Abstract
In T cells, anergy can be induced after T cell receptor engagement in the absence of costimulation. Under these conditions, the expression of a specific set of anergy-associated genes is activated. Several lines of evidence suggest that nuclear factor of activated T cells (NFAT) proteins may regulate the expression of many of those genes; however, the nature of the complexes responsible for the induction of this new program of gene expression is unknown. Here, we show that transcriptional complexes formed by NFAT homodimers are directly responsible for the activation of at least two anergy-inducing genes, Grail and Caspase3. Our data shows that Grail expression is activated by direct binding of NFAT dimers to the Grail promoter at two different sites. Consequently, a mutant NFAT protein with impaired ability to dimerize is not able to induce an unresponsive state in T cells. Our results not only identify a new biological function for NFAT dimers but also reveal the different nature of NFAT-containing complexes that induce anergy versus those that are activated during a productive immune response. These data also establish a basis for the design of immunomodulatory strategies that specifically target each type of complex.
T cell anergy is a mechanism of peripheral immune tolerance responsible for the inactivation of self-reactive T cells. Anergy occurs as a consequence of a partial or suboptimal T cell activation (1, 2). In vitro, clonal anergy can be induced by the engagement of the TCR in the absence of costimulation (e.g., CD28) or by stimulation with a weak agonist antigen in the presence of costimulation (3, 4). The TCR in anergic T cells becomes uncoupled from downstream signaling pathways, thus preventing proliferation and cytokine expression in response to subsequent antigen encounter (5–7).
Engagement of the TCR in the absence of costimulation results in the preferential activation of calcium signaling with only partial activation of those pathways that require costimulation (e.g., Ras/MAPK, PKC, and IKK) (8, 9). One of the main targets of calcium signaling in T cells is the phosphatase calcineurin. Calcineurin is responsible for the dephosphorylation and activation of the NFAT transcription factors (10). Three of the four calcineurin-regulated members of the NFAT family are expressed in T cells: NFAT1 (NFATp, NFATc2), NFAT2 (NFATc, NFATc1), and NFAT4 (NFATc3) (11, 12). NFAT proteins contain three distinct domains: the NFAT homology region (NHR), the Rel-homology region (RHR), and the C-terminal domain. The NHR is conserved in all calcium-regulated NFAT proteins and contains a strong transactivation domain and a regulatory domain, containing several serine-rich regions and the docking sites for calcineurin and the NFAT kinases (11, 12). The RHR is highly conserved within all NFAT proteins and shares structural homology with the Rel domain of the NF-κB family of transcription factors. This is the region that binds DNA and it is also involved in mediating protein–protein interactions with other transcriptional partners (11, 12). The RHR of NFAT proteins contains two functionally distinct domains, the N-terminal (RHR-N) and the C-terminal (RHR-C). The RHR-N makes most of the base-specific DNA contacts and is also involved in interactions with other transcription factors (13–18). In resting T cells, NFAT proteins reside in the cytosol in a highly phosphorylated state. After antigen encounter, activated calcineurin dephosphorylates several serine residues located in the N-terminal regulatory domain of NFAT proteins leading to their translocation into the nucleus (19). In response to different stimuli and cell conditions, the integration of calcium signaling with other signaling pathways determines NFAT interactions with distinct transcriptional partners that result in the activation of specific cellular programs. In activated T cells, NFAT cooperates with AP-1 transcription factors to form cooperative ternary complexes at NFAT–AP-1 composite sites that induce the expression of activation-induced genes (20, 21). NFAT can also synergize with lineage-specific factors (e.g., T-bet and GATA3) and contribute to the programs of differentiation that generate Th type 1 or 2 cells (22, 23). Similarly, cooperation between NFAT and Foxp3 seems to be essential for regulatory T (T reg) cell function and development (24).
In response to tolerizing stimuli, T cells activate a program of gene expression that is responsible for the uncoupling of the TCR from downstream signaling and for the suppression of cytokine expression (25). Anergy-associated genes are different from those genes activated during a productive immune response, and therefore their expression must be regulated by different transcriptional complexes (9, 26, 27). These genes include, among others, several E3 ubiquitin ligases, such as Cblb, Itch, and Grail; transcription factors, such as Egr2, Egr3, and Ikaros; Caspase3; and diacylglycerol kinase α (Dgka) (9, 28–35). The expression of many of these genes seems to be controlled by NFAT, as their expression is drastically reduced in T cells that lack NFAT1 (9). The mechanisms that control the regulation and the nature of the NFAT-containing transcriptional complexes involved remain largely unknown.
Here, we show that transcriptional complexes formed by NFAT dimers regulate the induction of T cell anergy. We present evidence that these complexes regulate the expression of anergy-inducing genes, including Grail and Caspase3. We show that, under anergizing conditions, NFAT dimers bind to two κB-like sites in the proximal Grail promoter and directly induce its expression, making NFAT dimer formation necessary to induce T cell unresponsiveness in vitro and in vivo. Our data support a model in which NFAT dimers regulate the establishment of anergy in T cells, and suggest that it should be possible to independently disrupt the surfaces involved in NFAT–NFAT or NFAT–AP-1 interactions to modulate the activation or inhibition of T cell responses.
RESULTS
Mutations of critical residues in the RHR-C interfere with NFAT homodimerization
The RHR of NFAT1 had been previously shown to have the ability to form homodimers on specific DNA sites containing two NFAT core binding sites separated by 1 or 2 nt (16, 17). To test the cooperative nature of these complexes, we analyzed binding of the NFAT1 RHR to probes containing two minimal consensus NFAT sites (GGAA) separated by 1 or 6 nt. As expected, recombinant NFAT1 RHR homodimers formed on the probe with a 1-nt spacer. Incubation of this probe with increasing amounts of recombinant NFAT1 RHR yielded increasing amounts of dimers (Fig. 1 A). However, dimer formation was almost completely absent when a 6-nt spacer was inserted between both NFAT monomer sites (Fig. 1 B). These data suggested that the binding of two NFAT1 RHR molecules to κB-like sites might be cooperative, and therefore disruption of the protein–protein contacts that maintained both NFAT molecules together should prevent binding of dimers onto the DNA.
Figure 1.
