Abstract
Many pathogens that cause human disease infect only humans. To identify the mechanisms of immune protection against these pathogens and also to evaluate promising vaccine candidates, a small animal model would be desirable. We demonstrate that primary T cell responses in mice with reconstituted human immune system components control infection with the oncogenic and persistent Epstein-Barr virus (EBV). These cytotoxic and interferon-γ–producing T cell responses were human leukocyte antigen (HLA) restricted and specific for EBV-derived peptides. In HLA-A2 transgenic animals and similar to human EBV carriers, T cell responses against lytic EBV antigens dominated over recognition of latent EBV antigens. T cell depletion resulted in elevated viral loads and emergence of EBV-associated lymphoproliferative disease. Both loss of CD4+ and CD8+ T cells abolished immune control. Therefore, this mouse model recapitulates features of symptomatic primary EBV infection and generates T cell–mediated immune control that resists oncogenic transformation.
Mice are a preferred species for many avenues of immunological research in vivo. Because of the evolutionary divergence of mouse and man 65 million years ago, however, these two species have inhabited different ecological niches and have been challenged with minimally overlapping groups of pathogens. The human and mouse immune systems, evolving to meet these challenges, have therefore accumulated many differences (1), making genes related to immunity, together with genes involved in reproduction and olfaction, the most divergent between the two species (2).
Pathogens that drive this divergence are those that exclusively infect either humans or mice with a high frequency of population penetration and life-threatening pathology. One such example is EBV, a human γ-herpesvirus that infects >90% of the human adult population. Humans are the only known natural reservoir for EBV, and this virus is associated with malignancies of lymphocyte and epithelial cell origin (3, 4). EBV is thought to cause or contribute to these life-threatening tumors by its ability to induce proliferation and to protect infected cells from apoptosis (5). All EBV-associated lymphomas and carcinomas express latent EBV antigens, whereas lytic EBV particle production does not cause significant pathology. Latent EBV antigen expression differs between the various EBV-associated malignancies. Burkitt lymphoma expresses only the nuclear antigen 1 of EBV (EBNA1) as the sole latent gene product (latency I). In contrast, Hodgkin lymphoma and nasopharyngeal carcinoma express, in addition, one or both latent membrane proteins (LMPs), LMP1 and 2 (latency II). Only in immune-compromised patients, like HIV-infected individuals or transplant recipients, do B cell lymphomas occur that express the five additional EBNA proteins 2, 3A, B, C, and LP (latency III). In addition, all EBV-infected B cells express small nontranslated virally encoded RNAs, including the EBV-encoded RNAs (EBERs). All three types of EBV latencies can also be found in healthy EBV carriers, and this graded EBV protein expression depends on the B cell differentiation stage of the infected cell (6, 7). Healthy virus carriers therefore have already established tumor-associated latent EBV protein expression. Hence, the increased risk of immune-compromised individuals to develop EBV-associated tumors indicates loss of EBV-specific immune control that normally prevents viral tumorigenesis.
T cells constitute the decisive component of EBV-specific immune control against virus-associated malignancies because adoptive transfer of EBV-specific T cell lines can eradicate EBV-associated posttransplant lymphomas (8). During primary infection, CD8+ T cells targeting epitopes of EBV lytic antigens make up the majority of EBV-specific T cells, but T cells targeting latent epitopes can also be detected. Different latency patterns confer different degrees of immunogenicity for recognition by cytotoxic CD8+ and helper CD4+ T cells (9, 10). In healthy EBV carriers, the EBNA3 proteins are the dominant targets of CD8+ T cell responses, whereas EBNA1, 2, and 3C are the most consistently recognized CD4+ T cell antigens. In contrast, the LMPs are subdominant T cell antigens. At least a subset of investigated CD4+ and CD8+ T clones, however, recognize both dominant and subdominant specificities directly on EBV-infected cells. Both CD4+ and CD8+ T cells therefore contribute to immune control against virus-infected cells (11), in addition to maintenance of functional CD8+ T cell memory by CD4+ T cells (12).
The lack of an animal model of EBV infection prevents assignment of a protective value to the known T cell reactivities. Design and evaluation of vaccines against EBV-associated tumors and symptomatic primary infection, however, require knowledge about the degree of protection conferred by different EBV-specific immune responses. Mouse models that partially reconstitute human immune system components after engraftment of hematopoietic progenitor cells (HPCs) are of particular interest to study vaccine candidates and EBV-specific immune responses in vivo. In this respect, three novel models of human immune system reconstitution are being considered. The first one uses BALB/c Rag2−/− γc−/− mice, which reconstitute macrophages, T cells, B cells, natural killer cells, and dendritic cells after neonatal intrahepatic HPC transfer (13, 14). The second model reconstitutes NOD-scid γc−/− mice by intravenous injection of human HPCs (15, 16), which also leads to significant development of human myeloid and lymphoid cells. Finally, the most labor-intensive model is the BLT mouse, which requires implantation of human fetal liver and thymus pieces under the kidney capsule of NOD-scid mice in addition to intravenous HPC injection (17). Immune compartment reconstitution in peripheral blood of BLT mice is very similar to human. However, although signs of primary immune responses were reported in all three of these current mouse models of human immune system reconstitution, the protective value of this immunocompetence and, thus, the potential of these in vivo systems as challenge models for vaccine development against pathogens with exclusive tropism for humans has not been evaluated. Therefore, we analyzed the ability of mice with reconstituted immune system components to exert antigen-specific T cell–mediated control of EBV infection and EBV-associated lymphomas.
RESULTS
Immune reconstitution of NOD-scid γc−/− mice injected with human CD34+ HPCs
To generate mice susceptible to EBV infection and, hence, potentially capable of generating EBV-specific human immune responses in vivo, we compared previously described mouse models for human immune system reconstitution (NOD-scid γc−/− and Rag2−/− γc−/− mice) (13–16). For this purpose we engrafted irradiated newborn Rag2−/− γc−/− and NOD-scid γc−/− mice with human fetal liver–derived CD34+ HPCs. We observed higher levels of overall reconstitution and, in particular, human T and NK cell reconstitution in NOD-scid γc−/− mice (unpublished data), and therefore pursued this model, which we will refer to as hu-NSG mice in the remainder of the text.
We consistently achieved reconstitution of 20–50 hu-NSG mice from the same graft with similar B and T cell frequencies in peripheral blood by 3 mo (Fig. S1 A). Frequencies of human CD45+ cells routinely exceeded 60% of total splenocytes after 4 mo of reconstitution (Fig. S1 B), with the majority (80–90%) being B and T cells (ratios of 2:1 for B to T cells, and 3:2 for CD4+ to CD8+ T cells). Additionally, CD123+ plasmacytoid cells (1–2%) and CD11c+ conventional dendritic cells (1–2%), monocytes (3–5%), and NK cells (2–5%) also engrafted (Fig. S1 B and not depicted). Furthermore, human CD45+ cells and all major subsets of human immune cells were detected in the thymus, mesenteric lymph node, bone marrow, liver, and lung of hu-NSG mice (unpublished data). Compared with the structured architecture in human secondary lymphoid organs like lymph nodes and tonsils, immune cells in the spleens of hu-NSG mice showed a primitive organization into white and red pulp (Fig. S1 C). In line with previous findings, we therefore successfully achieved human multilineage reconstitution in NOD-scid γc−/− mice after neonatal HPC transfer, thus generating a small animal model with the potential to generate human immune responses in vivo.
