requirement of transcription for hypermutation and the possibility that AID may associate with the transcription complex, we systematically mapped the position of RNA pol II molecules across the Sμ locus using nuclear run-on and chromatin immunoprecipitation (ChIP) assays. The former measures RNA polymerase activity in a region for a given time, whereas the latter measures the amount of RNA pol II molecules bound to DNA at the instant of cell lysis. B cells from Ung−/− mice were studied because the entire Sμ region can be analyzed without deletions caused by switching (15). The 5-kb Sμ region was divided into a series of contiguous 500-bp probes for run-on analysis (Fig. 1 A, S1–9). However, the 1-kb region containing highly repetitive G:C-rich DNA could not be amplified by our PCR techniques, although we tried a variety of DNA polymerases and conditions (unpublished data). We prepared nuclei from naive B cells or cells stimulated ex vivo with LPS and IL-4 for 2 d and initiated de novo transcription by the addition of ribonucleotides and radioactive UTP for 30 min. The labeled nascent RNA molecules were hybridized to membranes containing DNA probes of the Sμ region, and membrane-bound radioactivity was quantified by phosphorimager analysis (Fig. 1 B). Data were normalized to β-actin transcription and corrected for dTTP content because radiolabeled UTP was incorporated. In nuclei from days 0 and 2, the strongest signals were obtained from probes S5 and S7, which flank the repetitive region and are within the R-loop region. Hybridizations were also performed with single-strand DNA probes made in M13 vectors. There was hybridization to transcribed strand probes but no detectable hybridization to nontranscribed strand probes (unpublished data), which indicates that the majority of RNA transcripts are complementary to the transcribed strand, as expected.
Figure 1.
Location of RNA polymerases by nuclear run-on. (A) A map of the fragments that were analyzed is shown; the Sμ repetitive region (RR) was not analyzed. Position 1 corresponds to nt 135,091 in GenBank/EMBL/DDBJ under accession no. AC073553. The restriction sites ScaI (S), EcoRI (E), and HindIII (H) are depicted. The bent arrow shows the position of the Iμ promoter. The region forming the proposed R-loops is indicated. For graphs in B–D, representative blots for each graph are shown on the right. The intensity of hybridization of labeled nascent RNA to each of the fragments was graphed based on the value of β-actin, which was set as 1. γ-Actin and CD3δ were positive and negative controls, respectively. (B) Ung−/− splenic B cells before and after stimulation with LPS and IL-4. The error bars represent the SD of the values from triplicate blots from one experiment on days 0 and 2 with five mice. Three independent experiments with five mice each were performed and showed similar patterns and intensities. (C) Aid−/−Ung−/− cells after stimulation. Data are from triplicate blots from one experiment with three mice, with error bars showing SD. Three independent experiments with three mice each were performed with similar results. (D) Sμ del cells after stimulation. The ΔS probe corresponded to the 5′ and 3′ ends flanking the 3.7-kb deletion (16). Data are from triplicate blots from one experiment with three mice. Error bars indicate SD.
To determine if the accumulation of polymerases around the repetitive region was caused by mutations initiated by AID, nuclear run-ons were performed in B cells from Aid−/−Ung−/− mice 2 d after stimulation. As seen in Fig. 1 C, there was still an accumulation of polymerases in S5 and S7 in the absence of AID. We also saw a similar pattern of hybridization in nuclei from hybridoma cells that do not express AID (unpublished data). There were twofold fewer transcripts in S5 and S7 in day-2 cells from Ung−/− mice, which express the AID protein, compared with day-0 and day-2 cells from Ung−/− and Aid−/−Ung−/− cells, respectively. This implies that AID affects the level of transcription, but the molecular basis for this is unknown. To see if the pileup of polymerases was directly caused by the Sμ DNA sequence, we performed run-on assays in cells from mice which had a 3.7-kb deletion in Sμ (Sμ del) (16) and have no detectable R-loop formation (11). As shown in Fig. 1 D, after 2 d of stimulation, there was no accumulation of RNA pol II when the Sμ sequence was virtually deleted.