Mutations of critical residues in the NFAT1 dimer interface disrupt dimer formation without affecting DNA binding affinity or cooperation with AP-1. (A) EMSAs were performed using a probe containing a κB-like site and increasing amounts of recombinant NFAT1 RHR. Two complexes are identified: NFAT1 monomers (N) and dimers (N–N). Graph shows the relative percentage of dimer complexes compared with the total RHR bound to DNA. FP, free probe. (B) EMSAs were performed using recombinant NFAT1 RHR and either a probe containing a consensus κB-like site (left) or a probe in which the two core NFAT binding sites in a κB-like site were separated by 6 nt. (C) Representation of the crystal structure of RHR-C domains in the NFAT1 dimer. Residues involved in the dimer interface are highlighted. (D) Binding of recombinant WT NFAT1 RHR or the dimerization mutants ΔCC′ and ΔCC′QQ-AA was assayed using a probe containing a κB-like dimer site incubated with increasing concentration of these proteins. (E) Increasing amounts of recombinant c-Fos and c-Jun proteins were incubated with constant amounts of recombinant WT NFAT1 RHR or the ΔCC′QQ-AA mutant to analyze their ability to form ternary complexes on a probe containing a NFAT–AP-1 composite site. N, RHR monomer; N-A, RHR–cFos–cJun complex. (F–H). Nuclear extracts prepared from HEK293 cells transfected with plasmids expressing GFP (-), WT NFAT1, or the ΔCC′QQ-EE mutant were analyzed by EMSA to determine the ability of these proteins to form dimers (F), using a probe containing a κB-like site; to bind DNA (G); and to form complexes with AP-1 (H), using a probe containing an NFAT–AP-1 composite site. N-A, NFAT1–cFos–cJun complex. *, unspecific bands.
Analysis of the crystal structure of the NFAT1 RHR dimers (16, 17) allowed us to identify a series of residues located in the dimer interface that, when mutated, should impair protein–protein interactions (Fig. 1 C). Deletion of the CC′ loop (aa616-623) in the RHR-C generated a mutant with reduced capacity to form dimers, although these could still be detected with high concentrations of the CC′ loop-deleted NFAT1 RHR (Fig. 1 D). Additional mutations on Q671 and Q673 produced a protein with a complete impairment of its ability to bind κB-like sites as a dimer (Fig. 1 D).
All mutations were performed in residues located in the RHR-C and, in theory, should not affect NFAT1's ability to form complexes with Fos and Jun proteins on composite sites because these interactions occur predominantly through the RHR-N (Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20082731/DC1). To test this hypothesis, recombinant RHR was incubated with increasing amounts of c-Fos and c-Jun, and a probe containing the ARRE2 composite site of the murine Il2 promoter (see Supplemental materials and methods). Binding reactions showed that the mutant lacking the CC′ loop and bearing Q to A substitutions on Q671 and Q673 (ΔCC′QQ–AA), had the same ability to form cooperative complexes with Fos and Jun as the WT RHR (Fig. 1 E).
Next, we tested whether a full-length NFAT1 protein bearing those mutations would also show dimer formation impairment. Nuclear extracts prepared from HEK293 cells transfected with plasmids encoding WT NFAT1 or a ΔCC′QQ-EE mutant were analyzed by EMSA using a probe containing a κB-like dimer site. Glutamines were substituted for glutamic acid in this mutant to introduce electrostatic repulsion between monomers. This mutant showed almost no capacity to form dimers, but retained its ability to bind DNA and to form complexes with AP-1 (Fig. 1, F–H). Direct targeting of the dimer interface can, thus, prevent dimer formation without affecting NFAT interactions with AP-1 proteins.
Disruption of NFAT homodimer formation results in lack of transcriptional activity from κB-like dimer sites
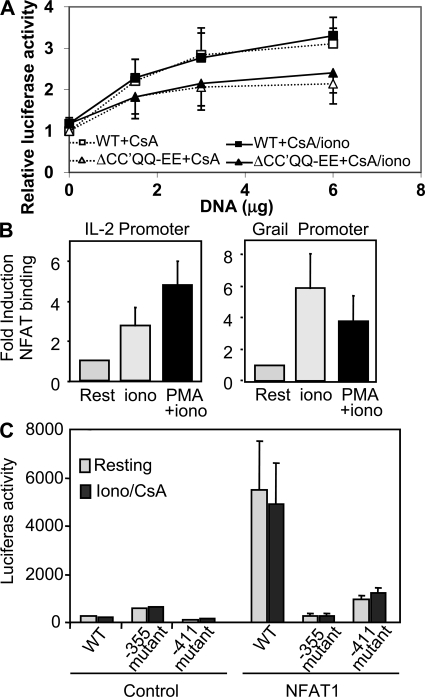
Next, we determined if mutations in the dimer interface would inhibit NFAT-induced transcription from a promoter containing κB-like elements. Jurkat cells were transfected with vectors encoding constitutively active forms of WT or the ΔCC′QQ-EE NFAT1 and a reporter vector containing two tandem κB-like sites. We have previously shown that these active NFAT proteins, which contain several serine-to-alanine substitutions in key residues located in the regulatory region of the NHR domain, are constitutively nuclear and insensitive to the inhibitory effect of cyclosporine A (CsA) (19). Therefore, to assess the effect of the plasmid-encoded NFAT proteins, transfected cells were treated with CsA to block endogenous NFAT activation. WT NFAT1 was transcriptionally active on κB-like sites. Its transcriptional activity was similar in cells treated with or without ionomycin, suggesting that NFAT1 did not require cooperation with other calcium-induced transcription factors to activate that promoter (Fig. 2 A). Cooperative dimer formation was clearly necessary to induce transcription from κB-like sites, as the ΔCC′QQ-EE mutant showed almost no transcriptional activity (Fig. 2 A). All NFAT1 proteins tested were, however, equally active when a reporter containing three tandem ARRE2 sites from the Il2 promoter was used (Fig. 2 B), confirming that disruption of the dimer interface did not affect NFAT interactions with Fos or Jun.
Figure 2.
The NFAT1 ΔCC′QQ-EE mutant cannot transactivate from dimer sites, but conserves full activity on NFAT–AP1 composite sites. Jurkat T cells were cotransfected with reporter plasmids in which the expression of the luciferase gene was driven by enhancers containing tandem κB-like sites (A), composite NFAT–AP-1 sites (B), or NFAT monomer binding sites (C), as well as an empty control vector or plasmids expressing an HA-tagged constitutively active NFAT1 (WT) or the ΔCC′QQ-EE dimerization mutant protein. Cells were then treated with CsA, CsA and ionomycin (A and C), or CsA+ionomycin+PMA (B) for 8 h, and luciferase activity was measured. Normalized values (to the activity of the stimulated cells expressing WT NFAT1) are presented from three to eight independent experiments. Error bars are the SEM. (inset) Immunoblot of NFAT1 WT and ΔCC′QQ-EE expression in transfected cells using an anti-HA antibody.
To exclude that NFAT proteins could bind as monomers and activate transcription, the activity of constitutively active NFAT1 proteins was measured using a reporter plasmid containing 6 tandem copies of the core “GGAA” NFAT binding site. Neither the WT nor the ΔCC′QQ-EE mutant proteins showed any activity on this vector (Fig. 2 C). A reporter containing 6 9-bp κB-like sites was used as control and, as expected, only the WT protein could activate its transcription (Fig. 2 C).
NFAT dimers activate the expression of anergy-inducing genes
Having shown that NFAT1 dimers were transcriptionally active, we determined whether they would control the transcription of anergy-associated genes in T cells. For that purpose, we transduced primary Th1 cells with retroviruses expressing constitutively active forms of WT or ΔCC′QQ-EE NFAT1 proteins and analyzed the expression of four of the genes that had previously been suggested to be NFAT dependent (9, 30). Confirming previous data, quantitative real-time PCR (qPCR) analysis revealed that the expression of those genes was up-regulated in Th1 cells in response to increases in intracellular calcium caused by ionomycin, whereas CsA blocked their expression (Fig. 3 A).