EBV infection of humanized mice
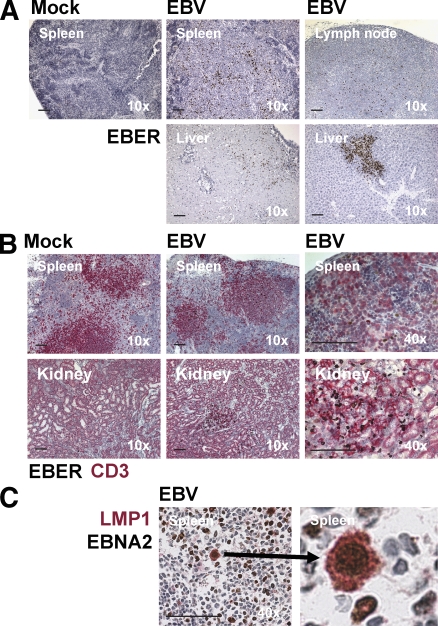
To test the reconstituted human immune system's ability to generate pathogen-specific immune control, we infected hu-NSG mice with EBV, an oncogenic human virus that is efficiently controlled by infected individuals with normal immune function. We chose an infectious dose that would approximate the number of virus particles in 100 µl of saliva from a symptomatic EBV converter (18). After injection of hu-NSG mice with 105 Raji-infecting units (RIU) of purified viral particles intraperitoneally, EBV-infected cells were readily detectable in the spleen 4 wk after infection by in situ hybridization for EBERs. EBER+ cells were not detected in mock-infected animals (Fig. 1 A). At later time points, we also detected EBER+ cells in the lymph nodes and livers of most infected animals (Fig. 1 A). In the spleens of infected animals, EBER+ cells were consistently surrounded by CD3+ T cells (Fig. 1 B). Furthermore, when EBER+ cells were detected in nonlymphoid organs such as liver and kidney, these infected cells were again in close proximity to CD3+ T cells, suggesting that T cells can home to similar peripheral sites as EBV-infected cells in hu-NSG mice (Fig. 1 B). To characterize the latency type of EBV-infected cells, we stained spleen sections for the two EBV-encoded proteins EBNA2 and LMP1. We did not detect any LMP1 single-positive cells, but we found similar frequencies of EBNA2+ compared with EBER+ cells, some of which also expressed LMP1, indicating latency III–type EBV infection (Fig. 1 C). These results indicate that hu-NSG mice establish latent EBV infection, which constitutes the basis of B cell transformation by EBV.
Figure 1.
EBV-infected cells are detected in hu-NSG mice in multiple organs and express EBNA2 and LMP1. (A) EBV-infected cells could be detected in the indicated organs by EBER hybridization. (B) EBER+ cells were surrounded by T cells (CD3+) both in the spleen and after migration to the kidney. (C) EBNA2+ cells coexpressed LMP1 in the spleen (the right panel is a magnification of the left panel). These analyses were performed 4–6 wk after EBV infection. Data are representative of three independent experiments. Bars, 100 µm.
Development of HLA-restricted EBV-specific human T cell responses in infected hu-NSG mice
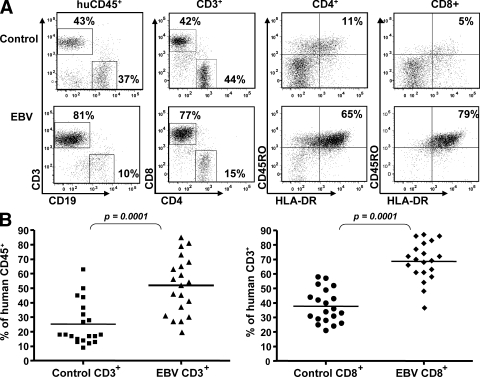
Because we rarely observed tumors after 6 wk of infection in hu-NSG mice, we investigated whether they developed EBV-specific T cell responses, such as those that protect healthy human carriers from EBV-associated malignancies (9, 10). For this purpose, we first analyzed the splenic lymphocyte composition of the infected animals by flow cytometry. We found dramatic expansions of CD3+ T cells among the human CD45+ leukocytes after EBV infection, which correlated with an increase in the percentage of CD8+ cells among the human T cells (Fig. 2 A). On average, there was a statistically significant twofold expansion of splenic CD3+ CD8+ T cells in 10 independent experiments with a total of 40 mice (Fig. 2 B). In addition, in both the CD8+ and the CD4+ T cell compartments, there was a marked-up regulation of HLA-DR and CD45RO surface expression, indicating an activated memory phenotype of the expanded T cells, similar to that seen in humans during symptomatic primary EBV infection (9, 10).
Figure 2.
Expansion of human CD3+ T cells after EBV infection. (A) Splenocytes from control or EBV-infected animals were harvested 6 wk after infection. Frequencies of lymphocyte subsets were determined by flow cytometry. Activation and memory phenotypes of both the CD4+ and CD8+ T cells were monitored by measuring the up-regulation of the HLA-DR and CD45RO surface markers, respectively. Representative data from 10 experiments are shown. (B) Summary of CD3+CD8+ T cell expansion for 40 mice in 10 different experiments. Horizontal bars represent means.
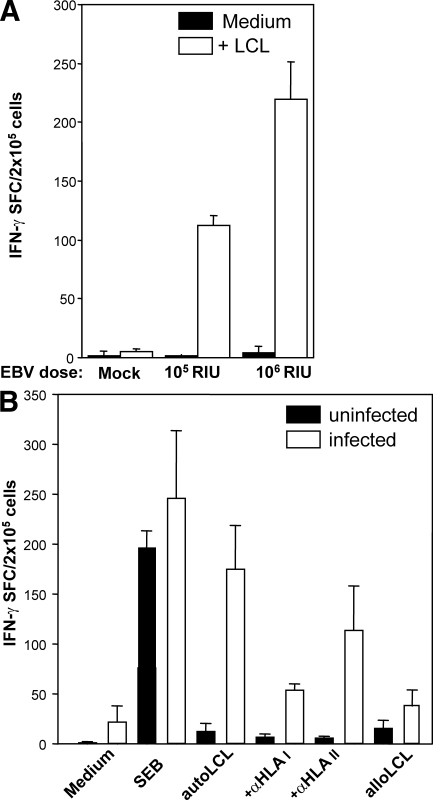
We next examined whether T cells from infected animals would respond to autologous EBV-transformed B cells presenting viral antigens by human MHC molecules. For this purpose, we first established lymphoblastoid cell lines (LCLs) by in vitro infection of B cells derived from a littermate mouse reconstituted with cells from the same HPC donor used for reconstitution of the EBV-infected animals. B cell–depleted splenocytes isolated from control and infected animals were incubated with these autologous LCLs, and IFN-γ secretion was monitored by ELISPOT assays. Autologous LCLs stimulated significant amounts of IFN-γ production, whereas responses to allogeneic LCLs were comparable to background IFN-γ production (Fig. 3, A and B). Animals infected with higher doses of EBV also developed more vigorous EBV-specific T cell responses (Fig. 3 A), indicating that T cells can be primed in a dose-dependent manner in hu-NSG mice. Pretreatment of LCLs with antibodies against HLA-A/B/C, HLA-DR/DP/DQ, or both blocked their recognition (Fig. 3 B and not depicted). These data clearly demonstrate that HLA-restricted, EBV-specific CD8+ and CD4+ human T cells were primed in hu-NSG mice upon infection with EBV.
Figure 3.
Dose-dependent induction of HLA-restricted T cell responses against autologous EBV-transformed B cells in infected hu-NSG mice. (A) Reconstituted NSG mice were infected with 105 or 106 RIU EBV. 6 wk after infection, human B cell–depleted splenocytes were incubated with autologous EBV-transformed B cells (LCLs) to measure EBV-specific IFN-γ secretion using ELISPOT assays. IFN-γ–specific spots per 105 cells are shown for a representative experiment with three mice in each group. One representative out of six experiments is shown. (B) Humanized NSG mice were infected with 106 RIU EBV. 6 wk after infection, splenocytes were harvested from control and infected animals, and T cell reactivity was evaluated by IFN-γ ELISPOT assays under similar conditions as described in A. Staphylococcal enterotoxin B (SEB) superantigen and allogenic LCLs were used as positive and negative controls, respectively. Human HLA restriction was determined using inhibitory antibodies against HLA I and II as indicated. One representative out of two experiments is shown. Data represent means + SD. SFC, spot-forming cells.
