Abstract
Invariant natural killer T (iNKT) cells constitute a distinct subset of T lymphocytes exhibiting important immune-regulatory functions. Although various steps of their differentiation have been well characterized, the factors controlling their development remain poorly documented. Here, we show that TGF-β controls the differentiation program of iNKT cells. We demonstrate that TGF-β signaling carefully and specifically orchestrates several steps of iNKT cell development. In vivo, this multifaceted role of TGF-β involves the concerted action of different pathways of TGF-β signaling. Whereas the Tif-1γ branch controls lineage expansion, the Smad4 branch maintains the maturation stage that is initially repressed by a Tif-1γ/Smad4-independent branch. Thus, these three different branches of TGF-β signaling function in concert as complementary effectors, allowing TGF-β to fine tune the iNKT cell differentiation program.
Natural killer T cells (NKT) are a unique T cell lineage with the capacity to regulate immune responses. NKT cells are multifunctional. They have been described as being important in a variety of immune responses, including the response to tumors, infectious agents, the maintenance of self-tolerance, and the prevention of autoimmunity (1, 2). In contrast to conventional T lymphocytes and other regulatory T cells, NKT cells do not recognize peptide antigen presented by classical MHC-encoded class I or II molecules; instead, their TCR interacts with glycolipid antigen presented by the MHC class I–like molecule CD1d (3). These T lymphocytes are mainly CD4+CD8−, but a subset can be CD4−CD8−. NKT cells branch away from T cell lineage at the double-positive (DP) CD4+CD8+ stage in the thymus (4).
Invariant NKT (iNKT) cells represent a subclass of NKT cells. They derive from DP thymocytes that express a TCR with a CDR3α region formed by a semiinvariant rearrangement of Vα14-Jα18. iNKT cells are positively selected upon recognition of CD1d expressed on DP thymocytes loaded with glycolipids (1). iNKT react to a glycosphingolipid α-galactosylceramide (αGalCer) presented by CD1d (5). CD1d-αGalCer tetramers, which uniformly stain all iNKT cells, have been used to characterize some NKT cell developmental steps and precursors. As CD1d-αGalCer tetramer+ precursors progress to mature HSAlow cells, three differentiation stages have been reported. iNKT cells go from the CD44low NK1.1− stage, through the CD44high NK1.1− stage, and finally reach the CD44high NK1.1+ stage. This iNKT differentiation sequence is accompanied by a massive cellular expansion occurring between the CD44low NK1.1− stage and the CD44high NK1.1− stage. A majority of CD44high NK1.1− cells emigrate to the peripheral tissues, where they cease to proliferate and rapidly express a mature phenotype, becoming CD44high NK1.1+ and producing both IL-4 and IFN-γ (6). iNKT cells represent ∼2.5% of splenic T lymphocytes and 30% of liver T cells (5). Interestingly, a fraction of the CD44high NK1.1− cells do not emigrate to the periphery, but instead proceed to their terminal maturation inside the thymus, where they acquire the NK1.1 marker and other NK-related markers that characterize the lineage (e.g., Ly49 and NKG2 family members). The CD44high NK.1.1+ population constitutes long-lived resident cells for several months (7). Although the importance of peripheral iNKT cells in the regulation of immune system has been widely reported, the significance of the iNKT intrathymic long-lived residents remains unknown (5).
Although it is known that CD1d expression on DP thymocytes is required for iNKT cell selection and lineage differentiation (6), and that both IL-15 and CD122 (IL-15Rβ) expression are involved in the transition between the CD44high NK1.1− and CD44high NK1.1+ stages, factors that control iNKT cell differentiation are poorly documented (8, 9). Some studies have proposed a role for two cytokines, GM-CSF and lymphotoxin, in iNKT cell development (10, 11). However, a recent investigation has reported that GM-CSF does not affect iNKT cell numbers in the thymus or in the periphery, but rather regulates their ability to secrete cytokines (12). Moreover, a role for lymphotoxin has been ascribed in iNKT cell emigration from the thymus rather than in iNKT cell development (13). Recently, it has been suggested, by us and others, that TGF-β could influence iNKT differentiation (14, 15).
TGF-β is a widely expressed cytokine involved in the control of proliferation and differentiation of numerous cell types both inside and outside the immune system. The three isoforms of TGF-β (TGF-β1, TGF-β2, and TGF-β3) signal through a common serine–threonine kinase receptor complex composed of the TGF-βRI and TGF-βRII subunits. Binding of TGF-β to its receptor leads to the activation of the intracellular kinase domain of TGF-βRII, which in turn phosphorylates the kinase domain of the TGF-βRI subunit. The latter then phosphorylates the receptor-associated Smads (R-Smads), Smad2 and/or Smad3, which bind the common Smad (C-Smad), Smad4. Complexes of Smad2–Smad4 and Smad3–Smad4 then migrate to the nucleus and regulate the transcription of various genes. Interestingly, Smad-independent pathways, such as MEK/MAP kinase, have also been reported (16). For many years Smad4 had been considered to be an obligate partner for the R-Smads Smad2 and Smad3 and the mediation of the TGF-β response through the Smad branch. However, the differences in phenotype observed between R-Smad-null and Smad4-null organisms suggested that Smad2 and Smad3 could have Smad4-independent biological functions. A role has recently been proposed for the transcriptional intermediary factor 1γ (Tif-1γ) in the TGF-β signaling pathway (17, 18). This ubiquitous protein (also known as TRIM33, RFG7, PTC7, and ectodermin) selectively binds receptor-phosphorylated Smad2/Smad3 in competition with Smad4. Although the Smad2–Tif-1γ interaction has biological effects independent of the Smad3–Tif-1γ complex in vitro, the importance of the Tif-1γ branch in TGF-β signaling in vivo remains unknown.
TGF-β signaling plays a crucial role on T cell biology because mice selectively deprived of TGF-βRI or -βRII on their T cells develop massive autoimmunity and die by 3 wk of age (15, 19). Thymic development of conventional CD4 and CD8 T cells does not appear to be under TGF-β control, but TGF-β seems clearly involved in the peripheral differentiation of selective T cell subsets such as Foxp3+ regulatory T cells and Th17 cells (15, 20, 21, 22). Interestingly, the complete deprivation of TGF-β signaling in α/β T lymphocytes, obtained by the crossing of a strain of floxed TGF-βRII mice with CD4-Cre transgenic mice (CD4-Cre x TGF-βRIIfl/fl), leads to a sharp defect in CD1d-αGalCer tetramer+ NK.1.1+ cells in both the thymus and the spleen (14, 15). These observations suggest that TGF-β could control the thymic development of the iNKT cells. However, given that the ablation of TGF-β signaling in T cells is associated with strong autoimmune disorders, T cell activation and acute cytotoxicity (15), it has been difficult to decipher whether the defect of iNKT cells in CD4-Cre x TGF-RIIfl/fl mice is a direct effect of TGF-β on iNKT cell precursors or the consequence of autoimmune disorders that affect the capacity of the thymus environment to differentiate iNKT cells. Whether TGF-β constitutes a specific factor directly controlling the thymic differentiation of iNKT precursor thus remains totally unknown.
In this study, we demonstrate that TGF-β signaling directly orchestrates several steps of iNKT precursor differentiation, ascribing a crucial and specific role for this cytokine in iNKT thymic development. We provide evidence that three branches of TGF-β signaling, Smad4, Tif-1γ, and the Tif-1γ–Smad4-independent signaling pathway, function in concert. Each of these TGF-β signaling branches is selectively dedicated to the control of a specific differentiation step of iNKT cells, allowing TGF-β to fine tune the iNKT cell differentiation program.