Figure 3.
Expression of the anergy-associated genes Grail and Caspase3 is regulated by NFAT1 dimers. (A) Primary mouse Th1 cells were transduced with retroviral RV-IRES-GFP empty vector (GFP) or retroviruses expressing the constitutively active WT NFAT1 (WT) or the ΔCC′QQ-EE mutant. Cells were sorted for GFP expression (infected cells) and left resting or treated with ionomycin or CsA+ionomycin for 4 h. RNA was prepared, and the expression of four anergy-associated genes was quantified by qPCR. Values are normalized to control untreated Th1 cells. The data represents the mean of seven independent experiments ± SEM. *, P < 0.05. (B) Primary Nfat1+/+ or Nfat1−/− CD4+ T cells were transfected with plasmids expressing WT NFAT1 or the ΔCC′QQ-EE mutant and an Il2 promoter–luciferase reporter vector. 24 h after transfection, cells were left resting or activated with PMA and ionomycin for 8 h. Luciferase activity (mean + SEM) from three independent experiments is shown. (C) Th1 cells from Nfat1+/+ or Nfat1−/− mice were stimulated with 1 µm ionomycin for 6 h. RNA was obtained, and expression of Grail and Caspase3 was measured by qPCR. Values (mean + SEM) show fold induction of the expression of those genes compared with the untreated cells from three independent experiments. (D) NIH-3T3 cells were transfected with plasmids expressing NFAT1 (N1), NFAT2 (N2), or NFAT4 (N4), and reporter plasmids in which the expression of luciferase was controlled by the Grail promoter (pGrail) or by enhancers containing three tandem copies of the ARRE2 site of the Il2 promoter. Luciferase values are the mean + SEM of three different experiments.
Transcription of those genes was clearly NFAT dependent, as it was induced in cells expressing an active WT NFAT1 at similar levels to those detected in ionomycin-treated cells (Fig. 3 A). As in previous experiments, these cells were always treated with CsA to block endogenous NFAT activation. Whereas the expression of Grg4 and Dgka was also up-regulated by the ΔCC′QQ-EE mutant, this protein failed to induce the expression of Grail or Caspase3 (Fig. 3 A). These results indicated that the transcription of some anergy-inducing genes, such as Grail or Caspase3, was directly or indirectly regulated by transcriptional complexes containing NFAT dimers. Transcription of other anergy-associated genes, such as Grg4 or Dgka, although still dependent on NFAT proteins, would be activated by other NFAT-containing transcriptional complexes. To confirm that the ΔCC′QQ-EE mutant was still active on promoters containing nondimer sites, primary Nfat1+/+ or Nfat1−/− Th1 cells were cotransfected with plasmids expressing WT or ΔCC′QQ-EE NFAT1 proteins and a reporter vector in which the expression of luciferase was regulated by the murine proximal Il2 promoter. In these assays, the ΔCC′QQ-EE mutant showed similar transactivational activity as the WT NFAT1 protein (Fig. 3 B).
Given that we had previously shown that Nfat1−/− T cells were less susceptible to anergizing stimuli and showed a markedly reduced ability to up-regulate the expression of most anergy-associated genes, we focused our study on the role that this NFAT protein had on the regulation of the expression of those genes. To determine if other NFAT proteins might also form dimers and be able to compensate for the lack of NFAT1, we analyzed the induction of the expression of Grail and Caspase3 in Nfat1+/+ and Nfta1−/− T cells. Our results showed that T cells that lacked NFAT1 were not able to up-regulate the expression of these dimer-dependent genes in response to calcium signaling (Fig. 3 C). To further study if, in any other circumstances, dimers containing other NFAT family members could activate the expression of those genes, we cotransfected NIH3T3 cells, which express very low levels of endogenous NFAT proteins, with plasmids expressing NFAT1, NFAT2, or NFAT4 (the three NFAT proteins that can be found in T cells) and a reporter vector in which the promoter of Grail directed the expression of luciferase. As expected, NFAT1 induced transcription from the Grail promoter (Fig. 3 D). Similar results were obtained for NFAT4, but NFAT2 did not show any transactivational activity on the Grail promoter, even though it was perfectly capable of inducing the expression of a reporter controlled by the Il2 promoter ARRE2 site, which contains composite NFAT–AP-1 sites (Fig. 3 D).
NFAT1 induces Grail expression by binding to κB-like sites located in the Grail promoter
To determine whether the expression of Grail and Caspase3 could be induced by the NFAT dimer-mediated activation of their promoters, we measured the capacity of WT NFAT1 and the ΔCC′QQ-EE dimerization mutant to activate two reporter vectors where the expression of the luciferase gene was controlled by those promoters. We did not detect any NFAT-mediated activation of the Caspase3 proximal promoter (unpublished data). However, expression of an active form of NFAT1 was able to activate the Grail promoter (Fig. 4 A). Levels of activation were almost identical in cells treated with CsA and in cells treated with CsA and ionomycin, which suggested that, under the conditions used in the assay, NFAT did not require additional calcium-activated proteins to transactivate the Grail promoter (Fig. 4 A). Supporting the results obtained in primary Th1 cells, the ΔCC′QQ-EE dimerization mutant NFAT1 protein had a markedly reduced ability to activate transcription from the Grail promoter (Fig. 4 A), confirming that Grail expression was dependent on NFAT dimers.
Figure 4.
NFAT1 dimers activate the transcription of Grail by binding two κb-like sites in the Grail promoter. (A) Jurkat T cells were cotransfected with a luciferase reporter vector containing the Grail promoter (89 to −880), and a control empty plasmid or increasing concentration of plasmids expressing a constitutively active WT NFAT1 or the ΔCC′QQ-EE mutant. 36 h after transfection, cells were treated with CsA or CsA+ionomycin for 8 h, and luciferase activity was measured. Values are normalized to luciferase levels of activated cells transfected with the control empty vector. Graph shows the mean of three independent experiments ± SEM. (B) Primary mouse Th1 cells were treated with ionomycin (iono) or PMA+ionomycin for 3 h. ChIP was performed using an anti-NFAT1 antibody. NFAT binding to DNA was quantified by qPCR analysis using primers for the proximal regions of the Il2 promoter (left) and the Grail promoter (right). Bars show the mean + SEM of values for fold induction (relative to resting cells) from three independent experiments. (C) Jurkat T cells were transiently cotransfected with a luciferase reporter vector containing the Grail promoter with or without mutations in the NFAT dimer sites located at positions −355 or −455, and a control empty plasmid or a plasmid expressing the constitutively active NFAT1. 36 h after transfection, cells were treated with CsA/ionomycin for 8 h and luciferase activity was measured. The graph shows the mean of three independent experiments + SEM.
