Abstract
Cancer development is often associated with the lack of specific and efficient recognition of tumor cells by the immune system. Natural killer (NK) cells are lymphocytes of the innate immune system that participate in the elimination of tumors. We report the identification of a tumor cell surface molecule that binds NKp30, a human receptor which triggers antitumor NK cell cytotoxicity and cytokine secretion. This previously unannotated gene belongs to the B7 family and, hence, was designated B7-H6. B7-H6 triggers NKp30-mediated activation of human NK cells. B7-H6 was not detected in normal human tissues but was expressed on human tumor cells, emphasizing that the expression of stress-induced self-molecules associated with cell transformation serves as a mode of cell recognition in innate immunity.
NK cells are large granular lymphocytes that produce chemokines and cytokines and, so, participate in the shaping of the inflammatory and adaptive immune response (Vivier et al., 2008; Yokoyama, 2008). They are also an important part of the innate immune system as they directly kill transformed and virally infected cells (Smyth et al., 2002; Lanier, 2005; Yokoyama, 2008). NK cells survey potential targets for expression of MHC class I via cell surface inhibitory receptors (Parham, 2005). Engagement of these inhibitory receptors by MHC class I prevents NK cell activation, thereby protecting the target cells from NK cell attack. NK cell effector function thus arises in part from recognition of “missing self” (Kärre et al., 1986). The discovery of NK-activating receptors, including the natural cytotoxicity receptor (NCR) family, NKG2D, and DNAM-1, revealed that in addition to an absent inhibitory signal, activating signals were also necessary for NK activation and tumor cell lysis (Pende et al., 1999; Moretta et al., 2001). The NCR family includes NKp46 (NCR1, CD335), NKp44 (NCR2, CD336), and NKp30 (NCR3, CD337). NCR association with CD3ζ, KARAP/DAP12, and Fcrγ immunoreceptor tyrosine-based activation motif–bearing transducing polypeptides is reminiscent of the architecture of other pivotal immune receptor complexes, such as the T, B, and Fc receptors, and makes them very potent activating receptors (Bryceson et al., 2006). Although multiple ligands for NKG2D and DNAM-1 have been identified (Bottino et al., 2005), tumor cell surface ligands for the NCR family have remained elusive, hindering a complete understanding of their role in tumor surveillance. For example, despite the involvement of NKp30 in the lysis of various tumor cell lines and primary tumor cells, including carcinomas, neuroblastomas, and myeloid and lymphoblastic leukemias (Pende et al., 1999, 2002; Sivori et al., 2000; Castriconi et al., 2004; Nowbakht et al., 2005; Byrd et al., 2007), its tumor ligands are still unknown. The HLA–B-associated transcript 3 (BAT3) and the pp65 proteins have been shown to bind NKp30, but they do not correspond to tumor cell surface ligands because pp65 is a human cytomegalovirus tegument protein (Arnon et al., 2005) and BAT3 is a nuclear protein released upon heat shock treatment (Pogge von Strandmann et al., 2007). In this paper, we report that human NKp30 directly interacts with a novel member of the B7 family that we refer to as B7-H6. Using NKp30 reporter cells, human IL-2–driven NK cell lines, and freshly isolated human blood NK cells, we show that the expression of B7-H6 on tumor cells induces NKp30-dependent cell activation and cytotoxicity. Furthermore, B7-H6 was absent from all normal cells tested but was selectively expressed on tumor cells. These results show the first instance of a tumor-specific B7 family member that can directly trigger NK cell activation.
RESULTS AND DISCUSSION
Identification of B7-H6 as a ligand for NKp30
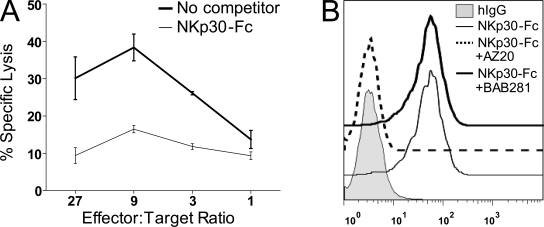
The human NK-92 cell line expresses surface NKp30 (not depicted) and mediates killing of the human chronic myelogenous leukemia cell line K562 (Fig. 1 A). To search for molecules that bind to NKp30 and trigger NKp30-mediated cytolysis in NK cells, we generated soluble NKp30-Fc fusion proteins. Soluble NKp30-Fc proteins bound to K562 cells, and this binding was abolished by blocking anti-NKp30mAb (AZ20) but not by blocking control anti-NKp46 mAb (BAB281; Fig. 1 B). In addition, soluble NKp30 reduced the ability of NK-92 cells to kill K562 targets (Fig. 1 A). Collectively, these experiments revealed K562 cells and soluble NKp30-Fc proteins as practicable tools for the identification of a cell surface ligand for NKp30.
Figure 1.
Soluble NKp30-Fc inhibits NK-92 cytolysis and binds K562 cells. (A) Soluble NKp30-Fc inhibited the killing of K562 cells by NK-92 cells. A 4-h cytoxicity assay was performed in the presence (thin line) or absence (bold line) of 2 µg/ml NKp30-Fc. Data indicate the mean of triplicates ± SD and are representative of three independent experiments. (B) Soluble NKp30-Fc binds K562. The binding of NKp30-Fc to K562 cells was measured by flow cytometry in the presence of blocking anti-NKp30 F(ab′)2 mAb (AZ20, IgG1) or a negative control anti-NKp46 F(ab′)2 mAb (BAB281, IgG1). Data are representative of at least three independent experiments.
