Abstract
The t(14;18) translocation constitutes the initiating event of a causative cascade leading to follicular lymphoma (FL). t(14;18) translocations are present in blood from healthy individuals, but there is a trend of increased prevalence in farmers exposed to pesticides, a group recently associated with higher risk of t(14;18)+ non-Hodgkin's lymphoma development. A direct connection between agricultural pesticide use, t(14;18) in blood, and malignant progression, however, has not yet been demonstrated. We followed t(14;18) clonal evolution over 9 yr in a cohort of farmers exposed to pesticides. We show that exposed individuals bear particularly high t(14;18) frequencies in blood because of a dramatic clonal expansion of activated t(14;18)+ B cells. We further demonstrate that such t(14;18)+ clones recapitulate the hallmark features of developmentally blocked FL cells, with some displaying aberrant activation-induced cytidine deaminase activity linked to malignant progression. Collectively, our data establish that expanded t(14;18)+ clones constitute bona fide precursors at various stages of FL development, and provide a molecular connection between agricultural pesticide exposure, t(14;18) frequency in blood, and clonal progression.
Agricultural pesticide use has been repeatedly, but not consistently, associated with risk of non-Hodgkin's lymphoma (NHL; Blair et al., 1992; Dreiher and Kordysh, 2006). Over the past years, it has become clear that NHL heterogeneity is setting many hurdles to this connection and that pertinent biomarkers must be identified according to malignant subtypes (Harris et al., 1994). Among those, t(14;18) translocation constitute the genetic hallmark and early initiating event of follicular lymphoma (FL), a germinal center (GC)–derived malignancy representing one of the most common adult NHLs (Cleary et al., 1986; Kuppers, 2005). However, the t(14;18) is also present in the peripheral blood of a significant proportion of healthy individuals (HI; Limpens et al., 1995; Dolken et al., 1996; Roulland et al., 2003). Although the significance of t(14;18) in blood from HI is as yet unclear, we have previously reported trends to higher t(14;18) prevalence among farmers occupationally exposed to pesticides (Roulland et al., 2004). Since then, recent epidemiological data have established that pesticide exposure is strongly associated with the risk of t(14;18)+ NHL development but not with t(14;18)-negative cases, suggesting that pesticides may act through a t(14;18)-dependant pathway (Chiu et al., 2006). Nonetheless, a direct mechanistic link between environmental exposure and the t(14;18) during FL pathogenesis remains to be conclusively demonstrated. Because securing this correlation between a biomarker of risk and a carcinogen exposure could have major impact on cancer prevention and public health, we sought to determine the molecular links of this connection.
RESULTS AND DISCUSSION
To address this issue, we used a prospective agricultural cohort representing a total of 753 individuals, including adult family members living/working on a farm. We performed a longitudinal cohort study in a sample of 128 farmers from this cohort defined as pesticide users on crops at enrollment (the “exposed” population). As a reference, an independent cohort of 25 unexposed nonfarmers recruited from the same geographical area (the “control” population) was similarly followed, as detailed previously (Roulland et al., 2003; Roulland et al., 2004).
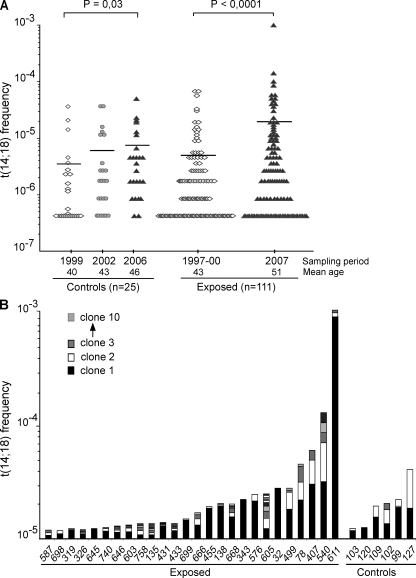
The evolution of t(14;18) frequency was followed over 9 yr in both cohorts. Strikingly, although t(14;18) frequency slowly increased in the control population (+87%; P = 0.03), mostly as the result of aging (Roulland et al., 2006a), a dramatic increase was observed for the exposed cohort (+253%; P < 0.0001 using the paired t test; Fig. 1 A). Contrary to controls, neither age nor aging effects were found in the exposed cohort at enrollment or follow up (P = 0.8 or 0.47, respectively, using the Spearman rank test). However, the relation with age was restored in farmers who stopped all agricultural activity (P = 0.05 using the Spearman rank test; Fig. S1). Collectively, our data firmly establish that cumulative agricultural pesticide exposure is associated with a dramatic increase of t(14;18) frequency in blood.
Figure 1.
Agricultural exposure drives a dramatic increase of t(14;18)+ clones in blood. (A) Evolution of t(14;18) frequency in PBMCs from control and exposed populations over time. The mean age at enrollment was 40 yr (±9 yr) for the controls and 43 yr (±9 yr) for the exposed individuals, with an average follow-up time of 7 and 9 yr, respectively. t(14;18) frequency was assessed at each time point by fluctuation SR-PCR. Horizontal lines indicate mean frequencies. p-values were calculated using the Mann-Whitney test. (B) Clonality analysis. Exposed (n = 26/51, 51%) and control (n = 6/46, 13%) samples with a high t(14;18) frequency (>10−5) in the last sampling period are shown and ordered according to increasing t(14;18) frequencies (the total number of clones in each individual for which more than one positive amplicon was found is summarized in Table S1). BCL2/JH breakpoints (Table I and unpublished data) were identified by the cloning/sequencing of SR-PCR products (n ≈ 1,000) amplified from PBMCs, and were used to monitor the extent of polyclonality within t(14;18)+ cells in each individual. For each individual, independent BCL2/JH clones are pictured in distinct shades and are represented from bottom to top according to increasing frequency.
We next sought to define how environmental exposure might be causally connected to this increase. Pesticides are often regarded as genotoxic chemicals generating DNA breaks, thereby favoring the accumulation of chromosomal translocations (Garry et al., 1996). We thus monitored the extent of polyclonality of t(14;18) breaks by analyzing BCL2/JH junctions. Surprisingly, although some exposed individuals indeed displayed an unusual accumulation of distinct t(14;18) translocations, most individuals carried one to two dominant clones (Fig. 1 B and Table S1). Furthermore, upon cell sorting of the peripheral B cell subsets, most clones displayed markers of GC transit: sCD27 and/or class switch recombination (CSR)/somatic hypermutation (SHM; Klein and Dalla-Favera, 2008), including the previously described “allelic paradox” (Roulland et al., 2006b) that stands as a hallmark of FL (simultaneous presence of sIgM+D+ on the productive allele and of CSR on the nonfunctional allele; Table I; and Figs. S2 and S3). Note that the presence of this allelic paradox is in sharp contrast with the features of the peripheral IgM+IgD+CD27+ B cell subset (being devoid of CSR both on the productive and the nonproductive alleles), and adds to the evidence of GC-expanded clones, distinct from a putative marginal zone B cell origin (Weller et al., 2004; Roulland et al., 2006b). This indicates that the t(14;18) increase observed in exposed farmers results far more from an immunogenic effect of agricultural pesticide exposure, leading to an extensive clonal expansion of activated t(14;18)+ B cells, than from a genotoxic effect. Importantly, expansions of activated t(14;18)+ B clones (>10−5) could also be observed among a few control individuals (Fig. 1 B and Table I). Although the expansions were more moderate, this suggests that a complex set of environmental factors (possibly including infectious agents such as the hepatitis C virus) can give rise to the activation of t(14;18)+ B clones (Giannelli et al., 2003; Roulland et al., 2008), with agricultural pesticide exposure provoking a particularly pronounced immunogenic effect.