Isolation of EBV-specific T cell clones
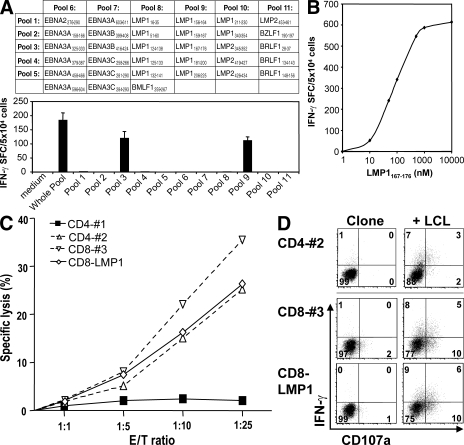
To analyze the peptide epitope specificity and effector functions of the in vivo–primed EBV-specific T cell responses, we isolated splenocytes from hu-NSG mice reconstituted with HLA-A2+ HPCs 10 wk after EBV infection. After labeling with CFSE, these splenocytes were stimulated with either autologous LCLs or a pool of 33 peptides derived from lytic and latent EBV antigens (Fig. 4 A). To enrich for LCL- or peptide-specific cells, CFSElow T cells were sorted after 6 d by flow cytometry, cloned by limiting dilution, and finally retested with either autologous LCLs or the pool of 33 EBV-derived peptides in IFN-γ ELISPOT assays. Three CD8+ T cell clones recognized the library of 33 peptides, and subsequent testing against a matrix of smaller peptide libraries identified the individual peptides recognized by the clones (Fig. 4 A). All three clones recognized the LMP1167-176 peptide (Fig. 4 A), which is recognized by CD8+ T cells in EBV-infected HLA-A2+ individuals (10). Titration of the cognate peptide demonstrated high affinity recognition by the clones (Fig. 4 B).
Figure 4.
Isolation of EBV-specific T cell clones from infected hu-NSG mice. (A) T cell clones were established by limiting dilution cloning from sorted CFSElow T cells of spleens from animals 6 wk after EBV infection. These T cells had proliferated in response to EBV-transformed B cells (LCLs) or EBV-derived peptides. The library of 33 EBV peptides that was used for the initial T cell proliferation was divided into the indicated matrix of peptide pools and used to assess the fine specificity of obtained T cell clones. Reactivity of one out of three CD8+ T cell clones specific for the HLA-A2–restricted peptide LMP1167-176 in IFN-γ ELISPOT is shown. (B) Epitope affinity was determined by cognate peptide titration on LMP1167-176–specific CD8+ T cells in IFN-γ ELISPOT assays. One representative out of two experiments is shown. (C) The cytotoxicity of LMP1167-176–specific (CD8-LMP1) and LCL-specific (#1–3) CD4+ and CD8+ T cell clones against autologous EBV-transformed B cells (LCLs) was assessed by flow cytometric TO-PRO-3-iodide exclusion assays at the indicated effector/target ratios (E/T). One representative out of three experiments is shown. (D) Degranulation and IFN-γ production were evaluated after co-culture with autologous LCLs by flow cytometric surface staining for CD107a and intracellular IFN-γ staining (percentages are shown). One representative out of three experiments is shown.
We also identified CD4+ and CD8+ T cell clones that recognized the autologous LCLs in IFN-γ ELISPOT assays. Subsequently, we compared the cytotoxicity of three of these LCL-specific clones (#1–3) and one of the LMP1-specific CD8+ T cell clones (CD8-LMP1) against autologous LCLs, and found that one of the two CD4+ T cell clones, the LCL-specific CD8+ T cell clone, and the LMP1-specific CD8+ T cell clone lysed autologous LCLs at similar effector/target ratios (Fig. 4 C). These T cells also degranulated and secreted IFN-γ upon LCL recognition (Fig. 4 D). These data indicate that multifunctional cytotoxic EBV-specific T cells are primed in hu-NSG mice and can kill EBV-transformed B cells.
Improved detection of dominant EBV peptide–specific CD8 T cell responses in HLA-A2 transgenic hu-NSG mice
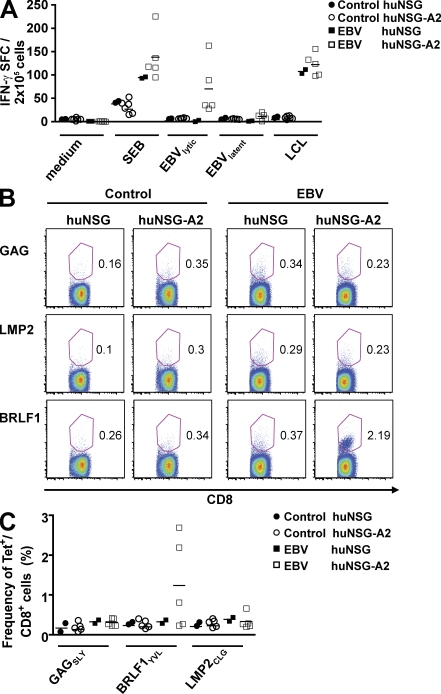
Because we were able to identify EBV-derived peptide epitopes for our isolated T cell clones only occasionally from EBV-infected hu-NSG mice, we aimed to bias EBV-specific T cell recognition to peptide epitopes that dominate EBV-specific immune control in humans by introducing a HLA-A2 transgene into NSG mice. We reconstituted a group of regular NSG mice and NSG mice transgenic for HLA-A2 (NSG-A2) with CD34+ cells from the same HLA-A2+ donor. Human immune cells and, in particular, human CD4+ and CD8+ T cells developed in both groups of mice with similar frequencies and distributions (Fig. S2 A). Furthermore, transgenic expression of HLA-A2 did not influence homeostatic proliferation and survival of T cells (Fig. S2, B and C). We infected hu-NSG and hu-NSG-A2 mice with EBV, and 5 wk after infection we stimulated splenocytes from these mice with two pools of EBV-derived HLA-A2–restricted peptides, one containing 8 lytic epitopes and one containing 12 latent epitopes (Table S1). We detected IFN-γ secretion by ELISPOT after stimulation with the lytic pool in all five EBV-infected hu-NSG-A2 mice, but only in four out of five mice after stimulation with the latent pool (Fig. 5 A). Notably, these latent responses were significantly lower than the lytic responses (11 vs. 70 SFU/2 × 105 cells; P = 0.04). Moreover, no IFN-γ–secreting cells were detected in any of the control mice or in EBV-infected hu-NSG mice reconstituted with HLA-A2+ matching (n = 2) or nonmatching (n = 8) CD34+ cells against the used peptide pools ex vivo. Interestingly, after stimulation with autologous LCLs, splenocytes from both EBV-infected hu-NSG and hu-NSG-A2 mice responded similarly (107 vs. 124 SFU/2 × 105 cells). To detect EBV-specific CD8+ T cells directly, splenocytes were stained with control tetramers (HIV GAGSLY), and two well-characterized EBV tetramers derived from a lytic epitope (BRLF1YVL) and a latent epitope (LMP2CLG), respectively (Fig. 5 B). Although we did not detect any tetramer-positive cells in control hu-NSG and hu-NSG-A2 mice as well as in EBV-infected hu-NSG mice, in three out of five EBV-infected hu-NSG-A2 mice up to 2.7% of the CD8+ T cells stained positive with the BRLF1YVL tetramer (Fig. 5 C). These tetramer-positive cells had an activated phenotype that was exclusively CD45RO+HLA-DR+ (unpublished data). But, in these mice we also did not observe any cells positive for the LMP2CLG tetramer, possibly because their frequency was too low. These experiments suggest that HLA transgenic hu-NSG mice bias primary T cell responses of reconstituted human immune system components toward recognition of EBV-derived peptide epitopes that are dominant during EBV infection in humans.
Figure 5.
Enhanced priming of CD8+ T cell responses against dominant EBV peptides in HLA-A2 transgenic hu-NSG mice. (A) HLA-A2 transgenic and nontransgenic NSG mice were reconstituted with HLA-A2+ CD34+ HPCs from the same donor. 4 wk after EBV infection or mock treatment, splenocytes were restimulated for IFN-γ ELISPOT assays with medium alone, staphylococcal enterotoxin B (SEB) as a positive control, the autologous EBV-transformed B cell line (LCLs), 8 lytic EBV antigen–derived dominant CD8+ T cell epitopes, and 12 latent EBV antigen–derived CD8+ T cell epitopes, which had been defined as dominant CD8+ T cell epitopes in human EBV carriers. The data summarize two independent experiments. (B and C) In parallel, tetramer staining on splenocytes of EBV-infected or control mice was performed ex vivo. Tetramers of HLA-A*0201 with the HIV gag aa 77–85 (GAGSLY), EBV LMP2 aa 426–434 (LMP2CLG), or EBV BRLF1 aa 109–117 (BRLF1YVL) peptides were used in costaining with anti-CD8 and analyzed by flow cytometry (percentages are shown). B shows a representative experiment and C shows the summary of two independent experiments. Horizontal bars represent means.