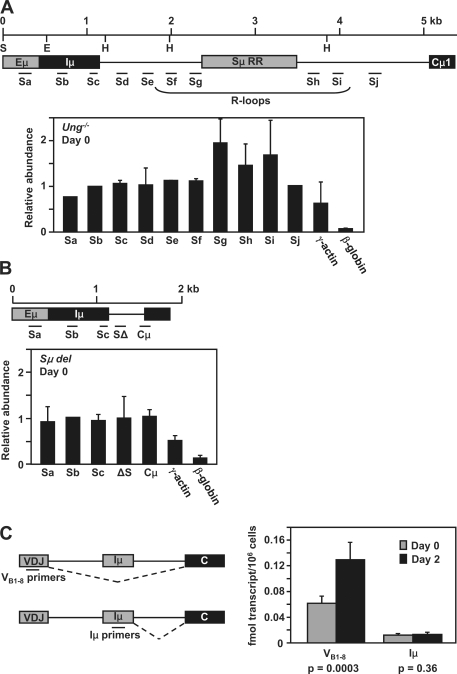
To confirm the nuclear run-on data, the density of RNA pol II molecules was directly assayed by ChIP with anti-RNA pol II antibodies, which were used to enrich polymerase-bound chromatin. After purification, the associated genomic DNA was amplified by PCR with 10 primer sets located across the Sμ region (Fig. 2 A, Sa–j). The polymerase distribution across the region was the same in day-0 and day-2 activated cells. The highest levels of RNA pol II were detected in the amplicons that were closest to the repetitive region, Sg, Sh, and Si, which showed a twofold increase compared with adjacent amplicons. In contrast, there was no accumulation of RNA pol II in mice devoid of the Sμ region (Fig. 2 B). Therefore, both the nuclear run-on and ChIP assays indicate that RNA pol II accumulates close to the repetitive region. This pattern is directly related to the DNA sequence encoding Sμ and is not related to the activation status of the cells or to the presence of AID-dependent mutation. It should be noted that the quantity of RNA pol II flanking the repetitive region differs between the two assays in that the ChIP assay shows a twofold increase, whereas the run-on assay detects a four- to eightfold increase. This could happen if the ChIP assay depicts the loading of RNA pol II at a given instant, whereas the run-on assay shows the activity of the polymerases during a 30-min incubation time, which may produce a more intense signal, as demonstrated in Fig. S1. In both cases, the increase in polymerase density around the repetitive region supports the notion that DNA structure retards the movement of the polymerases.
Figure 2.
RNA analyses by ChIP and absolute qPCR. (A) A map of the position of PCR primers for the ChIP analysis is diagrammed. Data from Ung−/− splenic B cells at day 0 is shown below the map. The abundance of amplified sequences was graphed relative to the input DNA. The data are shown compared with a positive signal from γ-actin and negative signal from β-globin. Error bars represent the SD of values from two independent experiments with three mice each. (B) ChIP analysis in Sμ del splenic B cells on day 0. The position of primers is shown with the data below. Error bars represent the SD of values from one experiment with three mice. (C) Absolute qPCR analysis of VB1-8 and Iμ RNA spliced transcripts from B1-8 cells before and after stimulation. The position of primers and splicing patterns are shown to the left. The data on the right is expressed as fmols of transcripts per million cells. Error bars represent the SD of values from two independent experiments with three mice each. Statistical significance comparing days 0 and 2 was determined by the student's t test.
