Abstract
The molecular mechanisms underlying the development of hepatocellular carcinoma are not fully understood. Liver-specific signal transducer and activator of transcription (STAT) 5A/B–null mice (STAT5-LKO) were treated with carbon tetrachloride (CCl4), and histological analyses revealed liver fibrosis and tumors. Transforming growth factor (TGF)–β levels and STAT3 activity were elevated in liver tissue from STAT5-LKO mice upon CCl4 treatment. To define the molecular link between STAT5 silencing and TGF-β up-regulation, as well as STAT3 activation, we examined STAT5-null mouse embryonic fibroblasts and primary hepatocytes. These cells displayed elevated TGF-β protein levels, whereas messenger RNA levels remained almost unchanged. Protease inhibitor studies revealed that STAT5 deficiency enhanced the stability of mature TGF-β. Immunoprecipitation and immunohistochemistry analyses demonstrated that STAT5, through its N-terminal sequences, could bind to TGF-β and that retroviral-mediated overexpression of STAT5 decreased TGF-β levels. To confirm the in vivo significance of the N-terminal domain of STAT5, we treated mice that expressed STAT5 lacking the N terminus (STAT5-ΔN) with CCl4. STAT5-ΔN mice developed CCl4-induced liver fibrosis but no tumors. In conclusion, loss of STAT5 results in elevated TGF-β levels and enhanced growth hormone–induced STAT3 activity. We propose that a deregulated STAT5–TGF-β–STAT3 network contributes to the development of chronic liver disease.
Hepatocellular carcinomas (HCC) are a major cause of cancer death and mostly develop as a result of advanced liver fibrosis. Studies have linked the development of HCC with a methylation-induced silencing of SOCS (suppressor of cytokine signaling) genes, which in turn leads to elevated activation of signal transducer and activator of transcription (STAT) 3 (1) or mutations of Axin1, a negative regulator of Wnt signaling (2). However, the mechanisms by which liver fibrosis induces cancer remain elusive.
It is well known that TGF-β is an important cytokine in these fibrotic processes (3, 4) and that excess TGF-β production is a key problem for the treatment of liver diseases as it triggers liver fibrosis (4). Three different gene products, TGF-β1, -β2, and -β3, have been cloned from mammalian tissues (5, 6) and mature TGF-β proteins are composed of two 12.5-kD polypeptides. They are derived from 55-kD polypeptides, which dimerize upon synthesis followed by processing and secretion (7). Most studies describing biological effects of TGF-β have examined how it affects targeted cells. In this paper, we have focused on intracellular processing of the TGF-β complex before secretion. Transformation of hepatocytes, but not of hepatic stellate cells (HSC) or Kupffer cells (KC), leads to cancer as a result of liver fibrosis. Although hepatocytes have not been considered a source of TGF-β, we can hypothesize that intracellular synthesis and modification of TGF-β in these cells is involved in cancer development.
The transcription factors STAT5 and STAT3 promote cell cycle progression and suppress apoptosis, thus contributing to cellular transformation. Notably, activated STAT3 can mediate cellular transformation (8) and has been detected in primary tumors including HCC (1, 9–11). Although STAT5 has been linked to tumorigenesis (12–15), it can also act as a tumor suppressor by inhibiting expression of NPM1-ALK (16). As for solid tumors, the role for STAT5 is controversial (17–19). To explore the role of cytokine signaling through STAT5 in liver fibrosis and cancer, we deleted the Stat5 locus specifically in hepatocytes using Cre-mediated recombination. Using this experimental approach, we discovered a relationship between STAT5, TGF-β, and STAT3 in the pathogenesis of chronic liver disease.
RESULTS
Progression of liver fibrosis in hepatocyte-specific STAT5-null mice
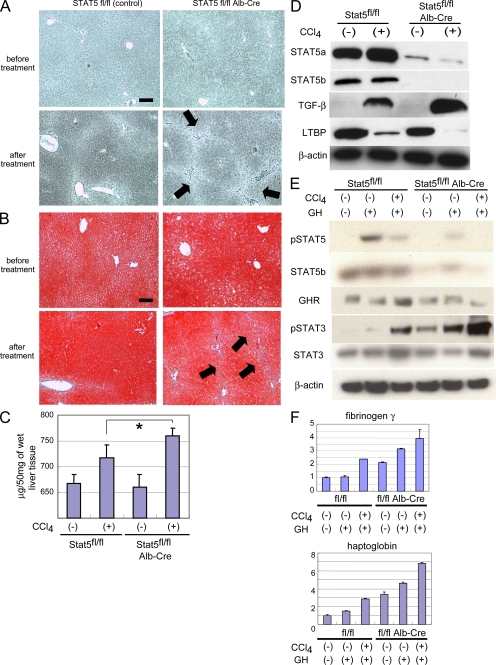
Treatment of mice with carbon tetrachloride (CCl4) induces liver fibrosis and is an accepted model to mimic human disease. To examine the significance of cytokine-STAT5 signaling in the development of liver fibrosis, control mice and mice from which the Stat5 locus had been deleted in hepatocytes using Cre-mediated recombination (STAT5-LKO) were treated for 8 wk with CCl4. Loss of STAT5 did not affect the sensitivity of primary hepatocytes to CCl4 and the slight increase of cell death was not significant (Fig. S1, A and B). Before treatment, there were no histological differences between mice from both groups based on silver and Azan staining (Fig. 1, A and B, top). The reticular fibers (Fig. 1, A and B, arrows) extended and developed into the interlobular septum in STAT5-LKO mice, whereas few reticular fibers deposited in the periportal tissue space in controls (Fig. 1 A, bottom). To evaluate fibrotic changes, we also performed Azan staining to detect collagen accumulation and identified the formation of nodules (Fig. 1 B, arrows) in STAT5 mutant mice (Fig. 1 B, bottom). In agreement with these observations, the content of hepatic collagen was higher in STAT5-LKO mice after treatment (Fig. 1 C). These histological analyses and hepatic collagen assays revealed that STAT5 mutant mice displayed more advanced liver fibrosis than control mice.
Figure 1.
Progression of liver fibrosis in hepatocyte-specific Stat5A/B KO mice. (A and B) Silver staining (A) and Azan staining (B) of liver sections from STAT5A/Bf/f (control) mice and STAT5A/Bf/f;Alb-Cre (STAT5-LKO) mice before and after CCl4 treatment for 8 wk. Arrows indicate reticular fibers. All pictures are shown in low magnification (100×). Bars, 200 µm. (C) Quantification of total acid pepsin-soluble collagens in liver tissue of STAT5A/Bf/f and STAT5A/Bf/f;Alb-Cre before and after CCl4 treatment. Data are mean ± SD. The asterisk indicates a significant difference (P = 0.023) between the control and KO mice (n = 5–7). (D) Expression levels of STAT5A, STAT5B, TGF-β, and LTBP in liver from control and KO mice before and after the treatment as detected by Western blot analysis. (E) Activation levels of STAT5 and STAT3 and expression levels of STAT5, STAT3, and GH receptor (GHR) in liver from control and KO mice before and after CCl4 treatment with or without GH stimulation as detected by Western blot analysis. (F) Expression levels of fibrinogen γ and Haptoglobin in liver from control and STAT5-LKO mice before and after CCl4 treatment with or without GH stimulation as detected by real-time RT-PCR analysis. The relative expression level of the untreated sample of control mice was set as 1, and the fold expression level of each sample was calculated (n = 3 in each group). Data are mean ± SD. Three independent experiments were performed in triplicate and representative data were shown.
TGF-β levels and STAT3 activity are elevated in STAT5-LKO mice after CCl4 treatment
The deletion of the Stat5a/b locus in hepatocytes was confirmed by Western blot analyses and STAT5A/B levels were reduced by ∼80% (Fig. 1 D). TGF-β (mature TGF-β) levels were more profoundly induced in STAT5-LKO mice after CCl4 treatment compared with controls. Latent TGF-β binding protein (LTBP) levels were reduced in STAT5-LKO mice after CCl4 treatment and to a lesser extent in controls. This reduction of LTBP might be the result of an increased secretion of mature TGF-β (20, 21). The expression levels of other fibrogenesis-related factors, such as platelet-derived growth factor α/β and TGF-β pseudo receptor Bambi, were examined and no differences were observed between control and STAT5-LKO mice (Fig. S2).