RESULTS
TGF-β signaling directly and specifically controls iNKT precursor differentiation
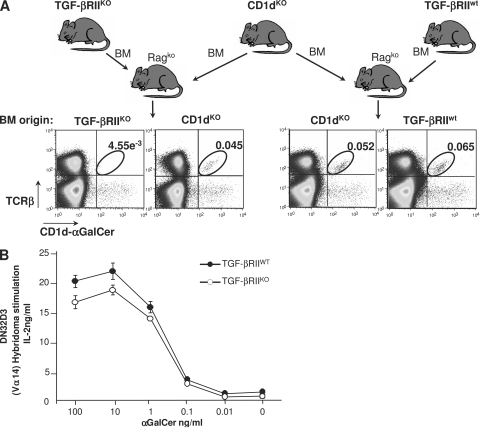
We have previously reported that thymic development of conventional CD4, CD8 T lymphocytes is not affected by TGF-β signaling (14, 15, 21). In the thymus, iNKT cells branch away from the T cell lineage at the CD4+CD8+ (DP) stage and are positively selected upon recognition of CD1d expressed on DP cells that are themselves loaded with glycolipids. Thus, a putative effect of TGF-β on the thymic development of iNKT cells could affect iNKT precursors alone, the capacity of DP to select iNKT precursors, or both. To test these three hypotheses, we have performed mixed BM transfers in reconstituted, sublethally irradiated B cell– and T cell–deficient recipients (rag2ko). BM cells from CD1dko mice, which DP cells can lead to iNKT precursors but cannot positively select, were transferred into rag2ko recipients with congenic Ly9.1-expressing BM cells from either CD4-Cre x TGF-βRIIfl/fl mice or CD4-Cre x TGF-βRIIwt littermate controls (Fig. 1 A). In CD4-Cre mice, the Cre transgene is expressed at the DP stage of thymocyte development. Consistent with this, DP thymocytes and their progeny from CD4-Cre x TGF-βRIIfl/fl BM cells escape TGF-β control. 6–7 wk after BM transfers, we analyzed the capacity of CD4-Cre x TGF-βRIIfl/fl DP thymocytes to complement the deficiency of DP from CD1dko BM to select iNKT cells. Strikingly, iNKT cells from CD1dKO donors were found in the spleens of all chimeric mice. However, iNKT cells from CD4-Cre x TGF-βRIIfl/fl BM were barely detectable compared with iNKT cells from CD4-Cre x TGF-βRIIwt BM (Fig. 1 A). In addition, from the CD4-Cre x TGF-βRIIfl/fl/CD4-Cre x TGF-βRIIwt mixed BM transfer, only iNKT cells from wild-type mouse BM were detectable (unpublished data). Thus, in the absence of TGF-β control, DP thymocytes remain a suitable environment for iNKT differentiation. In agreement with this, the expression of CD1d at the surface of DP thymocytes from CD4-Cre x TGF-βRIIfl/fl mice was similar to that of DP cells from littermate controls (unpublished data) and co-culture of Vα14 hybridoma cell lines with DP thymocytes from either CD4-Cre x TGF-βRIIfl/fl mice or littermate controls showed no significant difference in the capacity of DP to present αGalCer antigen (Fig. 1 B).
Figure 1.
Functional thymic environment to select iNKT cells in the absence of TGF-β signaling. (A) Flow cytometric analysis of splenocytes in mixed BM chimeras. T cell–depleted BM from 2-wk-old CD4-Cre x TGF-βRIIfl/fl mice or wild-type littermate mice were mixed with BM from CD1dKO mice (ratio 1:1) and transferred into rag2KO animals. 6–7 wk later, the presence in the spleen of iNKT cells was investigated using CD1d-αGalCer tetramers and TCR-β staining. The results are representative of three different experiments with three animals per group. (B) Proliferation analysis of CD1d-restricted Vα14-expressing DN32D3 hybridoma in the presence of thymocytes from either wild-type animals or CD4-Cre x TGF-βRIIfl/fl mice and different concentration of αGalCer. Mean ± SD. n = 2, from two independent experiments.
Collectively, this first set of experiments reveals that TGF-β directly affects thymic iNKT precursor cells and not the thymic environment mandatory for their selection, suggesting a specific role for TGF-β in the thymus that is confined to the control of iNKT precursor development.
TGF-β signaling controls several steps of iNKT precursor differentiation
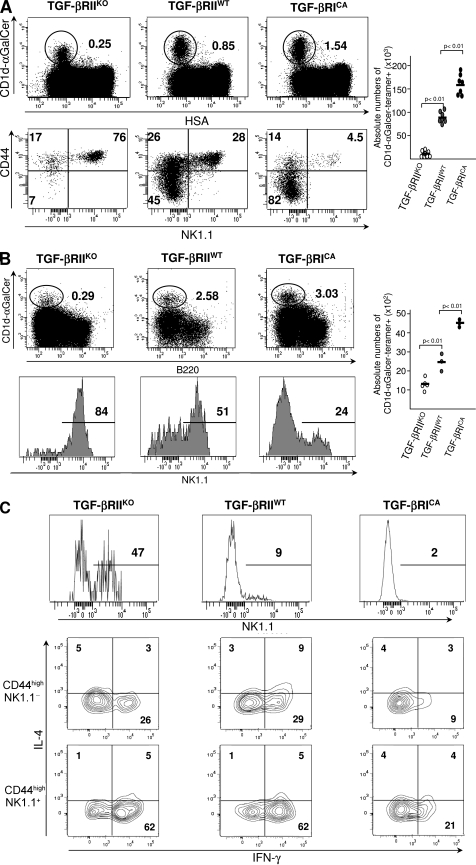
Thymic iNKT precursor differentiation goes through several established chronological steps defined by CD44 and NK1.1 expression. To determine the effects of TGF-β signaling on iNKT precursor differentiation, CD44 and NK1.1 stainings were performed on CD1d-αGalCer tetramer+ thymocytes, purified either from CD4-Cre x TGF-βRIIfl/fl mice or CD4-Cre x Stopfl/fl TGF-βRICA mice, the latter exhibiting a constitutively active (CA) TGF-β signaling in their T cells, and a normal thymic development of CD4 and CD8 conventional T lymphocyte (21). Few CD1d-αGalCer-tetramer+ thymocytes were observed in CD4-Cre x TGFβ-RIIfl/fl mice and their repartition among the different steps of differentiation was greatly affected compared with those from littermate controls (Fig. 2 A). Whereas in wild-type thymus, iNKT cell precursors were mainly NK1.1− at 2 wk of age, CD4-Cre x TGF-βRIIfl/fl iNKT cell precursors from littermate animals exhibited a repartition among the different steps of differentiation that was more similar to what is usually observed in adult mice. The early stages of development were rarely represented because the majority of the precursors were NK1.1+ and also express NK markers such as NKG2D, NKG2A/C/E, and Ly49C/I (Fig. S1). In clear contrast with the aforementioned observations, our analysis on CD1d-αGalCer-tetramer+ thymocytes purified from CD4-Cre x Stopfl/fl TGF-βRICA mice revealed an overrepresentation of the early differentiation stage CD44low NK1.1− and a large deficiency in both CD44high NK1.1− precursors and the mature CD44high NK1.1+ cell subset (Fig. 2 A). To exclude the possibility that the rare iNKT cells observed in CD4-Cre x TGF-βRIIfl/fl animals were caused by a potential defect of Cre-mediated recombination, CD4-Cre x TGF-βRIIfl/fl mice were bred onto a Cre recombination reporter allele, i.e., YFP flanked by LoxP sites were knocked into the ROSA26 locus (Rosa26 YFPfl/fl). In these animals, all the CD1d-αGalCer tetramer+ cells were both YFP+ and deficient for TGF-βRII (unpublished data). In agreement with the lack of thymic iNKT cell precursors, few CD1d-αGalCer tetramer+ T cells were detectable in both the liver and spleen from CD4-Cre x TGF-βRIIfl/fl mice compared the controls (Fig. 2 B and not depicted). Moreover, as in the thymus, the few iNKT cells observed at the periphery of CD4-Cre x TGF-βRIIfl/fl mice were mostly NK1.1+ (Fig. 2, B and C), but produced both IFN-γ and IL-4, which is similar to iNKT cells from wild-type animals (Fig. 2 C), suggesting that these cells are fully functional. In clear contrast, the overexpression of TGF-β signaling in T cells lead to an increase of peripheral CD1d-αGalCer tetramers+ T cells, but these cells presented a defect of maturation with both a defective NK.1.1 expression and a weak IFN-γ production compared with iNKT cells from wild-type animals (Fig. 2, B and C).