To determine if NFAT-dimers would bind directly to the Grail promoter and regulate its expression, we followed two different approaches. First, we assessed the recruitment of NFAT1 to the Grail promoter by chromatin immunoprecipitation. We used an anti-NFAT1 antibody to pull down NFAT–DNA complexes in murine Th1 cells, and we quantified binding of NFAT1 by qPCR using specific primers for the proximal promoters of the Il2 and Grail genes (see Supplemental materials and methods). As expected, NFAT1 bound the Il2 promoter. Although some binding was already detected in cells treated with ionomycin, it was in fully activated T cells where NFAT1 binding to the Il2 promoter was clearly induced (Fig. 4 B). These results were consistent with the binding of NFAT–AP-1 complexes to composite sites on the Il2 promoter in activated T cells. NFAT1 was also able to bind to the Grail promoter; however, the pattern of binding was different from the one detected on the Il2 promoter. Consistent with the role of NFAT1 in the transcription of anergy-inducing genes, NFAT1 bound the Grail promoter preferentially in cells receiving an anergizing stimulus (ionomycin), whereas full activation of T cells with PMA and ionomycin reduced NFAT1 binding (Fig. 4 B).
To determine if two putative dimer sites we had identified in the Grail promoter (−355 gtaacgtttcc and −411 ggattcttcc) were binding sites for NFAT, and to simultaneously measure their contribution to the regulation of Grail expression, reporter vectors containing the Grail promoter with mutations that disrupted those sites were constructed (see Supplemental materials and methods). Analysis of Jurkat cells transiently transfected with these plasmids showed that disruption of any of those two sites resulted in an almost complete loss of NFAT-dependent transcriptional activity (Fig. 4 C).
The ΔCC′QQ-EE dimerization mutant has an impaired ability to induce unresponsiveness in T cells
Expression of an active form of NFAT1 is able to induce unresponsiveness in T cells (9). To determine the contribution of NFAT dimer complexes to the induction of the anergic phenotype, we transduced murine Th1 cells with retroviral vectors expressing a constitutively active WT NFAT1 or the ΔCC′QQ-EE mutant and analyzed the ability of these proteins to induce unresponsiveness. Western blot analysis demonstrated that equivalent amounts of both proteins were expressed in the sorted infected Th1 cells (Fig. 5 A). As previously reported (9), Th1 cells transduced with a constitutively active NFAT1 became resistant to stimulation, and activation-induced IL-2 expression decreased almost 12-fold when compared with control cells. However, the expression of the NFAT1 dimerization mutant, although still able to cause some inhibition of IL-2 expression, was much less efficient than the WT protein in blocking T cell responses to stimulation (3.5-fold compared to 11.8-fold; Fig. 5 A). Similar results were obtained when Nfat1−/− T cells were transduced with retrovirus expressing nonconstitutively active forms of NFAT1 or the ΔCC′QQ-EE dimerization mutant. As previously shown (9), Nfat1−/− cells were less susceptible to anergizing stimuli than Nfat1+/+ T cells (Fig. 5 B). Restoring the expression of NFAT1 by retroviral infection in these cells also restored their ability to become anergic in response to ionomycin; however, no effect could be detected when cells were transduced with a retrovirus that expressed the ΔCC′QQ-EE mutant protein (Fig. 5 B).
Figure 5.
Disruption of NFAT1 dimerization ability compromises its capacity to induce unresponsiveness in T cells. (A) Primary mouse Th1 cells were infected with retrovirus expressing an active form of WT NFAT1 (WT) or the ΔCC′QQ-EE mutant. Infected cells were sorted and stimulated with anti-CD3 and anti-CD28 for 8 h. IL-2 production was quantified by ELISA. The mean + SEM of seven experiments is presented (left). *, P < 0.05. (inset) Immunoblot showing protein expression using an anti-HA antibody. Fold reduction of IL-2 expression compared with control infected T cells (anergy index) is also shown (right). (B) Nfat1−/− Th1 cells were transduced with the retroviral RV-IRES-GFP vector or retrovirus expressing a WT NFAT1 or the ΔCC′QQ-EE mutant. GFP+ cells were sorted and left resting (open bars) or anergized with 1 µm ionomycin (filled bars). Cells were then washed, rested for 4 h, and stimulated with plate-bound anti-CD3 and anti-CD28 for 24 h. IL-2 expression was then measured by ELISA. Values are the mean + SEM of three independent experiments. Uninfected Nfat1+/+ Th1 cells were used as controls. *, P < 0.05. Fold reduction of IL-2 expression in anergic T cells (anergy index) compared with control untreated nonanergic cells is also shown (right). (C) CD4+ T cells from DO11.10 mice were transduced with retroviral vectors expressing GFP or GFP and a WT constitutively active NFAT1 or the ΔCC′QQ-EE dimerization mutant, sorted for GFP expression and transferred into BALB/c recipients. Host mice were then challenged subcutaneously with CFA or CFA plus OVA323-339 peptide. 7 d after challenge, cells were collected from draining lymph nodes and the presence of GFP+CD4+ T cells was determined by FACS. A representative experiment is shown. Graph shows the mean + SEM of the numbers of GFP+ cells from 4 independent experiments. *, P < 0.05 .
To confirm these results in vivo, adoptive transfer experiments were performed. CD4+ T cells isolated from DO11.10 mice were adoptively transferred to BALB/c recipients and challenged with CFA or OVA323-339 peptide in CFA. Upon analysis of the draining lymph nodes 7 d after challenge, proliferative responses could be observed only in mice challenged with the antigenic peptide (Fig. S2, available at http://www.jem.org/cgi/content/full/jem.20082731/DC1). When transduced T cells expressing GFP alone or GFP and constitutively active forms of WT NFAT1 or the ΔCC′QQ-EE mutant were transferred to BALB/c recipients, increased numbers of GFP+ T cells in response to OVA323-339 challenge could be readily detected in control GFP-expressing cells, but they were almost completely absent in mice receiving cells expressing an active WT NFAT1 protein. When cells that expressed the ΔCC′QQ-EE dimerization mutant NFAT1 protein were transferred, it was evident that this protein had lost most of its ability to induce unresponsiveness in T cells and clear proliferative responses induced by OVA323-339 challenge could be detected in those mice (Fig. 5 C).
DISCUSSION
Tolerizing stimuli induce the expression of a specific set of genes that are different from those expressed during a productive immune response (9, 27). In various models of immune tolerance, an elevation of intracellular Ca2+ can be consistently detected (8, 36, 37). In T cells, weakly agonistic antigens that induce T cell anergy also elicit sustained increases in the levels of cytoplasmic Ca2+ (8). Thus, tolerizing stimuli that cause suboptimal activation of T cells may only induce low levels of sustained increase in intracellular Ca2+ that have been shown to be sufficient to activate NFAT, but not AP-1 or NF-κB, proteins (38, 39). Several lines of evidence support the involvement of Ca2+ and NFAT in the expression of anergy-associated genes: cells treated with a Ca2+ ionophore become unresponsive and up-regulate the expression of anergy-associated genes, which can be blocked by the calcineurin inhibitor CsA; T cells from Nfat1−/− mice are hyperactive and more resistant to becoming anergic; and, finally, expression in Th1 cells of a constitutively active NFAT1 induces a state of unresponsiveness and up-regulates many anergy-associated genes (9).
