Abstract
Plasmodium and Toxoplasma are parasites of major medical importance that belong to the Apicomplexa phylum of protozoa. These parasites transform into various stages during their life cycle and express a specific set of proteins at each stage. Although little is yet known of how gene expression is controlled in Apicomplexa, histone modifications, particularly acetylation, are emerging as key regulators of parasite differentiation and stage conversion. We investigated the anti-Apicomplexa effect of FR235222, a histone deacetylase inhibitor (HDACi). We show that FR235222 is active against a variety of Apicomplexa genera, including Plasmodium and Toxoplasma, and is more potent than other HDACi's such as trichostatin A and the clinically relevant compound pyrimethamine. We identify T. gondii HDAC3 (TgHDAC3) as the target of FR235222 in Toxoplasma tachyzoites and demonstrate the crucial role of the conserved and Apicomplexa HDAC-specific residue TgHDAC3 T99 in the inhibitory activity of the drug. We also show that FR235222 induces differentiation of the tachyzoite (replicative) into the bradyzoite (nonreplicative) stage. Additionally, via its anti-TgHDAC3 activity, FR235222 influences the expression of ∼370 genes, a third of which are stage-specifically expressed. These results identify FR235222 as a potent HDACi of Apicomplexa, and establish HDAC3 as a central regulator of gene expression and stage conversion in Toxoplasma and, likely, other Apicomplexa.
Apicomplexa are unicellular eukaryotes that multiply intracellularly in their hosts. They include parasites of major medical importance like Plasmodium species, the causative agent of malaria, and Toxoplasma gondii, an opportunistic parasite of immunosuppressed individuals and a cause of congenital disease. In the mammalian host, Plasmodium differentiate and multiply inside host erythrocytes, whereas in the intermediate host, T. gondii alternates between two developmental forms: the tachyzoite, the proliferative form that rapidly divides and disseminates in the host, and the bradyzoite, the cystic form responsible for persistence in host tissues (1-3).
Stage conversion in Apicomplexa is associated with global modifications of mRNA contents, suggesting that developmental switches are transcriptionally regulated (4-6). The mechanisms by which Apicomplexa regulate expression of their genes are still poorly understood. They lack many of the typical eukaryotic transcription factors, with one exception being the plant-like AP2 DNA binding family, the major lineage-specific expansion of transcriptional regulators in the phylum (7). In contrast, these parasites possess a rich repertoire of enzymes involved in histone modification and chromatin remodeling (8). This suggests that Apicomplexa may be unusually reliant on epigenetic mechanisms to control developmental gene expression and cellular identity (2, 8).
In yeast and metazoa, acetylases and histone deacetylases (HDACs) play a major role in controlling gene expression by switching between the acetylated and deacetylated states of chromatin (9). In T. gondii, histone acetylation affects gene expression (10, 11) and correlates with tachyzoite to bradyzoite differentiation (11), and HDAC inhibitors (HDACi's) modify the abundance of developmentally regulated gene transcripts (12). Thus, acetylases and HDACs likely play an important role in the control of stage-specific gene expression during parasite differentiation.
The effect of HDACi's on Apicomplexa has been previously documented with the discovery of apicidin, a cyclic tetrapeptide having broad-spectrum antiparasitic activity (13). Despite this promising discovery, the mechanism of action of apicidin against Apicomplexa has not been documented so far. To date, various HDACi's have been isolated, and each chemical compound displays different properties in terms of the class of HDAC inhibited and downstream cellular effect on human cell lines (14, 15).
In this work, we examined the effect and investigated the mode of action of FR235222, a novel cyclic tetrapeptide HDACi isolated from the fermentation broth of Acremonium species (16). We first show that the drug is active against a wide range of Apicomplexa, blocks the growth and differentiation of P. falciparum and berghei parasites in red blood cells, and induces T. gondii tachyzoite to bradyzoite differentiation. Using a genetic approach, we identify HDAC3 as the target of the drug in T. gondii, and show that the drug inhibitory activity depends on a two-residue insertion within the catalytic site of the enzyme (A98T99 in T. gondii HDAC3 [TgHDAC3]), which is present exclusively in the HDAC3 family of proteins in Apicomplexa and is absent from any other HDAC identified so far in other organisms. Finally, using chromatin immunoprecipitation (ChIP) combined with DNA microarray (ChIP-on-chip) assays, we identify 369 Toxoplasma gene upstream regions containing hyperacetylated nucleosomes upon FR235222 treatment, one third of which are mainly expressed in the sporozoite and/or bradyzoite stage of parasite. Collectively, our results confirm that histone acetylation plays a significative role in the control of parasite differentiation and that TgHDAC3 is a regulator acting within the regulatory pathway leading to parasite differentiation.