Figure 2.
Control of iNKT cell precursor development by TGF-β signaling. Flow cytometric analysis of the presence of iNKT cells in the thymus (A), liver (B), or spleen (C) of 14-d-old CD4-Cre x TGF-βRIIfl/fl mice (TGF-βRIIKO), CD4-Cre x Stopfl/fl TGF-βRICA mice (TGF-βRICA), or wild-type mice (TGF-βRIIwt). Absolute numbers of CD1d-αGalCer tetramer+ cells among either HSAlow cells or B220+ cells are illustrated by graphs and absolute numbers for each development stage are illustrated in supplemental table 1. CD44, NK1.1 staining on purified CD1d-αGalCer tetramers2 thymocytes (A) and on CD1d-αGalCer tetramers+ B220− cells from either the liver (B) or spleen (C). IL-4, IFN-γ, staining on either NK1.1− or NK1.1+ CD1d-αGalCer tetramer+ B220− cells from spleen (C). The results are representative of three or four different experiments with three animals per group.
Overall, these results reveal that TGF-β signaling controls several key steps of iNKT cell differentiation ranging from the early to late precursor differentiation stage. TGF-β signaling affects the maturation of both the thymic resident iNKT cells and iNKT precursors that have reached the periphery.
TGF-β signaling controls apoptosis in early iNKT precursors
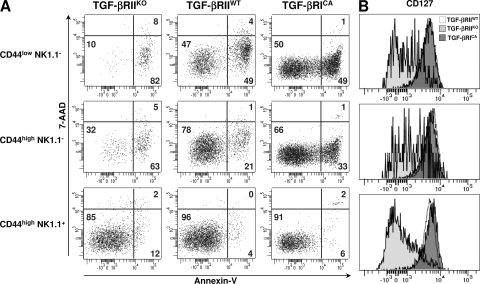
Because the iNKT differentiation step between the CD44low NK1.1− and the CD44high NK.1.1− stage is accompanied by a massive cellular expansion (6), it is possible that the defect of both CD44low NK1.1− and CD44high NK.1.1− cells observed in CD4-Cre x TGF-βRIIfl/fl mice, results from an impairment of their proliferation capacity. Surprisingly, Ki-67 staining of both CD44low NK1.1− and CD44high NK.1.1− cells from 14-d-old CD4-Cre x TGF-βRIIfl/fl mice and CD4-Cre x Stopfl/fl TGF-βRICA mice was similar to that observed for all differentiation stages in the littermate controls (Fig. S2). We thus postulated that TGF-β could affect the survival of iNKT cell precursors. When CD1d-αGalCer tetramer+ thymocytes were stained with both anti–annexin V and 7-amino-actinomycin-D (AAD), two to three times more apoptotic CD44low NK1.1− cells and CD44high NK.1.1− cells were observed in CD4-Cre x TGF-βRIIfl/fl mice compared with the littermate controls, demonstrating an antiapoptotic effect of TGF-β signaling on early iNKT precursors (Fig. 3 A). Because IL-7 has been established as a crucial cytokine for the survival of thymocytes, particularly for iNKT cells (23, 24), we asked whether TGF-β could affect CD127 (IL-7-Rα) expression on early iNKT cell precursors (Fig. 3 B). Consistent with their apoptosis, CD44low NK1.1− and CD44high NK1.1− cells from CD4-Cre x TGF-βRIIfl/fl mice expressed lower levels of CD127. Interestingly, the overexpression of TGF-β signaling (TGF-βRICA) affects neither CD127 expression nor the survival capacity of iNKT precursors, suggesting that in wild-type animals, endogenous TGF-β signaling levels appear to be sufficient to maintain early iNKT survival.
Figure 3.
Control iNKT cell precursor apoptosis by TGF-β signaling. Flow cytometric analysis of CD1d-αGalCer tetramer+ enriched from thymus from 14-d-old CD4-Cre x TGF-βRIIfl/fl mice (TGF-βRIIKO), CD4-Cre x Stopfl/fl TGF-βRICA mice (TGF-βRICA), or wild-type mice (TGF-βRIIwt) and stained for either CD44, NK1.1, or annexin V and incubated with 7-AAD (A) or CD44, NK1.1, and CD127 (B). These results are representative of three different experiments with two or three mice per group.
TGF-β signaling is thus crucial to both maintain a high level of CD127 expression on early iNKT cell precursors and prevent the apoptotic death of these highly dividing cells.
TGF-β signaling controls T-bet and CD122 (IL-15Rβ) in iNKT precursors
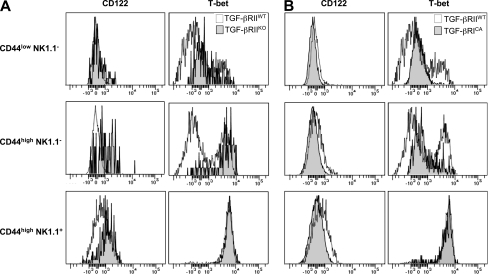
The deprivation of TGF-β signaling leads to a massive representation of CD44high NK1.1+ precursors among the few CD1d-αGalCer tetramers+ T cells. Moreover the overexpression of TGF-β signaling is associated with a sharp reduction of this same late stage of iNKT differentiation (Fig. 2), suggesting that TGF-β signaling represses the transition between CD44high NK1.1− and CD44high NK1.1+ cells and that the few mature cells observed have escaped TGF-β signaling. The transition between CD44high NK1.1− and CD44high NK1.1+ cells is known to be dependent on the cell surface expression of the IL-2/IL-15 receptor common β subunit (CD122). Interestingly, although CD122 is not highly expressed on CD44high NK1.1− and CD44high NK1.1+ cells, its modest expression has been shown to play a key role on iNKT maturation (25). To further investigate the mechanisms by which TGF-β controls the progression of CD44high NK1.1− to CD44high NK1.1+ cells, we analyzed CD122 (IL-15Rβ) expression on CD1d-αGalCer tetramer+ thymocytes. In agreement with the representation of the mature stage observed in Fig. 2, CD122 was expressed early on iNKT precursors escaping TGF-β regulation as compared with cell controls (Fig. 4 A). In clear contrast, CD122 cell-surface expression was impaired on all precursors from CD4-Cre x Stopfl/fl TGF-βRICA mice compared with precursors from their littermate controls (Fig. 4 B). Because the transcription factor T-bet has been shown to be a key factor for the maturation of iNKT cell precursors and to control CD122 cell surface expression (26), we proceeded to investigate whether TGF-β signaling modulates T-bet expression in iNKT cell precursors. The abrogation of TGF-β signaling in CD4-Cre x TGF-βRIIfl/fl mice lead to early expression of T-bet in precursors, whereas T-bet expression was highly repressed in the same iNKT precursors from CD4-Cre x Stopfl/fl TGF-βRICA mice (Fig. 4, A and B). Interestingly, CD44high NK1.1+ cells from all three kinds of mice, even when rare, expressed similar levels of T-bet, suggesting that these cells are fully mature.
Figure 4.
Control of T-bet and CD122 expression by TGF-β signaling. (A and B) Flow cytometric analysis of CD1d-αGalCer tetramer+ cells enriched from thymocytes from either CD4-Cre x TGF-βRIIfl/fl mice (TGF-βRIIKO; A) or CD4-Cre x Stopfl/fl TGF-βRICA mice (TGF-βRICA; B) stained for CD44, NK1.1, T-bet, and CD122. These results are representative of four different experiments with two mice per group.
Altogether, these results reveal that TGF-β signaling directly or indirectly represses T-bet and CD122 expression in both CD44low NK1.1− and CD44high NK1.1− precursors, and prevents the latter from massively maturing.