Table I.
Phenotypic and genotypic characterization of BCL2/IGH junctions in isolated B cell subsets from both exposed and control HI
| B cell subsets | PCRa | RAG-mediated activity | AID-mediated activity | GC transitb | |||||||
| IgDc | CD10 | CD27d | Nb clones | BCL2-mbr (5′ to 3′)e | N nucleotides (5′ to 3′) | JH | CSR | SHMf | Rate (%) | ||
| Exposedg | |||||||||||
| 78 | + | + | − | SR (1) | CAGTGGTGCTTACGCTCCACCA | CCCCTGAGGCAGAT | J6 | n.a. | n.a. | – | – |
| n.d. | + | + | SR(3)/LR(1) | AAGAAAGCAGGAAe | GAACGTTACCTCAGGGGACTTCCCCGCGCCCCCCG | J6 | γ1 | 9/821 | 10.9 | yes | |
| + | + | − | SR (1) | CAGACCTCCCCCGGC | ∅ | J6 | n.a. | n.a. | – | ||
| + | − | + | LR (1) | CAGTGGTGCTTACGCTCC | TTCGGGTGTGATCGGTCGG | J6 | μ | 3/1,262 | 2.4 | yes | |
| + | − | + | LR (1) | AAGAAAGCAGGAAe | GAACGTTACCTCAGGGGACTTCCCCGCGCCCCCCG | J6 | γ1 | 8/821 | 9.8 | yes | |
| + | − | + | LR (1) | CAGTGGTG | TTGTAGTAGGAAA | J6 | μ | 1/1,262 | 0.8 | yes | |
| + | − | + | LR (1) | CAGTGGTG | TTGTAGTAGGAAA | J6 | γ1 | 1/474 | 2.1 | yes | |
| 138 | + | + | − | SR (1) | CAGTGGTGCTTACGCT | ATCGCGACGGGCTA | J3-6 | n.a. | n.a. | ||
| + | − | − | LR (1) | ACAAGTCCTTTA (*ICR) | GATCGGGGTCCCCCCGGGA | J6b | γ2 | 4/900 | 4.4 | yes | |
| +/−h | − | + | LR (3) | CAGACCTC | TTGTGAGGGGCCATGGTTATCCCAGGACGTCGTCGCCGA | J6b | γ1 | 44/4,176 | 10.5 | yes | |
| 326 | + | + | − | SR (1) | CAGTGGe | ATACTTGGGACAG | J4 | n.a. | n.a. | – | |
| + | + | − | SR (1) | CAGTGGTGCTTA | TCAGGTAGGGGG | J6 | n.a. | n.a. | – | ||
| − | − | + | LR (1) | CAGTGGe | ATACTTGGGACAG | J4 | γ1 | 1/687 | 1.5 | yes | |
| + | − | + | LR (2) | CCTCCTGCCCTCC | GGTAGACTAGACCC | J4 | μ | 6/2,524 | 2.4 | yes | |
| 431 | + | + | − | SR(7)/LR(2) | CAGTGGTGCTTAe | AATTTAGTCT | J6 | μ | 6/2,524 | 2.4 | yes |
| + | − | + | LR (2) | CAGTGGTGCTTAe | AATTTAGTCT | J6 | μ | 3/2,276 | 1.3 | yes | |
| 433 | + | + | − | SR (1) | CAGACCTCCCCCGGCGG | TTTTTCGAAGACGATATCAGGGT | J6 | n.a. | n.a. | – | |
| + | − | + | LR (1) | CAGACCTCCCCC | CCGAGCG | J4 | μ | 10/753 | 13.5 | yes | |
| 576 | +/− | − | + | LR (3) | CCTCCTGCCCTC | GTACGATTTTTGGAGTGGTCT | J6 | μ | 4/2,046 | 1.9 | yes |
| 605 | + | + | − | SR (1) | CCTCCTGCCCTCCTTCe | GGGTTTACTCCCCGGGTTTT | J5 | n.a. | n.a. | – | |
| +/− | + | + | SR(1)/LR(1) | TGCTTTACGT | TGCTC | J6 | μ | 1/1,262 | 0.8 | yes | |
| + | + | + | SR (1) | AAGAAAGCAGGAe | CCCCCACGGAG | J6 | n.a. | n.a. | yes | ||
| + | + | − | LR (1) | TCAGGGAACAGAA | CCCCTAGATAGTGGGAGCTCCAT | J6b | μ | 0/1,262 | 0 | – | |
| − | − | + | LR (1) | CAGTGGTGCTTA | AGGAAGAAGTCG | J4 | μ | 2/1,262 | 1.6 | yes | |
| + | − | + | LR (1) | AAGAAAGCAGGAe | CCCCCACGGAG | J6 | μ | n.d. | n.d. | yes | |
| + | − | + | LR (1) | CCTCCTGCCCTCCTTCCe | TCGAGAGAAG | J5 | μ | 5/1,262 | 4 | yes | |
| 611 | + | + | − | LR (3) | CAGTGGTGCe | CGAATGG | J6 | μ | 20/3,500 | 5.7 | yes |
| n.d. | + | + | SR (3) | CAGACCTCCCCCGGe | TTAGAGTTTGTGGGGCCCGTTGGGACG | J6 | n.a. | n.a. | yes | ||
| n.d. | + | + | SR (1) | CAGTGGTGCTTACG | TGCCGACCAAA | J3 | n.a. | n.a. | yes | ||
| + | + | − | SR (2) | AAGAAAGCAG | AGGGCCCCACCCTG | J6 | n.a. | n.a. | – | ||
| n.d. | + | + | SR (4) | AAGAAAGCA | AGGCAGGGCTTCGAGGG | J6 | n.a. | n.a. | yes | ||
| +/− | − | + | LR (4) | CAGTGGTGCe | CGAATGG | J6 | μ | 21/5,048 | 4.2 | yes | |
| +/− | − | + | LR (3) | CAGTGGTGCe | CGAATGG | J6 | γ1 | 2/95 | –i | yes | |
| − | − | + | LR (1) | CAGACCTCCCCCGGe | TTAGAGTTTGTGGGGCCCGTTGGGACG | J6 | γ3 | 23/1,109 | 20.7 | yes | |
| − | − | + | LR (1) | CAGACCTCCC | GAGATAGGGTTCAGGGGC | J6 | γ3 | 11/667 | 16.5 | yes | |
| + | − | + | LR (1) | CAGTGGTGCTTAC | CGCCCTAGCTGC | J6 | γ1 | 7/857 | 8.1 | yes | |
| Anonymous blood donorsf | |||||||||||
| 33 | n.d. | + | n.d. | LR (1) | CTTTACGTGGCCTe | TAGTACCAGCTGCCCCCTA | J6 | μ | n.d. | n.d. | – |
| n.d. | + | n.d. | SR (3) | CAGTGGTGCe | CAGCTGCCCCCCCCCGCCTGGCGGTGGGTTGTGCGGCCCC | J1 | n.a. | n.a. | – | ||
| n.d. | + | n.d. | LR (1) | CAGTGGTGC | CCTCGCCATATGTTCGTAACCCCGTCACCACC | J6 | γ3 | 2/521 | 3.8 | yes | |
| n.d. | − | + | SR (1) | CTTTACGTGGCCTe | TAGTACCAGCTGCCCCCTA | J6 | n.a. | n.a. | yes | ||
| n.d. | − | + | SR (7) | CAGTGGTGCe | CAGCTGCCCCCCCCCGCCTGGCGGTGGGTTGTGCGGCCCC | J1 | n.a. | n.a. | yes | ||
| n.d. | − | + | SR(13)/LR(3) | CAGTGG | GGGT | J6 | μ | 5/1,262 | 4 | yes | |
| 37 | n.d. | + | n.d. | LR (1) | CCTCCTGCCCTCCTT | AAACCA | J4 | μ | 13/1,282 | 11.1 | yes |