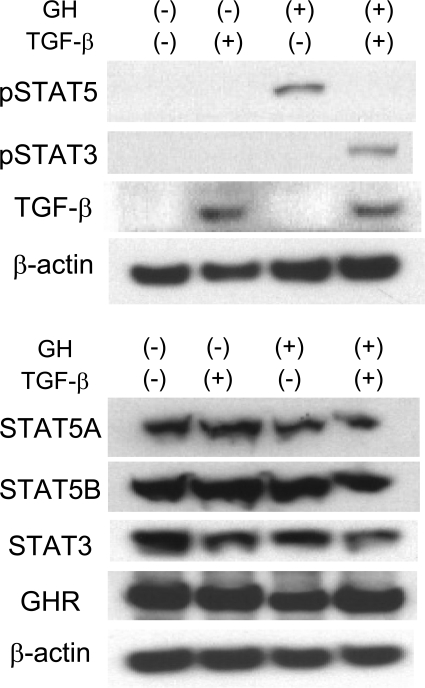
Liver fibrosis is reported to associate with acquired growth hormone (GH) resistance because of low GH receptor levels (22, 23). Although strong activation of STAT5 was observed in hepatocytes of control mice upon stimulation with GH, reduced activation was detected after CCl4 treatment (Fig. 1 E), suggesting that these cells loose their hormonal response during early stages of fibrosis. STAT5 levels and activation were greatly reduced in STAT5-LKO mice before and after GH stimulation irrespective of CCl4 treatment. GH receptor levels were largely independent of GH and CCl4 treatment.
We have recently reported that in the absence of STAT5, other STAT members, especially STAT3 and STAT1, could be recruited to receptors normally engaged by STAT5, which in turn would result in their aberrant activation (24, 25). As shown in Fig. 1 E, GH weakly induced tyrosine phosphorylation of STAT3 (pSTAT3) in control mice, which was exacerbated in the combined presence of GH and CCl4. In STAT5-LKO mice, strong STAT3 activation was observed even before GH injection and was even more pronounced upon GH treatment. Combined CCl4 and GH treatment further enhanced STAT3 activation in STAT5-LKO mice (Fig. 1 E). STAT3 levels were similar in the different groups. Expression levels of the STAT3 target genes fibrinogen γ and haptoglobin were also increased in liver tissue from STAT5-LKO mice and further elevated upon GH and CCl4 treatment (Fig. 1 F and Fig. S3). STAT1 activation was largely independent of CCl4 treatment (unpublished data). These results demonstrate that in the absence of STAT5, GH can directly activate STAT3, and that this activation is strongly enhanced by CCl4.
Development of liver cancer in STAT5-LKO mice
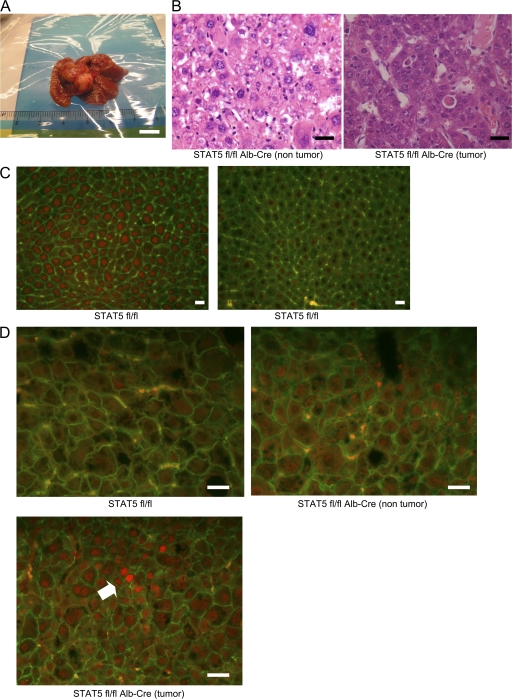
4 out of 20 STAT5-LKO mice had developed multiple tumors after 8 wk of CCl4 treatment, whereas none were detected in 26 control mice (P = 0.0199; Fig. 2 A). Histological analysis revealed that these tumors were mostly formed by moderately differentiated HCCs (Fig. 2 B). It has been established that most HCCs develop as a result of severe liver fibrosis. However, because STAT5-LKO mice had not developed severe fibrosis, we speculated these HCCs were induced through a different mechanism. We again confirmed an almost complete absence of nuclear STAT5B in STAT5-LKO mice, whereas it was present in hepatocytes from control mice (Fig. 2 C). Although little nuclear pSTAT3 was detected in control mice, STAT5-LKO mice exhibited slightly stronger staining of pSTAT3 in nontumor tissue and extensive staining in tumor tissue (Fig. 2 D). These results indicate that STAT3 is gradually activated in the fibrosis-cancer process in STAT5-LKO mice.
Figure 2.
Development of cancer in hepatocyte-specific STAT5A/B KO mice. (A) Appearance of a representative hepatic tumor from STAT5A/Bf/f;Alb-Cre mice after CCl4 treatment. Bar, 1 cm. (B) H&E staining of nontumor (left) and tumor (right) sections from STAT5A/Bf/f;Alb-Cre after CCl4 treatment. Bars, 40 µm. (C) Livers from STAT5f/f (left) and STAT5f/f;Alb-Cre (right) mice were harvested 30 min after GH injection and analyzed for STAT5B expression using by immunofluorescent staining with anti-STAT5B (red) and anti–β-catenin (green) antibodies. Bars, 40 µm. (D) Tissues from STAT5f/f (top) and STAT5A/Bf/f;Alb-Cre (nontumor, top right; tumor, bottom) after CCl4 treatment are analyzed for pSTAT3 activation using immunofluorescent staining with anti-pSTAT3 (red) and anti–β-catenin (green) antibodies. Strong activation of pSTAT3 is detected around the arrow. Bars, 40 µm.
TGF-β protein levels, but not messenger RNA (mRNA) levels, are elevated in STAT5-null mouse embryo fibroblasts and primary hepatocytes.
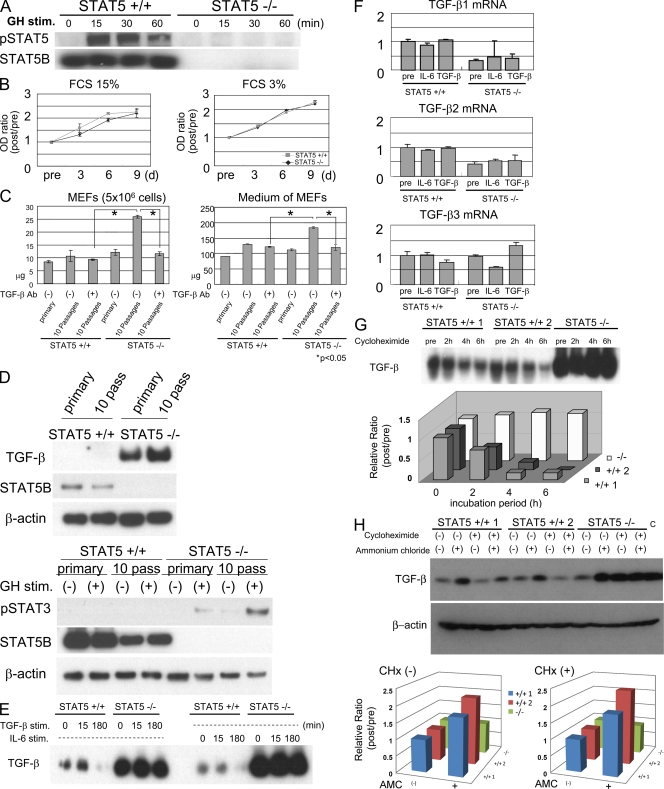
To define the molecular link between STAT5 silencing, STAT3 activation, and elevated TGF-β levels, we performed experiments with mouse embryonic fibroblasts (MEFs) from control and STAT5-null mice. We first confirmed the absence of STAT5 expression and activation in cell lysates from STAT5-null MEFs by Western blot analyses (Fig. 3 A). Proliferation of control and STAT5-null MEFs was similar both in 15 and 3% FCS (Fig. 3 B). Fibroblastic cells are known to produce TGF-β and to be stimulated by autocrine and paracrine pathways (26). Based on our experiments with hepatocytes, we hypothesized that TGF-β levels would be elevated in STAT5-null MEFs, which would lead to a paracrine activation and excess production of extracellular matrix protein. The intracellular content of collagen was similar in MEFs from STAT5-null and control mice immediately after their preparation (Fig. 3 C, left). In contrast, elevated collagen levels were observed in STAT5-null but not control MEFs after 10 passages. Similarly, collagen levels were higher in culture medium obtained from STAT5-null MEFs after 1 mo in culture (Fig. 3 C, right). Furthermore, collagen levels were reduced in MEFs of the 10th passage in the presence of anti–TGF-β antibody, suggesting that increased levels of TGF-β caused excess production of collagen. TGF-β levels were greatly elevated in cell lysates and medium of STAT5-null MEFs compared with control MEFs (Fig. 3 D, top; and Fig. S4). GH-induced STAT3 activation was also elevated in STAT5-KO MEFs (Fig. 3 D, bottom), which is similar to what was observed in liver section from treated mice. Increased TGF-β levels and GH-induced STAT3 activation were also observed in primary hepatocytes stimulated by CCl4 (Fig. S5).