The Smad4 branch of TGF-β signaling controls iNKT cell late differentiation
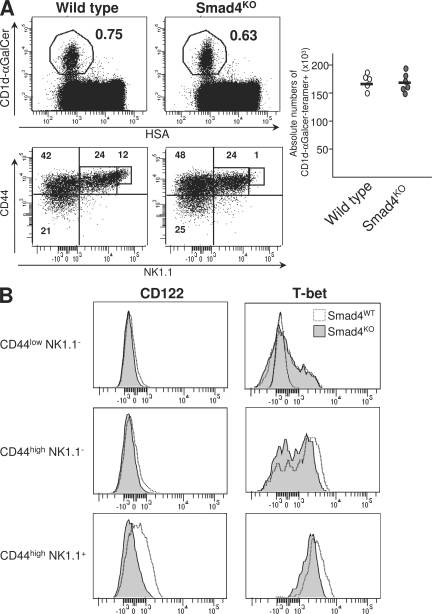
TGF-βR can signal through three different branches: the Smad4-dependent pathway, the Tif-1γ–dependent pathway, and the Tif-1γ/Smad4-independent pathway. The Smad4 branch has been reported to act as a mediator for many TGF-β effects in numerous cell types. Thus, we first investigated the role of Smad4 in iNKT cell differentiation. To do this, we used mice selectively deficient for Smad4 in their T cells from the DP stage. These animals were obtained by breeding of CD4-Cre transgenic animals with floxed smad4 mice, as previously described (27). Interestingly, the proportion of CD44low NK1.1− and CD44high NK1.1− precursors was similar to that observed in control littermate mice (Fig. 5 A), excluding a role for the Smad4 branch in the survival maintenance of early iNKT precursors. Moreover mature CD44high NK1.1+ cells were not overrepresented in the absence of Smad4-dependent signaling, contrary to what was observed when TGF-β signaling was totally abrogated (Fig. 5 A). This, again, excludes a role for the Smad4 branch in the transition of CD44high NK1.1− cells to CD44high NK1.1+ cells. However, the CD44high NK1.1+ cells, express a lower level of NK.1.1 marker in the absence of Smad4-dependent signaling (Fig. 5 A). Because NK1.1 expression has been shown to be dependent on CD122 and T-bet expression (25, 26), we next analyzed the expression of CD122 and T-bet. We found that the low level of NK1.1 expression observed in CD4-Cre x Smad4fl/fl was associated with the impairment of both CD122 and T-bet expression, in CD44high NK1.1+ precursors (Fig. 5 B).
Figure 5.
Role of Smad4 signaling in iNKT cell differentiation. (A) Flow cytometric analysis of the presence of iNKT cells in the thymuses from either 16-d-old CD4-Cre x Smad4 mice (Smad4KO) or wild-type mice and staining for CD44 and NK1.1 on CD1d-αGalCer tetramer+ cells from the same thymuses. Absolute numbers of CD1d-αGalCer tetramer+ cells among either HSAlow cells are illustrated by graphs and absolute numbers for each development stage are illustrated in Table S1. (B) Flow cytometric analysis on CD1d-αGalCer tetramer+ cells enriched from thymuses from either CD4-Cre x Smad4fl/fl mice (Smad4KO) or wild-type mice littermate stained for CD44, NK1.1, T-bet, and CD122. These results are representative of three different experiments with two mice per group.
The Smad4 branch of TGF-β signaling thus maintains both T-bet and CD122 expressions in mature iNKT precursors.
The Tif-1γ branch of TGF-β signaling controls early iNKT cell differentiation
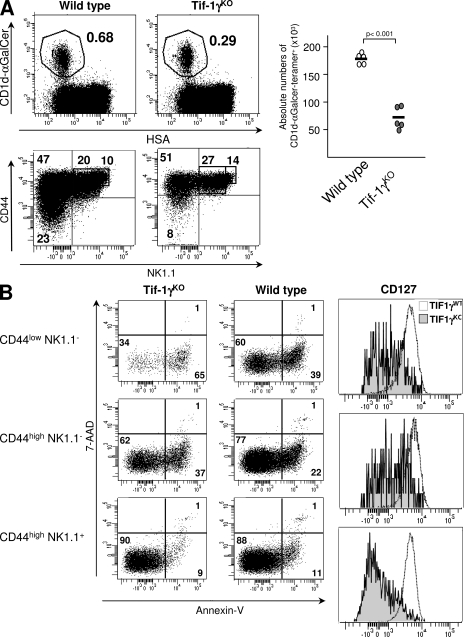
Given that the role of the Smad4 branch appears to be restricted to the late maturation steps of iNKT precursors, we proceeded by investigating the contribution of another TGF-β signaling pathway, the Tif-1γ branch, in iNKT cell development. We generated mice selectively deficient for Tif-1γ in their T cells, from the DP stage, by breeding CD4-Cre transgenic animals with floxed tif-1γ mice (Fig. S3 A). The CD4-Cre x Tif-1γfl/fl mice did not develop any sign of autoimmunity and inflammation (unpublished data). Moreover, in these animals, the conventional CD4/CD8 T cell development in the thymus was similar that observed in wild-type animals (Fig. S3 B). However, as observed in CD4-Cre x TGF-βRIIfl/fl animals, the deprivation of the Tif-1γ–dependent signaling pathway led to a large decrease in the proportion of CD1d-αGalCer–tetramer+ cells in the thymus. As in the absence of total TGF-β signaling, the deprivation of the Tif-1γ branch led to a large defect in CD44low NK1.1− precursors associated with their apoptosis and to a loss of CD127 surface expression (Fig. 6, A and B). However, contrary to what we observed in CD4-Cre x TGF-βRIIfl/fl mice, no effect, either at the CD44high NK1.1+ precursor stage or on CD122 and T-bet expression, was observed in CD4-Cre x Tif-1γfl/fl mice (unpublished data), excluding a potential role for the Tif-1γ branch in the transition of CD44high NK1.1− precursors to CD44high NK1.1+ precursors.
Figure 6.
Role of Tif-1γ signaling in iNKT cell differentiation. (A) Flow cytometric analysis of the presence of iNKT cells in thymuses from either 16 d-old CD4-Cre x Tif-1γfl/fl mice (Tif-1γKO) or wild-type mice and staining for CD44 and NK1.1 on CD1d-αGalCer tetramer+ cells from the same thymuses. Absolute numbers of CD1d-αGalCer tetramer+ cells among either HSAlow cells are illustrated by graphs and absolute numbers for each development stage are illustrated in supplemental table 1. B) Flow cytometric analysis on CD1d-αGalCer tetramer+ cells enriched from thymuses from either CD4-Cre x Tif-1γfl/fl mice (Tif-1γKO) or wild-type littermate mice and stained for CD44, NK1.1, Annexin V, 7-AAD, and CD127. Mean fluorescence intensity is reported on the CD127 histograms. These results are representative of three different experiments with two mice per group.
Altogether, these results demonstrate that the Tif-1γ branch of TGF-β signaling is responsible for the survival maintenance of the early precursors of iNKT cells.