To evaluate where the RNA pol II molecules initiated transcription, we measured the quantity of transcripts starting at the VDJ versus Iμ promoters. Expression of Iμ transcripts has been shown to be constitutive before and after mitogen stimulation (17), but a direct comparison to VDJ transcripts has not been performed. RNA was isolated from Ung−/− B cells and analyzed as described in the supplemental materials and methods. There was a threefold increase in RNA levels per cell 2 d after activation (Fig. S2 A), which is likely the result of increased transcription of many genes in cells preparing for G1 progression. For instance, transcription of the β-actin gene increased after stimulation as measured by PCR (unpublished data) and Northern assays (Fig. S2 B). Northern blots also showed that Cμ spliced transcripts originating from VDJ promoters were in much greater abundance compared with Iμ spliced transcripts before and after stimulation (Fig. S2 B). We then quantified the amount of transcripts per cell by absolute quantitative PCR (qPCR; Fig. 2 C) using B1-8 mice, which have a homozygous knockin of a rearranged VHJ558 gene (18). Total cellular RNA from cells before and after ex vivo stimulation was measured with VB1-8 and Iμ primers. Although these cells can switch, the primers should detect transcripts from any rearranged CH gene. The results show that VDJ transcripts increased twofold after ex vivo stimulation, whereas Iμ transcript levels remained constant. The data also show that spliced VDJ transcripts were 5–13-fold higher than Iμ transcripts before and after stimulation, respectively. Thus, transcripts initiating from the VDJ promoter may contribute substantially to forming secondary structure in the Sμ region.
Mutations increase before the Sμ repetitive region and decrease afterward
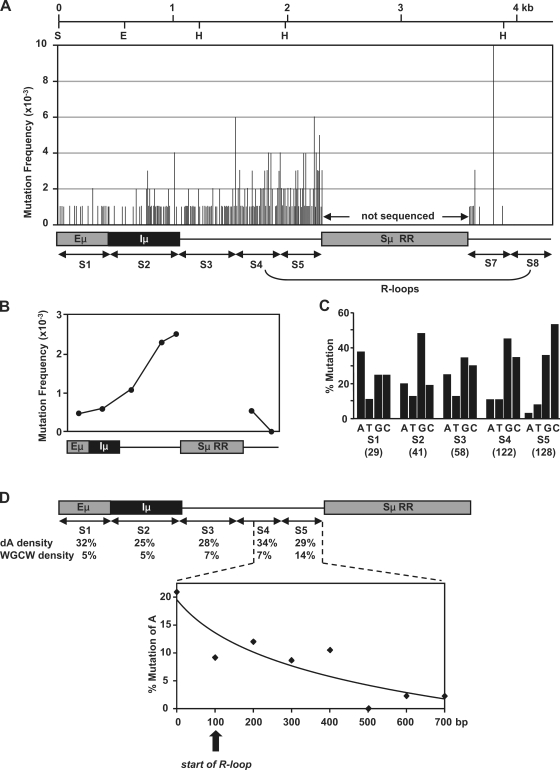
To evaluate the relationship between DNA structure and somatic hypermutation, we sequenced DNA in the Sμ region. This region has been previously sequenced in Ung−/−Msh2−/− mice, and it showed that the DNA sequence upstream of the repetitive region was targeted for deamination by AID, as measured by G:C to A:T transitions (5). However, somatic hypermutation also consists of mutations of A:T bp, which are made during error-prone processing of uracils by MSH2-MSH6 and DNA pol η. To assess where the A:T mutations occurred, we analyzed the pattern in Ung−/− mice, which have a normal frequency of A:T mutations (15), using DNA from Peyer's patch and immunized splenic B cells. A 4.3-kb region spanning Eμ to downstream of the repetitive region was amplified in ∼500-bp segments that corresponded to the location of probes used for the run-on analysis. As shown in Fig. 3 A and Fig. S3, mutations were sparse around the Eμ-Iμ region and increased sequentially up to the repetitive region, with the highest frequency in S5 (Fig. 3 B). Although we could not sequence the repetitive region because of difficulties in PCR amplification, a high frequency of mutations is likely to be found there as well. After the repetitive region, the frequency fell precipitously, with the exception of a hot spot at T in an AGGCTGGGA motif in S7. Because no mutations were seen in S8, we did not analyze sequences further downstream.
Figure 3.