| n.d. | + | n.d. | SR(5)/LR (1) | CCTCCTGCCCTCCTTCCe | CCTTGGGGGGCT | J6 | μ | 3/1,448 | 2 | yes | |
| n.d. | + | n.d. | SR (1) | CAGTGGTGCTTe | GATTAGAGG | J5 | n.a. | n.a. | – | ||
| n.d. | + | n.d. | LR (1) | CAGACCTCCCCGe | AGAACCCCGCATAAA | J6 | μ | 1/1,526 | 0.7 | – | |
| n.d. | − | + | SR(4)/LR (1) | CCTCCTGCCCTCCTTCCe | CCTTGGGGGGCT | J6 | μ | 8/2,793 | 2.9 | yes | |
| n.d. | − | + | SR(1)/LR (1) | CAGTGGTGCTTe | GATTAGAGG | J5 | μ | 2/2,966 | 0.7 | yes | |
| n.d. | − | + | SR (4) | CAGACCTCCCCGe | AGAACCCCGCATAAA | J6 | n.a. | n.a. | yes | ||
| Controlsf | |||||||||||
| 118 | + | n.d. | + | LR (1) | CTCCTTCCGCGGG | A | J4 | μ | 5/2,620 | 1.9 | yes |
| + | n.d. | + | LR (1) | CAGTGGTGCTTA | GTGAATAGCGGGGCTC | J3 | μ | 3/1,262 | 2.4 | yes | |
| 92 | + | n.d. | + | LR (1) | GCTTTCTCATGG | TTCTTTTTCGGTAGGGTTGAGCACG | J5 | μ | 0/1,262 | 0 | yes |
| LR (1) | CAGTGGTGCTTAC | TGACCTCGGTCGA | J4 | μ | 2/1,262 | 1.6 | |||||
| 111 | + | n.d. | + | LR (2) | CAGTGGT | TTCTTTCGGGG-(DH3-9/DH3-10)-GTTAGG | J5 | μ | 17/2,524 | 6.8 | yes |
| 100 | + | n.d. | + | LR (1) | CAGTGGTG | GATATTTTCGGGGGCAACGACCGGGAGGATTGTCTCAAAA | J4 | μ | 1/1,262 | 0.8 | yes |
| 103 | + | n.d. | + | LR (1) | CAGACCTCCCCGGC | CCCCCCCCAGAA | J6 | μ | 2/1,262 | 1.5 | yes |
| LR (4) | CAGACCTCCCCGGC | CCCCCCCCAGAA | J6 | γ1 | 7/5,048 | 1.4 | yes | ||||
| 102 | + | n.d. | + | LR (4) | CCTCCTGCCCTCCTTCCG | TTCCTGTCCAAAAAG | J6 | μ | 5/5,048 | 0.9 | yes |
| + | n.d. | + | LR (1) | CAGACCTCCCCGGC | TGGGTTAGG | J5 | γ1 | 3/645 | 4.6 | yes | |
| − | n.d. | + | LR (1) | CCTCCTGCCCTCCTTCCGCGG | CCGTTGTGGGGACTCA | J6 | μ | 5/1,262 | 4 | yes | |
| 127 | + | n.d. | + | LR (2) | CAGACCTCCCCGG | TATCGCTAAACACGAT | J6 | γ2 | 20/1,932 | 10.3 | yes |
| − | n.d. | + | LR (3) | CAGACCTCCCCGG | TATCGCTAAACACGAT | J6 | γ2 | 110/7,329 | 15 | yes | |
| − | n.d. | + | LR (3) | CAGTGGTGCTTA | AGGTGGTC | J6 | γ1 | n.d. | n.d. | yes | |
n.a., not applicable; n.d., not determined.
Type of PCR assay in which the fragment was obtained (number of positive amplicons of the given immunogeno-/phenotype; Fig. S6 B).
Based on CD27+ and/or CSR+ and/or SHM+.
See Fig. S3 for evaluation of the IgM status.
CD27 is a pertinent GC marker only when present, as the loss/absence of CD27 after GC transit has been shown to occur in recently identified memory B cell subsets (Ehrhardt et al., 2005; Wirths and Lanzavecchia, 2005; Tangye and Good, 2007) and is variably expressed on FL cells.
BCL2/JH junctions detected in both CD10+ and CD10− fractions.
SHM corresponds to the number of somatic mutations/total length of Sµ analyzed (bp). Background mutation levels were calculated to be 0.55‰ (Fig. S2 A).
Sorted according to CD10, CD27, and IgD for exposed samples (Fig. S6 A, top); according to CD10 and CD27 into CD10+ and CD10−CD27+ for anonymous blood donors (Fig. S6 A, bottom); and according to CD27 and IgD for controls (only the CD27+ is shown). Eight additional anonymous blood donors were screened and excluded from the analysis due to the absence of the t(14;18) detection.
+/− indicates PCR fragments detected in both positive and negative sorted B cell fractions.
The mutation rate was not estimated due to the partial deletion of the Sµ region.
Is this clonal expansion of activated t(14;18)+ cells connected to a high risk of malignant transformation? Upon antigen encounter, normal activated B cells reach the GCs, where the activation-induced cytidine deaminase (AID)–mediated program of affinity maturation randomly modifies their receptors through CSR and SHM (Muramatsu et al., 2000). In the vast majority of cases, this process leads to a decrease in receptor affinity and B cells die by neglect in situ. In FL, three successive steps lay out the lymphomagenesis pathway: (a) t(14;18)-enforced ectopic expression of BCL2 in GC-activated B cells, leading to their illegitimate rescue from cell death (McDonnell et al., 1989; Kuppers, 2005; Klein and Dalla-Favera, 2008); (b) consequent maturation arrest and accumulation as centrocyte/centroblast-like cells with ongoing AID activity (Aarts et al., 2000; Smit et al., 2003); and (c) consequent accumulation of complementary oncogenic hits, partly because of AID-mediated genomic instability (Ramiro et al., 2006; Bende et al., 2007; Pasqualucci et al., 2008). We thus sought to determine where the activated t(14;18)+ B cells stood in this cascade of events leading to FL genesis.