In activated T cells, NFAT cooperates with other transcription factors on multiple cytokine promoters (21, 40). However, during anergy induction, many of the NFAT transcription partners, such as AP-1, are not fully activated. Thus, other NFAT-containing transcriptional complexes must regulate the expression of anergy-associated genes. Recently, two groups characterized the dimer structure of the NFAT1 RHR bound to the κB-like sites of the human IL-8 promoter and the HIV-1 LTR (16, 17). Our results show that NFAT dimers are able to induce transcription from κB-like sites and regulate the expression of at least two genes responsible for blocking TCR signaling in anergic T cells. Whereas NFAT1 can induce the expression of Grail and Caspase3, a mutant NFAT1 protein unable to form dimers fails to transactivate those two genes. Transient reporter assays indicate that the NFAT-dependent regulation of Caspase3 expression does not occur through its proximal promoter. It is possible, though, that distal control elements containing NFAT sites are responsible for the regulation of the expression of this gene, as it has been described for several cytokines (41–43). Grail, however, is regulated by direct binding of NFAT dimers to at least two κB-like sites located in its promoter. Grail is also posttranslationally regulated by two isoforms of the ubiquitin-specific protease Otubain 1 (44). After T cell activation, Otubain1 binds Grail and inhibits its deubiquitination, leading to Grail degradation. During anergy induction, a different spliced form, Otubain1-ARF, is expressed and binds and stabilizes Grail by recruitment of USP8 (44). It is still unknown how the expression of these isoforms of Otubain is regulated, and it would be interesting to determine if NFAT might also be involved.
Our data indicates that NFAT homodimers are cooperative. Increasing the distance between the NFAT binding hemi-sites in a κB-like site abrogates binding of NFAT dimers. Similarly, mutations of the NFAT1 dimer interface that inhibit protein–protein interactions result in a markedly reduced ability of NFAT1 to bind and transactivate from κB-like sites. Most of the interactions between NFAT and DNA or AP-1 occur through the RHR-N (Fig. S1) (15), which explains why mutations in the RHR-C do not affect the formation of NFAT–AP-1 complexes. NFAT-containing transcriptional complexes activated during a productive immune response are different from those formed during the induction of T cell tolerance, and involve different protein surfaces on NFAT. Molecules could thus be designed to specifically manipulate these cellular responses. For instance, targeting the critical residues needed for NFAT–AP-1 interactions should interfere with the formation of these complexes without affecting dimer formation, thus skewing T cell responses toward tolerance. Conversely, targeting the dimer interface should not affect AP-1 interactions, and should selectively disrupt dimer transcriptional complexes, tilting T cell responses toward activation.
The NFAT1 residues involved in the dimer interface are conserved in NFAT2 and NFAT4 (16, 17). Thus, structurally, it is likely that other NFAT proteins can also homodimerize or even heterodimerize. It has been well characterized that redundancy within the NFAT family exists. In general, knocking out one NFAT protein results only in a mild phenotype, whereas knocking out two or more members leads to striking defects (45–53). The phenotypes of these mice seem to indicate that although NFAT1 and NFAT2 proteins regulate the expression of activation-induced genes, it is NFAT1 and NFAT4 that might be specifically involved in controlling cellular programs that lead to the negative regulation of T cell responses. As suggested by the phenotypes of those mice, NFAT1 and NFAT4 had a much higher activity on the promoter of Grail than NFAT2. NFAT4 dimers are unlikely to compensate for NFAT1, as T cells that lack NFAT1 showed an almost complete lack of up-regulation of Grail and Caspase3 when anergized. Although we cannot rule out a possible role for NFAT1–NFAT4 heterodimers, recent reports have indicated that, because of differences in the residues involved in dimer interaction between NFAT1 and NFAT4, these complexes are not likely to occur (16).
We have also observed that the regulation of other anergy-inducing genes, such as Grg4 and Dgka, is NFAT dependent but dimer independent. How are these genes regulated? Is it possible that NFAT monomers may drive their expression? The crystal structure of the NFAT1 RHR bound to a single NFAT cognate DNA site shows the formation of four different DNA–NFAT binary complexes (18). In all four, the RHR-N recognizes the DNA and provides a relatively rigid domain to interact with other transcriptional factors. A flexible linker exists between the RHR-N and RHR-C that permits the RHR-C to acquire different conformational changes, providing a dynamic surface for protein–protein interactions. This flexibility should facilitate the formation of higher order NFAT transcriptional complexes. Our experimental data supports this conclusion and shows that NFAT1 is not able to induce any significant transcriptional activity in a reporter vector driven by NFAT monomer sites. Overall, the data suggests that NFAT1 must cooperate with other transcription factors to activate gene expression. It is also possible that NFAT may indirectly regulate the expression of some anergy-inducing genes. It has recently been shown that anergic T cells up-regulate transcription of Cblb by inducing the expression of the transcription factors Egr2 and Egr3 in a NFAT-dependent way (28).
Could NFAT dimers be involved in other cellular functions as well? T reg cells are also intrinsically anergic; they do not produce IL-2 and proliferate poorly upon stimulation in vitro. A hallmark of T reg cells is the expression of the Foxp3 transcription factor, which seems to be necessary for T reg cell development and suppressor activity (54–56). Recently, it has been reported that T reg cells express up to 10 times more Grail than naive CD4+ T cells. Grail may thus be necessary to maintain the anergic status of T reg cells and to acquire suppressor activity, as T cells overexpressing Grail gain suppressor capacity (57). The mechanism of action and the regulation of Grail in T reg cells are unknown. Recent evidence indicates that NFAT–Foxp3 complexes are crucial for the development of T reg cell suppressor function (24). However, we cannot rule out that NFAT dimers might also be involved in the regulation of the program of gene expression that leads to the establishment of a regulatory phenotype.
The identification of the transcriptional complexes that activate distinct programs of gene expression in T cells should allow us to understand the mechanisms that regulate specific cellular responses. At the same time, this information should provide valuable insight to design tools that may interfere with the formation or functioning of these complexes, and therefore may modulate T cell function.
MATERIALS AND METHODS
Mice.
WT or Nfat1−/− C57BL6/J, BALB/c, and DO11.10 mice were maintained in pathogen-free conditions. All animal work was approved by the Animal Institute Committee of the Albert Einstein College of Medicine.
Cell culture.
Primary CD4+ T cells were isolated from lymph nodes and spleen of 4–6-wk-old mice using anti-CD4–coupled magnetic beads, stimulated with 0.5 µg/ml plate-bound anti-CD3ϵ and anti-CD28, and differentiated for 7 d in the presence of 10 ng/ml IL-12, 10 µg/ml anti–IL-4, and 10 U/ml of recombinant hIL-2. Cells were cultured in DME supplemented with 10% fetal calf serum, 2 mM l-glutamine, nonessential amino acids, and essential vitamins, 550 nM l-arginine, 240 nM l-asparagine, 14 nM folic acid, and 50 µM β-mercaptoethanol. Jurkat, HEK293, and NIH-3T3 cells were cultured in DME supplemented with 10% fetal calf serum and 2 mM l-glutamine.