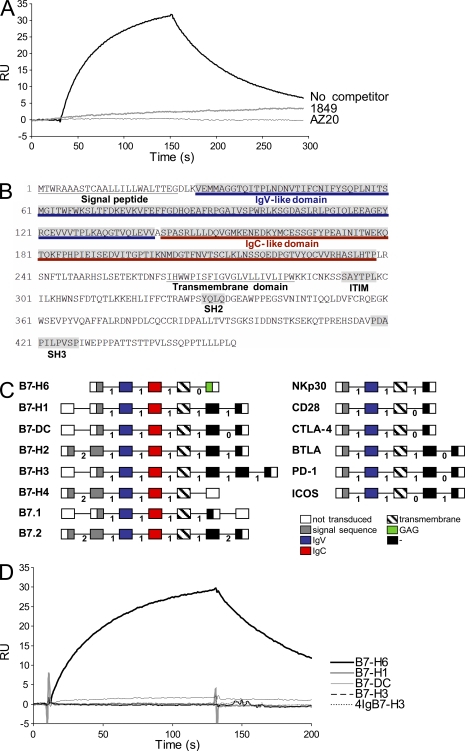
A proteomic approach was used to search for this counter structure. Biotinylated NKp30-Fc was used as a probe in chemical cross-linking studies to K562 cells. Proteins cross-linked to the NKp30-Fc protein were precipitated, separated by gel electrophoresis, and subjected to in-gel tryptic digestion. The resulting peptides were analyzed by mass spectrometry. Substractive proteomic analysis was used to identify candidate NKp30 binding proteins that were unique to K562 cell lysates and were identified by two or more peptides. Two of these candidates were predicted to have transmembrane domains, and their complementary DNAs were obtained for subsequent studies. A recombinant Fc fusion form of one of these candidates (hypothetical protein DKFZp686O24166) gave specific binding to soluble NKp30 as judged by surface plasmon resonance (SPR) experiments (Fig. 2 A). NKp30 interaction with DKFZp686O24166 was inhibited by anti-NKp30 mAbs (1849 and AZ20) that block NKp30-dependent NK cell activation (Fig. 2 A).
Figure 2.
Characterization of B7-H6. (A) Direct interaction of B7-H6 with NKp30 using SPR. The data show binding of soluble DKFZp686O24166-Fc to immobilized NKp30-Fc in the presence or absence of blocking anti-NKp30 mAb (1849 and AZ20). Data are representative of three independent experiments. (B) Protein sequence encoded by the B7-H6 gene. Predicted signal peptide, IgV, IgC, and transmembrane domains are indicated. (C) Schematic representation of the B7-H6 and the NKp30 gene orgnaization compared with members of the B7 and the CD28 families, respectively. Each domain is represented as a box and is encoded by exons that are separated by phase 0, 1, or 2 introns as indicated. The scheme refers to the “canonical” sequence as defined by www.uniprot.org. (D) The data show SPR experiments using soluble B7-H1-Fc, B7-H3-Fc, 4IgB7-H3-Fc, B7-H6-Fc, and B7-DC-Fc and immobilized NKp30-Fc. RU, relative units. Data are representative of two independent experiments.
DKFZp686O24166 codes for a 454-aa-long type I transmembrane protein (51 kD predicted molecular mass) with an intracytoplasmic domain that is homologous to GAG polyprotein. Interestingly, this intracytoplasmic domain also contains various signaling motifs, such as an immunoreceptor tyrosine-based inhibition motif (SaYtpL), an SH2 (Src homology 2)-binding domain (YqlQ), and an SH3-binding motif (PdaPilPvsP), suggesting the possibility that DKFZp686O24166 could provide signals when engaged (Fig. 2 B). A BLAST query revealed that two of the most related proteins are members of the B7 family (B7-H1 and B7-H3). Like all known B7 family members (B7.1, B7.2, B7-DC, and B7-H1 to B7-H4/B7h.5; Zou and Chen, 2008), DKFZp686O24166 has two Ig domains with adjacent phase 1 introns in the extracellular region (Fig. 2 C). TPSNR (tapasin-related protein), PVR (poliovirus receptor; CD155), CADM3 (cell adhesion molecule 3), and CXADR (Coxsackie virus and adenovirus receptor) also showed amino acid similarity, but their gene and predicted domain organization were different from those of DKFZp686O24166. Further protein homology analysis revealed that the percentage of DKFZp686O24166 identity with other B7 family members is comparable to the percentages of identity observed within this group of distantly related molecules (Table S1). In addition, a BLAST query revealed that the closest homologue of NKp30 was CTLA-4, a receptor for B7.1 and B7.2. Like other members of the CD28 family, NKp30 has a single predicted IgV domain in its extracellular region, whereas nearly all other Ig domain–containing immunoreceptors have multiple Ig domains. Moreover, the gene structure of the extracellular region is identical between NKp30 and all other CD28 family genes. Specifically, there is an exon containing the leader peptide, followed by a phase 1 intron, then the exon containing the IgV domain, and another phase 1 intron, followed by a short exon containing the transmembrane region (Fig. 2 C). The only other proteins identified to date that have this exact gene structure are the known members of the CD28 family including CD28, CTLA-4 (CD152), BTLA (CD272), ICOS (CD278), and PD-1 (CD279). NKp30 could thus be considered as a member of the CD28 family whose members do not share significant sequence similarity to each other but do share a similar gene structure. Thus, because DKFZp686O24166 had a gene and protein structure similar to that of other members of the B7 family, two of its closest homologues were members of that group, and its putative counter structure, NKp30, is related to the CD28 family, we designated the gene B7-H6. B7-H6 was present in genomic clone AC124798 (available at GenBank/EMBL/DDBJ) which maps to chromosome region 11p15.1. A short region corresponding to the first exon of human B7-H6 was found in the mouse genome (near bp 47,000 of AC146610.4, available at GenBank/EMBL/DDBJ), but a full-length gene sequence was not evident. This mouse exon appears to have intact splice sites at either end, but no other nearby regions of the mouse genome have other exons related to B7-H6. Finally, this isolated exon from the mouse does not have any related ESTs from mouse libraries, and we conclude that the Mus musculus genome is missing a full-length gene corresponding to human B7-H6.