Figure 3.
TGF-β protein levels are enhanced in STAT5A/B-null MEFs. (A) Changes in the pSTAT5 and STAT5B levels in control and STAT5-null MEFs before and after GH stimulation as detected by Western blot analysis. (B) Control and STAT5-null MEFs were seeded on a 96-well culture plate, maintained in 15% FCS (left) or 3% FCS (right), and viable cells were assessed by the WST colorimetric assay. Data are mean ± SD. (C) Quantification of total collagen in cell lysates of MEFs (left) or in culture medium (right). Primary MEFs (bars 1 and 4) or MEFs after 10 passages in the absence (bars 2 and 5) or presence (bars 3 and 6) of TGF-β antibody were used. Neutralization antibody was added to the medium every passage (final concentration, 100 ng/ml). Data are mean ± SD. The asterisks indicate significant differences (left, P = 0.024 [left] or P = 0.011 [right]; right, P = 0.021 [left] or P = 0.024 [right]). (D) Changes in TGF-β levels and STAT3 activation in primary MEFs and MEFs after 10 passages before and after GH stimulation as detected by Western blot analysis. (E) Changes in TGF-β levels of cell lysates in control and STAT5-null MEFs before and after TGF-β or IL-6 stimulation as detected by Western blot analysis. (F) Expression levels of TGF-β1, β2, and β3 mRNA in both cells before and after stimulation as detected by real-time RT-PCR analysis. The relative expression level of untreated sample of control MEFs was considered as 1, and the fold expression level of each sample was calculated. Data are mean ± SD. (G and H) Changes in TGF-β levels in control (two lines were used) and STAT5-null MEFs before and after CHx or ammonium chloride treatment. The intensity of protein bands was measured by AlphaImager (Alpha Innotech). The relative intensity level of the untreated sample of each MEF was considered as 1, and the intensity level of each sample was calculated. Three independent experiments were performed and representative data are shown. Recombinant murine TGF-β protein (Wako Chemicals USA, Inc.) was used as a positive control (H, right).
To confirm that excessive collagen production was linked to an increase of TGF-β, protein and mRNA levels in cell lysates were examined. Mature TGF-β levels were ∼20-fold elevated in STAT5-null MEFs, and these differences were independent of IL-6 or TGF-β stimulation (Fig. 3 E). However, mRNA levels for TGF-β were similar in both cells irrespective of IL-6 or TGF-β stimulation (Fig. 3 F).
Loss of STAT5 enhances the stability of mature TGF-β
Because TGF-β mRNA levels were similar in control and STAT5-null MEFs, we examined the effects of STAT5 on TGF-β protein stability and monitored TGF-β levels after inhibiting protein synthesis with cycloheximide (CHx). Serum-starved wild-type and STAT5-null MEFs were treated with CHx for 0-6 h, harvested, and protein was extracted. Although the levels of mature TGF-β remained fairly stable in STAT5-null cells, they rapidly declined in control cells. Quantitative analysis confirmed an elevated half-life of mature TGF-β in STAT5-null MEFs (Fig. 3 G). To understand the underlying mechanism, we tested the effects of the lysosomal inhibitor ammonium chloride on the loss of mature TGF-β. Pretreatment with this inhibitor restored the levels of mature TGF-β in wild-type MEFs in the presence and absence of CHx, whereas it did not change levels in STAT5-null MEFs (Fig. 3 H). These results demonstrate that loss of STAT5 enhances the stability of mature TGF-β protein.
Interaction of the STAT5 N terminus with TGF-β
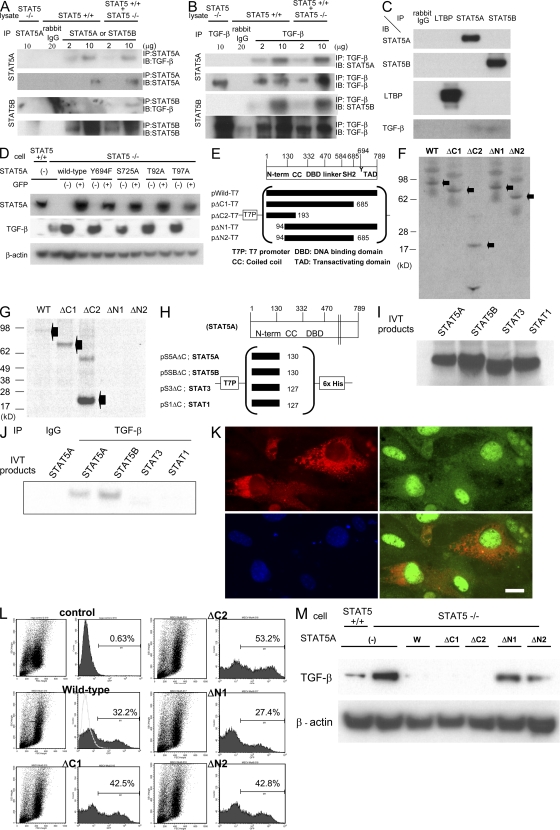
We next used a combination of immunoprecipitation (IP) and immunoblotting to explore the possibility of a physical interaction between STAT5 and TGF-β (Fig. 4, A and B). Cell lysates from STAT5-null and control MEFs, as well as a mixture of the two lysates, were applied to IP followed by immunoblotting. TGF-β was found to interact with STAT5A in extracts from wild-type cells (Fig. 4 A, top). STAT5-null cells served as a negative control. Binding of TGF-β to STAT5B was detected in mixed lysates (Fig. 4 A, bottom). To confirm the interaction between TGF-β and STAT5, we performed the reverse assay performing the IP with antibody against TGF-β (Fig. 4 B). Again, binding of STAT5A and STAT5B to TGF-β was established.
Figure 4.
STAT5 binds to TGF-β and controls its levels. (A and B) Cellular lysates from wild-type and/or STAT5-null MEFs were immunoprecipitated with nonspecific γ-globulin and antibodies against STAT5A/B or TGF-β, and the immunoprecipitates were subjected to Western blotting. (C) Cellular lysates from STAT5 wild-type MEFs were immunoprecipitated with nonspecific γ-globulin and antibodies against LTBP or STAT5A/B. The immunoprecipitates were subjected to Western blotting. (D) Expression levels of STAT5A and TGF-β in wild-type MEFs, STAT5-null MEFs, and STAT5-null MEFs infected with various kinds of retroviral STAT5-expressing vector. After infection, GFP-negative/positive cells were sorted and applied to Western blot analysis. (E and H) Schematic presentation of protein-expressing plasmids. (F and I) Aliquots of in vitro translation products from each plasmid were fractionated by SDS-PAGE. The proteins were labeled with l-[35S]methionine. (G and J) In vitro–synthesized wild-type and mutant STAT5A and the N-terminal domain of each STAT protein were mixed with cellular lysates and immunoprecipitated with TGF-β antibody. (K) STAT5-null MEFs were infected with STAT5A-GFP–expressing retrovirus and transfected with pUB6–TGF-β–V5, which expresses a TGF-β–V5 fusion protein. Cells were analyzed for TGF-β and STAT5A using immunofluorescent staining with anti-V5 (red) and anti-GFP (green) antibodies. Nuclei were stained with DAPI (blue). Bar, 10 µm. (L) Flow cytometry detection of STAT5-GFP–infected cells from STAT5 KO MEFs infected with various kinds of STAT5-expressing retrovirus. The ratio of GFP-positive cells is shown in each graph. (M) TGF-β levels in wild-type, STAT5-null MEFs, and STAT5-null MEFs infected with different STAT5-expressing retroviruses. After infection, GFP-positive cells were sorted and subjected to Western blotting.