Disseminated EBV-associated malignancies in T cell–depleted hu-NSG mice
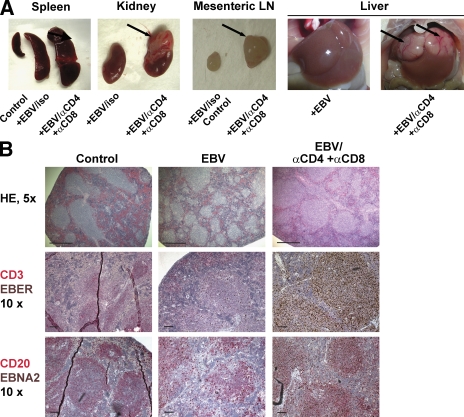
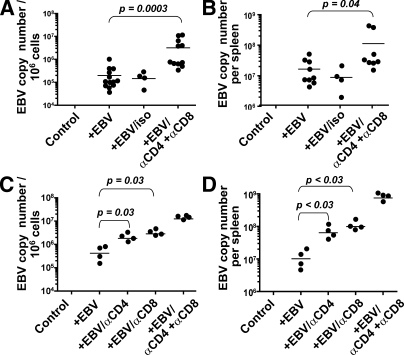
To study the role of these in vivo–primed EBV-specific T cells to control EBV infection and EBV-associated malignancies in hu-NSG mice, we depleted T cells before EBV infection using antibodies against CD4 and CD8 (Fig. S3). 4–5 wk after infection, mice were analyzed for the development of tumors as well as for EBV viral loads. Although we observed small splenic tumors in only 3 out of the 17 EBV-infected animals with or without isotype control antibody injection, all 11 T cell–depleted animals developed disseminated EBV-positive tumors in the spleen, mesenteric lymph node, kidney, and/or liver (Fig. 6 A). Histological analysis showed expansion of white pulp regions in the enlarged spleens of T cell–depleted and EBV-infected mice, which contained almost exclusively EBER+ CD20+ B cells (Fig. 6 B and not depicted). Morphologically these proliferations destroyed the underlying architecture of the tissue. They consisted of a polymorphous cell population composed of atypical large transformed cells (some of which resembled Reed-Sternberg variants), plasmacytoid cells, small lymphocytes, and histiocytes. Areas of coagulative necrosis were often also present. Overall, these proliferations resembled the EBV-associated polymorphic lesions seen in bone marrow and solid organ transplant recipients. The majority of these cells expressed EBNA2, once again indicating EBV latency III (Fig. 6 B). EBER+ cells located in the lymph node, liver, and kidney showed a similar phenotype (Fig. S4 and not depicted). In addition, lytic EBV replication, which was barely detectable during controlled primary EBV infection in vivo, as assessed by immunohistochemical staining for the immediate early EBV antigen BZLF1, was also increased in T cell–depleted hu-NSG mice 4 wk after EBV infection (Fig. S5). Furthermore, viral DNA loads increased significantly in T cell–depleted mice (3.2 × 106 vs. 1.9 × 105 per 106 splenocytes [P = 0.003] and 1.1 × 108 vs. 1.6 × 107 per spleen [P = 0.04]), indicating uncontrolled EBV infection after T cell depletion (Fig. 7, A and B). Notably, viral loads did not significantly differ between EBV-infected hu-NSG and hu-NSG-A2 mice (Fig. S6). To compare the contributions of CD4+ and CD8+ T cells to the T cell–mediated immune control of EBV infection, we then depleted T cell subsets separately (Fig. 7, C and D). Viral titers increased significantly both after the depletion of CD4+ and CD8+ T cells compared with EBV-infected animals (EBV, 6.4 × 105; αCD4, 1.9 × 106; αCD8, 3.2 × 106; and αCD4+αCD8, 1.3 × 107 per 106 splenocytes [P < 0.03]; and EBV, 1.5 × 107; αCD4, 7.2 × 107; αCD8, 1.2 × 108; and αCD4+αCD8, 8.6 × 108 per spleen [P < 0.03]). However, neither the separate depletion of CD4 or CD8 T cells increased the viral titers as much as the depletion of both CD4+ and CD8+ T cells in these experiments. Notably, we observed the occurrence of tumors in the spleens and mesenteric lymph nodes in all four pan–T cell–depleted and in three out of four CD8-depleted animals, but only in one out of four CD4-depleted animals. These results therefore demonstrate that hu-NSG mice are able to prime human EBV-specific T cell responses that protect mice against uncontrolled EBV infection and EBV-associated malignancies. Furthermore, both CD4+ and CD8+ T cells contribute to the successful immune control of EBV in this model.
Figure 6.
Development of EBV-associated tumors in EBV-infected hu-NSG mice after T cell depletion. (A) Disseminated tumors in EBV-infected animals after T cell depletion. T cell depletion in hu-NSG mice, which had been infected with EBV for 4–5 wk, resulted in splenomegaly and EBV-positive tumors either in the kidney, mesenteric lymph node, or liver (arrows). T cell–depleted and EBV-infected hu-NSG mice (EBV/αCD4+αCD8; n = 11) were compared with EBV-infected hu-NSG mice (EBV; n = 13), EBV-infected hu-NSG mice treated with isotype control antibodies (EBV/iso; n = 4), and uninfected hu-NSG mice (control; n = 11). Representative images are shown. Data summarize three independent experiments. (B) Immunohistological characterization of representative spleen sections of T cell–depleted and EBV-infected, EBV-infected and mock-treated, or uninfected hu-NSG mice. Splenic architecture was assessed by hematoxylin and eosin staining (HE), EBV-infected cells were identified by either EBER in situ hybridization (EBER) or staining with EBNA2-specific antibodies (EBNA2), and T and B cell content was characterized by CD3- and CD20-specific antibody staining, respectively. Bars: (HE) 500 µm; (CD3/EBER and CD20/EBNA2) 100 µm.
Figure 7.
Elevated viral loads in T cell–depleted and EBV-infected hu-NSG mice. (A) Splenic EBV loads were determined by quantitative real-time PCR 4 wk after EBV infection. Viral titers were calculated from three independent experiments with a total of 39 animals. No EBV titers were detected in uninfected hu-NSG mice (control). Samples were analyzed at least in duplicates, and statistical significance was calculated using the Mann-Whitney U test. (B) Total splenic EBV loads were determined by multiplying splenic viral loads determined as in A with total splenocyte numbers determined by counting. Results are shown for seven control mice, nine EBV-infected mice, four EBV-infected and isotype control antibody–treated mice, and eight T cell–depleted and EBV-infected mice. Statistical significance was calculated using the Mann-Whitney U test. (C) EBV episome copy numbers in 106 splenocytes were determined in EBV-infected hu-NSG mice after CD4+ (αCD4) and CD8+ (αCD8) T cell single depletions, as well as double depletion (αCD4+αCD8). Composite data of two independent experiments are shown and were statistically analyzed using the Mann-Whitney U test. (D) Total viral loads per spleen were calculated from the viral copy numbers per 106 cells multiplied by the total counted splenocyte numbers. Statistical significance was assessed with the Mann-Whitney U test. Horizontal bars represent means.
DISCUSSION
In this paper, we describe the first mouse model with protective immune control of the human persistent and oncogenic EBV. We demonstrate that primary T cell responses stabilize EBV load at high levels and prevent the development of EBV-associated malignancies. We propose that NOD-scid γc−/− mice with reconstituted human immune system components provide a challenge model for testing the efficacy of vaccines against this human oncogenic virus, and for the characterization of innate and adaptive immune responses during primary EBV infection.
Although no similarly oncogenic γ-herpesviruses of the EBV-containing genus Lymphocryptoviridae (lymphocryptoviruses) has been identified in rodents to date (19), priming of immune responses to human (EBV) or closely related monkey lymphocryptoviruses has been observed after infection in rhesus macaques (20, 21), cottontop tamarins (22), and in the two other novel models of immunocompetent human immune system reconstitution in mice (13, 17). Cottontop tamarins were able to prime MHC class II–restricted CD4+CD8+ T cells with cytotoxicity against EBV-transformed B cells (22). In rhesus macaques, strong cytotoxic and IFN-γ–secreting T cell responses against the monkey virus homologues of the EBNA1 antigen and the immediate early lytic EBV antigen BZLF1 were consistently detected in infected animals (20, 21). In BLT mice, EBV infection elicited low levels of IFN-γ–secreting, HLA-restricted T cell responses against autologous EBV-transformed B cells (17), and in reconstituted BALB/c Rag2−/− γc−/− mice, T cell proliferation against autologous EBV-transformed B cells was detected after EBV infection (13). None of these animal models, however, has investigated the protective value of EBV-specific primary T cell responses in vivo, and the level of IFN-γ–producing EBV-specific T cell responses was also about 5- to 10-fold lower than in the present study. In addition, we report that EBV infection in hu-NSG mice elicited cytotoxic- and epitope-specific primary T cell responses with protective effector functions (9).