RESULTS
FR235222 is an efficient inhibitor of the intracellular multiplication of Apicomplexa parasites
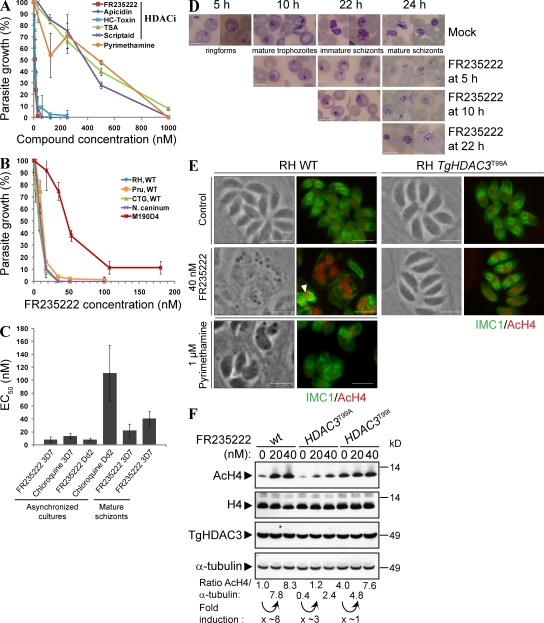
We first assessed the effect of FR235222 on the growth of T. gondii in human foreskin fibroblasts (HFFs). FR235222 and the other cyclopeptide HDACi's, such as HC-toxin and apicidin, inhibited T. gondii intracellular growth at low nanomolar concentrations (EC50 ≈ 10 nM; Fig. 1 A and Table I). In contrast, hydroxamic acid HDACi compounds (trichostatin A [TSA] and Scriptaid), which affect human cell proliferation by inducing cell-cycle arrest and/or stimulating apoptosis of certain cancer cells, were less efficient in inhibiting T. gondii proliferation (EC50 = 400 nM; Fig. 1 A). Note that cyclopeptide HDACi's more efficiently inhibit T. gondii growth than pyrimethamine (pyrimethamine EC50 = 300 nM; Fig. 1 A and Table I), a compound currently used clinically. FR235222 displayed comparable effects on T. gondii types I (RH), II (Prugniaud), and III (CTG), as well as on Neospora caninum, a closely related Apicomplexan parasite (Fig. 1 B). Low nanmolar concentrations of FR235222 also inhibited Plasmodium intraerythrocytic cycle in vitro. It is noteworthy that FR235222 EC50s are equivalent on P. falciparum 3D7 and Dd2 clones, which are sensitive and resistant, respectively, to the chloroquine (Fig. 1 C).
Figure 1.
In vitro antiprotozoal activity of FR235222 and other HDACi's. In vitro inhibitory concentrations for FR235222 and other compounds against Apicomplexan parasite growth were determined by measuring the incorporation of [3H]uracil by intracellular T. gondii tachyzoites and of [3H]thymidine by Plasmodium species, as described in Materials and methods. (A) Effect of FR235222 and other HDACi's on T. gondii RH strain growth in HFF monolayer. Pyrimethamine (non-HDACi compound) was used as a clinically relevant control. Means ± SD of parasite growth (percentages) are shown (n = 3 experiments). (B) Effect of FR235222 on N. caninum; T. gondii types I (RH), II (Pru), and III (CTG) wild-type strains; and the type I FR235222-resistant mutant M190D4. The data are plotted as the percentage of the control in the absence of drug. Means ± SD of parasite growth (percentages) are shown (n = 3 experiments). (C) Effects of FR235222 on P. falciparum clones 3D7 and Dd2. Effectiveness of FR235222 was compared with Chloroquine. EC50 values are plotted. Means ± SD of EC50 (nanomolar) are shown (n = 4 experiments for each set of data). (D) Effect of FR235222 on P. berghei blood stage development. Intraerythrocyte P. berghei GFP@hsp70 ANKA parasites were synchronized as described in Materials and methods. Synchronized in vitro cultures were treated with 54 nM FR235222 after 5, 11, and 22 h of intracellular growth. The development of the parasites was assessed at the indicated time points by Giemsa staining. Bars, 8 µm. (E) Effects of FR235222 on histone H4 acetylation in intracellular T. gondii parasites. Confluent monolayers of HFF cells were infected with T. gondii RH WT and R20D9 (TgHDAC3T99A) strains in the presence of 40 nM FR235222 and 0.1% DMSO as a control. As a control, parasites were treated with 1 µM pyrimethamine. After 24 h of growth, cells were fixed and stained for AcH4 (red) and IMC1 (green). The arrowhead indicates aberrant progeny. A representative set of data is shown. Bars, 5 µm. (F) Extracellular T. gondii parasites (RH WT, R20D9 [TgHDAC3T99A], and M3135E11 [TgHDAC3T99I]) were treated with the indicated concentrations of FR235222 for 4 h and lysed. Total cell lysates were analyzed by immunoblot with anti-AcH4, anti-H4, anti-TgHDAC3, and anti–α-tubulin antibodies as indicated.