Location of hypermutation in the Sμ region from Ung−/− mice. The region was sequenced in ∼500-bp segments (S1–8). DNA from Peyer's patch and splenic B cells was analyzed; the Sμ repetitive region (RR) was not sequenced. (A) Mutation frequency (mutations/bp) at each nucleotide position. (B) Total mutation frequency for each segment. (C) The indicated number of mutations in parentheses was corrected for nucleotide composition of each fragment, and the percentage of mutation at each base is shown for S1–5. The detailed mutation analysis is recorded in Fig. S4. (D) Mutations of A bases decreased in S4 and S5. The density of dA and WGCW motifs is calculated for each segment. Data for S4 and S5 are then plotted below in 100-bp increments. The vertical arrow shows the proposed start of R-loop formation (11).
Sμ DNA structure may limit activity of DNA pol η
As expected in Ung−/− mice, the vast majority of G:C mutations were transitions (Fig. S4). After correction for nucleotide composition, it was noted that the frequency of mutations of C increased relative to mutations of G in S5 as recorded from the nontranscribed strand (Fig. 3 C), which has also been reported in previous studies of this region (19–21). This is consistent with R-loop structures exposing the nontranscribed strand for cytosine deamination. Furthermore, there was an unexpected decrease in the frequency of mutations of A bases in S4 and S5. As shown in Fig. 3 D, the frequency of mutations of A plummeted 10-fold to just 3% before the repetitive region, even though the germline dA density was high, and the WGCW density increased only twofold. We conclude that DNA pol η, which incorporates mutations of A during somatic hypermutation, may not be active in S4 and S5. The observation that the decrease in mutations of A was greater than the decrease in mutations of T is also consistent with a role for DNA pol η to introduce mutations on the nontranscribed strand (22, 23). The decline in A mutations begins at the proposed start of R-loops (11), implicating DNA structure as a reason for reduced recruitment of pol η. One way this could happen is by the inability of MSH2-MSH6, which interacts with pol η (2), to recognize single-strand DNA within R-loops and thereby not enlist pol η. Alternatively, pol η could be at the site but unable to copy in a repair patch if the template-transcribed strand is stably complexed to RNA. In either case, pol η would have less activity on R-loop structures. A potential benefit to preventing repair and synthesis by pol η would be to retain uracils for removal by UNG and subsequent nicking of the abasic site to induce switch recombination.
Setting up the Sμ locus for AID
Our interpretation is that the DNA sequence of the Sμ region influences the location of RNA pol II and hypermutation. Recently, Conticello et al. (24) identified a protein that interacts with AID and affects hypermutation, class switching, and gene conversion, possibly via the transcriptional spliceosome complex. One speculative model incorporating DNA structure, transcription, and mutation is presented in Fig. S5. RNA pol II molecules move down the DNA and then pause when they encounter the R-loop region. At the other end of the sequence, they are no longer stalled and proceed to the Cμ gene. Why would the polymerases pause? R-loops contain long RNA transcripts forming a stable hybrid with the transcribed DNA strand, and RNA polymerases may slow down when trying to unwind the tight RNA-DNA hybrid. Such blocks in transcription have been reported in vitro when the nontranscribed strand is G rich and the transcribed strand is C rich and can form R-loops (25, 26). If AID associates with the transcription complex, it would be paused as well and have the opportunity to bind to DNA. Along these lines, a recent study has described an in vitro system where AID-generated mutations were increased when T7 RNA polymerase was stalled (27). The decrease in mutation frequency after the repetitive region could occur if prolonged stalling caused AID to dissociate from the transcription complex in an unknown manner, resulting in fewer mutations. Alternatively, the local concentration of AID molecules may become depleted after being bound to DNA in the repetitive sequence. This model could be tested by detecting AID molecules bound to DNA before and after the repetitive region.