These primary immune responses control EBV infection at high levels of viral load, with massive expansion and activation of the CD8+ T cell compartment. These features are reminiscent of symptomatic primary EBV infection, called infectious mononucleosis (IM). Such a phenotype seems to be even more pronounced in hu-NSG mice, because they carry a 10-fold elevated viral load in their splenocytes, compared with ∼104 viral DNA copies per 106 peripheral blood mononuclear cells in IM patients (18, 23). Of course, this assumes that the frequency of EBV-infected cells is similar in peripheral blood and spleen during IM, as has been shown for healthy virus carriers (24). Therefore, the reconstituted human immune system in hu-NSG mice has difficulty controlling EBV infection, similar to chronic active EBV infection in bone marrow transplantation recipients. Indeed, approximately 105 viral DNA copies per 106 peripheral blood mononuclear cells have been observed in >50% of chronic active EBV patients, and these also occasionally develop tumors (25, 26). Nevertheless, the reconstituted human immune system protects most infected mice from EBV-associated malignancies. In contrast, T cell–depleted mice resemble, with their 10-fold increase in EBV viral loads and the dissemination of EBV-induced B cell lymphomas to various tissues, EBV-associated lymphoproliferative disease in transplant recipients (8, 27). In addition, in both immunocompetent and T cell–depleted mice, we primarily detect coexpression of EBER, EBNA2, and LMP1 in EBV-infected cells. This indicates latency III–type cells, which also form the basis of EBV-associated lymphoproliferative disease in immune-compromised patients. Furthermore, these lesions morphologically resemble those seen in the organ transplant patient population (28). Therefore, EBV-infected hu-NSG mice control EBV infection at high viral loads with hallmarks of IM and develop EBV-associated lymphoproliferative disease after T cell depletion.
Both CD8+ and CD4+ T cells contribute to the control of viral infection. In the case of EBV, in vitro studies have demonstrated that CD8+ T cells mainly kill infected cells, whereas CD4+ T cells provide help for CD8+ T cells but can also directly target infected cells (9). Based on the results of our in vivo experiments, we conclude that CD4+ T cells contribute to the control of early EBV infection but are not able to restrict viral replication efficiently in the absence of CD8+ T cells. However, viral titers are further elevated upon additional depletion of CD8+ T cells in addition to CD4+ T cells, suggesting that CD4+ T cells can mediate some immune control of EBV (11). The inability of CD8+ T cells to control EBV infection on their own might reflect the requirement for CD4+ T cell help during primary EBV infection (12). This analysis demonstrates the usefulness of our in vivo model, which allows for the first time to dissect protective mechanisms of human lymphocyte compartments in vivo.
Although we demonstrate protective primary immune responses against EBV in human immune system–reconstituted mice, there are several limitations to this model. The most striking is probably the lack of germinal center formation and difficulty in developing efficient as well as class-switched humoral immune responses. Although we found low concentrations of human IgG accumulating in the plasma of reconstituted mice over time, we were unable to detect EBV-specific IgG or IgM responses against the viral capsid antigen in EBV-infected mice (unpublished data). We nevertheless cannot exclude that humoral responses might develop against other EBV antigens and perhaps at later time points of infection, as we have observed for EBNA1 in an experimental vaccine study using the same mouse model (29). Indeed, IgM responses against the lytic EBV antigen BFRF3 were recently reported after 6 wk of EBV infection in a subset of NOD-scid γc−/− mice with similar reconstitution of human immune compartments (30). Human B cell responses seem to be weak and slow in their development in hu-NSG mice. In addition, because more restricted expression patterns of EBV latent antigens have only been found in germinal center B cells of healthy virus carriers (6) and because latency I/II tumors are thought to originate from EBV-infected centrocytes or centroblasts (31), the study of EBV-associated Burkitt and Hodgkin lymphomas and T cell responses against these malignancies might be difficult in our model. Latency II tumors, however, have been observed in EBV-infected and human B cell–reconstituted NOD-scid mice, suggesting that these hosts allow signaling for germinal center formation. Because these mice are unable to reconstitute human T cells after HPC transfer alone (32), immunological studies are not possible in this model. Encouragingly, however, latency I/II patterns were recently also described in EBV-infected BALB/c Rag2−/− γc−/− mice with reconstituted human immune system components (33), suggesting that the mode of EBV infection and/or of the developing EBV-specific immune control might allow different latent EBV infections even in mice with compromised germinal center development. Furthermore, the EBV-specific T cell response in hu-NSG mice seems to favor subdominant EBV-derived peptide epitopes (9), and we were unable to detect T cells of in humans dominant specificities ex vivo. A possible explanation for this might be the suboptimal selection of human T cells and their TCRs on mouse thymic epithelial cells and on human bone marrow–derived cells. Even so, others and we have reported that human T cells recognize EBV-infected B cells after EBV infection of hu-NSG mice (this study and reference 30), and this recognition can be blocked with antibodies against human MHC molecules, the selection of EBV-specific T cells on human bone marrow–derived cells and by H2 molecules on mouse stromal cells seems to favor different affinities and specificities than those observed in humans with matching MHC type. This shortcoming of dominant EBV epitope recognition during infection in NSG mice can be overcome by introducing HLA transgenes, such as HLA-A2 in our case, into this mouse background. Accordingly, we were able to detect latent and lytic EBV antigen–specific T cell responses against dominant peptide epitopes from EBV-infected hu-NSG-A2 mice ex vivo. Interestingly and similar to human EBV carriers, lytic EBV-specific T cells were detected with nearly 1 log higher frequencies than latent EBV antigen specificities (34). Therefore, HLA transgenes seem to overcome one of the limitations of human immune responses in hu-NSG mice, and allow this immunocompetent small animal model with human immune system components to develop protective T cell responses against EBV infection with similar specificities to human virus carriers.
Because EBV-specific T cells are considered to be the cornerstone of immune control against this oncogenic and persistent γ-herpesvirus (9, 10), we propose to further characterize the innate and adaptive immune responses that lead to this T cell–based immune control. We also plan to evaluate vaccine candidates for eliciting these protective T cell responses against EBV and other pathogens with exclusive tropism for the human hematopoietic lineage. This includes HIV, which has been shown to establish infection in mice with reconstituted human immune system components (35–42).
MATERIALS AND METHODS
Preparation of humanized mice.
NOD/LtSz-scid IL2Rγnull (NOD-scid γc−/−) mice were obtained from the Jackson Laboratory and raised under specific pathogen-free conditions. The NOD.Cg-Prkdcscid Il2rgtm1Wjl Tg(HLA-A2.1)1Enge/Gck/Roly (NSG-A2) mouse was derived from crossing NOD.Cg-Prkdcscid Tg(HLA-A2.1) Enge/Dvs mice (a gift from D. Serreze, The Jackson Laboratory, Bar Harbor, ME) with NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice at Memorial Sloan-Kettering Cancer Center in the laboratory and with the generous support of R. O'Reilly (Memorial Sloan-Kettering Cancer Center, New York, NY). Brother and sister mating from the F2 offspring was performed with mice identified by PCR to be positive for both HLA-A*0201(A2+) and the IL-2Rγ KO. Both heterozygous and homozygous NSG-A2 mice were used in these experiments. Human fetal liver was obtained from Advanced Bioscience Resources. The tissue was minced and treated with 2 mg/ml collagenase D (Roche) in HBSS with CaCl2/MgCl2 for 30 min at room temperature, followed by filtering through 70-µm nylon cell strainers (BD). CD34+ human hematopoietic stem cells (HSCs) were isolated using the Direct CD34+ Progenitor Cell Isolation Kit (Miltenyi Biotec). 2–5-d-old NSG mice were irradiated with 100 cGy and injected intrahepatically with 1–3 × 105 CD34+ HSCs 6 h after irradiation. The mice were bled 10–12 wk after engraftment, and peripheral lymphocytes were analyzed by FACS, as described previously (15, 16) to check for the reconstitution of the human immune system. Animal protocols were approved by the Institutional Animal Care and Use Committee of the Rockefeller University.