Given the structure of B7-H6, we also tested it for possible interactions with the other known CD28 family members but only observed binding with soluble NKp30 (unpublished data). Moreover, B7-H6 did not bind to other NK-activating receptors including NKp44, NKp46, and NKG2D (unpublished data). No binding of NKp30-Fc to B7.1, B7-DC, B7-H3, 4IgB7-H3, or B7-H1 could be detected (Fig. 2 D). These results indicated that the extracellular domain of NKp30 directly and selectively interacts with the extracellular domain of B7-H6, a novel member of the B7 family.
Functional significance of B7-H6 surface expression
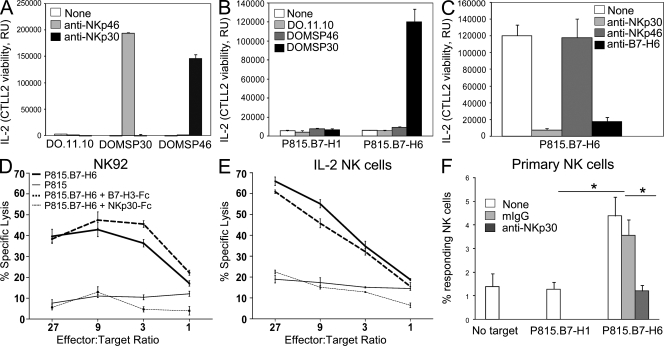
The IL-2–producing DO11.10 mouse T cell hybridoma expressing a chimeric receptor formed by NKp30 extracellular domain fused to CD3ζ (DOMSP30 cells; Schleinitz et al., 2008) was used as a reporter cell system to evaluate whether B7-H6 was able to induce signaling directly through NKp30. DOMSP30 reporter cells were activated by plate-bound anti-NKp30 mAbs, which is in contrast to DOMSP46 reporter cells expressing a NKp46-CD3ζ chimeric receptor which were selectively responsive to NKp46 cross-linking (Fig. 3 A). DOMSP30 reporter cells produced IL-2 in the presence of P815 expressing B7-H6 (P815.B7-H6), whereas P815 expressing a control protein (P815.B7-H1) failed to elicit such a response (Fig. 3 B). Parental DO11.10 cells and DOMSP46 reporter cells were unresponsive to P815.B7-H6 cells (Fig. 3 B). IL-2 production was markedly reduced by addition of either blocking anti-NKp30 mAb F(ab′)2 or affinity-purified B7-H6 antiserum (Fig. 3 C). No inhibition was observed with blocking anti-NKp46 mAb F(ab′)2 controls. Together, these results show that B7-H6 engagement of the NKp30/CD3ζ chimera leads to cell activation. To further examine the functional significance of the interaction of NKp30 with B7-H6, parental P815 cells and P815.B7-H6 cells were used as targets for the NK-92 cell line (Fig. 3 D). Although parental P815 cells were lysed poorly by NK-92 cells, P815.B7-H6 cells were lysed effectively (Fig. 3 D). This susceptibility to cytolysis was impaired by addition of soluble NKp30-Fc but not by addition of soluble B7-H3-Fc control proteins (Fig. 3 D). P815.B7-H6 cells were also lysed by IL-2–activated primary human NK cells and this cytotoxicity was blocked by addition of soluble NKp30-Fc (Fig. 3 E). These data indicate that B7-H6 expression on tumor cells can confer sensitivity to NKp30-specific cytolysis. Consistent with these data, freshly isolated blood NK cells were activated by P815.B7-H6 cells but not by P815.B7-H1 cells, as assessed by cell surface expression of CD107a (LAMP-1) and intracytoplasmic detection of IFN-γ (Fig. 3 F). That a tumor ligand for the activating receptor NKp30 was a member of the B7 family was an unexpected finding. Indeed, recent studies have shown that human and rodent cancer cells up-regulate expression of B7 molecules, such as B7-H1, 4IgB7-H3, and B7-H4, but this expression has been proposed to contribute to tumor immune evasion by engaging inhibitory receptors (Castriconi et al., 2004; Zou and Chen, 2008).
Figure 3.