Transfections and reporter assays.
Jurkat cells were transfected by electroporation in serum-free medium, and murine T cells were transfected by electroporation using a Nucleofector electroporator (Amaxa) following the manufacturer's recommendations. 24 h after transfection, cells were stimulated with 1 µM ionomycin and/or 20 nM PMA in the presence or absence of 1 µM CsA. 8 h after stimulation, cells were lysed and assayed for luciferase activity using the Promega Dual-Luciferase Reporter Assay System. HEK293 and NIH-3T3 cells were transfected using PolyFect transfection reagent (QIAGEN) following the manufacturer's recommendations.
Retroviral infection of primary T cells.
The retroviral vectors RV-IRES-GFP and RV-CA-HA-NFAT1-IRES-GFP have been previously described (9). Phoenix Ecotropic cells (a gift from G. Nolan, Stanford University, Palo Alto, CA) were transfected with retroviral vectors. Supernatants were collected 48 h after transfection, supplemented with 8 µg/ml polybrene, and used to infect CD4+ T cells 24 h after stimulation. Infected cells were sorted for GFP expression. NFAT expression was assessed by immunoblot using an anti-HA antibody.
PCR site-directed mutagenesis.
The QuickChange Site-Directed Mutagenesis kit (Stratagene) was used to remove the CC′ loop and substitute Q671 and Q673 for A or E in the NFAT dimer interface. The two NFAT dimer sites at position −355 and −411 on the Grail promoter were mutated from GTAACGTTTCC to GTCTCGTAACC and from GGATTCTTCC to TAATTCAACC, respectively.
Electrophoretic mobility shift assays (EMSAs).
Recombinant proteins were expressed fused to hexahistidine tags and purified using Ni-NTA agarose. Nuclear extracts were prepared from transfected HEK293 cells, and equal amounts of recombinant or HEK293-expressed NFAT proteins were incubated with [32P]-labeled probes containing κB-like dimer sites with different 1-, 2-, or 6-bp spacers or a composite NFAT–AP-1 site in a buffer containing 10 mM Hepes, pH 7.0, 125 mM NaCl, 10% glycerol, 0.25 mM DTT, 100 ng/µl of poly dI-dC, 0.8 mg/µl of BSA for 30 min, and then resolved in 4% polyacrylamide gels.
Ionomycin-induced anergy.
Th1 cells were treated with 1 µM ionomycin for 16 h. In some experiments, CsA at 1 µM was also added 30 min before the ionomycin treatment. Cells were then washed and rested for 2–4 h in fresh medium before stimulation.
ELISA.
25–50 × 103 Th1 cells were stimulated with 0.5 µg/ml plate-bound anti-CD3 and anti-CD28 in 96-well plates. Supernatants were collected 8 h after stimulation, and IL-2 levels were measured in a sandwich ELISA.
qPCR.
Total RNA samples were prepared from sorted GFP+ retrovirally transduced mouse Th1 cells. cDNA was synthesized using Superscript II reverse transcription. Gene expression was analyzed using SYBR Green in a Smart Cycler II thermocycler. A threshold was set in the linear range of the amplification curve (fluorescence = f[cycle]), and the number of cycles needed to reach it was calculated for every sample. Expression of each gene was normalized to levels of actin, and the fold induction was calculated relative to the resting values.
Chromatin immunoprecipitation (ChIP) assays.
NFAT1 binding was assayed using a ChIP assay kit (Millipore). Nuclear lysates from 107 Th1 cells untreated or treated with 1 µM ionomycin or 1 µM ionomycin/20 nM PMA for 2 h were subjected to immunoprecipitation overnight at 4°C with an anti-NFAT1 antibody (ABR). Taqman probes were used to analyze the ChIP products by qPCR. Results were expressed as fold induction over values in resting cells and were normalized by input values.
Adoptive transfers.
5 × 105 retrovirally transduced, GFP+ DO11.10 T cells were transferred into BALB/c recipients by tail vein injection. 1 d after the transfer, mice received a subcutaneous injection of 200 µl of complete Freund's adjuvant in the presence or absence of 400 µg of OVA323-339 peptide. On day 7, draining lymph nodes were collected. Cell suspensions were prepared and labeled with a PE-coupled anti-CD4 antibody, and the number of CD4+GFP+ T cells was measured by FACS.
Statistical analysis.
Differences in the inhibition of IL-2 production and gene expression in anergic T cells were analyzed using a Student's t test.
Online supplemental material.
Fig. S1 shows an amino acid sequences comparison of the of the RHRs of NFAT1, 2, and 4, highlighting the residues involved in contacts with Fos and Jun, those involved in NFAT-NFAT dimer interactions, and the amino acids mutated in this study. Fig. S2 shows the proliferative responses of adoptively transferred DO11.10 TCR transgenic T cells in BALB/c mice in response to stimulation with OVA323-339 peptide. The Supplemental materials and methods describes the sequences of the DNA probes and primers used in this study. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20082731/DC1.
Acknowledgments
We thank members of our laboratory for helpful discussions. We also thank Drs. Lin Chen, at the University of Southern California, Stephen Harrison, at Harvard Medical School, and Patrick Hogan, at the Immune Disease Institute, for helpful suggestions and discussion. Human IL-2 and antibodies against murine IL-4 were obtained from the Biological Resources Branch preclinical repository.
This work was supported by National Institutes of Health (NIH) grants AI059738 (F. Macian), CA42471 (A. Rao), and AI48213 (A. Rao); NIH training grants GM007288 (N. Soto-Nieves) and GM007288 (B.T. Abe and I. Baine); and the Irene Diamond Foundation (F. Macian).
The authors have no conflicting financial interests.
Footnotes
Abbreviations used: ChIP, chromatin immunoprecipitation; CsA, cyclosporine A; Dgka, diacylglycerol kinase α; EMSA, electrophoretic mobility shift assay; NHR, NFAT homology region; qPCR, quantitative real-time PCR; RHR, Rel-homology region.