EBV infection of mice and depletion of T cells.
Reconstituted mice were infected with EBV at different infectious doses ranging from 105 to 106 RIU by intraperitoneal injection. In select experiments, human CD4+ and CD8+ cells were depleted before EBV infection by intraperitoneal injection of 100 µg OKT-4 and 50 µg OKT-8 antibodies (BioLegend) on three consecutive days. To deplete T cells for the duration of the experiment, the same injection regimen was repeated 2 wk later.
Analysis of EBV-specific T cell responses.
4–10 weeks after infection, mice were sacrificed and EBV-specific T cell responses were analyzed using an IFN-γ ELISPOT assay, as previously described (43). In brief, splenocytes were depleted of human CD19+ cells and mouse CD45+ using anti-CD19 and anti-CD45 microbeads (Miltenyi Biotec). The negative fraction after the depletion was stimulated with autologous LCLs at a ratio of 1:4 for 18 h. Spots were counted with an ELISPOT reader (Autoimmun Diagnostika GmbH). In blocking experiments, LCL recognition by T cells was blocked by preincubation of the target cells with 10 µg/ml anti–HLA-A/B/C (clone W6/32; eBioscience), anti–HLA-DR/DP/DQ (clone Tü39; BD), and a combination thereof.
Cloning of peptide- and LCL-specific T cells.
10 wk after infection, mice were sacrificed and splenocytes were isolated. Cells were then labeled with 0.5 µM CFSE (Invitrogen) and cultured in complete medium containing a library of 33 EBV peptides (1 µM each) or autologous irradiated LCLs at a ratio of 5:1, respectively. On day 6, CFSElow CD3+ cells were sorted by flow cytometry and cloned by limiting dilution (44). After initial expansion, individual clones were screened by IFN-γ ELISPOT assays after restimulation with the library of 33 EBV peptides or autologous irradiated LCLs, respectively. Individual clones that recognized the library were further tested against a matrix of peptide subpools (Fig. 4 A) to identify the specific cognate peptides.
Cytotoxicity assay.
To evaluate the cytotoxic activity of T cell clones against the autologous LCLs, cytotoxicity assays were performed as previously described (45). In brief, target cells were labeled with PKH26 (Sigma-Aldrich) and incubated with T cell clones at the effector/target ratios. Cells were harvested after 4 h; TO-PRO-3-iodide (Invitrogen), a membrane-impermeable DNA stain, was added to each culture (0.5 µM final concentration); and cells analyzed by flow cytometry. Background and maximum TO-PRO-3-iodide stainings were obtained by incubation of target cells with medium and detergent, respectively. The percent specific lysis was calculated as (% TO-PRO-3-iodide+PKH26+ cells in effector/target cell co-culture − TO-PRO-3-iodide+PKH26+ cells in medium)/(% TO-PRO-3-iodide+PKH26+ cells in detergent − TO-PRO-3-iodide+PKH26+ cells in medium) × 100%.
Degranulation assay.
To characterize the multifunctionality of EBV-specific T cell clones, we stimulated T cell clones at an effector/target ratio of 5:1 with autologous LCLs for 6 h in the presence of anti-CD107 antibody. To detect spontaneous degranulation and cytokine production, a control without LCL was included. After 1 h, 1 µg/ml monensin (Sigma-Aldrich) was added to all samples. At the end of the incubation, cells were fixed, permeabilized, stained with an anti–IFN-γ antibody, and analyzed by flow cytometry.
Analysis of homeostatic T cell proliferation and survival using 5-ethynyl-2′-deoxyuridine (EdU) and annexin-V.
Reconstituted mice were injected once intraperitoneally with 100 µg EdU and sacrificed 24 h later. For EdU staining, cells were stained first for cell-surface antigens and then analyzed for EdU incorporation using the Click-iT EdU AF488 Cell Proliferation Assay Kit (Invitrogen) according to the manufacturer's instruction. For annexin-V staining, cells were stained first for cell-surface antigens and then stained with annexin-V and 7-AAD using the PE Annexin V Apoptosis Detection Kit (BD) according to the manufacturer's instructions.
Quantification of EBV viral loads by quantitative real-time PCR.
Splenic EBV viral DNA load was quantified by real-time PCR using a TaqMan PCR kit and a sequence detector (model 7900; Applied Biosystems). DNA was extracted using the Tissue and Blood DNA kit (QIAGEN) or the Wizard SV Genomic DNA purification system (Promega), according to the manufacturer's protocol. A region from the BamHI W fragment of EBV was amplified using primers 5′-GGACCACTGCCCCTGGTATAA-3′ and 5′-TTTGTGTGGACTCCTGGGG-3′, and detected with the fluorogenic probe FAM-TCCTGCAGCTATTTCTGGTCGCATCA-TAMRA. The human bcl-2 gene was amplified using primers 5′-CCTGCCCTCCTTCCGC-3′ and 5′-TGCATTTCAGGAAGACCCTGA-3′, and detected with the fluorogenic probe FAM-CTTTCTCATGGCTGTCC-TAMRA. The EBV BamHI W fragment copy number per cell was calculated using the formula N = 2 × W/B, where N is the EBV BamHI W copy number/cell, W is the EBV BamHI W copy number, and B is the bcl-2 copy number. All samples were tested in triplicates.
Immunohistochemistry.
Immunohistochemical staining was performed on formalin-fixed, paraffin-embedded tissue sections. The antibodies used in this study included CD20 (clone L26), CD8 (clone C8/144B), CD45 (clones 2B11 and PD7/26), CD68 (clone PGM1), EBNA2 (clone PE2), LMP1 (clone CS1-4; Dako), BZLF1 (clone BZ1; Santa Cruz Biotechnology, Inc.), CD3 (clone SP7; Thermo Fisher Scientific), CD21 (clone 2G9), and CD56 (ERIC-1; Novocastra). In situ hybridization for EBV was performed using an EBER probe (Vision Biosystems). Double immunohistochemical staining and dual in situ hybridization with immunohistochemical staining was performed using the Bond Max Autostainer (Leica). Formalin-fixed, paraffin-embedded tissue sections were deparaffinized, and endogenous peroxidase was inactivated. For the first antibody, antigen retrieval was performed using either the Bond Epitope Retrieval Solution 1 (ER1) or the Bond Epitope Retrieval Solution 2 (ER2) at 99–100°C for 20–30 min. After retrieval, the sections were incubated sequentially with the primary antibody for 25 min, the postprimary antibody for 15 min, and the polymer for 25 min (Bond Polymer Detection System; Vision Biosystems), followed by colorimetric development with diaminobenzidine (DAB; Vision Biosystems). For the subsequent staining with the second antibody, the sections were heated in either ER1 or ER2 at 99–100°C for 20–30 min, followed by blocking of endogenous alkaline phosphatase using Dual Endogenous Enzyme Block (Dako). The sections were then sequentially incubated with the second primary antibody, biotinylated link, and streptavidin–alkaline phosphatase (LSAB 2 System-AP; Dako) for 25, 15, and 30 min, respectively, followed by red chromagen development with permanent red (Dako). With respect to dual in situ hybridization–immunohistochemistry, in situ hybridization was performed first according to the manufacturer's instructions (Vision Biosystems) with colorimetric development using DAB, followed by immunostaining as described for the second antibody. Single immunohistochemical staining was performed as described for the first primary antibody.
Production of GFP-expressing EBV.
GFP-expressing EBV was produced as previously described (46). In brief, the EBV lytic cycle was induced in EBV-positive AGS cells by addition of PMA and sodium butyrate. EBV was further purified by ultracentrifugation over a 25% sucrose step gradient and subsequently titered on Raji cells. GFP+ cells were counted 2 d later and titers were calculated in RIU.
Tetramer staining.
To stain CD8+ T cells, soluble tetrameric complexes of HLA-A*0201–peptide were produced using the methods of Busch et al. (47) with the following peptides: HIV gag aa 77–85 (SLYNTVATL), EBV LMP2 aa 426–434 (CLGGLLTMV), and EBV BRLF1 aa 109–117 (YVLDHLIVV). For staining with these reagents, hu-NSG splenocytes were incubated for 30 min at 37°C with 0.5 µg of HLA-A*0201–peptide tetramers (PE-conjugated) and washed. The cells were stained with anti-CD8 mAb conjugated with FITC (BD) for 30 min at 4°C and washed. The samples were analyzed in a flow cytometer.