Mature TGF-β binds to LTBP, forming latent TGF-β, which is subsequently secreted. We examined which part of the TGF-β complex binds to STAT5 (Fig. 4 C). STAT5A/B were not detected in immunoprecipitates using LTBP antibodies and LTBP was also not identified in immunoprecipitates using STAT5A/B antibodies. TGF-β was found to bind to STAT5A/B but not to LTBP. Furthermore, these results indicate that the TGF-β complex is not detected inside cells after it binds to LTBP, whereas TGF-β is present in the cytoplasm after it binds to STAT5A/B.
To determine whether known functional amino acids in STAT5 are required for the interaction with TGF-β, we generated retroviral vectors encoding wild-type or mutant STAT5. First we prepared retroviruses carrying the wild-type STAT5a gene as well as mutations in amino acids, which are crucial for the function of STAT5A (27–29). None of these mutations adversely affected the interaction between STAT5A and TGF-β, and their presence led to the suppression of TGF-β levels in STAT5-null MEFs, whereas mRNA levels for TGF-β were similar in these cells (Fig. 4 D and Fig. S6).
To further determine which region of STAT5 interacts with TGF-β, we generated expression vectors carrying either wild-type STAT5 (pWild-T7) or various deletion mutants (pΔC1-T7, pΔC2-T7, pΔN1-T7, and pΔN2-T7; Fig. 4 E). Using an in vitro transcription–translation system with l-[35S]methionine, similar levels of wild-type and mutant STAT5A proteins were synthesized (Fig. 4 F). Subsequently, these STAT5 proteins were incubated with cellular lysates from STAT5-null MEFs and immunoprecipitated with antibody against TGF-β (Fig. 4 G). STAT5AΔC1 and ΔC2, as well as wild-type STAT5A, were detected after SDS-PAGE fractionation, whereas STAT5AΔN1 and ΔN2 did not bind to TGF-β. We conclude that the 94 N-terminal amino acids of STAT5 are critical for the interaction with TGF-β.
Because STAT1 and STAT3 share similarities with STAT5 (30), we asked whether their N termini were also able to interact with TGF-β. Using in vitro expression vectors encoding the N-terminal domains of STAT5A/B, STAT3, and STAT1 we determined that the N-terminal domains of STAT5A/B, but not those of STAT3 and STAT1, interacted with TGF-β (Fig. 4, H–J).
To determine the intracellular localization of each protein, we generated vectors expressing TGF-β with a V5 tag and STAT5A coexpressing GFP. STAT5-null MEFs were infected with the STAT5A-expressing retrovirus and subsequently transfected with the TGF-β–V5 plasmid. GFP, an indicator of STAT5A expression, was found preferentially in the nucleus but also in the cytoplasm (Fig. 4 K, top right). In contrast, TGF-β was detected preferentially in the cytoplasm (Fig. 4 K, top left). Cytoplasmic colocalization of STAT5A and TGF-β was observed (Fig. 4 K, bottom right).
TGF-β levels are controlled by STAT5
We have demonstrated an interaction between STAT5 and TGF-β and that elevated TGF-β levels in STAT5-null cells are a result of an enhanced stability. To further establish a causal relationship between STAT5 and TGF-β levels, we expressed wild-type STAT5 and different mutants in STAT5-null MEFs and analyzed TGF-β levels. The efficiency of retroviral infection was equivalent for all constructs (Fig. 4 L). GFP-positive cells were isolated, and subjected to Western blot analysis. TGF-β levels were elevated in STAT5-null MEFs compared with wild-type cells. Infection with retroviruses expressing wild-type, ΔC1, and ΔC2 STAT5A decreased TGF-β to almost undetectable levels. In contrast, TGF-β levels remained unaltered upon expression of STAT5ΔN1 and ΔN2 (Fig. 4 M).
To address the question of whether the interaction with STAT5 would prevent TGF-β from shuttling to lysosomes, we purified lysosomes and analyzed the presence of STAT5-TGF-β complexes. This complex was present in lysosomes in a STAT5A dose-dependent manner (Fig. S7), suggesting that binding to STAT5 does not prevent TGF-β shuttling to lysosomes.
GH-induced STAT3 and STAT5 activation is modulated by TGF-β
To explore the physiological consequences of the STAT5–TGF-β interactions on STAT5 activity, we infected control MEFs with a retrovirus expressing TGF-β/GFP. After treatment with or without GH, GFP-positive cells were isolated and protein extracts were analyzed. Phosphorylated STAT5 was detected upon GH stimulation in the absence of exogenous TGF-β but not in its presence (Fig. 5). Intriguingly, strong GH-induced STAT3 activation was detected only in the presence of exogenous TGF-β. The levels of STAT5A/B, STAT3, and GH receptor were equivalent before and after GH stimulation irrespective of TGF-β expression. These findings demonstrate that TGF-β levels determine whether GH activates STAT3 or STAT5.
Figure 5.
GH-induced STAT3 and STAT5 activation is modulated by TGF-β. Phosphorylation levels of STAT5 and STAT3 and expression levels of TGF-β, STAT5A, STAT5B, STAT3, and GH receptor in STAT5 wild-type MEFs before and after GH stimulation were determined by Western blot analysis. STAT5 wild-type MEFs were infected with a retrovirus based on pMSCV–TGF-β–GFP vector, sorted, and applied to Western blot analysis.
The STAT5 N terminus is critical for the development of liver fibrosis
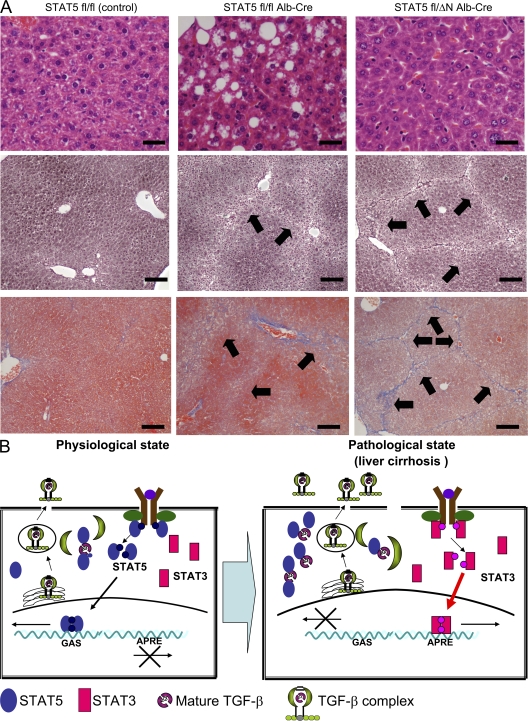
To further explore the in vivo role of the STAT5 N-terminal domain in the process of liver fibrosis, we took advantage of mice that express an N-terminal truncated form of STAT5 (31). We generated mice that carry one allele encoding STAT5A/B lacking the first 90 amino acids, one Stat5 floxed allele, and the Alb-Cre transgene (STAT5fl/ΔN;Alb-Cre mice). Western blot analyses confirmed the presence of STAT5A/B-ΔN in liver tissue (unpublished data). Control mice (STAT5fl/fl), STAT5fl/fl;Alb-Cre (STAT5-LKO), and STAT5fl/ΔN;Alb-Cre (STAT5-ΔN) mice were treated for 8 wk with CCl4. Hematoxylin-eosin (H&E) staining revealed an accumulation of fat droplets in STAT5-LKO but not in control or STAT5-ΔN mice (Fig. 6 A, top). Upon treatment with CCl4, we observed extended reticular fibers that developed similarly in STAT5-ΔN and STAT5-LKO mice but to a lesser extent in controls (Fig. 6 A, middle). Azan staining revealed the formation of nodules and collagen accumulation in STAT5-ΔN and STAT5-LKO mice (Fig. 6 A, bottom). In agreement with these histological findings, increased levels of hepatic collagen were detected after CCl4 treatment (unpublished data). However, no tumors were detected in 17 STAT5-ΔN mice. These results indicate that the STAT5 N terminus is dispensable for the tumor suppressor function of STAT5. Furthermore, it strongly suggests that fat accumulation in STAT5-LKO mice has almost no effect on the fibrotic process because STAT5-ΔN and STAT5-LKO mice had a similar degree of liver fibrosis after treatment.
Figure 6.