B7-H6 cell surface expression triggers NKp30-dependent cell activation. (A–C) DO.11.10, DOMSP30, and DOMSP46 cells were stimulated using the indicated plate-bound mAbs (A) or P815.B7-H1 versus P815.B7-H6 cells in the absence (B) or presence of indicated F(ab′)2 mAbs or B7-H6 mouse antiserum (C). Data indicate the mean of triplicates + SD and are representative of three independent experiments. (D and E) NK-92 cells (D) and IL-2–stimulated primary NK cells (E) were used in a cytolytic assay against P815 cells (thin line) or P815.B7-H6 cells (bold line). Soluble NKp30-Fc (thin dashed line) and control Fc-fusion protein (B7-H3-Fc; bold dashed line) were used at 2 µg/ml. (F) Freshly isolated primary blood NK cells in PBMCs were tested for their activation induced by P815.B7-H1 or P815.B7-H6 cells in the presence or absence of anti-NKp30 F(ab′)2 mAbs or control mouse Ig (mIg). 500,000 PBMCs were added to 100,000 tumor cells. The fraction of reactive NK cells (percentage of responding NK cells) was assessed by adding the percentage of CD107+IFN-γ−, CD107+IFN-γ+, and CD107−IFN-γ+ NK cells. Data are representative of three independent experiments. Error bars indicate SD. Statistical analysis was performed using a Mann-Whitney U test. *, P < 0.05.
Cellular distribution of B7-H6
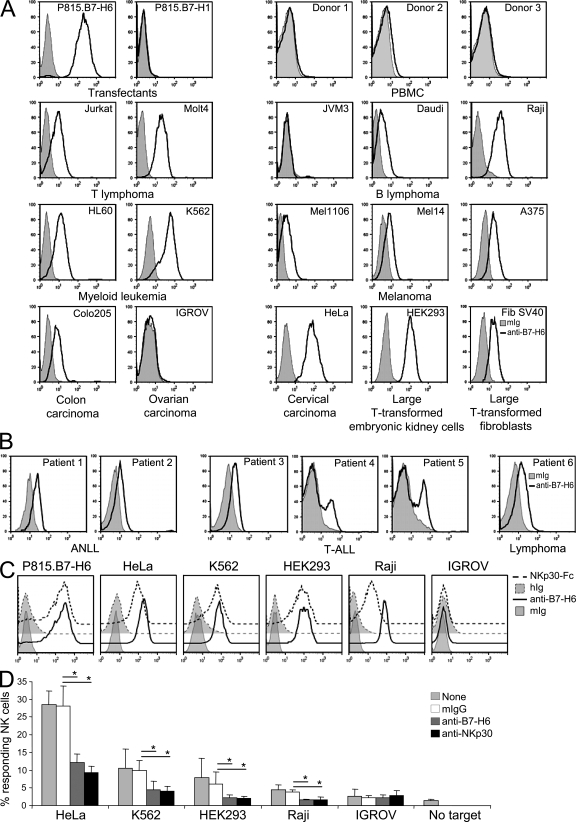
Having established that B7-H6 was a functionally relevant counter structure for NKp30, we next analyzed its expression pattern. Quantitative reverse transcription polymerase chain reaction analysis of B7-H6 messenger RNA (mRNA) on a normal tissue array showed no detectable expression in any of the 48 normal tissues assayed (unpublished data). This paucity of B7-H6 mRNA in normal tissues was consistent with microarray analysis of the gene (Shyamsundar et al., 2005; and not depicted). Collectively with the fact that few expressed sequence tags for B7-H6 have been identified, these data suggest that B7-H6 is not abundantly expressed in any normal tissues examined. This was also supported by the failure to detect B7-H6 at the surface of freshly isolated PBMCs from all donors tested using affinity-purified B7-H6 antiserum (Fig. 4 A) or mAbs directed against B7-H6 (not depicted). Moreover, no B7-H6 cell surface staining was observed on phytohemagglutinin-stimulated PBMCs (unpublished data). It is of note that although NKp30 has a role in NK cell–DC cross-talk (Ferlazzo et al., 2002; Vitale et al., 2005), no B7-H6 mRNA or protein expression was detected when assaying resting or activated DCs generated from PBMCs with IL-4 and GM-CSF (unpublished data). It is therefore possible that BAT3 serves as a ligand for NKp30 on DCs (Simhadri et al., 2008), indicating that NKp30 might interact with several ligands, as this is the case for NKG2D and DNAM-1 (Raulet, 2003; Soriani et al., 2009). Along this line, it has been shown that NKp30 and other NCRs can recognize heparan sulfate (Hecht et al., 2009). Heparan sulfates are found in the commonest linkage type with xylose O-glycosidically linked to serine. No potential O-glycosylation sites could be found in B7-H6. There are also studies where heparan sulfate has been generated as N-glycans (Sundblad et al., 1988), and the extracytoplasmic domain of B7-H6 harbors nine potential N-glycosylation sites. It thus remains to be tested whether the glycosylation of B7-H6 might contribute to its interaction with NKp30, although we could not alter DOMSP30 cell activation by addition of heparan sulfate (unpublished data).
Figure 4.
B7-H6 is not detected on blood cells but is expressed on tumor cells. (A and B) Assessment of B7-H6 cell surface staining on PBMCs, P815 cell transfectants, and tumor cells was performed by flow cytometry using anti–B7-H6 mouse antiserum. (C) NKp30-Fc cell surface staining on tumor cells is also represented. HIg, control human Ig; mIg, control mouse Ig. (D) Freshly isolated primary blood NK cells in PBMCs were tested for their activation induced by the indicated cell lines in the presence or absence of anti-NKp30 F(ab′)2 mAbs, B7-H6 mouse antiserum, or control mouse Ig (mIg). 500,000 PBMCs were added to 100,000 tumor cells. The fraction of reactive NK cells (percentage of responding NK cells) was assessed by adding the percentage of CD107+IFN-γ−, CD107+IFN-γ+, and CD107−IFN-γ+ NK cells (mean + SD; n = 3 independent experiments). Statistical analysis was performed using a Mann-Whitney U test. *, P < 0.05. Open histograms, B7-H6 staining; filled histograms, control mIg staining.