We also show that secondary structure affects the activity of DNA pol η because mutations of dA decrease at the start of R-loop formation. Therefore, both the pileup of RNA polymerases and the reduction of dA mutations provide independent and functional measures of DNA secondary structure in Sμ. In contrast, frog Sμ regions, which do not have G-clusters or R-loops, do not switch as efficiently as mouse S regions (28). Thus, the mouse Sμ structure appears to be the perfect substrate to amass mutations and associated strand breaks for switching. The pattern of RNA pol II accumulation preexists before B cell stimulation at day 0, which is most likely caused by R-loop formation from RNA polymerases transcribing membrane IgM. Both the stalling of transcription complexes and presence of single-strand DNA on the nontranscribed strand would give AID molecules optimal access to initiate the mutagenic cascade. This secondary structure may also explain why mutations readily arise in the Sμ region but not in nearby variable regions in B cells stimulated ex vivo.
MATERIALS AND METHODS
Mice.
Ung+/− mice on a mixed C57BL/6J × 129SV background were obtained from H. Krokan (Norwegian University of Science and Technology, Trondheim, Norway). Aid+/− mice on a C57BL/6 background were provided by T. Honjo (Kyoto University, Kyoto, Japan), courtesy of M. Scharff (Albert Einstein College of Medicine, Bronx, NY). B1-8 mice on a C57BL/6 background were obtained from R. Casellas (National Institute of Arthritis and Musculoskeletal and Skin Diseases, National Institutes of Health, Bethesda, MD). The mice were bred in our animal facilities and used at 3–6 mo of age. Spleens from Sμ del mice were provided by A. Khamlichi (16). All animal procedures were reviewed and approved by the National Institute on Aging Animal Care and Use Committee.
Stimulation of splenic B cells.
Spleen cells were treated with ACK lysing buffer (Quality Biological, Inc.) to lyse red blood cells, and B cells were purified by negative selection using anti-CD43 and anti-CD11b antibodies coupled to magnetic beads (Miltenyi Biotec). Cells were cultured at 106 cells/ml and stimulated with 5 µg/ml of Escherichia coli lipopolysaccharide serotype 0111:B4 (Sigma-Aldrich) and 5 ng/ml of recombinant mouse IL-4 (BD). The cells were harvested on days 0 and 2 after stimulation.
Nuclear run-on.
Isolation of nuclei and preparation of nuclear RNA were performed as previously described (29) with some modifications. In brief, ∼5 × 108 cells were pelleted, washed with PBS, resuspended in ice-cold lysis buffer (20 mM Tris-HCl, pH 7.4, 20 mM NaCl, 5 mM MgCl2, and 0.25% vol/vol NP-40), and incubated on ice for 10 min. Nuclei were spun down and resuspended in storage buffer (50 mM Tris-HCl, pH 8.0, 5 mM MgCl2, 0.1 mM EDTA-NaOH, pH 8.0, and 45% vol/vol glycerol). The nuclei in 200-µl aliquots were then mixed with 200 µl of reaction buffer (300 mM KCl, 10 mM MgCl2, and 1 mM each of ATP, CTP, and GTP) plus 500 µCi α-[32P]UTP (3,000 Ci/mmol, 10 mCi/ml; PerkinElmer) and incubated for 30 min at 30°C. Samples were then incubated with 100 U of RNase-free DNase I (Applied Biosystems) for 30 min at 37°C and 20 µg/ml of proteinase K for 45 min at 37°C. Finally, the labeled nascent RNA was purified by Sephadex G-25 column filtration (GE Healthcare) and hybridized to DNA probes as described in the following section.
Preparation of DNA probes and hybridization.
The Sμ region was amplified from genomic DNA with the primers listed in Table S1 and Taq DNA polymerase (Takara Bio Inc.). 1 μg PCR products was then printed on Hybond N+ positively charged nylon membranes (GE Healthcare) as described in the manufacturer's protocol (Bio-Dot Microfiltration apparatus; Bio-Rad Laboratories). The membranes were prehybridized overnight with 3 ml MicroHyb hybridization solution (Invitrogen), 10 µg of Cot DNA (Invitrogen), and 8 µg of poly A DNA (Invitrogen). The labeled RNA was then added in 1 ml of hybridization solution for 24 h in a rotisserie-style incubator at 62°C. The membranes were rinsed in 2× SSC and 0.1% SDS at 62°C, followed by washes in SSC and 0.1% SDS twice at 62°C. Membranes were exposed for 2 d and scanned using a Phosphorimager (GE Healthcare). ImageQuant software (GE Healthcare) was used to convert the hybridization signals into raw intensity values which were analyzed.