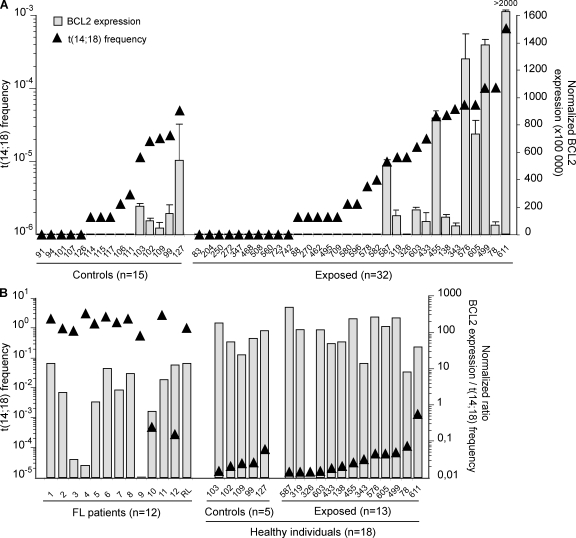
Although t(14;18) carried by HI are morphologically undistinguishable from those found in FL tumors (Dolken et al., 1996; Jager et al., 2000; Roulland et al., 2006b), the expression of BCL2/IGH fusion transcripts (Cleary et al., 1986; Seto et al., 1988) has to date never been functionally demonstrated in HI, partly because of the experimentally challenging issue of detecting transcripts at such low frequencies. We thus set up a sensitive RT-PCR assay (Fig. S4) and studied a set of control (n = 15) and exposed (n = 32) individuals with low (<10−5) or high (>10−5) frequencies (Fig. 2 A, triangles). BCL2/IGH quantification clearly shows that transcription levels closely parallel translocation frequencies (Fig. 2 A, histograms). Furthermore, in purified B cell subsets from a given individual, only t(14;18)-positive fractions allowed BCL2/IGH detection (Fig. S5). We next assessed how BCL2/IGH transcription levels compared with those in FL patients. To account for the large difference in t(14;18) frequency between FL and HI (Fig. 2 B, triangles), we calculated a normalized ratio of transcription level per t(14;18)+ cell. Our data clearly show that BCL2/IGH expression is comparable in the t(14;18)+ cells of FL and HI (Fig. 2 B, histograms). Collectively, these findings demonstrate that similar to tumor cells in FL patients (Seto et al., 1988), t(14;18)+ cells in HI are actively transcribing BCL2 from the translocated allele, allowing their rescue from apoptosis in the GCs.
Figure 2.
t(14;18)+ cells in HI are actively transcribing BCL2 from the translocated allele. (A) Superposition of t(14;18) frequency (triangles; left y axis) and the relative BCL2/JH expression levels (histograms; right y axis) in PBMC samples from controls (n = 15) and exposed individuals (n = 32) evaluated by real-time quantitative PCR. The absolute BCL2/JH expression data were normalized to ABL, and the value is arbitrarily set as ×100,000. Each bar represents the mean of replicate wells. The results shown were obtained from two independent PCRs performed in duplicate (B) BCL2/JH expression levels in FL biopsies (n = 12) compared with samples from HI (n = 18) with respect to their t(14;18) frequency. A normalized ratio of BCL2 expression per t(14;18)+ cell was calculated by dividing the normalized BCL2/JH transcription level of a given sample by the corresponding t(14;18) frequency (as determined by real-time PCR).
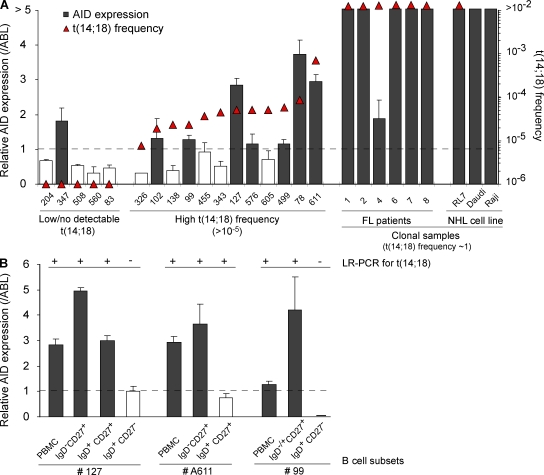
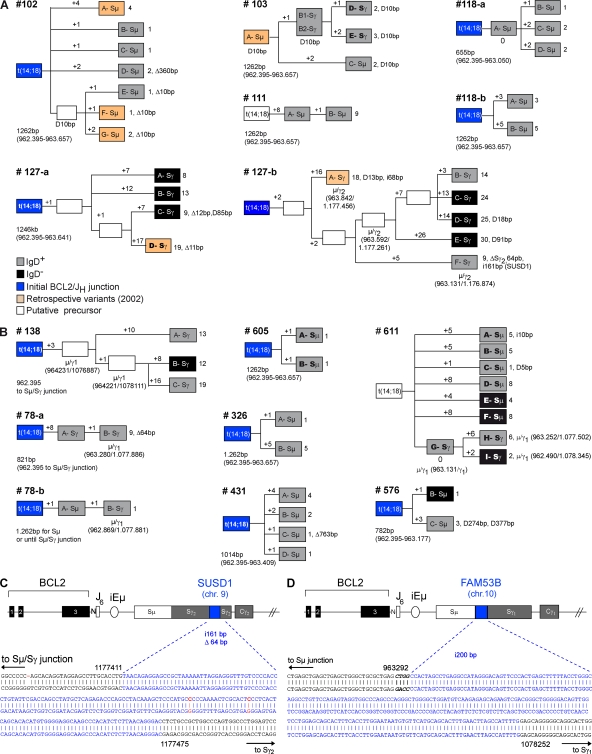
Malignant FL cells are frequently released from the founder follicle to the bloodstream, and undergo extensive trafficking between organs, thereby favoring tumor dissemination (Oeschger et al., 2002). However, consistent with their status as frozen centrocyte/centroblast-like cells, most nodal and at least a fraction of circulating FL tumor cells retain some GC markers such as CD10 (CD10+/−, REAL classification; Harris et al., 1994). To determine if t(14;18)+ clones from HI also represent released centrocyte/centroblast-like cells, CD10+ and CD10− cell populations were sorted (Fig. S6 A). In all individuals, t(14;18) could be found in both the CD10+ and CD10− subsets (50 vs. 66 amplicons, respectively; Table I and Fig. S6 C). In most cases, CD10+ and CD10− clones (with identical BCL2/IGH) coexisted in peripheral blood after GC transit (Table I, note e). This indicates that similar to FL patients, at least a fraction of the circulating t(14;18)+ cells from HI also retained the CD10 marker normally lost upon GC exit (Kuppers, 2005). One additional seminal feature of FL tumor cells that stands as a direct consequence of their status as frozen GC B cells is the sustained AID expression (Smit et al., 2003; Bende et al., 2007), normally down-regulated upon GC exit. We thus first attempted to evaluate AID expression in t(14;18)+ cells. Because AID transcripts are not different in translocated and normal cells, we sought to assess an expression difference of AID between individuals with low (<10−5) and high (>10−5) t(14;18) frequencies. Results show that detection of AID expression in total PBMCs correlates remarkably well with increasing t(14;18)+ frequencies (Fig. 3 A, shaded histograms). Moreover, AID levels could be enriched through fractionating total blood into t(14;18)-positive B cell subsets (Fig. 3 B). A similar tendency (but less significant because of high background levels; Ye et al., 1997) was observed with BCL6 (Fig. S7). To next functionally assess if, similar to FL, t(14;18)+ clones from HI also maintained AID activity and kept evolving with time, we sought to analyze filiations between t(14;18)+ cells. The prediction was that if AID were off during clonal expansion, identical clones (in terms of CSR and SHM) should be generated, whereas if AID were on, clonal expansion should lead to a population of clonal SHM/CSR variants. We thus obtained large blood samples from control (n = 7) and exposed (n = 8) individuals, and analyzed switched versus unswitched purified B cell subsets for both SHM and CSR events on the translocated allele (Fig. S6 B). For most BCL2/IGH clones issued from clonal expansions, we were able to build genealogical trees based on intraclonal variation (ICV) of the Sµ region on the translocated allele (Nagaoka et al., 2002), taking into account both CSR and SHM (Fig. 4). Strikingly, the vast majority of subclones displayed ICV: out of 69 amplicons (and despite the fact that the expected rate of SHM is much lower in the Sµ region than in the IgVH region), only 2 were found to be strictly identical (case #103, B1/B2). This quasisystematic presence of ICV clearly indicates that cell division is constantly associated with SHM and/or CSR events, and therefore that t(14;18)+ clones have maintained AID expression over time. Together with the expression of CD10, the presence of ongoing AID-mediated activity demonstrates that t(14;18)+ cells in HI indeed represent circulating counterparts of “frozen” GC B cells.