Table I.
Summary of EC50 of FR235222 on Apicomplexa and HFF primary host cells
| Cells | FR235222 EC50 (nM ± SD) | Apicidin EC50 (nM ± SD) | Pyrimethamine EC50 (nM ± SD) |
| T. gondii RH WT | 7.6 ± 0.6 | 15 ± 2 | 285 ± 61 |
| T. gondii RH M190D4 | 62 ± 5 | 67 ± 16 | 300 ± 11 |
| T. gondii Prugniaud | 10 ± 1 | – | 195 ± 1 |
| P. falciparum | 8 ± 4 | – | – |
| N. caninum | 8 ± 2 | – | – |
| HFFs | 107 ± 18 | 165 ± 33 | – |
Apicidin (HDACi) and pyrimethamine (non-HDACi) were used as controls.
FR235222 treatment also blocked the development of synchronized P. berghei cultures from the ring to the trophozoite stages, and from the trophozoite to the schizont stages, although schizonts were somewhat less sensitive to drug treatment (Fig. 1 D and not depicted).
Histone H4 hyperacetylation in FR235222-treated T. gondii
T. gondii multiplies intracellularly by endodyogeny. Antibodies directed against the inner membrane complex protein 1 (IMC1) allow monitoring of the formation of the internal daughter cells inside the parental dividing parasites (Fig. 1 E). Unlike parasites grown in the absence of drug, intracellular parasites treated with FR235222 were vacuolated and lacked IMC1-delineated daughter cells, or contained aberrant progeny (Fig.1 E, arrowhead). Also, the majority of the drug-treated parasites (∼80%) displayed massive DNA overreplication (>1N DNA content per cell), indicating that FR235222 interferes directly or indirectly with T. gondii cell-cycle progression.
To confirm the anti-HDAC activity of FR235222 (16-18) on T. gondii, the nuclei of FR235222-treated parasites were stained using antibodies directed against acetylated histone H4 (AcH4; Fig. 1 E and Fig. S1). As expected, AcH4 signals were higher in the nuclei of treated parasites than with the DMSO control. The increase in AcH4 signals as well as the DNA overreplication were not observed in parasites treated with pyrimethamine (non-HDACi compound; inhibitor of the dihydrofolate reductase) nor in the hxgprt− RH parasites treated with mycophenolic acid and xanthine (non-HDACi compound; inhibitor of guanine nucleotide synthesis pathway; Fig. S1), indicating that histone hyperacetylation and the abnormal cell cycle were specifically caused by FR235222 and were not the consequence of dying parasites.
To ascertain that the drug acted directly on the parasites, and not via host cells, extracellular tachyzoites were incubated in the presence of the drug and histone H4 acetylation levels were analyzed by immunoblotting (Fig. 1 F). The AcH4 signals increased approximately eightfold in the FR235222-treated parasites, confirming that the drug induces histone hyperacetylation in the absence of host cells. In agreement with a direct action of FR235222 on the parasites, pretreatment of HFF cells with 180 nM FR235222 for 12 h had no detectable effect on T. gondii growth after removal of the drug (unpublished data). In addition, immunoblot analysis showed that acetylation of histones H2B (K12 and K15) and H3K9 was not affected upon FR235222 treatment (Fig. S2), indicating that FR235222 specifically increases acetylation of the histone subunit H4.
FR235222 targets HDAC3 in T. gondii
The T. gondii genome (available from ToxoDB 5.0 at http://toxodb.org/toxo/home.jsp) contains five putative nicotinamide adenine dinucleotide–independent HDAC-encoding genes (11). Given that T. gondii growth inhibition by FR235222 correlates with increased levels of AcH4 (Fig. 1 F), the targeted enzymes are likely to function in the parasite nucleus. TgHDAC3, which acts on histone proteins (11) and localizes to the parasite nucleus (Fig. S3 A), appeared as a possible target of FR235222.