The Tif-1γ/Smad4-independent branch of TGF-β signaling controls iNKT cell intermediate differentiation
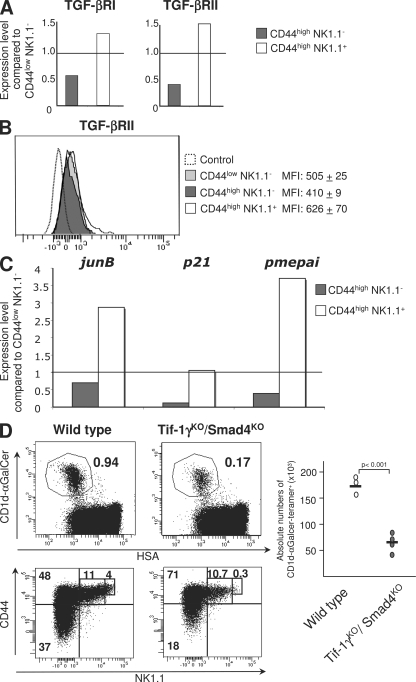
Because we found that TGF-β signaling represses both T-bet and CD122 expression in both CD44high NK1.1− and CD44high NK1.1+ precursors, and hence impairs their maturation in CD44high NK1.1+ cells, TGF-β signaling in wild-type mice should be repressed, at least at the intermediate stage (CD44high NK1.1−) of differentiation, to allow these precursors to proceed to maturity. The analysis of both tgf-βrI and tgf-βrII expression revealed a down-regulation of the expression of both TGF-βR genes in CD44high NK1.1− precursors compared with early and late precursors (Fig. 7 A). This down-regulation was confirmed at the protein level by analysis of the surface expression of TGF-βR (Fig. 7 B). In addition, this lack of TGF-βR expression at the CD44high NK1.1− stage was associated with the repression of several genes known to be up-regulated by TGF-β signaling (Fig. 7 C), confirming an impairment of TGF-β responsiveness in the CD44high NK1.1− precursors compared with the other differentiation stages.
Figure 7.
Role of Tif-1γ/Smad4-independent signaling in the control of iNKT maturation. (A) tgf-b receptor expression analysis by RT-PCR performed on purified CD1d-αGalCer thymocytes from 16-d-old C57BL/6 mice. Relative gene expression are normalized on both hprt and g3pdh expression. (B) Flow cytometric analysis of CD1d-αGalCer tetramer+ cells from iNKT cell-enriched thymocytes of C57BL/6 mice, stained for TGF-βRII. Means of fluorescence intensity (MFI) are reported with their SD. (C) jun-b, p21, pme-pai expression analysis by RT-PCR performed on purified CD1d-αGalCer thymocytes from C57BL/6 mice. Relative gene expression was normalized on βactin expression. The results are representative of three different experiments from five to eight mice. (D) Flow cytometric analysis of the presence of iNKT cells in thymuses from either 13–14-d-old CD4-Cre x Tif-1γfl/fl x Smad4fl/fl mice or wild-type littermate mice and staining for CD44 and NK1.1 on CD1d-αGalCer tetramer+ cells from the same thymuses. Absolute numbers of CD1d-αGalCer tetramer+ cells among HSAlow cells are illustrated by graphs, and absolute numbers for each development stage are illustrated in Table S1. The results are representative of three different experiments with at least three mice per group.
Interestingly, neither the ablation of the Tif-1γ branch nor the ablation of the Smad4 branch by itself can explain the overrepresentation of mature CD44high NK1.1+ precursor cells observed in the complete absence of TGF-β signaling (CD4-Cre x TGF-βRIIfl/fl mice; Fig. 2). We thus analyzed iNKT cell development in the absence of both the Tif-1γ and Smad4 branches, using CD4-Cre x Tif-1γfl/fl x Smad4fl/fl animals. Interestingly, the deprivation of these two branches of TGF-β signaling did not mimic the total ablation of the TGF-β signaling pathway (CD4-Cre x TGF-βRIIfl/fl mice; Fig. 2), but led to the addition of the phenotypes observed when the Tif-1γ and Smad4 branches were switched off separately (Fig. 7 D). We observed a large defect of CD44low NK1.1− precursors associated with their apoptosis, as in the absence of the Tif-1γ signaling pathway, and a low expression of NK1.1 on CD44high NK1.1+ cells associated with the impairment of both CD122 and T-bet expression in CD44high NK1.1+ as reported in the absence of the Smad4 signaling pathway (Fig. 7 D and not depicted). CD44high NK1.1+ precursors were not overrepresented, suggesting that the transition of CD44high NK1.1− precursors to CD44high NK1.1+ precursors is repressed by the Tif-1γ/Smad4-independent branch of TGF-β signaling, which is regulated by the level of expression of TGF-βR.
DISCUSSION
iNKT cells constitute a unique thymus-dependent T cell subset, characterized by the expression of a semiinvariant (Vα14-Jα18 in mice and Vα24-Jα18 in human) TCR-α chain and markers traditionally associated with the NK cell lineage. iNKT cells are developmentally and functionally distinct from the mainstream CD4 and CD8 T cells. iNKT cells have been associated with a broad array of disorders, ranging from transplant rejection to tumors, various forms of autoimmunity, atherosclerosis, allergy, and infection (5). Because of the importance of iNKT cells in immune regulation during various pathological processes (28), it is critical to define the factors that govern their development. Our findings reveal TGF-β to be a key factor for thymic iNKT cell development. Although thymic precursors of both iNKT cells and conventional T cells require interaction with their respective self-ligands in the thymus environment for positive selection, the HSAlow iNKT cell precursors subsequently undergo a complex series of activation and differentiation events. We have clearly demonstrated that TGF-β directly orchestrates the sequence of thymic events that iNKT precursors undergo. This sequence includes a lineage expansion, acquisition of the CD44high phenotype before either their migration to the periphery or their long stay in the thymus, and their maturation to CD44high NK1.1+ (25, 29). Unlike IL-15, whose action is restricted to the maturation step, we revealed that TGF-β controls several key steps of iNKT cell differentiation, ascribing a crucial role for this cytokine iNKT cell development. Moreover, although iNKT cells and conventional α/β T cells are derived from a common lineage at the DP stage, neither the ablation nor the overexpression of TGF-β signaling in DP cells affects the thymic development of conventional α/β T cells (15, 21). Thus, TGF-β appears as a key factor whose action in the thymus is confined to the control of several steps of iNKT cell differentiation.
iNKT cell lineage expansion has been regarded as the consequence of multiple rounds of cell division occurring between the CD44low NK1.1− and CD44high NK1.1− stages (5). Our results suggest that when they escape from TGF-β control, the CD44low NK1.1− and CD44high NK1.1− precursors keep dividing, but a large proportion of them undergo apoptosis, which dramatically reduces the pool of cells for the next iNKT differentiation steps, and thus reduces the total number of iNKT cells. Our observations strongly indicate that the iNKT lineage expansion is not only restricted to precursor cell proliferation, but also results from the survival maintenance of these dividing precursors by TGF-β. This TGF-β antiapoptotic effect is correlated with the maintenance of IL-7Rα (CD127) expression at the cell surface of iNKT precursors, suggesting a role for the IL-7/IL-7R signaling pathway in early iNKT precursors' survival. In agreement with this, a large decrease in iNKT precursors has been reported in IL-7KO mice (24). Furthermore, our findings demonstrate that both the survival of iNKT early precursors and the maintenance of CD127 expression are under the control of the Tif-1γ branch of TGF-β signaling. However, whether Tif-1γ directly controls CD127 expression remains to be investigated. Tif-1γ is a ubiquitous nuclear protein that has been recently shown to be involved in hematopoietic cell fate via the TGF-β pathway in vitro (17). Interestingly, in zebrafish, a mutation in mon, which is a gene identified as being an orthologous gene of tif-1γ in mammals, leads to massive apoptosis of hematopoietic cells (30). Moreover, that TGF-β has largely been reported to play an antiapoptotic role (31) supports our results, suggesting that the Tif1-γ branch could be a key signaling pathway for the TGF-β antiapoptotic effect. In addition to the control of apoptosis, Tif-1γ could regulate CD44 expression, and thus control the progression from CD44low to CD44high precursors.