Figure 3.
AID expression correlates with increasing t(14;18) frequencies in HI. (A) Superposition of t(14;18) frequency (triangles; left y axis) and the relative AID expression levels (histograms; right y axis) in PBMC samples from HI (n = 18), FL patient samples (n = 6), and lymphoma cell lines (RL7, FL; Daudi and Raji, Burkitt lymphoma) evaluated by real-time quantitative PCR. The expression data were normalized to ABL. As AID expression is assumed to be low/negative in naive cells, RT-PCR data are represented relative to the levels obtained in the sorted naive (CD19+CD27−IgD+) B cell fraction and are considered positive (shaded histograms) above this arbitrary threshold of 1 (dashed line). The results shown were obtained from two independent PCRs performed in duplicate (except for clonal FL samples). (B) AID expression is enriched in t(14;18)+ fractions. Comparison of t(14;18) detection (+/−, as determined by LR-PCR) and AID expression levels (shaded histograms) in isolated B cell subsets (based on CD27 and IgD markers) from two HI. The results shown were obtained from one PCR performed in duplicate (except for PBMC samples analyzed two times in duplicate). Data were normalized as in Fig. 3 A.
Figure 4.
AID mediates the evolution of t(14;18)+ activated clones in HI. (A and B) Genealogic trees generated from healthy controls (n = 5) and exposed individuals (n = 7) for which several molecular subclones derived from the same BCL2/JH junction (labeled A to G in boxes) were obtained. Trees are organized based on ICV, including SHM in the Sµ region and CSR on both the expressed (gray boxes, sIgD+; black boxes, sIgD−) and the translocated (indicated as Sµ or Sγ in boxes) alleles. For most trees, the BCL2/JH junction was already identified at enrollment (blue boxes, ICV information not available). Six LR-PCR amplicons were obtained from the retrospective PBMC samples (orange boxes) and could be branched in four distinct trees. Stepwise accumulation of mutations is indicated by numbers above the branches (+1 to +26). Total mutations, insertions (i), duplications (D), and deletions (▵) are summarized at the end of each branch. Shared mutations were used to define putative intermediate or precursor cells (open boxes). The length of the analyzed Sµ region is indicated below each tree and varied slightly in case of CSR. (C) Sequence analysis of an AID-mediated foreign DNA insertion (200 bp from the FAM53B gene, chr 10, in blue) into an Sµ/Sγ switch junction of an activated t(14;18)+ clone from sample #138. (D) Sequence analysis of a foreign DNA insertion (161 bp from the SUSD1 gene, chr 9, in blue) into an intra-Sγ2 deletion (▵64 bp) from an activated t(14;18)+ clone from sample #127-b (see clone F-Sγ in A). Numbers refer to germline IGH available from GenBank/EMBL/DDBJ under accession no. NG_001019.
In lymphomas, sustained AID activity provides a high propensity for the acquisition of further oncogenic aberrations. This mostly includes CSR-mediated translocations (Ramiro et al., 2006), aberrant CSR (Lenz et al., 2007), and the occurrence of SHM in non-Ig target genes (Pasqualucci et al., 2001; Takizawa et al., 2008). Remarkably, we found that almost 30% (19 out of 69) of the BCL2/IGH clones tested in the Sµ region display unusually large intra-Sµ deletions (up to 763 bp) or Sµ tandem duplications interspersed with mutations in AID hotspots (Fig. 4; and Fig. S2 A, note e), suggesting that CSR is functionally impaired in these clones (Reina-San-Martin et al., 2003; Lenz et al., 2007). Furthermore, the insertion of an irrelevant DNA fragment from a distinct chromosome into a Sµ deletion site has been recently observed in a diffuse large B cell lymphoma sample, and proposed as a surrogate marker of AID-mediated genomic instability and malignant progression (Lenz et al., 2007). Strikingly, two of such rare insertion events were found in our samples (Fig. 4, C and D). Although probably not directly relevant to tumor progression, such events demonstrate the occurrence of illegitimate AID-mediated events and highlight the propensity of some t(14;18)+ clones to withstand a complementary oncogenic hit over time.
Collectively, our results clearly demonstrate that the expanded t(14;18)+ clones, which are particularly prominent in farmers exposed to pesticides, constitute bona fide FL precursors standing at various stages of tumor progression. Together with the recent findings that pesticide exposure is specifically associated with a higher risk of t(14;18)+ NHL development (Chiu et al., 2006), our data provide the first direct connection between t(14;18) frequency in blood and malignant progression. Remarkably, the dynamics of clonal evolution observed in HI are strikingly similar to those observed in FL (Ruminy et al., 2008) and transformed-FL in diffuse large B cell lymphoma (Carlotti et al., 2009), suggesting that the selection process operating in malignant clones is already at work at early development stages (Fig. 4 and Fig. S8).
The most pressing question in our report is how to assess which of those individuals are more likely to develop FL in the near future. Some individuals displayed unusual t(14;18) frequencies (e.g., # 611, 1 in 1,000 PBMCs) reaching levels observed in some FL patients, and others displayed aberrant CSR events (e.g., #127-b, ectopic insertions) that indicate a predisposition to chromosomal translocations (Lenz et al., 2007). Our current challenge is to determine how the combined assessment of t(14;18) frequency and surrogate markers of malignant progression could be helpful in the identification of individuals at “high” risk for transformation to overt FL. Besides the identification of at-risk individuals, the precise understanding of the immunogenic effect described in this paper on t(14;18)+ cells might also provide important targets for its disruption in preventive approaches (Staudt, 2007; Roulland et al., 2008).
MATERIALS AND METHODS
Human samples.