Recent reports have revealed that the induction of stress-induced self is a common mechanism that leads to the initiation of innate immune response (Raulet, 2003). In preliminary experiments, no induction of B7-H6 cell surface expression was detected under various conditions of cellular stress such as heat shock, genotoxicity (γ radiation or sodium butyrate treatment), or inhibition of proteasomal degradation (MG-132 treatment). In contrast, B7-H6 was expressed at the surface of several tumor cell lines including T and B lymphomas, myeloid leukemias, melanomas, carcinomas, and large T SV40 antigen-transformed cells (Fig. 4 A). In a broader survey, B7-H6 expression was detected on 24 of 119 cancer cell lines (unpublished data). The surface expression of B7-H6 was also tested on primary tumor blood and bone marrow cells obtained at diagnosis from 43 individuals with a variety of hematological malignancies, including acute nonlymphoblastic leukemia (ANLL; n = 20), acute lymphoblastic leukemia (ALL; n = 11), and non-Hodgkin's and Hodgkin's lymphoma (n = 12). B7-H6 expression was observed on circulating tumor cells from three patients with ANLL, two patients with T-ALL, and one patient with marginal zone lymphoma (Fig. 4 B). B7-H6 cell surface expression correlated with the binding of NKp30-Fc to the cell surface and with the capacity of tumor cell lines to activate resting blood NK cells (Fig. 4, C and D). Collectively, these data indicated that B7-H6 is a major NKp30 ligand on tumor cells of various origins, including K562, a prototypical model of tumor NK cell target.
The absence of B7-H6 mRNA in normal tissues, coupled with its relative abundance among tumor cells, indicates that its expression is up-regulated by tumor transformation. These findings highlight the role of tumor-induced self-molecules in alerting innate immunity and prompt further investigations on whether B7-H6 expression correlates with tumor prognosis in a large cohort of patients and on the regulation of B7-H6 expression.
MATERIALS AND METHODS
Soluble recombinant proteins.
In-frame fusion proteins of the extracellular domains of NKp30 (aa 1–132) or of B7-H6 (aa 1–267) and an effector-negative version of the mouse IgG2a Fc region (aa 216 through the C-terminal lysine) were constructed in a mammalian expression vector (NKp30-mFc). Secreted Fc fusion proteins from stable Chinese hamster ovary transfectants were purified by affinity chromatography on Protein A Sepharose (GE Healthcare). For the generation of mouse Fc fusion protein, mutations in mIgG2a were introduced as follows. The hinge residue corresponding to EU Index position 219 was changed from Gly in the wild-type γ2a sequence to Ser to mimic the amino acid substitution made in human Fc4, Fc5, and Fc10. For CH2, one amino acid substitution was introduced in mFc2 relative to mouse wild-type γ2a at EU Index position 235 (Leu to Glu) to inactivate binding to FcγRI and FcγRII (Duncan et al., 1988; Zheng et al., 1999). Three additional changes were made at the complement C1q binding site to reduce complement fixation at EU Index positions 318, 320, and 322 (Duncan and Winter, 1988; Duncan et al., 1988). A soluble recombinant NKp30 molecule fused to mutated human IgG1 (NKp30-hFc) was also generated and purified from HEK293 transfected cells (provided by C. Cantoni, University of Genova, Genova, Italy). NKp30-hFc was used for the experiments represented in all the figures, with the exception of Fig. 1 A and Fig. 3 (D and E), where NKp30-mFc was used. Other Fc fusion proteins were obtained from R&D Systems.
Proteomics.
Human erythroleukemic K562 cells (NKp30-Fc+) or mouse pro–B Ba/F3 cells (NKp30-Fc−) were incubated with 2 mg/ml NKp30-Fc-biotin, washed, and then incubated with 3 mg/ml bis(sulfosuccinimidyl) suberate (Thermo Fisher Scientific) for in situ chemical cross-linking. After cell lysis, NKp30-Fc-biotin–bound materials were purified using streptavidin agarose. A coomassie-stained gel of these streptavidin agarose–purified lysates was divided into 16 sections across experiment and controls based on Western blot band laddering. Proteins were reduced with 25 mM TCEP (Tris[2-carboxyethyl] phospine) for 15 min at 80°C and free cysteines were capped with 100 mM iodoacetamide for 2 h at 25°C. Samples were digested with porcine trypsin (V5111; Promega) at 20 µg/ml in 25 mM NH4HCO3 for 18 h at 37°C. Peptides were extracted from the gel with 60% vol/vol acetonitrile in 1% formic acid, dried under vacuum, and reconstituted in 20 µl of 0.1% formic acid in H20. The resulting peptide mixture was separated on a Magic C18AQ (Michrom Bioresources, Inc.) 3-µm 200-Å resin packed into 10 cm of 50 µm of fused silica. Eluted peptides were analyzed by nano–liquid chromatography tandem mass spectrometry using an ion-trap mass spectrometer (Thermo Fisher Scientific) interfaced with nanoflow RP-HPLC. Proteins were identified by an X!Tandem search of peptide mass spectra against the human and mouse IPI database for K562 cells and BaF3 cells (used as a negative control), respectively.
Affinity-purified B7-H6 antiserum production and purification.