The N-terminal domain of STAT5 is critical for the development of liver fibrosis. (A) H&E staining of liver sections from control, STAT5-LKO, and STAT5-ΔN mice before CCl4 treatment (top) and silver (middle) and azan (bottom) staining of liver sections from these mice after CCl4 treatment. Arrows indicate reticular fibers. H&E images are shown at high magnification (400×; bars, 50 µm), and others are at low magnification (100×; bars, 200 µm). n = 3–5 in each group. (B) Model for the development of liver fibrosis and cancer through elevated expression of TGF-β and activation of STAT3 in hepatocytes. Under physiological conditions, TGF-β is present at low or nondetectable levels and some is secreted after binding to LTBP in a constitutive fashion. STAT5 binds to TGF-β and interferes with the formation of TGF-β complex, resulting in suppression of TGF-β protein stability. At this level of TGF-β, STAT5 levels and activation are not influenced. Upon development of liver fibrosis, TGF-β levels increase and LTBP levels decrease. Under these conditions, STAT5 is sequestered by TGF-β and can no longer be activated by GH. Instead, GH activates STAT3.
DISCUSSION
GH resistance and low serum levels of insulin-like growth factor 1 have been identified in a large percentage of cirrhotic patients (23, 32), suggesting a link between STAT5 signaling and liver fibrosis and possibly HCC. In this paper, we have demonstrated that TGF-β directly interacts with STAT5 and that elevated levels of TGF-β abrogate GH-induced STAT5 activation. Instead, elevated levels of TGF-β result in GH activation of STAT3, and constitutive STAT3 activity is commonly observed in HCC. Elevated phosphorylation of STAT3 has been associated with epithelial cancers and linked to the induction of STAT3 target genes. In support of a STAT5–TGF-β–STAT3 mechanism, deletion of STAT5 from hepatocytes in mice resulted in elevated levels of TGF-β, an activation of STAT3, and, finally, the development of HCC. We hypothesize that a TGF-β–mediated abrogation of STAT5 signaling results in the activation of STAT3, which is a key step in the development of HCC (Fig. 6 B).
Most likely, the development of liver cancer in this model is the result of more than one molecular insult. Notably, excessive STAT3 activation upon GH and CCl4 treatment in STAT5-LKO mice could result in unscheduled cell proliferation and transformation. TGF-β binds to the N terminus of STAT5, and loss of STAT5 led to elevated TGF-β levels as a result of increased stability. The absence of the STAT5 N terminus in STAT5-ΔN mice did not abrogate the development of liver fibrosis, but no tumors were observed in these mice. This suggests that an N-terminally truncated STAT5 still contains tumor suppressor functions similar to that seen with wild-type STAT5. Several studies that compared STAT5-ΔN mice and mice completely missing STAT5A/B established functional activity of the N-terminally truncated STAT5. For example, although the complete loss of STAT5 results in perinatal lethality, STAT5-ΔN mice are viable (33). Similarly, the complete loss of STAT5 causes more severe hematological defects (34, 35). A recent study demonstrated that the N-terminal domain plays an important role in maintaining normal balance of lymphoid and myeloid cells (36). Although it is clear that loss of STAT5 N terminus, which is required for the tetramerization of STAT5, does not result in increased liver cancer induced by CCl4, its contribution to oncogenesis might be cell specific. Moriggl et al. (37) established that loss of the N-terminal region protected the development of certain leukemia. At this point, the cell specificity of STAT5 in normal development and oncogenesis is poorly understood.
TGF-β and STAT3 are not only critical in the promotion of tumor progression and survival but they also contribute to tumor-immune escape (38, 39). Notably, increased TGF-β secretion might lead to the expansion of CD4+CD25+FoxP3+ regulatory T (T reg) cells that would impact liver fibrosis and tumor development (40, 41). Another possibility for the promotion of liver fibrosis is that the deletion of STAT5 might have an effect on HSC or KC and increase their sensitivity to TGF-β. However, the CD4/CD8 ratio and TH1- and TH2-cytokine production were similar in control and mutant mice (Fig. S8, A–C), and no selective expansion of T reg cells was observed after CCl4 treatment (Fig. S8, D and E). Moreover, the sensitivity of HSC to TGF-β in isolated HSC was identical in both mice (Fig. S9). These results contributed to a nonbiased analysis of the role of STAT5 in liver fibrosis and cancer development.
In the absence of inflammation, liver TGF-β is secreted from HSC and KC but not from hepatocytes. However, upon inflammation or injury hepatocytes gradually become the major source of TGF-β (42–45). As hepatic fibrosis develops, TGF-β levels increase, followed by the accumulation of extracellular matrix (4). Collectively, hepatocytes secrete an excess of TGF-β in the late stage of liver fibrosis when most cancer develops. It has been shown that TGF-β1 induces an epithelial-to-mesenchymal transition in mature hepatocytes, resulting in type I collagen synthesis (46, 47). Because embryonic fibroblasts secrete TGF-β, MEFs are reminiscent of myofibroblasts, which are activated in liver fibrosis state. TGF-β levels were highly elevated in STAT5-null MEFs and reintroduction of STA5 resulted in the suppression of TGF-β. Based on our studies, we propose that binding of STAT5 to TGF-β reduces its half-life. STAT5 has been reported to interact with other molecules, such as the glucocorticoid receptor (GR) and HNF (hepatocytes nuclear factor) 4α (48, 49). Although the GR can act as a transcriptional coactivator of STAT5 and enhances STAT5-dependent transcription, the GR–STAT5 complex diminishes the glucocorticoid response of promoters harboring glucocorticoid response elements. As for HNF-4α, it strongly inhibits GH-induced STAT5B transcriptional activity. Although STAT5 decreases TGF-β stability, resulting in a reduction of TGF-β, excess levels of TGF-β inhibit GH-induced activation of STAT5. In this context, GH preferentially activates STAT3, which in turn activates genes linked to cell proliferation and survival. Thus, STAT5 transcriptional functionality can be modulated by activating and suppressing cofactors (50).
The unscheduled ability of STAT3 to be activated by cytokines that normally only activate STAT5 is no artifact of the STAT5-KO mouse model. Recently, several inactivating STAT5B mutations have been identified in humans (51, 52) and linked to dwarfism and immunological disorders. Stimulation of primary cells from these patients with GH and prolactin leads to an unscheduled activation of STAT3 (52), and it is conceivable that some of the immunological disorders observed in these patients can be linked to the aberrant activation of STAT3 rather than the loss of STAT5B. With respect to liver fibrosis and HCC, we propose that TGF-β–mediated down-regulation of STAT5 levels in hepatocytes leads to an aberrant activation of STAT3.
MATERIALS AND METHODS
Mice and treatment.
Through breeding, we generated STAT5A/Bfl/fl;Alb-Cre, STAT5A/Bfl/fl, and STAT5A/Bfl/ΔN;Alb-Cre mice, which are in a mixed background (STAT5-ΔN mice were provided by J.N. Ihle, St Jude's Children's Research Hospital, Memphis, TN) (30, 33, 53). Only 10–18-wk-old female mice were used in the experiment unless otherwise indicated. Primers for genotyping STAT5 were described elsewhere (33). Hepatic fibrosis in mice was induced by i.p. injection with 2 ml/kg body weight of 10% CCl4 (Sigma-Aldrich) dissolved in olive oil (Sigma-Aldrich) three times a week for 8 wk. As for GH stimulation, mice were injected with 2 µg/g body weight of GH i.p., and 30 min after injection they were sacrificed. We treated the animals humanely and performed procedures according to the protocol approved by the Animal Use and Care committee at the National Institute of Diabetes and Digestive and Kidney Diseases.
Isolation of MEF cells.
MEFs were isolated from embryonic day–14 embryos of wild-type and STAT5A−/−STAT5B−/− mice by first mincing the embryos, digesting in 0.05% trypsin/0.02% EDTA for 30 min at 37°C, pelleting the tissue, and resuspending in growth medium consisting of DMEM containing 10% FCS (31). MEFs were maintained in high-glucose DMEM supplemented with 15% FCS, 50 µg/ml streptomycin sulfate, 50 units/ml of penicillin G sodium, and nonessential amino acid in 5% CO2 at 37°C.
Antibodies, immunostaining, and immunoblotting.