A population of 753 farmers/farm workers were included in an agricultural cohort that has been previously described (Roulland et al., 2004). After enrollment (1997–2000), a subset was selected to control for potential confounding factors based on the following criteria: males, no smoking history, and current pesticide applicators on open-field crops. Among the 144 individuals matching these criteria, 128 provided DNA/blood samples (∼50 ml) for analysis at enrollment (Roulland et al., 2004), and 111 participated in the average 9-yr follow up (response rate = 89%; Table S2). Large blood samples (∼250 ml) were obtained from eight of them at follow up. As controls, blood samples (∼50–250 ml) were taken from a group of 25 healthy adults issued from a followed-up cohort (Roulland et al., 2006a), coming from the same geographical area as the exposed group and defined as “subjects who have never lived/worked on a farm and were not exposed to pesticides.” Large blood samples were also collected from 10 anonymous bank donors. 2 out of the 10 were screened highly positive for t(14;18) frequency and subjected to cell sorting and molecular analysis (see B cell isolation…). In addition, 20 tumor biopsies were obtained from FL patients at diagnosis (Jager et al., 2000; Ruminy et al., 2008). This study was approved by the local ethics committee (Comité Consultatif de Protection des Personnes dans la Recherche Biomédicale #99-07, Caen, France), and all participants of the study provided their written informed consent.
Pesticide exposure assessment.
A face-to-face comprehensive standardized questionnaire was administrated to all enrolled subjects at each time point. The interviews included detailed information on agricultural practices and pesticide exposure, including the timing and duration of farming activities, the years of exposure, the personal handling of specific pesticides, the average annual number of days of pesticide use on crops, the type of crops and average acres grown, the use of personal protective equipment, the nature of chemical classes, and the spraying equipment (Table S2). Exposed and control individuals (excluding anonymous blood blank donors) were also asked to provide information about sociodemographic characteristics, tobacco and alcohol use, dietary habits, medical history, and lifetime occupational exposure (Roulland et al., 2004).
B cell isolation, cell sorting, and FACS analysis.
PBMCs were obtained by Ficoll-density gradient centrifugation. For large blood samples, enriched B cells were isolated from PBMCs by negative selection using a B cell isolation kit (Invitrogen and/or Miltenyi Biotec). The resulting purified B cells were then stained for CD19-allophycocyanin (APC) or PE-Cy7 (clone HIB-19; BD or eBioscience), IgD-FITC (Invitrogen), IgM-APC (G20-127; BD), and CD27-PE (clone O323; eBioscience) mAbs. For control samples, the naive (IgD+CD27−) and memory (IgD+CD27+, IgD−CD27+) B cell subsets were isolated by FACS sorting, as previously performed (Roulland et al., 2006b). 10 large blood samples from HI (8 from the exposed cohort and 2 from anonymous blood donors) showing translocation frequencies >10−5 were further stained with CD10-PeCy5 (clone HI10a; BD) and used to sort the CD19+CD10+ (4–7% of B cells) and CD19+CD10−CD27+ (25–35% of B cells) B cell fractions. In the exposed cohort, the CD19+CD10+ fraction was further split according to CD27 and IgD expression (Fig. S6 A). Cell sorting experiments were performed on a FACSAria (BD), and each sorted fraction was reanalyzed on a flow cytometer (LSR; BD) with FlowJo software (Tree Star, Inc.) showing at least 95% purity.
Fluctuation PCR amplification of BCL2/IGH translocations.
Genomic DNA was extracted from PBMCs, FL biopsies, and/or sorted fractions using DNA blood or a tissue mini kit (QIAGEN). A fluctuation two-step short-range PCR (SR-PCR) assay was used to amplify BCL2/JH junctions in HI, as previously described (Roulland et al., 2006b). Amplification of BCL2/Cμ and BCL2/Cγ fragments by long-range (LR-PCR) was performed using the same fluctuation approach and as previously described (Roulland et al., 2006b). In brief, nested primers flanking the BCL2–major breakpoint region (mbr) were used in combination with 3′ primers from the Cμ/Sµ region or the consensus Cγ/Sγ regions (Fig. S6 B). The PCR products were visualized by electrophoresis on a 0.8% agarose gel and ethidium bromide staining. All amplicons were gel purified from each independent PCR replicate, cloned into pGEMT-easy vector (Promega), and sequenced. Primers and probes are listed in Table S3.
Quantitative RT-PCR.
RNA was extracted from PBMCs, FL biopsies, and/or sorted fractions with the RNAqueous-4 PCR kit (Applied Biosystems), and 1 µg RNA was converted into cDNA using the high capacity RT kit (Applied Biosystems). To prevent the potential contamination of the RNA samples with genomic DNA, all preparations were DNaseI treated. To accurately measure BCL2/JH and ABL gene expression from different origins, we developed an absolute quantitative PCR strategy (Fig. S4). During the translocation process, most BCL2 breakpoints arise in the mbr, located in the 3′ untranslated region (UTR) of BCL2 exon III. This generates a long BCL2/IGH fusion transcript carrying the complete BCL2 open reading frame (Cleary et al., 1986; Seto et al., 1988). Recombinant RNA (recRNA) was generated (Fig. S4, left) to build standard calibration curves. RT- and real-time PCR steps were performed in parallel on recRNA standards and human RNA samples in a PCR System (ABI PRISM 7500; Applied Biosystems). Quantitative PCR was performed with 1/10th of the RT-PCR product (∼100 ng cDNA) using primers flanking the 5′ BCL2-mbr (3′ UTR region) in combination with the 3′ JH consensus reverse sequence from the IGH locus (Fig. S4, right) and a TaqMan MGB probe (Applied Biosystems) matching the mbr region. For absolute quantification, the samples were analyzed in two independent PCR reactions, each performed in duplicate and normalized to the signal obtained for the ABL copy number. PCR for AID and BCL-6 transcripts was performed with specific TaqMan gene expression assays (Hs00757808_m1 and Hs00277037_m1, respectively; Applied Biosystems) and normalized to the expression of the endogenous ABL. The results were analyzed according to the ΔΔCt comparative method, normalizing to 1 the expression of AID and BCL6 in the naive IgD+CD27− fraction of the R127 control.
Sµ SHM analysis.
The presence and patterns of mutations accumulating on the 5′ Sµ region (Nagaoka et al., 2002) from the IGH locus on the der(14) translocated allele were examined by sequencing cloned BCL2/Sµ and BCL2/Sγ LR-PCR amplicons. The analysis was performed in an ∼1,250-bp sequence upstream of and overlapping the repetitive Sµ region using two consecutive primers (label 4; Fig. S6 B). To evaluate the PCR error rate introduced by the Expand Taq DNA polymerase (Roche) and exclude polymorphic variants from analysis, the germline Sµ region of each individual was amplified using primers 3+5 and sequenced (Fig. S6 B). Mutations corresponding to polymorphic variants were excluded, and the mean baseline mutation rate was evaluated to be 0.55‰ (Fig. S2). Sµ deletions, mutations, duplications, and insertions were then organized into genealogical trees of the stepwise accumulation of these events into individual t(14;18)+ molecular clones (Fig. 4).
Statistical analysis.
To examine the relationships between t(14;18) and agricultural pesticide use, we first compared the evolution of t(14;18) frequency over time among exposed and control groups using the paired t test. Normality was tested and rejected because of the skewness of the distribution of t(14;18) frequencies. Relationships between t(14;18) frequency and age were made with the Spearman rank test. For mutation analysis, Fisher's exact test was used to compare Sµ mutation frequencies between various B cell subpopulations as compared with FL cells, and the χ2 tests were used to compare mutation distribution patterns.
Online supplemental material.