After the expansion phase, precursors that become CD44high NK1.1− can either stay in the thymus or colonize peripheral organs and undergo a maturation phase accompanied by the cell-surface expression of both CD122 (IL-15βR chain) and NK1.1. This maturation step has been reported to be dependent on the expression of T-bet, a transcription factor originally implicated in the Th1 cell lineage commitment (26). T-bet has been shown to induce CD122 in iNKT cell precursors (32). We have demonstrated a role for TGF-β in the repression of both T-bet and CD122 expression in iNKT cell precursors. Interestingly, the ectopic expression of T-bet in immature iNKT cell precursors has been shown to be sufficient to induce their full maturation and function (32). Given the central role of T-bet in iNKT cell maturation, by controlling the expression of T-bet in iNKT cells, TGF-β is a key regulator for iNKT cell maturation. The absence of overrepresentation of CD44high NK1.1+ mature stage in both single- and double-knockout mice for Tif-1γ and Smad4 revealed that a Tif-1γ/Smad4-independent branch of TGF-β signaling represses iNKT precursor maturation. Interestingly, in wild-type mice, TGF-β signaling in CD44high NK1.1− precursors is impaired, leading to iNKT precursor maturation. Several arguments suggest the MEK/MAPK branch as a putative candidate of the Tif1-γ/Smad4-independent pathway to explain how TGF-β represses both T-bet and iNKT precursor maturation. TGF-β has been proposed to repress T-bet during Th1/Th2 lineage commitment in vitro. Recent studies have demonstrated that this repression is under the MEK signaling pathway (33). Moreover, in wild-type animals, we found that the down-regulation of TGF-β signaling at the CD44high NK1.1− stage was associated with the repression of mek1, whereas the expression of other actors of the MAPK, such as p38 and Erk, remained constant during iNKT cell differentiation (Fig. S4). Additional experiments should be carried out to confirm the role of MEK in iNKT cell differentiation. By repressing the maturation of both highly dividing CD44low NK1.1− and CD44high NK1.1− cells, the Tif-1γ/Smad4-independent pathway allows precursors to stay longer in highly divided stages, and thus could also contribute to promote iNKT lineage expansion along with the Tif-1γ branch. Interestingly, the TGF-β signaling effects on the control of iNKT maturation are not restricted to the thymus because iNKT peripheral maturation was similarly affected. The large defect of mature NK1.1+ cells at the periphery in CD4-Cre x Stopfl/fl TGF-βRICA mice is also associated with their incapacity to produce IFN-γ, underlining the role of TGF-β in the repression of iNKT cell maturation. Moreover, the rare but fully mature, peripheral iNKT cells generated in the absence of TGF-β control produce both IL-4 and IFN-γ, which is similar to iNKT cells from wild-type mice, suggesting that TGF-β is not mandatory for iNKT cell function.
Maturing CD44high NK1.1+ precursors express high levels of TGF-βR at their cell surface, and their responsiveness to TGF-β is highly increased. This reactivation of TGF-β signaling in CD44high NK1.1+ precursors is not associated with a repression of T-bet expression. This apparent discrepancy can be explained by the relay role of the Smad4 branch of TGF-β signaling. The Smad4 branch is responsible for the maintenance of both T-bet and CD122 expression in the CD44high NK1.1+ precursors, and allows them to mature and acquire a high level of NK1.1. Our data suggest that once precursors have turned on their maturation program, the Smad4 branch thwarts the TGF-β inhibitory effect on maturation driven by the Tif-1γ/Smad4-independent branch. Thus, TGF-β has a double-edged sword effect on iNKT cell differentiation, through the Tif-1γ/Smad4-independent branch, which represses their maturation, and through the Smad4 branch, which continues the maturation process. In CD4-Cre x TGF-βRIIfl/fl mice, all TGF-β signaling branches are turned off, thus the maturation is not repressed by Tif-1γ/Smad4-independent branch, and the iNKT cell precursor matures fully.
Therefore, the multifaceted role of TGF-β during iNKT cell differentiation can be explained not only by the concerted action of different signaling branches, such as the Tif-1γ branch and the Tif-1γ/Smad4-independent branch, but also by the interaction, or competition, between branches, such as the Smad4 branch and the Tif-1γ/Smad4-independent branch. Interestingly, the analysis of TGF-β branches during the differentiation of several cell types has also suggested an interaction between the TGF-β signaling pathways. In numerous cancers, at the early stage of tumor development, TGF-β represses tumor-growth, whereas at later stages TGF-β facilitates tumor progression. The activation of the Tif-1γ/Smad4-independent branch (MAPK) by TGF-β has been proposed to facilitate tumor progression by TGF-β. The loss of tumor growth inhibition by TGF-β and the acquisition of oncogenic capacity correlates with the loss of the Smad4 branch function (34). Competition between the Smad4 branch and the Tif-1γ branch has also been suggested. During the blastula stage of Xenopus, a role for Tif-1γ in the targeting of Smad4 for ubiquitination and degradation has been reported, suggesting a potential regulation of the Smad4 branch by Tif-1γ protein (18). However, the interactions between TGF-β signaling branches appear to be specific for a given cell type, because during human hematopoiesis, although two distinct roles have been ascribed to the Tif-1γ branch and the Smad4 branch, no variation in Smad4 protein levels have been reported after overexpression or deletion of Tif-1γ in human hematopoietic cells (17). In iNKT cell differentiation, the expression levels of both Tif-1γ and Smad4 stay constant over the different stages of iNKT cell development (unpublished data) suggesting that the Tif-1γ branch and the Smad4 branch do not interfere with one another, but work in concert during a cellular differentiation program, each branch dedicated to a specific task.
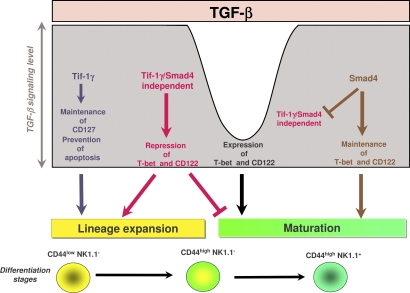
In sum, our results propose that TGF-β is a key regulator of iNKT cell differentiation. We have revealed that through different branches of its signaling pathway, TGF-β can fine tune the complex iNKT cell differentiation cell program. These different signaling pathways function as complementary effector arms that, in association with a modulation of TGF-β responsiveness between the differentiation stages, can explain the multiple roles of TGF-β during iNKT cell development. TGF-β assures lineage expansion via the Tif-1γ branch and maturation acquisition via the Tif-1γ/Smad4-independent branch, whereas the maintenance of the maturation process involves the Smad4-dependent pathway. At an early stage, the Tif-1γ/Smad4-independent branch represses precursor maturation and allows iNKT lineage expansion in concert with Tif-1γ branch. Next, the TGF-β responsiveness is impaired, allowing precursor maturation. Finally, maturing precursors restore their capacity to fully respond to TGF-β and the Smad4 branch assures the continuation of the maturation program by maintaining high level of both T-bet and CD122 (Fig. 8). The action of each branch is conditioned by the action of the others. These sequential roles of the TGF-β signaling branches during iNKT cell development ascribe a crucial role for TGF-β in the iNKT cell differentiation.
Figure 8.
Schematic model summarizing the effects of the three branches of TGF-β signaling on iNKT cell differentiation. The Tif-1γ–dependent branch prevents the apoptosis and allows lineage expansion. The Tif-1γ/Smad4-independent branch prevents maturation, and thus allows lineage expansion. The Smad4-dependent branch allows maturation.
MATERIALS AND METHODS
Mice.
CD4-Cre x TGF-βRIIfl/fl, and CD4-Cre x Smad4fl/fl mice were generated as previously described (15). Stopfl/fl TGF-βRICA transgenic animals have been generated by insertion at the HPRT locus of the Lox-Stop-Lox cassette positioned upstream of the TGF-βRICA-HA fusion gene (21). Tif1-γfl/fl mice harboring floxed exon 2–4 were generated by K.-P. Yan. The selective deletion of the Tif1-γ in Tif1-γ cells is illustrated in Fig. S3. R26RYFP (Rosa26-YFPfl/fl) mice were provided by Frank Constantini (Columbia University, New York, NY) and CD1dKO mice by Luc Van Kaer (Vanderbilt University, Nashville, TN). Beside the Stopfl/fl TGF-βRICA mice which were backcrossed 6 times in C57BL/6, each mouse stain was backcrossed 8–10 times in a C57BL/6 (B6) background, and only littermates were used as controls. C57BL/6 mice were purchased from Charles Rivers Laboratories. Mice were maintained in a specific pathogen–free animal facility at the Plateau de Biologie Expérimentale de la Souris (Lyon, France) and handled in the accordance with the institutional guidelines, and the protocols were approved by Comité Régional d'Ethique pour l'Expérimentation Animale.