The rabbit polyclonal anti-STAT5A (L-20), anti-STAT5B (C-17), and anti-STAT3 (C-20) antibodies were purchased from Santa Cruz Biotechnology, Inc. The rabbit phospho-STAT5 (Tyr694), phospho-STAT3 (Tyr705), and TGF-β antibodies were obtained from Cell Signaling Technology. Mouse monoclonal β-actin antibody was purchased from Millipore and Santa Cruz Biotechnology, Inc. Anti-LTBP, anti-GH receptor, and anti–TGF-β neutralization antibodies were obtained from R&D Systems, and anti-GFP and anti-V5 antibodies were obtained from Invitrogen. Immunohistochemistry and immunoblotting were performed using standard procedures according to the manufacturers' instructions. Some blots are shown in larger areas (Fig. S10).
Protein–protein interaction analysis.
To examine the binding of the STAT5 protein to TGF-β protein, the IP/Western blot analyses were applied in this study. The detailed procedure is described elsewhere (54). In short, the cellular protein was extracted from control and STAT5−/− MEFs and precleared by incubation with protein A–Sepharose beads at 4°C for 1 h. Next, the sample was incubated with the beads coupled to the TGF-β or STAT5A/B antibody, or with the rabbit nonspecific γ-globulin (Dako) for 18 h. The immune complex was eluted by being boiled for 5 min. Finally, the supernatant was used for Western blot to detect the TGF-β or STAT5A/B. To confirm the binding site of STAT5A protein to TGF-β protein, several STAT5A proteins possessing the wild-type or some deleted amino acid sequences were synthesized from pΔC1-T7, pΔC2-T7, pΔN1-T7, pΔN2-T7 (indicated in Fig. 4 E), pS5AΔC, p5SBΔC, pS3ΔC, and pS1ΔC (indicated in Fig. 4 H) by the in vitro translation method. The reaction was performed with the TNT T7 coupled reticulocyte lysate system (Promega) and labeled with l-[35S]methionine (GE Healthcare). The cellular lysates from STAT5 wild-type MEFs were extracted and mixed with various types of the in vitro–synthesized STAT5A/B, STAT3, or STAT1 proteins. The mixture was then subjected to IP using antibody against TGF-β as described in the first half of this paragraph. Finally, the immunoprecipitate was separated with SDS-PAGE and the gel was dried and autoradiographed.
Plasmid constructs.
Retroviral-expressing vectors of wild-type STAT5A, partially deleted STAT5A, and TGF-β were constructed from pMSCV-IRES GFP new polylinker (provided by R. Moriggl, Ludwig Boltzmann Institute for Cancer Research, Vienna, Austria). First, pMSCV-STAT5A-GFP or pMSCV–TGF-β–GFP, carrying the entire STAT5a complementary DNA (cDNA) or TGF-β cDNA, was made up by inserting each gene into EcoRI and SacII sites. Four kinds of single amino acid–mutated STAT5a expression vectors were generated by site-directed mutagenesis (T92A, from aca to gca; T97A, from acg to gcg; Y694F, from tac to ttc; and S725A, from tcc to gcc). Other retroviral-expressing vectors of partly deleted STAT5a, such as ΔC1, ΔC2, ΔN1, and ΔN2, were also made up by inserting each PCR product (indicated in Fig. 4 E). For the synthesis of entire or partly deleted STAT5 protein in vitro, we also made five kinds of expressing plasmids, each of which carries the entire or deleted region of STAT5a downstream of the T7 promoter based on pET161/GW/D-TOPO (Invitrogen). To examine the intracellular localization of STAT5 and TGF-β proteins, we generated two expressing plasmids with a certain tag. One of them is pUB6-TGF-β-V5, which carries the entire TGF-β gene attached with V5 tag in the C terminus of TGF-β, and the other is pMSCV-GFP-STAT5A, which has the STAT5a gene with GFP in the N terminus of STAT5a.
Collagen quantitation.
Collagen concentration in liver was determined using the Sircol collagen assay (Biodye Science). In brief, 50 mg of liver tissue was homogenized, and total acid pepsin-soluble collagens were extracted overnight using 5 mg/ml pepsin in 500 ml of 0.5 M of acetic acid. 1 ml of Sircol dye reagent was added to 100 µl of each sample in duplicate at 25°C for 30 min. After centrifugation, the pellet was suspended in 1 ml of alkali reagent, and absorbance was read at 540 nm.
Production and infection of retrovirus.
For making retrovirus, 293T cells were transfected with various kinds of pMSCV-IRES-GFP and Ecopac (the plasmid containing packaging sequences, pIK6.1MCV.ecopac.UTd) using FuGENE (Roche). All supernatants were collected for 48–72 h after transfection and passed through a 0.45-µm filter before freezing at −80°C.
For the infection of retrovirus to MEFs, 106 cells were seeded on a 10-cm culture dish and were infected the next day with retrovirus in the presence of 8 µg/ml polybrene. 48–72 h after infection, the cells were lysed and subjected to the subsequent experiments.
FACS.
After infection with retrovirus, MEFs were trypsinized, washed twice in PBS, resuspended in 1 ml of PBS, and, finally, subjected to flow cytometry (FACSCalibur; BD). Nonfluorescent cells and GFP-expressing cells were isolated using the FACS Vantage (BD) and sorted directly into PBS. Protein was extracted from each population and applied to Western blot analyses.
Histology.
H&E, silver staining, and Azan staining were performed using standard procedures. In short, liver pieces were rapidly removed and fixed in 10% neutral buffered formalin, processed routinely, and embedded in paraffin wax. The sections were cut to appropriate thickness followed by each staining.
RNA extraction and real-time PCR analysis.
Total RNA was isolated (RNeasy mini kit; QIAGEN), and then 1-µg amounts of RNA were reverse transcribed (SuperScript III First-Strand Synthesis SuperMix; Invitrogen), and cDNAs were amplified using Taqman (for fibrinogen γ and haptoglobin) or SYBR Green master mix and probe (for TGF-β1, β2, and β3; Applied Biosystems). These assays were performed according to the manufacturers' instructions.
Analysis of cell growth.
To examine the cell growth curve, 5 × 103 wild-type and STAT5 KO MEFs were seeded on a 96-well culture plate. After 3, 6, and 9 d, the net number of viable cells was assessed colorimetrically using water-soluble tetrazolium (WST; 2-[2-methoxy-4-nitrophenyl]-3-[4-nitrophenyl]-5-[2, 4-disulfophenyl]-2H-tetrazolium monosodium salt; Roche). This assay is based on the cleavage of tetrazolium salt by mitochondrial dehydrogenase in viable cells. For the assay, 10 µl WST reagent was added to the 100-µl culture medium, followed by incubation at 37°C for 2 h. Then the optical density at 450 nm was measured. The assay was done in quadruplicate, and the values were expressed as the means ± SD.
Degradation of TGF-β after blockade of protein synthesis and effects of lysosome inhibitors on TGF-β stability.
STAT5 wild-type (two independent lines) and KO MEFs were grown to 80% confluence in 60-mm dishes and serum starved overnight. Cells were then incubated with 20 mg/ml CHx for 0–6 h or 100 mM ammonium chloride for 2 h followed by CHx or PBS treatment. Cells lysates were resolved by SDS-PAGE and blotted with anti–TGF-β antibody.
Statistics.
All statistical analyses were performed using a two-tailed unpaired Student's t test. A p-value of 0.05 or less was considered significant.
Online supplemental material.
Fig. S1 shows cell availability in isolated hepatocytes and the ratio of apoptotic cells in liver tissues. Fig. S2 shows expression levels of fibrosis-related gene in liver tissues. Fig. S3 shows expression levels of fibrinogen α. Fig. S4 shows serum TGF-β levels in MEF culture medium. Fig. S5 shows pSTAT3 and TGF-β levels in isolated hepatocytes before and after CCl4 treatment. Fig. S6 shows TGF-β1 expression levels in various kinds of MEFs. Fig. S7 shows the existence of the TGF-β–STAT5 complex in lysosome fraction. Fig. S8 shows the immunological analysis, CD4/CD8 ratios, B cell ratios, cytokine production, and T reg cell ratios. Fig. S9 shows pSMAD2 level in isolated HSCs. Fig. S10 shows a larger area of some Western blots. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20080003/DC1.
Acknowledgments
We are grateful to James N. Ihle (St. Jude Children's Research Hospital, USA) for providing STAT5-ΔN mice. We also thank Tatsuki Kataoka (National Institute of Allergy and Infectious Diseases, National Institutes of Health, USA) for histological assessment, Rie Hanayama (Osaka University Graduate School of Medicine, Japan) for statistical data analysis, and April R. Robbins (National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, USA) for helping us to isolate lysosomal fractions. Traudl Robinson is acknowledged for help with figures.