Female BALB/c mice (Charles River Laboratories) were immunized by subcutaneous injection with purified recombinant human B7-H6 tetramer in combination with Emulsigen-P adjuvant (MVP Laboratories, Inc.) as per the manufacturer's instructions. Mice were boosted subcutaneously every 2 wk over a 9-wk period. Serum from mice challenged with B7-H6 tetramer was pooled and affinity purified on B7-H6-Fc protein coupled to CNBr-activated Sepharose 4B (GE Healthcare). Specificity of the eluted antiserum was confirmed by FACS analysis on B7-H6–transfected P815 cells compared with untransfected P815.
SPR.
SPR analysis was performed on a Biacore T100 apparatus (GE Healthcare) at 25°C. In all Biacore experiments, HBS-EP+ buffer served as running buffer and sensorgrams were analyzed with Biaevaluation 4.1 and Biacore T100 Evaluation software. Recombinant protein was immobilized covalently to carboxyl groups in the dextran layer of a Biacore Sensor Chip CM5. Competition assays were performed in acidic buffer, pH 5.6. Flow rate was set to 10 µl/min. mAbs were injected at 50 µg/ml before the injection of B7-H6-Fc.
Primary NK cell preparation, culture, and activation.
PBMCs were isolated from healthy volunteer donors using Ficoll-Paque Plus (GE Healthcare) density centrifugation. For CD107 mobilization and IFN-γ production, PBMCs were incubated 4 h at 37°C in the presence of 5 µM monensin (Sigma-Aldrich), CD107 mAb, and the indicated stimuli. Cells were then washed in PBS with 2 mM EDTA and stained for extracellular markers. Thereafter, cells were fixed with 2% paraformaldehyde and permeabilized using Perm/Wash buffer (BD). NK cells were electronically gated as CD3−CD56+. Intracellular IFN-γ was detected using APC–IFN-γ (BD).
Immunofluoresence and flow cytometry.
Mouse isotype control antibodies (mIg) consisted of an equimolar mixture of commercially available isotype control mouse IgG1, IgG2a, IgG2b, and IgM (BD). Human isotype control antibodies (hIg) were a mixture of human Ig (TEGELIN, Laboratoire français du Fractionnement et des Biotechnologies). For NKp30-hFc stainings, 5 µg/ml NKp30-hFc was mixed to 5 µg/ml of PE-conjugated anti–human Ig (Jackson ImmunoResearch Laboratories) for 1 h at 4°C before incubation with cells for 30 min at 4°C. Fluorescent stainings were analyzed on a FACSCanto (BD) using the FlowJo software (Tree Star, Inc.).
DOMSP30 assay.
DOMSP30 and DOMSP46 reporter cell lines were generated by transduction of the DO.11.10 T cell hybridoma with retroviral particles encoding a chimeric protein in which the intracytoplasmic domain of mouse CD3ζ was fused either to the extracellular portion of NKp30 (DOMSP30) or NKp46 (DOMSP46). Engagement of these chimeric proteins at the cell surface triggers IL-2 secretion. DOMSP30, DOMSP46, or DO.11.10 (30,000 cells/well in 96-well plates) were incubated on anti-NKp30 or anti-NKp46 mAb-coated plates or with tumor cell lines (30,000 cells/well in 96-well plates). After 20 h, cell supernatants were assayed for the presence of mouse IL-2 in a standard CTLL-2 survival assay using Cell Titer-Glo Luminescent Cell Viability Assay (Promega).
Cytolytic assay.
Target cells were loaded with calcein fluorescent dye as previously described (Lichtenfels et al., 1994). In brief, cells were washed in Hank's Balanced Salt Solution (Invitrogen)/5% FBS (HBSSF) and labeled at 106 cells/ml in HBSSF with 10 mM Calcein AM (Invitrogen) for 1 h at 37°C. Cells were washed with HBSSF and then resuspended at 50,000 cells/ml. Labeled target cells (5,000 cells/100 µl/well) were plated with varying numbers of effector cells in a 96-well U-bottom plate. Cells were briefly spun down and incubated at 37°C for 4 h. After 4 h, cells were pelleted and 100 µl of supernatant was transferred to fresh 96-well plates and read on a fluorimeter using a 1-s read at 485-nm excitation and 535-nm emission. Specific lysis was calculated by subtracting mean spontaneous lysis from mean lysis divided by mean spontaneous lysis subtracted from mean total lysis (obtained by lysing targets in 0.1% Tween 20).
Human primary leukemic and lymphoma cells.
Blood and bone marrow samples were obtained during diagnostic procedures. Once all the diagnostic tests were done, the leftover was qualified for research use, upon obtainment of patient informed consent, as per French regulations. The mononuclear cell fraction from either blood or bone marrow samples was cryopreserved in 4% human albumin with 10% DMSO, according to standard operating procedures at the Institut Paoli-Calmettes biological resource center. The use of leukemia and lymphoma cryopreserved cells for this study was reviewed and approved by the Institutional Review Board (Comité d'Orientation Stratégique) at Institut Paoli-Calmettes. Human and mouse experimental protocols were approved by the Western Institutional Review Board (Seattle, WA) and Comité consultatif d'éthique pour les sciences de la vie et de la santé (UnivMed, Institut National de la Santé et de la Recherche Médicale, Centre National de la Recherche Scientifique, Marseille, France).
Online supplemental material.
Table S1 exemplifies homologies in the extracellular domain of B7 family members. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20090681/DC1.