Statistical analysis.
Statistical analyses were performed with the paired two-tailed Student t test or the Mann-Whitney U test, as indicated in the figures. The p-value of significant differences is reported. Plotted data represent means + SD unless otherwise stated.
Online supplemental material.
Fig. S1 shows that human immune cells reconstitute NOD-scid γc−/− mice injected with human fetal liver–derived CD34+ HPCs. Fig. S2 demonstrates that transgenic expression of HLA-A2 does not affect overall reconstitution, homeostatic T cell proliferation, and T cell survival. Fig. S3 documents antibody-mediated depletion of human T cells in hu-NSG mice. Fig. S4 shows the development of EBV-associated tumors in nonlymphoid and secondary lymphoid organs after depletion of human T cells in EBV-infected hu-NSG mice. Fig. S5 demonstrates detection of EBV lytic replication in human B cells in the spleen of hu-NSG mice. Fig. S6 documents that transgenic expression of HLA-A2 does not significantly influence immune control of EBV infection in humanized mice. Table S1 lists the EBV-derived peptides used in this study. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20081720/DC1.
Acknowledgments
We thank Dr. R. Steinman for critically reading the manuscript, and Dr. R. O'Reilly for his generous support to derive the NOD.Cg-Prkdcscid Il2rgtm1Wjl Tg(HLA-A2.1)1Enge/Gck/Roly mouse strain by breeding.
C. Münz is supported by the Arnold and Mabel Beckman Foundation, the Alexandrine and Alexander Sinsheimer Foundation, the Burroughs Wellcome Fund, the Dana Foundation's Neuroimmunology program, the National Cancer Institute (grants R01CA108609 and R01CA101741), and the National Institute of Allergy and Infectious Diseases (grant RFP-NIH-NIAID-DAIDS-BAA-06-19). C. Münz and C.M. Rice received research funds from the Foundation for the National Institutes of Health (Grand Challenges in Global Health), and an Institutional Clinical and Translational Science Award (to the Rockefeller University Hospital). C. Münz, C.M. Rice, A. Chadburn, and J.W. Young are supported by the Starr Foundation. J.W. Young is supported by the National Cancer Institute (grants R01CA083070 and P01CA23766), by Mr. W.H. and Mrs. A. Goodwin of the Commonwealth Cancer Foundation for Research, and by the Experimental Therapeutics Center of Memorial Sloan-Kettering Cancer Center. T. Strowig is a recipient of a predoctoral fellowship from the Boehringer Ingelheim Foundation. C.M. Rice and A. Ploss are also supported by the Greenberg Medical Research Institute, and A. Ploss is a recipient of a Kimberley Lawrence-Netter Cancer Research Discovery Fund award. J. Sashihara and J.I. Cohen are supported by the intramural research program of the National Institute of Allergy and Infectious Diseases, and J. Sashihara is also supported by the Japanese Herpesvirus Infection Forum.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used: EBER, EBV-encoded RNA; EBNA, nuclear antigen of EBV; HPC, hematopoietic progenitor cell; IM, infectious mononucleosis; LCL, lymphoblastoid cell line; LMP, latent membrane protein; RIU, Raji-infecting units.
References
- Mestas J., Hughes C.C. 2004. Of mice and not men: differences between mouse and human immunology.J. Immunol. 172:2731–2738 [DOI] [PubMed] [Google Scholar]
- Waterston R.H., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., et al. 2002. Initial sequencing and comparative analysis of the mouse genome.Nature. 420:520–562 [DOI] [PubMed] [Google Scholar]
- Young L.S., Rickinson A.B. 2004. Epstein-Barr virus: 40 years on.Nat. Rev. Cancer. 4:757–768 [DOI] [PubMed] [Google Scholar]
- Cohen J.I. 2000. Epstein-Barr virus infection.N. Engl. J. Med. 343:481–492 [DOI] [PubMed] [Google Scholar]
- Kelly G.L., Milner A.E., Baldwin G.S., Bell A.I., Rickinson A.B. 2006. Three restricted forms of Epstein-Barr virus latency counteracting apoptosis in c-myc-expressing Burkitt lymphoma cells.Proc. Natl. Acad. Sci. USA. 103:14935–14940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babcock G.J., Hochberg D., Thorley-Lawson A.D. 2000. The expression pattern of Epstein-Barr virus latent genes in vivo is dependent upon the differentiation stage of the infected B cell.Immunity. 13:497–506 [DOI] [PubMed] [Google Scholar]
- Hochberg D., Middeldorp J.M., Catalina M., Sullivan J.L., Luzuriaga K., Thorley-Lawson D.A. 2004. Demonstration of the Burkitt's lymphoma Epstein-Barr virus phenotype in dividing latently infected memory cells in vivo.Proc. Natl. Acad. Sci. USA. 101:239–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk S., Rooney C.M., Heslop H.E. 2005. Post-transplant lymphoproliferative disorders.Annu. Rev. Med. 56:29–44 [DOI] [PubMed] [Google Scholar]
- Hislop A.D., Taylor G.S., Sauce D., Rickinson A.B. 2007. Cellular responses to viral infection in humans: lessons from Epstein-Barr virus.Annu. Rev. Immunol. 25:587–617 [DOI] [PubMed] [Google Scholar]
- Khanna R., Burrows S.R. 2000. Role of cytotoxic T lymphocytes in Epstein-Barr virus-associated diseases.Annu. Rev. Microbiol. 54:19–48 [DOI] [PubMed] [Google Scholar]
- Heller K.N., Gurer C., Munz C. 2006. Virus-specific CD4+ T cells: ready for direct attack.J. Exp. Med. 203:805–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan M.J. 2004. Helping the CD8+ T-cell response.Nat. Rev. Immunol. 4:595–602 [DOI] [PubMed] [Google Scholar]
- Traggiai E., Chicha L., Mazzucchelli L., Bronz L., Piffaretti J.C., Lanzavecchia A., Manz M.G. 2004. Development of a human adaptive immune system in cord blood cell-transplanted mice.Science. 304:104–107 [DOI] [PubMed] [Google Scholar]
- Gimeno R., Weijer K., Voordouw A., Uittenbogaart C.H., Legrand N., Alves N.L., Wijnands E., Blom B., Spits H. 2004. Monitoring the effect of gene silencing by RNA-interference in human CD34+ cells injected into newborn RAG2−/− gamma common−/− mice: functional inactivation of p53 in developing T cells.Blood. 104:3886–3893 [DOI] [PubMed] [Google Scholar]
- Ishikawa F., Yasukawa M., Lyons B., Yoshida S., Miyamoto T., Yoshimoto G., Watanabe T., Akashi K., Shultz L.D., Harada M. 2005. Development of functional human blood and immune systems in NOD/SCID/IL2 receptor gamma chainnull mice.Blood. 106:1565–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultz L.D., Lyons B.L., Burzenski L.M., Gott B., Chen X., Chaleff S., Kotb M., Gillies S.D., King M., Mangada J., et al. 2005. Human lymphoid and myeloid cell development in NOD/LtSz-scid IL2R gamma null mice engrafted with mobilized human hemopoietic stem cells.J. Immunol. 174:6477–6489 [DOI] [PubMed] [Google Scholar]
- Melkus M.W., Estes J.D., Padgett-Thomas A., Gatlin J., Denton P.W., Othieno F.A., Wege A.K., Haase A.T., Garcia J.V. 2006. Humanized mice mount specific adaptive and innate immune responses to EBV and TSST-1.Nat. Med. 12:1316–1322 [DOI] [PubMed] [Google Scholar]
- Fafi-Kremer S., Morand P., Brion J.P., Pavese P., Baccard M., Germi R., Genoulaz O., Nicod S., Jolivet M., Ruigrok R.W., et al. 2005. Long-term shedding of infectious epstein-barr virus after infectious mononucleosis.J. Infect. Dis. 191:985–989 [DOI] [PubMed] [Google Scholar]
- Ehlers B., Dural G., Yasmum N., Lembo T., de Thoisy B., Ryser-Degiorgis M.P., Ulrich R.G., McGeoch D.J. 2008. Novel mammalian herpesviruses and lineages within the Gammaherpesvirinae: cospeciation and interspecies transfer.J. Virol. 82:3509–3516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogg M.H., Garry D., Awad A., Wang F., Kaur A. 2006. The BZLF1 homolog of an Epstein-Barr-related gamma-herpesvirus is a frequent target of the CTL response in persistently infected rhesus macaques.J. Immunol. 176:3391–3401 [DOI] [PubMed] [Google Scholar]