Radiation BM chimeras.
Radiation BM chimeras were generated upon transfer of 2–3 × 106 T cell–depleted BM cells into lethally irradiated Rag2KO recipients, as previously described (15).
Lymphocyte isolation.
Cell suspensions were prepared from the thymus, spleen, and liver. Red cells in spleen were lysed with 168 mM NH4Cl and liver lymphocytes were separated by gradient density using Percoll (GE Healthcare), as previously described (15).
NKT cell enrichment.
Thymuses were depleted of CD8+, CD16/32+, CD19+ cells using mAbs directed against CD8α (clone 53–6.7), CD16/32 (clone 24G2), and CD19 (clone 1D3; all from BD), respectively, along with mouse depletion Dynabeads (Invitrogen) following the manufacturer's instructions.
Antibodies and flow cytometry.
CD24-FITC (clone M1/69) mAbs, CD122-PE (clone TM-β1), Ki67-PE (clone B56), NK-1.1-PerCP-Cy5.5 (clone PK136), Ly9.1, TCR-β-FITC (H57-597) annexin V-APC and 7-AAD, NKG2A/C/E (20D5), NKG2D (C7), and Ly49C/I (5E6) were obtained from BD. CD127-PE (clone A7R34), NK-1.1-PE-Cy7 (clone PK136), T-bet-Alexa Fluor 647 (clone eBio4B10), CD44-Alexa Fluor 700 (clone IM7), IL-4 (11B11), and IFN-γ (XMG1.2) were purchased from eBioscience. CD1d-α-GalCer tetramers were produced with streptavidin-APC (BD) or streptavidin-PE-Cy7 (eBioscience) and used for staining as previously described (25). Anti–TGF-βRII-biot was purchased from R&D Systems. Anti-Smad4 (EP618Y) was purchased from Abcam and monoclonal and anti-Tif-1γ (5Tγ3E9) were gifts from Institute of Genetics and Molecular and Cellular Biology (Strasbourg, France). For intracellular staining, cells were activated with 500 ng/ml PMA and 5 ng/ml ionomycin (Sigma-Aldrich), and then resuspended in Cytofix/Cytoperm solution from a Fixation/permeabilization kit (BD) and stained according to the manufacturer's instructions. Flow cytometry and cell sorting was performed using a two-laser FACSAria (BD) cell sorter/analyzer and analyzed using either BD FACSDiva software v5.0.1 (BD) or FlowJo (Tree Star, Inc.).
In vitro stimulation.
5 × 105 mouse thymocytes were incubated for 48 h with the CD1d-restricted Vα14-expressing DN32D3 hybridoma at a 1/10 ratio in 100 µl total volume with increasing concentrations of αGalCer, ranging from 0.01 to 100 ng/ml, and culture supernatants were analyzed by ELISA for IL-2 content, as described previously (25).
Quantitative RT-PCR.
CD1d-αGalCer tetramer+ thymocytes were FACS-sorted into three groups: CD44low NK1.1−, CD44high NK1.1−, and CD44high NK1.1+. RNA was isolated using the RNeasy kit (QIAGEN) and reverse transcribed into cDNA with Superscript RT-PCR kit (Invitrogen). RT-PCR was performed with platinium SYBR Green kit (Invitrogen) on an Applied Biosystems 7000 machine with the primers listed. Relative gene expression was normalized to hprt and g3pdh/βactin levels in agreement with works from vandesompele et al. (35). The primers used are as follows: hprt, 5′-TCATTATGCCGAGGATTTGGA-3′ and 5′-CAGAGGGCCACAATGTGATG-3′; g3pdh, 5′-GCATGGCCTTCCGTGTCC-3′ and 5′-TGTCATCATACTTGGCAGGTTTCT-3′; tgfβRI, 5′-CAGACGAAGCAGACTGGACCAG-3′ and 5′-TGCTGCAATCAGGACCACTGC-3′; tgfβRII, 5′-GAAGAATACACCACCAGCAGTC-3′ ανδ 5′-ATGATGACAGCTATGGCAATCC-3′; p38, 5′-ACCTAAAGCCCAGCAACCTAGC-3′ and 5′-GGTAGCCACGTAGCCTGTCATC-3′; mek1, 5′-AACTGGGAGCTGGCAACGG-3′ and 5′-TGCGGGTTTGATCTCCAGGTG-3′; erk, 5′-GCTGACTCCAAAGCTCTGGATTTAC-3′ and 5′-CTCCTTAGGTAAGTCGTCCAACTCC-3′; p21, 5′-TTGCACTCTGGTGTCTGAGC-3′ and 5′-GGGCACTTCAGGGTTTTCTC-3′; junB, 5′-GACGACCTGCACAAGATGAA-3′ and 5′-TGCTGAGGTTGGTGTAGACG-3′; pmepai, 5′-CAGGAGGAGAGACGATGGAC-3′ and 5′-AGGTAGGGGTAGGTGGGTTG-3′; and βactin SuperArray commercial Q-PCR primers.
Statistics.
The unpaired, two-tailed Student's t test was used for statistical confirmation of cell number comparisons.
Online supplemental materials.
NK marker stainings, MEK/ERK/P38 mRNA expression, Tif-1γ expression, NKT proliferation, and NKT absolute numbers are available online. The online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20090127/DC1.
Acknowledgments
The expert assistance of S. Guignès, A. Lambert, A. Ruiz, P. Contard, and C. Angleraux is highly appreciated. We thank S. Karlsson, C.B Wilson, F. Constantini, L. Van Kaer, for providing TgfβRIIfl/fl, Smad4fl/fl, CD4-Cre, R26RYFP, CD1dKO mice, respectively. We also thank members of our laboratory for advice and helpful discussions, and both R. Buckland and J.P. Rasmussen for manuscript editing.
This work was supported by grants from ANR JCJC06_136846 (J.C. Marie), ARC 3989 (J.C. Marie), ARC 3891 (L. Bartholin) ligue contre le cancer (J.C. Marie and L. Bartholin) INSERM Groupe AVENIR RO4193KS (K. Benlagha), FRM INE20051105133 (K. Benlagha), ANR RO7119KS (K. Benlagha), and INCa (R. Rimokh and J.C. Marie). F. Cyprian and D. Vincent are supported by an MRT fellowship. L. Bartholin is supported by Ligue Nationale Contre le Cancer and INSERM AVENIR.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used: αGalCer, α-galactosylceramide; AAD, amino-actinomycin-D; CA, constitutively active; DP, double-positive; iNKT, invariant natural killer T; Tif-1γ, transcriptional intermediary factor 1γ.