Acknowledgments
The authors would like to thank Pallavur Sivakumar, Pierre Golstein, Jonathan Ewbank, and E.V. laboratory members for critically reviewing the manuscript, as well as Claudia Cantoni (Genova University), François Sigaux (Saint Louis Hospital, Paris), Louis Gastinel (Limoges University), Laurent Gauthier, Mathieu Bléry, Nicola Hugues, and Nicolas Viaud (Innate-Pharma) for their help and advice.
The E.V. laboratory is supported by Ligue Nationale contre le Cancer (Equipe labelisée La Ligue), Agence Nationale de la Recherche, Institut National de la Santé et de la Recherche Médicale, Centre National de la Recherche Scientifique, and Ministère de l'Enseignement Supérieur et de la Recherche. E. Vivier is an Institut Universitaire de France scholar. A. Moretta is supported by grants awarded by Associazione Italiana per la Ricerca sul Cancro, Istituto Superiore di Sanità, Ministero della Sanità, Ministero dell'Istruzione, and dell'Università e della Ricerca Scientifica e Tecnologica.
C.S. Brandt, E.C. Yi, J. Kennedy, Z. Gao, B. Fox, B. Haldeman, C.D. Ostrander, R. West, W. Xu, and S.D. Levin were all employees of ZymoGenetics, Inc. at the time the work described here was done and hence declare a competing financial interest. E. Vivier and A. Moretta are cofounders and shareholders of Innate-Pharma and hence declare a competing financial interest. The authors have no additional financial interests.
Footnotes
Abbreviations used: BAT3, B-associated transcript 3; mRNA, messenger RNA; NCR, natural cytotoxicity receptor; SPR, surface plasmon resonance.
References
- Arnon T.I., Achdout H., Levi O., Markel G., Saleh N., Katz G., Gazit R., Gonen-Gross T., Hanna J., Nahari E., et al. 2005. Inhibition of the NKp30 activating receptor by pp65 of human cytomegalovirus.Nat. Immunol. 6:515–523 [DOI] [PubMed] [Google Scholar]
- Bottino C., Castriconi R., Moretta L., Moretta A. 2005. Cellular ligands of activating NK receptors.Trends Immunol. 26:221–226 [DOI] [PubMed] [Google Scholar]
- Bryceson Y.T., March M.E., Ljunggren H.G., Long E.O. 2006. Activation, coactivation, and costimulation of resting human natural killer cells.Immunol. Rev. 214:73–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd A., Hoffmann S.C., Jarahian M., Momburg F., Watzl C. 2007. Expression analysis of the ligands for the natural killer cell receptors NKp30 and NKp44.PLoS One. 2:e1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castriconi R., Dondero A., Augugliaro R., Cantoni C., Carnemolla B., Sementa A.R., Negri F., Conte R., Corrias M.V., Moretta L., et al. 2004. Identification of 4Ig-B7-H3 as a neuroblastoma-associated molecule that exerts a protective role from an NK cell-mediated lysis.Proc. Natl. Acad. Sci. USA. 101:12640–12645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan A.R., Winter G. 1988. The binding site for C1q on IgG.Nature. 332:738–740 [DOI] [PubMed] [Google Scholar]
- Duncan A.R., Woof J.M., Partridge L.J., Burton D.R., Winter G. 1988. Localization of the binding site for the human high-affinity Fc receptor on IgG.Nature. 332:563–564 [DOI] [PubMed] [Google Scholar]
- Ferlazzo G., Tsang M.L., Moretta L., Melioli G., Steinman R.M., Munz C. 2002. Human dendritic cells activate resting natural killer (NK) cells and are recognized via the NKp30 receptor by activated NK cells.J. Exp. Med. 195:343–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht M.L., Rosental B., Horlacher T., Hershkovitz O., De Paz J.L., Noti C., Schauer S., Porgador A., Seeberger P.H. 2009. Natural cytotoxicity receptors NKp30, NKp44 and NKp46 bind to different heparan sulfate/heparin sequences.J. Proteome Res. 8:712–720 [DOI] [PubMed] [Google Scholar]
- Kärre K., Ljunggren H.G., Piontek G., Kiessling R. 1986. Selective rejection of H-2-deficient lymphoma variants suggests alternative immune defense strategy.Nature. 319:675–678 [DOI] [PubMed] [Google Scholar]
- Lanier L.L. 2005. NK cell recognition.Annu. Rev. Immunol. 23:225–274 [DOI] [PubMed] [Google Scholar]
- Lichtenfels R., Biddison W.E., Schulz H., Vogt A.B., Martin R. 1994. CARE-LASS (calcein-release-assay), an improved fluorescence-based test system to measure cytotoxic T lymphocyte activity.J. Immunol. Methods. 172:227–239 [DOI] [PubMed] [Google Scholar]
- Moretta A., Bottino C., Vitale M., Pende D., Cantoni C., Mingari M.C., Biassoni R., Moretta L. 2001. Activating receptors and coreceptors involved in human natural killer cell-mediated cytolysis.Annu. Rev. Immunol. 19:197–223 [DOI] [PubMed] [Google Scholar]