Fig. S1 shows the restoration of the age effect in farmers who stopped agricultural pesticide use at follow up. Fig. S2 shows the Sµ mutation patterns in normal and t(14;18)+ B cells from HI and FL patients. Fig. S3 shows that the majority of t(14;18)+ cells are IgD+M+. Fig. S4 illustrates the absolute quantitative RT-PCR strategy to accurately measure the BCL2/JH transcripts levels in t(14;18)+ B cells from HI as compared with FL. Fig. S5 shows that BCL2/JH transcript expression strictly correlates with t(14;18)+ fractions. Fig. S6 depicts a representative cell sorting experiment to isolate peripheral B cell subsets according to CD27, CD10, and IgD cell-surface markers, and a diagram showing the positions of primers used to characterize BCL2/IGH translocations by SR-PCR and/or LR-PCR assays. Fig. S7 shows BCL6 expression in PBMCs and B cell subsets from HI with various t(14;18) frequencies. Fig. S8 illustrates the two kinds of dynamics than can be modelized from circulating FL precursor cells in HI. Table S1 provides a summary of the BCL2/JH polyclonality in individuals for which more than one positive amplicon was found. Table S2 describes the demographic and exposure characteristics of the farmer population at enrollment and follow up. Table S3 list the primers used for PCR and quantitative RT-PCR reactions. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20082842/DC1.
Acknowledgments
We are grateful to Drs. E. Jaffe, E. Vivier, C. Schiff, and L. Leserman for critical reading of the manuscript. We are indebted to H. Tilly and C. Bastard (INSERM U918, Center H. Becquerel, Rouen, France) for the kind gift of FL samples. We also thank N. Brun and M. Barad for cell sorting assistance, and N. Leveque, A.S. Lacauve, and C. Meyer for data managing of the exposed cohort. We are also indebted to the Service de Médecine de Jour du Centre François Baclesse for help with the organization of the large blood sample collection. Finally, we are grateful to all of the study subjects for their participation and all of the nurses for helping us with blood collection from exposed individuals.
This work was funded by grants from the Institut National du Cancer (INCa; PL06-010), the Fondation pour la Recherche Médicale (FRM; INE2003114116), the Association pour la Recherche sur le Cancer (3618/6974), and Le Conseil Général des Bouches du Rhône, and institutional grants from the Institut National de la Santé et de la Recherche Médicale (INSERM) and the Centre National de la Recherche Scientifique. The cohort study was funded by grants from la Ligue Nationale Contre le Cancer (comité de la Manche), the Conseil Général du Calvados, the Fondation Weisbren Benenson, the Union des Industries de la Protection des Plantes, and the Université de Caen Basse-Normandie. Work in the B. Nadel laboratory was supported by the INSERM AVENIR2003 program. S. Roulland was supported by the INSERM Young Investigator Program and INCa, A.-C. Gac was supported by an FRM fellowship, and B. Nadel is a recipient of a Contrat d'Interface INSERM/AP-Hôpitaux de Marseille award.
The authors declare no conflicting financial interests.
Footnotes
Abbreviations used: AID, activation-induced cytidine deaminase; CSR, class switch recombination; FL, follicular lymphoma; GC, germinal center; HI, healthy individuals; ICV, intraclonal variation; LR-PCR, long-range PCR; mbr, major breakpoint region; NHL, non-Hodgkin's lymphoma; SHM, somatic hypermutation; SR-PCR, short-range PCR.
References
- Aarts W.M., Bende R.J., Steenbergen E.J., Kluin P.M., Ooms E.C., Pals S.T., van Noesel C.J. 2000. Variable heavy chain gene analysis of follicular lymphomas: correlation between heavy chain isotype expression and somatic mutation load.Blood. 95:2922–2929 [PubMed] [Google Scholar]
- Bende R.J., Smit L.A., van Noesel C.J. 2007. Molecular pathways in follicular lymphoma.Leukemia. 21:18–29 [DOI] [PubMed] [Google Scholar]
- Blair A., Zahm S.H., Pearce N.E., Heineman E.F., Fraumeni J.F. 1992. Clues to cancer etiology from studies of farmers.Scand. J. Work Environ. Health. 18:209–215 [DOI] [PubMed] [Google Scholar]
- Carlotti E., Wrench D., Matthews J., Iqbal S., Davies A., Norton A., Hart J., Lai R., Montoto S., Gribben J.G., et al. 2009. Transformation of follicular lymphoma to diffuse large B-cell lymphoma may occur by divergent evolution from a common progenitor cell or by direct evolution from the follicular lymphoma clone.Blood. 113:3553–3557 [DOI] [PubMed] [Google Scholar]
- Chiu B.C., Dave B.J., Blair A., Gapstur S.M., Zahm S.H., Weisenburger D.D. 2006. Agricultural pesticide use and risk of t(14;18)-defined subtypes of non-Hodgkin lymphoma.Blood. 108:1363–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary M.L., Smith S.D., Sklar J. 1986. Cloning and structural analysis of cDNAs for bcl-2 and a hybrid bcl-2/immunoglobulin transcript resulting from the t(14;18) translocation.Cell. 47:19–28 [DOI] [PubMed] [Google Scholar]
- Dolken G., Illerhaus G., Hirt C., Mertelsmann R. 1996. BCL-2/JH rearrangements in circulating B cells of healthy blood donors and patients with nonmalignant diseases.J. Clin. Oncol. 14:1333–1344 [DOI] [PubMed] [Google Scholar]
- Dreiher J., Kordysh E. 2006. Non-Hodgkin lymphoma and pesticide exposure: 25 years of research.Acta Haematol. 116:153–164 [DOI] [PubMed] [Google Scholar]
- Ehrhardt G.R.A., Hsu J.T., Gartland L., Leu C.M., Zhang S.Y., Davis R.S., Cooper M.D. 2005. Expression of the immunoregulatory molecule FcRH4 defines a distinctive tissue-based population of memory B cells.J. Exp. Med. 202:783–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garry V.F., Tarone R.E., Long L., Griffith J., Kelly J.T., Burroughs B. 1996. Pesticide appliers with mixed pesticide exposure: G-banded analysis and possible relationship to non-Hodgkin's lymphoma.Cancer Epidemiol. Biomarkers Prev. 5:11–16 [PubMed] [Google Scholar]
- Giannelli F., Moscarella S., Giannini C., Caini P., Monti M., Gragnani L., Romanelli R.G., Solazzo V., Laffi G., La Villa G., et al. 2003. Effect of antiviral treatment in patients with chronic HCV infection and t(14;18) translocation.Blood. 102:1196–1201 [DOI] [PubMed] [Google Scholar]
- Harris N.L., Jaffe E.S., Stein H., Banks P.M., Chan J.K., Cleary M.L., Delsol G., Wolf-Peeters C., Falini B., Gatter K.C. 1994. A revised European-American classification of lymphoid neoplasms: a proposal from the International Lymphoma Study Group.Blood. 84:1361–1392 [PubMed] [Google Scholar]