ChIP.
ChIP assays were performed as previously described (30), using anti-RNA pol II antibodies (Millipore). The DNA from immunoprecipitation was quantified using Pico green (Invitrogen) and amplified by qPCR using the primers listed in Table S2. Each primer set was examined for >90% amplification efficiency and for the lack of secondary products.
Absolute qPCR.
Total cellular RNA was collected from unstimulated (day 0) and stimulated (day 2) B cells from B1-8 mice. Complementary DNA (cDNA) was produced using 0.5 µg and 1.0 µg RNA and the iScript cDNA synthesis kit (Bio-Rad Laboratories). The Iμ standard was amplified using cDNA template and primers Iμ-5′ 51019 (5′-GCTTGAGTAGTTCTAGTTTCCCCAAACTTAAG-3′) and Iμ-3′ 50615 (5′-GAGTTGGTGGTTGGTCGTACAAGTTG-3′) and cloned into a pGEM-T-easy (Promega) cloning vector. Similarly, the VB1-8 standard was amplified using primers VB1-8-2F (5′-CTGAGCACACAGGACCTCACC-3′) and VB1-8-2R(5′-GGACTCACCTGAGGAGACTGTG-3′). For the standard curve, plasmid standards were linearized with ScaI-HF (New England Biolabs, Inc.) and purified by agarose gel electrophoresis and gel extraction (QIAGEN). qPCR was performed using IQ SYBR green supermix (Bio-Rad Laboratories) with primers Iμ-5′ 50827 (5′-CCAATACCCGAAGCATTTACAGTGAC-3′) and Iμ-3′ 50726 (5′-GTGAAGCCGTTTTGACCAGAATGTC-3′) and with primers VB1-8-3F (5′-GACGAGGCCTTGAGTGGATTG-3′) and VB1-8-3R (5′-CATGTAGGCTGTGCTGGAGG-3′). Each primer set was examined for >90% amplification efficiency and for the lack of secondary products. Reactions were performed using a 7900HT real-time instrument (Applied Biosystems) and data analyzed using SDS 2.3 software (Applied Biosystems).
Hypermutation.
Activated B cells were obtained from the Peyer's patches of three mice, and they were obtained from spleens of four mice immunized with sheep red blood cells for 1 mo and sacrificed 4 d after a boost. Cells were stained with phycoerythrin-labeled antibody to B220 (eBioscience) and fluorescein-labeled peanut agglutinin (PNA; E-Y Laboratories). The cells were separated by flow cytometry, and DNA was prepared from B220+PNA+ cells. Libraries were made by amplifying DNA using the primers listed in Table S1. 100 ng DNA was amplified using Pfx DNA polymerase and PCR enhancer (Invitrogen) in a 50-µl vol using the external primers for 35 cycles. Nested PCR was performed with 5 µl of the first reaction and the internal primers for another 35 cycles. The products were digested, cloned into pBluescript (Agilent Technologies), and sequenced. Only clones with unique patterns of mutation were recorded.
Online supplemental material.
The supplemental materials and methods describe RNA isolation and Northern assays. Fig. S1 presents a model to compare polymerase density by ChIP and run-on assays. Fig. S2 shows the promoter analysis of RNA by Northern blots. Fig. S3 shows the location of mutations in the Sμ region. Fig. S4 displays the spectrum of mutations for each base in the S1–7 regions. Fig. S5 shows a potential model for AID and DNA pol η activity in the Sμ region. Table S1 lists the primers used for sequencing and run-on assays. Table S2 lists the primers used for ChIP analysis. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20082514/DC1.