- Nowbakht P., Ionescu M.C., Rohner A., Kalberer C.P., Rossy E., Mori L., Cosman D., De Libero G., Wodnar-Filipowicz A. 2005. Ligands for natural killer cell-activating receptors are expressed upon the maturation of normal myelomonocytic cells but at low levels in acute myeloid leukemias.Blood. 105:3615–3622 [DOI] [PubMed] [Google Scholar]
- Parham P. 2005. MHC class I molecules and KIRs in human history, health and survival.Nat. Rev. Immunol. 5:201–214 [DOI] [PubMed] [Google Scholar]
- Pende D., Parolini S., Pessino A., Sivori S., Augugliaro R., Morelli L., Marcenaro E., Accame L., Malaspina A., Biassoni R., et al. 1999. Identification and molecular characterization of NKp30, a novel triggering receptor involved in natural cytotoxicity mediated by human natural killer cells.J. Exp. Med. 190:1505–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pende D., Rivera P., Marcenaro S., Chang C.C., Biassoni R., Conte R., Kubin M., Cosman D., Ferrone S., Moretta L., Moretta A. 2002. Major histocompatibility complex class I-related chain A and UL16-binding protein expression on tumor cell lines of different histotypes: analysis of tumor susceptibility to NKG2D-dependent natural killer cell cytotoxicity.Cancer Res. 62:6178–6186 [PubMed] [Google Scholar]
- Pogge von Strandmann E., Simhadri V.R., Von Tresckow B., Sasse S., Reiners K.S., Hansen H.P., Rothe A., Boll B., Simhadri V.L., Borchmann P., et al. 2007. Human leukocyte antigen-B-associated transcript 3 is released from tumor cells and engages the NKp30 receptor on natural killer cells.Immunity. 27:965–974 [DOI] [PubMed] [Google Scholar]
- Raulet D.H. 2003. Roles of the NKG2D immunoreceptor and its ligands.Nat. Rev. Immunol. 3:781–790 [DOI] [PubMed] [Google Scholar]
- Schleinitz N., Cognet C., Guia S., Laugier-Anfossi F., Baratin M., Pouget J., Pelissier J.F., Harle J.R., Vivier E., Figarella-Branger D. 2008. Expression of the CD85j (leukocyte Ig-like receptor 1, Ig-like transcript 2) receptor for class I major histocompatibility complex molecules in idiopathic inflammatory myopathies.Arthritis Rheum. 58:3216–3223 [DOI] [PubMed] [Google Scholar]
- Shyamsundar R., Kim Y.H., Higgins J.P., Montgomery K., Jorden M., Sethuraman A., Van De Rijn M., Botstein D., Brown P.O., Pollack J.R. 2005. A DNA microarray survey of gene expression in normal human tissues.Genome Biol. 6:R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simhadri V.R., Reiners K.S., Hansen H.P., Topolar D., Simhadri V.L., Nohroudi K., Kufer T.A., Engert A., Pogge Von Strandmann E. 2008. Dendritic cells release HLA-B-associated transcript-3 positive exosomes to regulate natural killer function.PLoS One. 3:e3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivori S., Parolini S., Marcenaro E., Castriconi R., Pende D., Millo R., Moretta A. 2000. Involvement of natural cytotoxicity receptors in human natural killer cell-mediated lysis of neuroblastoma and glioblastoma cell lines.J. Neuroimmunol. 107:220–225 [DOI] [PubMed] [Google Scholar]
- Smyth M.J., Hayakawa Y., Takeda K., Yagita H. 2002. New aspects of natural-killer-cell surveillance and therapy of cancer.Nat. Rev. Cancer. 2:850–861 [DOI] [PubMed] [Google Scholar]
- Soriani A., Zingoni A., Cerboni C., Iannitto M.L., Ricciardi M.R., Di Gialleonardo V., Cippitelli M., Fionda C., Petrucci M.T., Guarini A., et al. 2009. ATM-ATR dependent up-regulation of DNAM-1 and NKG2D ligands on multiple myeloma cells by therapeutic agents results in enhanced NK cell susceptibility and is associated with a senescent phenotype.Blood. 113:3503–3511 [DOI] [PubMed] [Google Scholar]
- Sundblad G., Holojda S., Roux L., Varki A., Freeze H.H. 1988. Sulfated N-linked oligosaccharides in mammalian cells. II. Identification of glycosaminoglycan-like chains attached to complex-type glycans.J. Biol. Chem. 263:8890–8896 [PubMed] [Google Scholar]
- Vitale M., Della Chiesa M., Carlomagno S., Pende D., Arico M., Moretta L., Moretta A. 2005. NK-dependent DC maturation is mediated by TNFalpha and IFNgamma released upon engagement of the NKp30 triggering receptor.Blood. 106:566–571 [DOI] [PubMed] [Google Scholar]
- Vivier E., Tomasello E., Baratin M., Walzer T., Ugolini S. 2008. Functions of natural killer cells.Nat. Immunol. 9:503–510 [DOI] [PubMed] [Google Scholar]
- Yokoyama W.M. 2008. Mistaken notions about natural killer cells.Nat. Immunol. 9:481–485 [DOI] [PubMed] [Google Scholar]
- Zheng X.X., Steele A.W., Hancock W.W., Kawamoto K., Li X.C., Nickerson P.W., Li Y., Tian Y., Strom T.B. 1999. IL-2 receptor-targeted cytolytic IL-2/Fc fusion protein treatment blocks diabetogenic autoimmunity in nonobese diabetic mice.J. Immunol. 163:4041–4048 [PubMed] [Google Scholar]
- Zou W., Chen L. 2008. Inhibitory B7-family molecules in the tumour microenvironment.Nat. Rev. Immunol. 8:467–477 [DOI] [PubMed] [Google Scholar]