References
- Macian F., Im S.H., Garcia-Cozar F.J., Rao A. 2004. T-cell anergy.Curr. Opin. Immunol. 16:209–216 [DOI] [PubMed] [Google Scholar]
- Schwartz R.H. 2003. T cell anergy.Annu. Rev. Immunol. 21:305–334 [DOI] [PubMed] [Google Scholar]
- Jenkins M.K., Schwartz R.H. 1987. Antigen presentation by chemically modified splenocytes induces antigen-specific T cell unresponsiveness in vitro and in vivo.J. Exp. Med. 165:302–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quill H., Schwartz R.H. 1987. Stimulation of normal inducer T cell clones with antigen presented by purified Ia molecules in planar lipid membranes: specific induction of a long-lived state of proliferative nonresponsiveness.J. Immunol. 138:3704–3712 [PubMed] [Google Scholar]
- Fields P.E., Gajewski T.F., Fitch F.W. 1996. Blocked Ras activation in anergic CD4+ T cells.Science. 271:1276–1278 [DOI] [PubMed] [Google Scholar]
- Fathman C.G., Lineberry N.B. 2007. Molecular mechanisms of CD4(+) T-cell anergy.Nat. Rev. Immunol. 7:599–609 [DOI] [PubMed] [Google Scholar]
- Li W., Whaley C.D., Mondino A., Mueller D.L. 1996. Blocked signal transduction to the ERK and JNK protein kinases in anergic CD4+ T cells.Science. 271:1272–1276 [DOI] [PubMed] [Google Scholar]
- Sloan-Lancaster J., Steinberg T.H., Allen P.M. 1996. Selective activation of the calcium signaling pathway by altered peptide ligands.J. Exp. Med. 184:1525–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macian F., Garcia-Cozar F., Im S.H., Horton H.F., Byrne M.C., Rao A. 2002. Transcriptional mechanisms underlying lymphocyte tolerance.Cell. 109:719–731 [DOI] [PubMed] [Google Scholar]
- Aramburu J., Rao A., Klee C.B. 2000. Calcineurin: from structure to function.Curr. Top. Cell. Regul. 36:237–295 [DOI] [PubMed] [Google Scholar]
- Hogan P.G., Chen L., Nardone J., Rao A. 2003. Transcriptional regulation by calcium, calcineurin, and NFAT.Genes Dev. 17:2205–2232 [DOI] [PubMed] [Google Scholar]
- Macian F. 2005. NFAT proteins: key regulators of T-cell development and function.Nat. Rev. Immunol. 5:472–484 [DOI] [PubMed] [Google Scholar]
- Huxford T., Huang D.B., Malek S., Ghosh G. 1998. The crystal structure of the IkappaBalpha/NF-kappaB complex reveals mechanisms of NF-kappaB inactivation.Cell. 95:759–770 [DOI] [PubMed] [Google Scholar]
- Jacobs M.D., Harrison S.C. 1998. Structure of an IkappaBalpha/NF-kappaB complex.Cell. 95:749–758 [DOI] [PubMed] [Google Scholar]
- Chen L., Glover J.N., Hogan P.G., Rao A., Harrison S.C. 1998. Structure of the DNA-binding domains from NFAT, Fos and Jun bound specifically to DNA.Nature. 392:42–48 [DOI] [PubMed] [Google Scholar]
- Giffin M.J., Stroud J.C., Bates D.L., von Koenig K.D., Hardin J., Chen L. 2003. Structure of NFAT1 bound as a dimer to the HIV-1 LTR kappa B element.Nat. Struct. Biol. 10:800–806 [DOI] [PubMed] [Google Scholar]
- Jin L., Sliz P., Chen L., Macian F., Rao A., Hogan P.G., Harrison S.C. 2003. An asymmetric NFAT1 dimer on a pseudo-palindromic kappa B-like DNA site.Nat. Struct. Biol. 10:807–811 [DOI] [PubMed] [Google Scholar]
- Stroud J.C., Chen L. 2003. Structure of NFAT bound to DNA as a monomer.J. Mol. Biol. 334:1009–1022 [DOI] [PubMed] [Google Scholar]
- Okamura H., Aramburu J., Garcia-Rodriguez C., Viola J.P.B., Raghavan A., Tahiliani M., Zhang X., Qin J., Hogan P.G., Rao A. 2000. Concerted dephosphorylation of the transcription factor NFAT1 induces a conformational switch that regulates transcriptional activity.Mol. Cell. 6:539–550 [DOI] [PubMed] [Google Scholar]
- Jain J., McCaffrey P.G., Miner Z., Kerppola T.K., Lambert J.N., Verdine G.L., Curran T., Rao A. 1993. The T-cell transcription factor NFATp is a substrate for calcineurin and interacts with Fos and Jun.Nature. 365:352–355 [DOI] [PubMed] [Google Scholar]
- Macian F., Lopez-Rodriguez C., Rao A. 2001. Partners in transcription: NFAT and AP-1.Oncogene. 20:2476–2489 [DOI] [PubMed] [Google Scholar]
- Lee D.U., Avni O., Chen L., Rao A. 2004. A distal enhancer in the interferon-gamma (IFN-gamma) locus revealed by genome sequence comparison.J. Biol. Chem. 279:4802–4810 [DOI] [PubMed] [Google Scholar]
- Avni O., Lee D., Macian F., Szabo S.J., Glimcher L.H., Rao A. 2002. T(H) cell differentiation is accompanied by dynamic changes in histone acetylation of cytokine genes.Nat. Immunol. 3:643–651 [DOI] [PubMed] [Google Scholar]
- Wu Y., Borde M., Heissmeyer V., Feuerer M., Lapan A.D., Stroud J.C., Bates D.L., Guo L., Han A., Ziegler S.F., et al. 2006. FOXP3 controls regulatory T cell function through cooperation with NFAT.Cell. 126:375–387 [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay S., Soto-Nieves N., Macian F. 2007. Transcriptional regulation of T cell tolerance.Semin. Immunol. 19:180–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoechel B., Lohr J., Zhu S., Wong L., Hu D., Ausubel L., Abbas A.K. 2006. Functional and molecular comparison of anergic and regulatory T lymphocytes.J. Immunol. 176:6473–6483 [DOI] [PubMed] [Google Scholar]
- Lechner O., Lauber J., Franzke A., Sarukhan A., von Boehmer H., Buer J. 2001. Fingerprints of anergic T cells.Curr. Biol. 11:587–595 [DOI] [PubMed] [Google Scholar]
- Safford M., Collins S., Lutz M.A., Allen A., Huang C.T., Kowalski J., Blackford A., Horton M.R., Drake C., Schwartz R.H., Powell J.D. 2005. Egr-2 and Egr-3 are negative regulators of T cell activation.Nat. Immunol. 6:472–480 [DOI] [PubMed] [Google Scholar]
- Jeon M.S., Atfield A., Venuprasad K., Krawczyk C., Sarao R., Elly C., Yang C., Arya S., Bachmaier K., Su L., et al. 2004. Essential role of the E3 ubiquitin ligase Cbl-b in T cell anergy induction.Immunity. 21:167–177 [DOI] [PubMed] [Google Scholar]