Acknowledgments
We thank Joshua Erlandsen, William Yang, Jinshui Fan, and Nicholas Durham for assistance and advice and the Comparative Medicine Section for mouse breeding.
This research was supported entirely by the Intramural Research Program of the National Institutes of Health National Institute on Aging.
The authors have no conflicting financial interests.
References
- Stavnezer J., Guikema J.E., Schrader C.E. 2008. Mechanism and regulation of class switch recombination.Annu. Rev. Immunol. 26:261–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson T.M., Vaisman A., Martomo S.A., Sullivan P., Lan L., Hanaoka F., Yasui A., Woodgate R., Gearhart P.J. 2005. MSH2–MSH6 stimulates DNA polymerase eta, suggesting a role for A:T mutations in antibody genes.J. Exp. Med. 201:637–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guikema J.E., Linehan E.K., Tsuchimoto D., Nakabeppu Y., Strauss P.R., Stavnezer J., Schrader C.E. 2007. APE1- and APE2-dependent DNA breaks in immunoglobulin class switch recombination.J. Exp. Med. 204:3017–3026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennon G.G., Perry R.P. 1985. Cμ-containing transcripts initiate heterogeneously within the IgH enhancer region and contain a novel 5′-nontranslatable exon.Nature. 318:475–478 [DOI] [PubMed] [Google Scholar]
- Xue K., Rada C., Neuberger M.S. 2006. The in vivo pattern of AID targeting to immunoglobulin switch regions deduced from mutation spectra in msh2−/− ung−/− mice.J. Exp. Med. 203:2085–2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A., Storb U. 1996. Somatic hypermutation of immunoglobulin genes is linked to transcription initiation.Immunity. 4:57–65 [DOI] [PubMed] [Google Scholar]
- Nambu Y., Sugai M., Gonda H., Lee C.G., Katakai T., Agata Y., Yokota Y., Shimizu A. 2003. Transcription-coupled events associating with immunoglobulin switch region chromatin.Science. 302:2137–2140 [DOI] [PubMed] [Google Scholar]
- Yu K., Chedin F., Hsieh C.L., Wilson T.E., Lieber M.R. 2003. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells.Nat. Immunol. 4:442–451 [DOI] [PubMed] [Google Scholar]
- Duquette M.L., Handa P., Vincent J.A., Taylor A.F., Maizels N. 2004. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA.Genes Dev. 18:1618–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H.M., Storb U. 2004. Activation-induced cytidine deaminase (AID) can target both DNA strands when the DNA is supercoiled.Proc. Natl. Acad. Sci. USA. 101:12997–13002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F.T., Yu K., Balter B.B., Selsing E., Oruc Z., Khamlichi A.A., Hsieh C.L., Lieber M.R. 2007. Sequence dependence of chromosomal R-loops at the immunoglobulin heavy-chain Sμ class switch region.Mol. Cell. Biol. 27:5921–5932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlot T., Li G., Alt F.W. 2008. Antisense transcripts from immunoglobulin heavy-chain locus V(D)J and switch regions.Proc. Natl. Acad. Sci. USA. 105:3843–3848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaoka H., Muramatsu M., Yamamura N., Kinoshita K., Honjo T. 2002. Activation-induced deaminase (AID)–directed hypermutation in the immunoglobulin Sμ region: implication of AID involvement in a common step of class switch recombination and somatic hypermutation.J. Exp. Med. 195:529–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reina-San-Martin B., Difilippantonio S., Hanitsch L., Masilamani R.F., Nussenzweig A., Nussenzweig M.C. 2003. H2AX is required for recombination between immunoglobulin switch regions but not for intra-switch region recombination or somatic hypermutation.J. Exp. Med. 197:1767–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada C., Williams G.T., Nilsen H., Barnes D.E., Lindahl T., Neuberger M.S. 2002. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice.Curr. Biol. 12:1748–1755 [DOI] [PubMed] [Google Scholar]