This work was supported by the intramural program of the National Institute of Diabetes and Digestive and Kidney Diseases/National Institutes of Health.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used: cDNA, complementary DNA; CHx, cycloheximide; GH, growth hormone; GR, glucocorticoid receptor; HCC, hepatocellular carcinoma; HSC, hepatic stellate cell; IP, immunoprecipitation; KC, Kupffer cell; LTBP, latent TGF-β binding protein; MEF, mouse embryonic fibroblast; mRNA, messenger RNA; STAT, signal transducer and activator of transcription; WST, water-soluble tetrazolium.
References
- Yoshikawa H., Matsubara K., Qian G.S., Jackson P., Groopman J.D., Manning J.E., Harris C.C., Herman J.G. 2001. SOCS-1, a negative regulator of the JAK/STAT pathway, is silenced by methylation in human hepatocellular carcinoma and shows growth-suppression activity.Nat. Genet. 28:29–35 [DOI] [PubMed] [Google Scholar]
- Satoh S., Daigo Y., Furukawa Y., Kato T., Miwa N., Nishiwaki T., Kawasoe T., Ishiguro H., Fujita M., Tokino T., et al. 2000. AXIN1 mutations in hepatocellular carcinomas, and growth suppression in cancer cells by virus-mediated transfer of AXIN1.Nat. Genet. 24:245–250 [DOI] [PubMed] [Google Scholar]
- Raghow R., Postlethwaite A.E., Keski-Oja J., Moses H.L., Kang A.H. 1987. Transforming growth factor-beta increases steady state levels of type I procollagen and fibronectin messenger RNAs posttranscriptionally in cultured human dermal fibroblasts.J. Clin. Invest. 79:1285–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaja M.J., Weiner F.R., Flanders K.C., Giambrone M.A., Wind R., Biempica L., Zern M.A. 1989. In vitro and in vivo association of transforming growth factor–β1 with hepatic fibrosis.J. Cell Biol. 108:2477–2482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derynck R., Jarrett J.A., Chen E.Y., Eaton D.H., Bell J.R., Assoian R.K., Roberts A.B., Sporn M.B., Goeddel D.V. 1985. Human transforming growth factor-beta complementary DNA sequence and expression in normal and transformed cells.Nature. 316:701–705 [DOI] [PubMed] [Google Scholar]
- de Martin R., Haendler B., Hofer-Warbinek R., Gaugitsch H., Wrann M., Schlusener H., Seifert J.M., Bodmer S., Fontana A., Hofer E. 1987. Complementary DNA for human glioblastoma-derived T cell suppressor factor, a novel member of the transforming growth factor-beta gene family.EMBO J. 6:3673–3677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyytiainen M., Penttinen C., Keski-Oja J. 2004. Latent TGF-beta binding proteins: extracellular matrix association and roles in TGF-beta activation.Crit. Rev. Clin. Lab. Sci. 41:233–264 [DOI] [PubMed] [Google Scholar]
- Bromberg J.F., Wrzeszczynska M.H., Devgan G., Zhao Y., Pestell R.G., Albanese C., Darnell J.E., Jr. 1999. Stat3 as an oncogene.Cell. 98:295–303 [DOI] [PubMed] [Google Scholar]
- Frank D.A. 1999. STAT signaling in the pathogenesis and treatment of cancer.Mol. Med. 5:432–456 [PMC free article] [PubMed] [Google Scholar]
- Grandis J.R., Drenning S.D., Chakraborty A., Zhou M.Y., Zeng Q., Pitt A.S., Tweardy D.J. 1998. Requirement of Stat3 but not Stat1 activation for epidermal growth factor receptor- mediated cell growth In vitro.J. Clin. Invest. 102:1385–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catlett-Falcone R., Landowski T.H., Oshiro M.M., Turkson J., Levitzki A., Savino R., Ciliberto G., Moscinski L., Fernandez-Luna J.L., Nunez G., et al. 1999. Constitutive activation of Stat3 signaling confers resistance to apoptosis in human U266 myeloma cells.Immunity. 10:105–115 [DOI] [PubMed] [Google Scholar]
- Migone T.S., Lin J.X., Cereseto A., Mulloy J.C., O'Shea J.J., Franchini G., Leonard W.J. 1995. Constitutively activated Jak-STAT pathway in T cells transformed with HTLV-I.Science. 269:79–81 [DOI] [PubMed] [Google Scholar]
- Shuai K., Halpern J., ten Hoeve J., Rao X., Sawyers C.L. 1996. Constitutive activation of STAT5 by the BCR-ABL oncogene in chronic myelogenous leukemia.Oncogene. 13:247–254 [PubMed] [Google Scholar]
- Moriggl R., Topham D.J., Teglund S., Sexl V., McKay C., Wang D., Hoffmeyer A., van Deursen J., Sangster M.Y., Bunting K.D., et al. 1999. Stat5 is required for IL-2-induced cell cycle progression of peripheral T cells.Immunity. 10:249–259 [DOI] [PubMed] [Google Scholar]
- Socolovsky M., Fallon A.E., Wang S., Brugnara C., Lodish H.F. 1999. Fetal anemia and apoptosis of red cell progenitors in Stat5a−/−5b−/− mice: a direct role for Stat5 in Bcl-X(L) induction.Cell. 98:181–191 [DOI] [PubMed] [Google Scholar]
- Zhang Q., Wang H.Y., Liu X., Wasik M.A. 2007. STAT5A is epigenetically silenced by the tyrosine kinase NPM1-ALK and acts as a tumor suppressor by reciprocally inhibiting NPM1-ALK expression.Nat. Med. 13:1341–1348 [DOI] [PubMed] [Google Scholar]
- Xi S., Zhang Q., Gooding W.E., Smithgall T.E., Grandis J.R. 2003. Constitutive activation of Stat5b contributes to carcinogenesis in vivo.Cancer Res. 63:6763–6771 [PubMed] [Google Scholar]
- Kannangai R., Sahin F., Torbenson M.S. 2006. EGFR is phosphorylated at Ty845 in hepatocellular carcinoma.Mod. Pathol. 19:1456–1461 [DOI] [PubMed] [Google Scholar]
- Lee T.K., Man K., Poon R.T., Lo C.M., Yuen A.P., Ng I.O., Ng K.T., Leonard W., Fan S.T. 2006. Signal transducers and activators of transcription 5b activation enhances hepatocellular carcinoma aggressiveness through induction of epithelial-mesenchymal transition.Cancer Res. 66:9948–9956 [DOI] [PubMed] [Google Scholar]
- Miyazono K., Olofsson A., Colosetti P., Heldin C.H. 1991. A role of the latent TGF-beta 1-binding protein in the assembly and secretion of TGF-beta 1.EMBO J. 10:1091–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drews F., Knobel S., Moser M., Muhlack K.G., Mohren S., Stoll C., Bosio A., Gressner A.M., Weiskirchen R. 2008. Disruption of the latent transforming growth factor-beta binding protein-1 gene causes alteration in facial structure and influences TGF-beta bioavailability.Biochim. Biophys. Acta. 1783:34–48 [DOI] [PubMed] [Google Scholar]
- Moller S., Becker U., Gronbaek M., Juul A., Winkler K., Skakkebaek N.E. 1994. Short-term effect of recombinant human growth hormone in patients with alcoholic cirrhosis.J. Hepatol. 21:710–717 [DOI] [PubMed] [Google Scholar]
- Shen X.Y., Holt R.I., Miell J.P., Justice S., Portmann B., Postel-Vinay M.C., Ross R.J. 1998. Cirrhotic liver expresses low levels of the full-length and truncated growth hormone receptors.J. Clin. Endocrinol. Metab. 83:2532–2538 [DOI] [PubMed] [Google Scholar]
- Cui Y., Hosui A., Sun R., Shen K., Gavrilova O., Chen W., Cam M.C., Gao B., Robinson G.W., Hennighausen L. 2007. Loss of signal transducer and activator of transcription 5 leads to hepatosteatosis and impaired liver regeneration.Hepatology. 46:504–513 [DOI] [PubMed] [Google Scholar]
- Hosui A., Hennighausen L. 2008. Genomic dissection of the cytokine controlled STAT5 signaling network in liver.Physiol. Genomics. 34:135–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses H.L., Coffey R.J., Jr., Leof E.B., Lyons R.M., Keski-Oja J. 1987. Transforming growth factor beta regulation of cell proliferation.J. Cell. Physiol. Suppl. 133:1–7 [DOI] [PubMed] [Google Scholar]