- Fogg M.H., Kaur A., Cho Y.G., Wang F. 2005. The CD8+ T-cell response to an Epstein-Barr virus-related gammaherpesvirus infecting rhesus macaques provides evidence for immune evasion by the EBNA-1 homologue.J. Virol. 79:12681–12691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A.D., Shooshstari M., Finerty S., Watkins P., Morgan A.J. 1996. Virus-specific cytotoxic T cell responses are associated with immunity of the cottontop tamarin to Epstein-Barr virus (EBV).Clin. Exp. Immunol. 103:199–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochberg D., Souza T., Catalina M., Sullivan J.L., Luzuriaga K., Thorley-Lawson D.A. 2004. Acute infection with Epstein-Barr virus targets and overwhelms the peripheral memory B-cell compartment with resting, latently infected cells.J. Virol. 78:5194–5204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laichalk L.L., Hochberg D., Babcock G.J., Freeman R.B., Thorley-Lawson D.A. 2002. The dispersal of mucosal memory B cells: evidence from persistent EBV infection.Immunity. 16:745–754 [DOI] [PubMed] [Google Scholar]
- Kimura H., Hoshino Y., Kanegane H., Tsuge I., Okamura T., Kawa K., Morishima T. 2001. Clinical and virologic characteristics of chronic active Epstein-Barr virus infection.Blood. 98:280–286 [DOI] [PubMed] [Google Scholar]
- Miyamura T., Chayama K., Wada T., Yamaguchi K., Yamashita N., Ishida T., Washio K., Morishita N., Manki A., Oda M., Morishima T. 2008. Two cases of chronic active Epstein-Barr virus infection in which EBV-specific cytotoxic T lymphocyte was induced after allogeneic bone marrow transplantation.Pediatr. Transplant. 12:588–592 [DOI] [PubMed] [Google Scholar]
- Kutok J.L., Wang F. 2006. Spectrum of Epstein-Barr virus-associated diseases.Annu. Rev. Pathol. 1:375–404 [DOI] [PubMed] [Google Scholar]
- Knowles D.M., Cesarman E., Chadburn A., Frizzera G., Chen J., Rose E.A., Michler R.E. 1995. Correlative morphologic and molecular genetic analysis demonstrates three distinct categories of posttransplantation lymphoproliferative disorders.Blood. 85:552–565 [PubMed] [Google Scholar]
- Gurer C., Strowig T., Brilot F., Pack M., Trumpfheller C., Arrey F., Park C.G., Steinman R.M., Münz C. 2008. Targeting the nuclear antigen 1 of Epstein Barr virus to the human endocytic receptor DEC-205 stimulates protective T-cell responses.Blood. 112:1231–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajima M., Imadome K., Nakagawa A., Watanabe S., Terashima K., Nakamura H., Ito M., Shimizu N., Honda M., Yamamoto N., Fujiwara S. 2008. A new humanized mouse model of Epstein-Barr virus infection that reproduces persistent infection, lymphoproliferative disorder, and cell-mediated and humoral immune responses.J. Infect. Dis. 198:673–682 [DOI] [PubMed] [Google Scholar]
- Küppers R. 2003. B cells under influence: transformation of B cells by Epstein-Barr virus.Nat. Rev. Immunol. 3:801–812 [DOI] [PubMed] [Google Scholar]
- Islas-Ohlmayer M., Padgett-Thomas A., Domiati-Saad R., Melkus M.W., Cravens P.D., Martin Mdel P., Netto G., Garcia J.V. 2004. Experimental infection of NOD/SCID mice reconstituted with human CD34+ cells with Epstein-Barr virus.J. Virol. 78:13891–13900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocco M., Bellan C., Tussiwand R., Corti D., Traggiai E., Lazzi S., Mannucci S., Bronz L., Palummo N., Ginanneschi C., et al. 2008. CD34+ cord blood cell-transplanted Rag2−/− gamma c−/− mice as a model for Epstein-Barr virus infection.Am. J. Pathol. 173:1369–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münz C. 2005. Immune response and evasion in the host-EBV interaction. Epstein-Barr Virus Robertson E.S., editor Caister Academic Press, Norfolk, VA: 197–231 [Google Scholar]
- Zhang L., Kovalev G.I., Su L. 2007. HIV-1 infection and pathogenesis in a novel humanized mouse model.Blood. 109:2978–2981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berges B.K., Akkina S.R., Folkvord J.M., Connick E., Akkina R. 2008. Mucosal transmission of R5 and X4 tropic HIV-1 via vaginal and rectal routes in humanized Rag2−/−gammac−/− (RAG-hu) mice.Virology. 373:342–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S., Ohta S., Yajima M., Terashima K., Ito M., Mugishima H., Fujiwara S., Shimizu K., Honda M., Shimizu N., Yamamoto N. 2007. Humanized NOD/SCID/IL2Rgammanull mice transplanted with hematopoietic stem cells under nonmyeloablative conditions show prolonged life spans and allow detailed analysis of human immunodeficiency virus type 1 pathogenesis.J. Virol. 81:13259–13264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baenziger S., Tussiwand R., Schlaepfer E., Mazzucchelli L., Heikenwalder M., Kurrer M.O., Behnke S., Frey J., Oxenius A., Joller H., et al. 2006. Disseminated and sustained HIV infection in CD34+ cord blood cell-transplanted Rag2−/−gamma c−/− mice.Proc. Natl. Acad. Sci. USA. 103:15951–15956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An D.S., Poon B., Ho Tsong Fang R., Weijer K., Blom B., Spits H., Chen I.S., Uittenbogaart C.H. 2007. Use of a novel chimeric mouse model with a functionally active human immune system to study human immunodeficiency virus type 1 infection.Clin. Vaccine Immunol. 14:391–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorantla S., Sneller H., Walters L., Sharp J.G., Pirruccello S.J., West J.T., Wood C., Dewhurst S., Gendelman H.E., Poluektova L. 2007. Human immunodeficiency virus type 1 pathobiology studied in humanized BALB/c-Rag2−/−gammac−/− mice.J. Virol. 81:2700–2712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z., Denton P.W., Estes J.D., Othieno F.A., Wei B.L., Wege A.K., Melkus M.W., Padgett-Thomas A., Zupancic M., Haase A.T., Garcia J.V. 2007. Intrarectal transmission, systemic infection, and CD4+ T cell depletion in humanized mice infected with HIV-1.J. Exp. Med. 204:705–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Duyne R., Cardenas J., Easley R., Wu W., Kehn-Hall K., Klase Z., Mendez S., Zeng C., Chen H., Saifuddin M., Kashanchi F. 2008. Effect of transcription peptide inhibitors on HIV-1 replication.Virology. 376:308–322 [DOI] [PubMed] [Google Scholar]
- Bickham K., Goodman K., Paludan C., Nikiforow S., Tsang M.L., Steinman R.M., Münz C. 2003. Dendritic cells initiate immune control of Epstein-Barr virus transformation of B lymphocytes in vitro.J. Exp. Med. 198:1653–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonteneau J.F., Larsson M., Somersan S., Sanders C., Münz C., Kwok W.W., Bhardwaj N., Jotereau F. 2001. Generation of high quantities of viral and tumor-specific human CD4+ and CD8+ T-cell clones using peptide pulsed mature dendritic cells.J. Immunol. Methods. 258:111–126 [DOI] [PubMed] [Google Scholar]
- Ferlazzo G., Thomas D., Lin S.L., Goodman K., Morandi B., Muller W.A., Moretta A., Münz C. 2004. The abundant NK cells in human lymphoid tissues require activation to express killer cell Ig-like receptors and become cytolytic.J. Immunol. 172:1455–1462 [DOI] [PubMed] [Google Scholar]
- Borza C.M., Hutt-Fletcher L.M. 2002. Alternate replication in B cells and epithelial cells switches tropism of Epstein-Barr virus.Nat. Med. 8:594–599 [DOI] [PubMed] [Google Scholar]
- Busch D.H., Pilip I.M., Vijh S., Pamer E.G. 1998. Coordinate regulation of complex T cell populations responding to bacterial infection.Immunity. 8:353–362 [DOI] [PubMed] [Google Scholar]