References
- Kronenberg M., Gapin L. 2002. The unconventional lifestyle of NKT cells.Nat. Rev. Immunol. 2:557–568 [DOI] [PubMed] [Google Scholar]
- Kronenberg M., Rudensky A. 2005. Regulation of immunity by self-reactive T cells.Nature. 435:598–604 [DOI] [PubMed] [Google Scholar]
- Godfrey D.I., MacDonald H.R., Kronenberg M., Smyth M.J., Van Kaer L. 2004. NKT cells: what's in a name? Nat. Rev. Immunol. 4:231–237 [DOI] [PubMed] [Google Scholar]
- Kronenberg M. 2005. Toward an understanding of NKT cell biology: progress and paradoxes.Annu. Rev. Immunol. 23:877–900 [DOI] [PubMed] [Google Scholar]
- Bendelac A., Savage P.B., Teyton L. 2007. The biology of NKT cells.Annu. Rev. Immunol. 25:297–336 [DOI] [PubMed] [Google Scholar]
- McNab F.W., Berzins S.P., Pellicci D.G., Kyparissoudis K., Field K., Smyth M.J., Godfrey D.I. 2005. The influence of CD1d in postselection NKT cell maturation and homeostasis.J. Immunol. 175:3762–3768 [DOI] [PubMed] [Google Scholar]
- Berzins S.P., McNab F.W., Jones C.M., Smyth M.J., Godfrey D.I. 2006. Long-term retention of mature NK1.1+ NKT cells in the thymus.J. Immunol. 176:4059–4065 [DOI] [PubMed] [Google Scholar]
- Lodolce J.P., Boone D.L., Chai S., Swain R.E., Dassopoulos T., Trettin S., Ma A. 1998. IL-15 receptor maintains lymphoid homeostasis by supporting lymphocyte homing and proliferation.Immunity. 9:669–676 [DOI] [PubMed] [Google Scholar]
- Ohteki T., Ho S., Suzuki H., Mak T.W., Ohashi P.S. 1997. Role for IL-15/IL-15 receptor beta-chain in natural killer 1.1+ T cell receptor-alpha beta+ cell development.J. Immunol. 159:5931–5935 [PubMed] [Google Scholar]
- Sato H., Nakayama T., Tanaka Y., Yamashita M., Shibata Y., Kondo E., Saito Y., Taniguchi M. 1999. Induction of differentiation of pre-NKT cells to mature Valpha14 NKT cells by granulocyte/macrophage colony-stimulating factor.Proc. Natl. Acad. Sci. USA. 96:7439–7444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elewaut D., Brossay L., Santee S.M., Naidenko O.V., Burdin N., De Winter H., Matsuda J., Ware C.F., Cheroutre H., Kronenberg M. 2000. Membrane lymphotoxin is required for the development of different subpopulations of NK T cells.J. Immunol. 165:671–679 [DOI] [PubMed] [Google Scholar]
- Bezbradica J.S., Gordy L.E., Stanic A.K., Dragovic S., Hill T., Hawiger J., Unutmaz D., Van Kaer L., Joyce S. 2006. Granulocyte-macrophage colony-stimulating factor regulates effector differentiation of invariant natural killer T cells during thymic ontogeny.Immunity. 25:487–497 [DOI] [PubMed] [Google Scholar]
- Franki A.S., Van Beneden K., Dewint P., Hammond K.J., Lambrecht S., Leclercq G., Kronenberg M., Deforce D., Elewaut D. 2006. A unique lymphotoxin {alpha}beta-dependent pathway regulates thymic emigration of V{alpha}14 invariant natural killer T cells.Proc. Natl. Acad. Sci. USA. 103:9160–9165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.O., Sanjabi S., Flavell R.A. 2006. Transforming growth factor-beta controls development, homeostasis, and tolerance of T cells by regulatory T cell-dependent and -independent mechanisms.Immunity. 25:455–471 [DOI] [PubMed] [Google Scholar]
- Marie J.C., Liggitt D., Rudensky A.Y. 2006. Cellular mechanisms of fatal early-onset autoimmunity in mice with the T cell-specific targeting of transforming growth factor-beta receptor.Immunity. 25:441–454 [DOI] [PubMed] [Google Scholar]
- Derynck R., Zhang Y.E. 2003. Smad-dependent and Smad-independent pathways in TGF-beta family signalling.Nature. 425:577–584 [DOI] [PubMed] [Google Scholar]
- He W., Dorn D.C., Erdjument-Bromage H., Tempst P., Moore M.A., Massague J. 2006. Hematopoiesis controlled by distinct TIF1gamma and Smad4 branches of the TGFbeta pathway.Cell. 125:929–941 [DOI] [PubMed] [Google Scholar]
- Dupont S., Zacchigna L., Cordenonsi M., Soligo S., Adorno M., Rugge M., Piccolo S. 2005. Germ-layer specification and control of cell growth by Ectodermin, a Smad4 ubiquitin ligase.Cell. 121:87–99 [DOI] [PubMed] [Google Scholar]
- Liu Y., Zhang P., Li J., Kulkarni A.B., Perruche S., Chen W. 2008. A critical function for TGF-beta signaling in the development of natural CD4+CD25+Foxp3+ regulatory T cells.Nat. Immunol. 9:632–640 [DOI] [PubMed] [Google Scholar]
- Marie J.C., Letterio J.J., Gavin M., Rudensky A.Y. 2005. TGF-β1 maintains suppressor function and Foxp3 expression in CD4+CD25+ regulatory T cells.J. Exp. Med. 201:1061–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholin L., Cyprian F.S., Vincent D., Garcia C.N., Martel S., Horvat B., Berthet C., Goddard-Leon S., Treilleux I., Rimokh R., Marie J.C. 2008. Generation of mice with conditionally activated transforming growth factor beta signaling through the TbetaRI/ALK5 receptor.Genesis. 46:724–731 [DOI] [PubMed] [Google Scholar]
- Li M.O., Flavell R.A. 2008. TGF-beta: a master of all T cell trades.Cell. 134:392–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard W.J., Shores E.W., Love P.E. 1995. Role of the common cytokine receptor gamma chain in cytokine signaling and lymphoid development.Immunol. Rev. 148:97–114 [DOI] [PubMed] [Google Scholar]
- Matsuda J.L., Gapin L., Sidobre S., Kieper W.C., Tan J.T., Ceredig R., Surh C.D., Kronenberg M. 2002. Homeostasis of V alpha 14i NKT cells.Nat. Immunol. 3:966–974 [DOI] [PubMed] [Google Scholar]
- Benlagha K., Kyin T., Beavis A., Teyton L., Bendelac A. 2002. A thymic precursor to the NK T cell lineage.Science. 296:553–555 [DOI] [PubMed] [Google Scholar]
- Townsend M.J., Weinmann A.S., Matsuda J.L., Salomon R., Farnham P.J., Biron C.A., Gapin L., Glimcher L.H. 2004. T-bet regulates the terminal maturation and homeostasis of NK and Valpha14i NKT cells.Immunity. 20:477–494 [DOI] [PubMed] [Google Scholar]
- Yang X.O., Nurieva R., Martinez G.J., Kang H.S., Chung Y., Pappu B.P., Shah B., Chang S.H., Schluns K.S., Watowich S.S., et al. 2008. Molecular antagonism and plasticity of regulatory and inflammatory T cell programs.Immunity. 29:44–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda J.L., Mallevaey T., Scott-Browne J., Gapin L. 2008. CD1d-restricted iNKT cells, the ‘Swiss-army knife’ of the immune system.Curr. Opin. Immunol. 20:358–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicci D.G., Hammond K.J., Uldrich A.P., Baxter A.G., Smyth M.J., Godfrey D.I. 2002. A natural killer T (NKT) cell developmental pathway involving a thymus-dependent NK1.1−CD4+ CD1d-dependent precursor stage.J. Exp. Med. 195:835–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransom D.G., Bahary N., Niss K., Traver D., Burns C., Trede N.S., Paffett-Lugassy N., Saganic W.J., Lim C.A., Hersey C., et al. 2004. The zebrafish moonshine gene encodes transcriptional intermediary factor 1gamma, an essential regulator of hematopoiesis.PLoS Biol. 2:E237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letterio J.J., Roberts A.B. 1998. Regulation of immune responses by TGF-beta.Annu. Rev. Immunol. 16:137–161 [DOI] [PubMed] [Google Scholar]
- Matsuda J.L., Zhang Q., Ndonye R., Richardson S.K., Howell A.R., Gapin L. 2006. T-bet concomitantly controls migration, survival, and effector functions during the development of Valpha14i NKT cells.Blood. 107:2797–2805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park I.K., Letterio J.J., Gorham J.D. 2007. TGF-beta 1 inhibition of IFN-gamma-induced signaling and Th1 gene expression in CD4+ T cells is Smad3 independent but MAP kinase dependent.Mol. Immunol. 44:3283–3290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javelaud D., Mauviel A. 2005. Crosstalk mechanisms between the mitogen-activated protein kinase pathways and Smad signaling downstream of TGF-beta: implications for carcinogenesis.Oncogene. 24:5742–5750 [DOI] [PubMed] [Google Scholar]
- Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes.Genome Biol. 3:RESEARCH0034 [DOI] [PMC free article] [PubMed] [Google Scholar]