- Wood T.J., Sliva D., Lobie P.E., Pircher T.J., Gouilleux F., Wakao H., Gustafsson J.A., Groner B., Norstedt G., Haldosen L.A. 1995. Mediation of growth hormone-dependent transcriptional activation by mammary gland factor/Stat 5.J. Biol. Chem. 270:9448–9453 [DOI] [PubMed] [Google Scholar]
- Beuvink I., Hess D., Flotow H., Hofsteenge J., Groner B., Hynes N.E. 2000. Stat5a serine phosphorylation. Serine 779 is constitutively phosphorylated in the mammary gland, and serine 725 phosphorylation influences prolactin-stimulated in vitro DNA binding activity.J. Biol. Chem. 275:10247–10255 [DOI] [PubMed] [Google Scholar]
- Gewinner C., Hart G., Zachara N., Cole R., Beisenherz-Huss C., Groner B. 2004. The coactivator of transcription CREB-binding protein interacts preferentially with the glycosylated form of Stat5.J. Biol. Chem. 279:3563–3572 [DOI] [PubMed] [Google Scholar]
- Darnell J.E., Jr. 1997. STATs and gene regulation.Science. 277:1630–1635 [DOI] [PubMed] [Google Scholar]
- Teglund S., McKay C., Schuetz E., van Deursen J.M., Stravopodis D., Wang D., Brown M., Bodner S., Grosveld G., Ihle J.N. 1998. Stat5a and Stat5b proteins have essential and nonessential, or redundant, roles in cytokine responses.Cell. 93:841–850 [DOI] [PubMed] [Google Scholar]
- Donaghy A., Ross R., Gimson A., Hughes S.C., Holly J., Williams R. 1995. Growth hormone, insulinlike growth factor-1, and insulinlike growth factor binding proteins 1 and 3 in chronic liver disease.Hepatology. 21:680–688 [PubMed] [Google Scholar]
- Cui Y., Riedlinger G., Miyoshi K., Tang W., Li C., Deng C.X., Robinson G.W., Hennighausen L. 2004. Inactivation of Stat5 in mouse mammary epithelium during pregnancy reveals distinct functions in cell proliferation, survival, and differentiation.Mol. Cell. Biol. 24:8037–8047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoelbl A., Kovacic B., Kerenyi M.A., Simma O., Warsch W., Cui Y., Beug H., Hennighausen L., Moriggl R., Sexl V. 2006. Clarifying the role of Stat5 in lymphoid development and Abelson-induced transformation.Blood. 107:4898–4906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z., Cui Y., Watford W.T., Bream J.H., Yamaoka K., Hissong B.D., Li D., Durum S.K., Jiang Q., Bhandoola A., et al. 2006. Stat5a/b are essential for normal lymphoid development and differentiation.Proc. Natl. Acad. Sci. USA. 103:1000–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Wang Z., Zhang Y., Kang Z., Haviernikova E., Cui Y., Hennighausen L., Moriggl R., Wang D., Tse W., Bunting K.D. 2007. STAT5 requires the N-domain to maintain hematopoietic stem cell repopulating function and appropriate lymphoid-myeloid lineage output.Exp. Hematol. 35:1684–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriggl R., Sexl V., Kenner L., Duntsch C., Stangl K., Gingras S., Hoffmeyer A., Bauer A., Piekorz R., Wang D., et al. 2005. Stat5 tetramer formation is associated with leukemogenesis.Cancer Cell. 7:87–99 [DOI] [PubMed] [Google Scholar]
- Thomas D.A., Massague J. 2005. TGF-beta directly targets cytotoxic T cell functions during tumor evasion of immune surveillance.Cancer Cell. 8:369–380 [DOI] [PubMed] [Google Scholar]
- Wang T., Niu G., Kortylewski M., Burdelya L., Shain K., Zhang S., Bhattacharya R., Gabrilovich D., Heller R., Coppola D., et al. 2004. Regulation of the innate and adaptive immune responses by Stat-3 signaling in tumor cells.Nat. Med. 10:48–54 [DOI] [PubMed] [Google Scholar]
- Unitt E., Rushbrook S.M., Marshall A., Davies S., Gibbs P., Morris L.S., Coleman N., Alexander G.J. 2005. Compromised lymphocytes infiltrate hepatocellular carcinoma: the role of T-regulatory cells.Hepatology. 41:722–730 [DOI] [PubMed] [Google Scholar]
- Seki E., De Minicis S., Osterreicher C.H., Kluwe J., Osawa Y., Brenner D.A., Schwabe R.F. 2007. TLR4 enhances TGF-beta signaling and hepatic fibrosis.Nat. Med. 13:1324–1332 [DOI] [PubMed] [Google Scholar]
- Takehara T., Tatsumi T., Suzuki T., Rucker E.B., III, Hennighausen L., Jinushi M., Miyagi T., Kanazawa Y., Hayashi N. 2004. Hepatocyte-specific disruption of Bcl-xL leads to continuous hepatocyte apoptosis and liver fibrotic responses.Gastroenterology. 127:1189–1197 [DOI] [PubMed] [Google Scholar]
- Canbay A., Friedman S., Gores G.J. 2004. Apoptosis: the nexus of liver injury and fibrosis.Hepatology. 39:273–278 [DOI] [PubMed] [Google Scholar]
- Gao C., Gressner G., Zoremba M., Gressner A.M. 1996. Transforming growth factor beta (TGF-beta) expression in isolated and cultured rat hepatocytes.J. Cell. Physiol. 167:394–405 [DOI] [PubMed] [Google Scholar]
- Jeong W.I., Do S.H., Yun H.S., Song B.J., Kim S.J., Kwak W.J., Yoo S.E., Park H.Y., Jeong K.S. 2004. Hypoxia potentiates transforming growth factor-beta expression of hepatocyte during the cirrhotic condition in rat liver.Liver Int. 24:658–668 [DOI] [PubMed] [Google Scholar]
- Kaimori A., Potter J., Kaimori J.Y., Wang C., Mezey E., Koteish A. 2007. Transforming growth factor-beta1 induces an epithelial-to-mesenchymal transition state in mouse hepatocytes in vitro.J. Biol. Chem. 282:22089–22101 [DOI] [PubMed] [Google Scholar]
- Iwano M., Plieth D., Danoff T.M., Xue C., Okada H., Neilson E.G. 2002. Evidence that fibroblasts derive from epithelium during tissue fibrosis.J. Clin. Invest. 110:341–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocklin E., Wissler M., Gouilleux F., Groner B. 1996. Functional interactions between Stat5 and the glucocorticoid receptor.Nature. 383:726–728 [DOI] [PubMed] [Google Scholar]
- Park S.H., Wiwi C.A., Waxman D.J. 2006. Signalling cross-talk between hepatocyte nuclear factor 4alpha and growth-hormone-activated STAT5b.Biochem. J. 397:159–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennighausen L., Robinson G.W. 2008. Interpretation of cytokine signaling through the transcription factors STAT5A and STAT5B.Genes Dev. 22:711–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernasconi A., Marino R., Ribas A., Rossi J., Ciaccio M., Oleastro M., Ornani A., Paz R., Rivarola M.A., Zelazko M., Belgorosky A. 2006. Characterization of immunodeficiency in a patient with growth hormone insensitivity secondary to a novel STAT5b gene mutation.Pediatrics. 118:e1584–e1592 [DOI] [PubMed] [Google Scholar]
- Kofoed E.M., Hwa V., Little B., Woods K.A., Buckway C.K., Tsubaki J., Pratt K.L., Bezrodnik L., Jasper H., Tepper A., et al. 2003. Growth hormone insensitivity associated with a STAT5b mutation.N. Engl. J. Med. 349:1139–1147 [DOI] [PubMed] [Google Scholar]
- Yakar S., Liu J.L., Stannard B., Butler A., Accili D., Sauer B., LeRoith D. 1999. Normal growth and development in the absence of hepatic insulin-like growth factor I.Proc. Natl. Acad. Sci. USA. 96:7324–7329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosui A., Ohkawa K., Ishida H., Sato A., Nakanishi F., Ueda K., Takehara T., Kasahara A., Sasaki Y., Hori M., Hayashi N. 2003. Hepatitis C virus core protein differently regulates the JAK-STAT signaling pathway under interleukin-6 and interferon-gamma stimuli.J. Biol. Chem. 278:28562–28571 [DOI] [PubMed] [Google Scholar]