- Anandasabapathy N., Ford G.S., Bloom D., Holness C., Paragas V., Seroogy C., Skrenta H., Hollenhorst M., Fathman C.G., Soares L. 2003. GRAIL: an E3 ubiquitin ligase that inhibits cytokine gene transcription is expressed in anergic CD4+ T cells.Immunity. 18:535–547 [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay S., Dure M., Paroder M., Soto-Nieves N., Puga I., Macian F. 2007. Interleukin 2 gene transcription is regulated by Ikaros-induced changes in histone acetylation in anergic T cells.Blood. 109:2878–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heissmeyer V., Macian F., Im S.H., Varma R., Feske S., Venuprasad K., Gu H., Liu Y.C., Dustin M.L., Rao A. 2004. Calcineurin imposes T cell unresponsiveness through targeted proteolysis of signaling proteins.Nat. Immunol. 5:255–265 [DOI] [PubMed] [Google Scholar]
- Olenchock B.A., Guo R., Carpenter J.H., Jordan M., Topham M.K., Koretzky G.A., Zhong X.P. 2006. Disruption of diacylglycerol metabolism impairs the induction of T cell anergy.Nat. Immunol. 7:1174–1181 [DOI] [PubMed] [Google Scholar]
- Zha Y., Marks R., Ho A.W., Peterson A.C., Janardhan S., Brown I., Praveen K., Stang S., Stone J.C., Gajewski T.F. 2006. T cell anergy is reversed by active Ras and is regulated by diacylglycerol kinase-alpha.Nat. Immunol. 7:1166–1173 [DOI] [PubMed] [Google Scholar]
- Puga I., Rao A., Macian F. 2008. Targeted cleavage of signaling proteins by caspase 3 inhibits T cell receptor signaling in anergic T cells.Immunity. 29:193–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauld S.B., Benschop R.J., Merrell K.T., Cambier J.C. 2005. Maintenance of B cell anergy requires constant antigen receptor occupancy and signaling.Nat. Immunol. 6:1160–1167 [DOI] [PubMed] [Google Scholar]
- Healy J.I., Dolmetsch R.E., Timmerman L.A., Cyster J.G., Thomas M.L., Crabtree G.R., Lewis R.S., Goodnow C.C. 1997. Different nuclear signals are activated by the B cell receptor during positive versus negative signaling.Immunity. 6:419–428 [DOI] [PubMed] [Google Scholar]
- Dolmetsch R.E., Lewis R.S., Goodnow C.C., Healy J.I. 1997. Differential activation of transcription factors induced by Ca2+ response amplitude and duration.Nature. 386:855–858 [DOI] [PubMed] [Google Scholar]
- Dolmetsch R.E., Xu K., Lewis R.S. 1998. Calcium oscillations increase the efficiency and specificity of gene expression.Nature. 392:933–936 [DOI] [PubMed] [Google Scholar]
- Jain J., McCaffrey P.G., Valge-Archer V.E., Rao A. 1992. Nuclear factor of activated T cells contains Fos and Jun.Nature. 356:801–804 [DOI] [PubMed] [Google Scholar]
- Agarwal S., Avni O., Rao A. 2000. Cell-type-restricted binding of the transcription factor NFAT to a distal IL-4 enhancer in vivo.Immunity. 12:643–652 [DOI] [PubMed] [Google Scholar]
- Johnson B.V., Bert A.G., Ryan G.R., Condina A., Cockerill P.N. 2004. Granulocyte-macrophage colony-stimulating factor enhancer activation requires cooperation between NFAT and AP-1 elements and is associated with extensive nucleosome reorganization.Mol. Cell. Biol. 24:7914–7930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawwari A., Burrows J., Vadas M.A., Cockerill P.N. 2002. The human IL-3 locus is regulated cooperatively by two NFAT-dependent enhancers that have distinct tissue-specific activities.J. Immunol. 169:1876–1886 [DOI] [PubMed] [Google Scholar]
- Soares L., Seroogy C., Skrenta H., Anandasabapathy N., Lovelace P., Chung C.D., Engleman E., Fathman C.G. 2004. Two isoforms of otubain 1 regulate T cell anergy via GRAIL.Nat. Immunol. 5:45–54 [DOI] [PubMed] [Google Scholar]
- Kiani A., Viola J.P., Lichtman A.H., Rao A. 1997. Down-regulation of IL-4 gene transcription and control of Th2 cell differentiation by a mechanism involving NFAT1.Immunity. 7:849–860 [DOI] [PubMed] [Google Scholar]
- Hodge M.R., Ranger A.M., Charles de la Brousse F., Hoey T., Grusby M.J., Glimcher L.H. 1996. Hyperproliferation and dysregulation of IL-4 expression in NF-ATp-deficient mice.Immunity. 4:397–405 [DOI] [PubMed] [Google Scholar]
- Xanthoudakis S., Viola J.P., Shaw K.T., Luo C., Wallace J.D., Bozza P.T., Luk D.C., Curran T., Rao A. 1996. An enhanced immune response in mice lacking the transcription factor NFAT1.Science. 272:892–895 [DOI] [PubMed] [Google Scholar]
- Yoshida H., Nishina H., Takimoto H., Marengere L.E., Wakeham A.C., Bouchard D., Kong Y.Y., Ohteki T., Shahinian A., Bachmann M., et al. 1998. The transcription factor NF-ATc1 regulates lymphocyte proliferation and Th2 cytokine production.Immunity. 8:115–124 [DOI] [PubMed] [Google Scholar]
- Ranger A.M., Hodge M.R., Gravallese E.M., Oukka M., Davidson L., Alt F.W., de la Brousse F.C., Hoey T., Grusby M., Glimcher L.H. 1998. Delayed lymphoid repopulation with defects in IL-4-driven responses produced by inactivation of NF-ATc.Immunity. 8:125–134 [DOI] [PubMed] [Google Scholar]
- Ranger A.M., Oukka M., Rengarajan J., Glimcher L.H. 1998. Inhibitory function of two NFAT family members in lymphoid homeostasis and Th2 development.Immunity. 9:627–635 [DOI] [PubMed] [Google Scholar]
- Peng S.L., Gerth A.J., Ranger A.M., Glimcher L.H. 2001. NFATc1 and NFATc2 together control both T and B cell activation and differentiation.Immunity. 14:13–20 [DOI] [PubMed] [Google Scholar]
- Starr T.K., Jameson S.C., Hogquist K.A. 2003. Positive and negative selection of T cells.Annu. Rev. Immunol. 21:139–176 [DOI] [PubMed] [Google Scholar]
- Rengarajan J., Tang B., Glimcher L.H. 2002. NFATc2 and NFATc3 regulate T(H)2 differentiation and modulate TCR-responsiveness of naive T(H)cells.Nat. Immunol. 3:48–54 [DOI] [PubMed] [Google Scholar]
- Fontenot J.D., Gavin M.A., Rudensky A.Y. 2003. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells.Nat. Immunol. 4:330–336 [DOI] [PubMed] [Google Scholar]
- Zheng Y., Rudensky A.Y. 2007. Foxp3 in control of the regulatory T cell lineage.Nat. Immunol. 8:457–462 [DOI] [PubMed] [Google Scholar]
- Sakaguchi S. 2004. Naturally arising CD4+ regulatory t cells for immunologic self-tolerance and negative control of immune responses.Annu. Rev. Immunol. 22:531–562 [DOI] [PubMed] [Google Scholar]
- MacKenzie D.A., Schartner J., Lin J., Timmel A., Jennens-Clough M., Fathman C.G., Seroogy C.M. 2007. GRAIL is up-regulated in CD4+ CD25+ T regulatory cells and is sufficient for conversion of T cells to a regulatory phenotype.J. Biol. Chem. 282:9696–9702 [DOI] [PubMed] [Google Scholar]