- Khamlichi A.A., Glaudet F., Oruc Z., Denis V., Le Bert M., Cogne M. 2004. Immunoglobulin class-switch recombination in mice devoid of any S mu tandem repeat.Blood. 103:3828–3836 [DOI] [PubMed] [Google Scholar]
- Li S.C., Rothman P.B., Zhang J., Chan C., Hirsh D., Alt F.W. 1994. Expression of I mu-C gamma hybrid germline transcripts subsequent to immunoglobulin heavy chain class switching.Int. Immunol. 6:491–497 [DOI] [PubMed] [Google Scholar]
- Shih T.A., Roederer M., Nussenzweig M.C. 2002. Role of antigen receptor affinity in T cell-independent antibody responses in vivo.Nat. Immunol. 3:399–406 [DOI] [PubMed] [Google Scholar]
- Zeng X., Negrete G.A., Kasmer C., Yang W.W., Gearhart P.J. 2004. Absence of DNA polymerase η reveals targeting of C mutations on the nontranscribed strand in immunoglobulin switch regions.J. Exp. Med. 199:917–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martomo S.A., Yang W.W., Wersto R.P., Ohkumo T., Kondo Y., Yokoi M., Masutani C., Hanaoka F., Gearhart P.J. 2005. Different mutation signatures in DNA polymerase eta- and MSH6-deficient mice suggest separate roles in antibody diversification.Proc. Natl. Acad. Sci. USA. 102:8656–8661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martomo S.A., Yang W.W., Gearhart P.J. 2004. A role for Msh6 but not Msh3 in somatic hypermutation and class switch recombination.J. Exp. Med. 200:61–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayorov V.I., Rogozin I.B., Adkison L.R., Gearhart P.J. 2005. DNA polymerase eta contributes to strand bias of mutations of A versus T in immunoglobulin genes.J. Immunol. 174:7781–7786 [DOI] [PubMed] [Google Scholar]
- Unniraman S., Schatz D.G. 2007. Strand-biased spreading of mutations during somatic hypermutation.Science. 317:1227–1230 [DOI] [PubMed] [Google Scholar]
- Conticello S.G., Ganesh K., Xue K., Lu M., Rada C., Neuberger M.S. 2008. Interaction between antibody-diversification enzyme AID and spliceosome-associated factor CTNNBL1.Mol. Cell. 31:474–484 [DOI] [PubMed] [Google Scholar]
- Tous C., Aguilera A. 2007. Impairment of transcription elongation by R-loops in vitro.Biochem. Biophys. Res. Commun. 360:428–432 [DOI] [PubMed] [Google Scholar]
- Tornaletti S., Park-Snyder S., Hanawalt P.C. 2008. G4-forming sequences in the non-transcribed DNA strand pose blocks to T7 RNA polymerase and mammalian RNA polymerase II.J. Biol. Chem. 283:12756–12762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canugovi C., Samaranayake M., Bhagwat A.S. 2009. Transcriptional pausing and stalling causes multiple clustered mutations by human activation-induced deaminase.FASEB J. 23:34–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrin A.A., Alt F.W., Chaudhuri J., Stokes N., Kaushal D., Du Pasquier L., Tian M. 2004. An evolutionarily conserved target motif for immunoglobulin class-switch recombination.Nat. Immunol. 5:1275–1281 [DOI] [PubMed] [Google Scholar]
- Cheadle C., Fan J., Cho-Chung Y.S., Werner T., Ray J., Do L., Gorospe M., Becker K.G. 2005. Control of gene expression during T cell activation: alternate regulation of mRNA transcription and mRNA stability.BMC Genomics. 6:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty T., Chowdhury D., Keyes A., Jani A., Subrahmanyam R., Ivanova I., Sen R. 2007. Repeat organization and epigenetic regulation of the DH-Cμ domain of the immunoglobulin heavy-chain gene locus.Mol. Cell. 27:842–850 [DOI] [PubMed] [Google Scholar]