- Jager U., Bocskor S., Le T., Mitterbauer G., Bolz I., Chott A., Kneba M., Mannhalter C., Nadel B. 2000. Follicular lymphomas' BCL-2/IgH junctions contain templated nucleotide insertions: novel insights into the mechanism of t(14;18) translocation.Blood. 95:3520–3529 [PubMed] [Google Scholar]
- Klein U., Dalla-Favera R. 2008. Germinal centres: role in B-cell physiology and malignancy.Nat. Rev. Immunol. 8:22–33 [DOI] [PubMed] [Google Scholar]
- Kuppers R. 2005. Mechanisms of B-cell lymphoma pathogenesis.Nat. Rev. Cancer. 5:251–262 [DOI] [PubMed] [Google Scholar]
- Lenz G., Nagel I., Siebert R., Roschke A.V., Sanger W., Wright G.W., Dave S.S., Tan B., Zhao H., Rosenwald A., et al. 2007. Aberrant immunoglobulin class switch recombination and switch translocations in activated B cell–like diffuse large B cell lymphoma.J. Exp. Med. 204:633–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limpens J., Stad R., Vos C., de Vlaam C., De Jong D., van Ommen G.J., Schuuring E., Kluin P.M. 1995. Lymphoma-associated translocation t(14;18) in blood B cells of normal individuals.Blood. 85:2528–2536 [PubMed] [Google Scholar]
- McDonnell T.J., Deane N., Platt F.M., Nunez G., Jaeger U., McKearn J.P., Korsmeyer S.J. 1989. bcl-2-immunoglobulin transgenic mice demonstrate extended B cell survival and follicular lymphoproliferation.Cell. 57:79–88 [DOI] [PubMed] [Google Scholar]
- Muramatsu M., Kinoshita K., Fagarasan S., Yamada S., Shinkai Y., Honjo T. 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme.Cell. 102:553–563 [DOI] [PubMed] [Google Scholar]
- Nagaoka H., Muramatsu M., Yamamura N., Kinoshita K., Honjo T. 2002. Activation-induced deaminase (AID)–directed hypermutation in the immunoglobulin Smu region: implication of AID involvement in a common step of class switch recombination and somatic hypermutation.J. Exp. Med. 195:529–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeschger S., Brauninger A., Kuppers R., Hansmann M.L. 2002. Tumor cell dissemination in follicular lymphoma.Blood. 99:2192–2198 [DOI] [PubMed] [Google Scholar]
- Pasqualucci L., Neumeister P., Goossens T., Nanjangud G., Chaganti R.S., Kuppers R., Dalla-Favera R. 2001. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas.Nature. 412:341–346 [DOI] [PubMed] [Google Scholar]
- Pasqualucci L., Bhagat G., Jankovic M., Compagno M., Smith P., Muramatsu M., Honjo T., Morse H.C., III, Nussenzweig M.C., Dalla-Favera R. 2008. AID is required for germinal center-derived lymphomagenesis.Nat. Genet. 40:108–112 [DOI] [PubMed] [Google Scholar]
- Ramiro A.R., Jankovic M., Callen E., Difilippantonio S., Chen H.T., McBride K.M., Eisenreich T.R., Chen J., Dickins R.A., Lowe S.W., et al. 2006. Role of genomic instability and p53 in AID-induced c-myc-Igh translocations.Nature. 440:105–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reina-San-Martin B., Difilippantonio S., Hanitsch L., Masilamani R.F., Nussenzweig A., Nussenzweig M.C. 2003. H2AX is required for recombination between immunoglobulin switch regions but not for intra-switch region recombination or somatic hypermutation.J. Exp. Med. 197:1767–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roulland S., Lebailly P., Roussel G., Briand M., Cappellen D., Pottier D., Hardouin A., Troussard X., Bastard C., Henry-Amar M., Gauduchon P. 2003. BCL-2/JH translocation in peripheral blood lymphocytes of unexposed individuals: lack of seasonal variations in frequency and molecular features.Int. J. Cancer. 104:695–698 [DOI] [PubMed] [Google Scholar]
- Roulland S., Lebailly P., Lecluse Y., Briand M., Pottier D., Gauduchon P. 2004. Characterization of the t(14;18) BCL2-IGH translocation in farmers occupationally exposed to pesticides.Cancer Res. 64:2264–2269 [DOI] [PubMed] [Google Scholar]
- Roulland S., Lebailly P., Lecluse Y., Heutte N., Nadel B., Gauduchon P. 2006a. Long-term clonal persistence and evolution of t(14;18)-bearing B cells in healthy individuals.Leukemia. 20:158–162 [DOI] [PubMed] [Google Scholar]
- Roulland S., Navarro J.M., Grenot P., Milili M., Agopian J., Montpellier B., Gauduchon P., Lebailly P., Schiff C., Nadel B. 2006b. Follicular lymphoma–like B cells in healthy individuals: a novel intermediate step in early lymphomagenesis.J. Exp. Med. 203:2425–2431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roulland S., Suarez F., Hermine O., Nadel B. 2008. Pathophysiological aspects of memory B-cell development.Trends Immunol. 29:25–33 [DOI] [PubMed] [Google Scholar]
- Ruminy P., Jardin F., Picquenot J.M., Parmentier F., Contentin N., Buchonnet G., Tison S., Rainville V., Tilly H., Bastard C. 2008. S(mu) mutation patterns suggest different progression pathways in follicular lymphoma: early direct or late from FL progenitor cells.Blood. 112:1951–1959 [DOI] [PubMed] [Google Scholar]
- Seto M., Jaeger U., Hockett R.D., Graninger W., Bennett S., Goldman P., Korsmeyer S.J. 1988. Alternative promoters and exons, somatic mutation and deregulation of the Bcl-2-Ig fusion gene in lymphoma.EMBO J. 7:123–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit L.A., Bende R.J., Aten J., Guikema J.E., Aarts W.M., van Noesel C.J. 2003. Expression of activation-induced cytidine deaminase is confined to B-cell non-Hodgkin's lymphomas of germinal-center phenotype.Cancer Res. 63:3894–3898 [PubMed] [Google Scholar]
- Staudt L.M. 2007. A closer look at follicular lymphoma.N. Engl. J. Med. 356:741–742 [DOI] [PubMed] [Google Scholar]
- Takizawa M., Tolarova H., Li Z., Dubois W., Lim S., Callen E., Franco S., Mosaico M., Feigenbaum L., Alt F.W., et al. 2008. AID expression levels determine the extent of cMyc oncogenic translocations and the incidence of B cell tumor development.J. Exp. Med. 205:1949–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tangye S.G., Good K.L. 2007. Human IgM+CD27+ B cells: memory B cells or “memory” B cells? J. Immunol. 179:13–19 [DOI] [PubMed] [Google Scholar]
- Weller S., Braun M.C., Tan B.K., Rosenwald A., Cordier C., Conley M.E., Plebani A., Kumararatne D.S., Bonnet D., Tournilhac O., et al. 2004. Human blood IgM “memory” B cells are circulating splenic marginal zone B cells harboring a prediversified immunoglobulin repertoire.Blood. 104:3647–3654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirths S., Lanzavecchia A. 2005. ABCB1 transporter discriminates human resting naive B cells from cycling transitional and memory B cells.Eur. J. Immunol. 35:3433–3441 [DOI] [PubMed] [Google Scholar]
- Ye B.H., Cattoretti G., Shen Q., Zhang J., Hawe N., de Waard R., Leung C., Nouri-Shirazi M., Orazi A., Chaganti R.S., et al. 1997. The BCL-6 proto-oncogene controls germinal-centre formation and Th2-type inflammation.Nat. Genet. 16:161–170 [DOI] [PubMed] [Google Scholar]