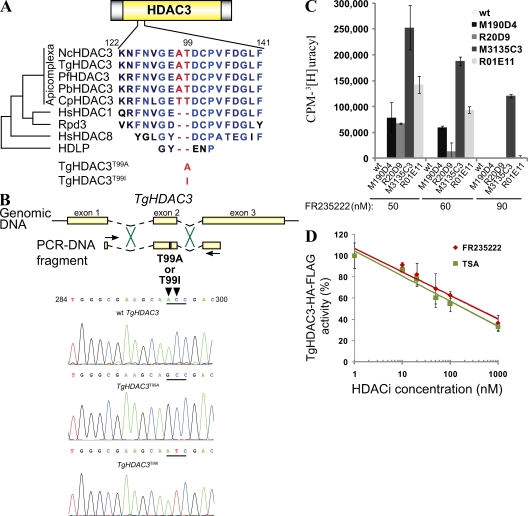
To further investigate the molecular basis of FR235222-induced growth inhibition of T. gondii, we generated FR235222-resistant parasite lines by chemical mutagenesis (see Materials and methods). Extracellular tachyzoites were mutagenized using N-nitroso-N-ethylurea (NEU), and resistant parasites were selected in the presence of 90 or 135 nM FR235222 (Table S1). Three resistant clones were isolated that grew normally in the presence of the drug. To determine whether these clones had mutated TgHDAC3, TgHDAC3 messenger RNA was amplified by RT-PCR and sequenced. Two clones, M190D4 and M3135C3, possessed T99A- and T99I-encoding mutations in exon 2 of TgHDAC3, respectively (Table S1). Interestingly, the T99 residue is part of a two-residue (A98T99) extension specific to HDAC3 of Apicomplexan parasites (Fig. 2 A and Fig. S4). The third clone, M3135D8, grew normally in the presence of the drug but not in its absence, and contains a WT TgHDAC3 gene. The molecular basis of the FR235222 resistance and dependence of this clone remains unknown.
Figure 2.
FR235222 induces acetylation of histones in T. gondii. (A) Sequence alignment of HDAC3 homologues in Apicomplexan parasites and other organisms. The region (from amino acids 122–141 of TgHDAC3) surrounding the point mutation identified in TgHDAC3-resistant mutants is shown. Rpd3 and HDLP are the HDAC homologues in Saccharomyces cerevisiae and the hyperthermophilic bacterium Aquifex aeolicus, respectively. Point mutations identified in the T. gondii FR235222-resistant mutants are shown at the bottom. The multiple sequence alignment was generated by the Clustal alignment method using the BLOSUM scoring matrix (CLC Free Workbench, available at http://www.clcbio.com). Regions identical to the three proteins (light blue) and amino acids conserved exclusively in Apicomplexan parasites (red) are shown. Cp, Cryptosporidium parvum; Hs, Homo sapiens; Nc, N. caninum; Pb, P. berghei; Pf, P. falciparum; Tg, T. gondii. (B) Schematic representation of the exon (yellow boxes)–intron (dashed lines) structure of TgHDAC3. DNA fragments encompassing the point mutations T99A and T99I were PCR amplified using the primers indicated (arrows), and genomic DNA from the resistant mutants M190D4 and M3135C3 as DNA template, respectively. T. gondii RH strain was transformed with the resulting PCR-DNA fragments, and FR235222 was used to select transformants harboring the allelic replacement of TgHDAC3 on the T. gondii genome. The mutated residues T99A and T99I (red) are shown. The corresponding chromatograms are shown at the bottom. (C) Comparison of T. gondii WT and FR235222-resistant mutant line growth in the presence of different concentrations of FR235222. T. gondii RH WT strain resistant lines from the mutagenesis screen carrying the HDAC3T99A (M190D4) and the HDAC3T99I (M3135C3) mutations, and the reconstructed mutants by allelic replacement for HDAC3T99A (R20D9) and HDAC3T99I (R01E11) were grown in HFF monolayer until numerous parasitophorous vacuoles were observed with the resistant parasites (∼6 d). Means ± SD of cpm are shown (n = 2 experiments). (D) FR235222 is a direct inhibitor of TgHDAC3. Partially purified TgHDAC3-HA-FLAG (tag in the C-terminal position) from T. gondii parasites was assayed for HDAC activity in vitro as described in Materials and methods. Equivalent amounts of immunoprecipitated protein were incubated with [3H]-labeled acetylated histones to determine enzymatic activity. Enzymatic activity is shown as a percentage of HDAC activity in the absence of inhibitor (set at 100%). Means ± SD are shown (n = 2 experiments).
To test whether the TgHDAC3 T99 mutations could account for T. gondii resistance to FR235222, mutations were introduced into the parental T. gondii RH strain. WT parasites were transfected with TgHDAC3 linear fragments encompassing an exon 2 that was either WT or contained the T99A- or T99I-encoding mutations (Fig. 2 B). Resistant parasites were selected in the presence of 90 nM FR235222 and emerged after transfection of the mutated but not WT TgHDAC3 fragments (unpublished data). Clones R20D9 and R01E11 were selected after parasite transfection with the fragments containing the T99A- or T99I-encoding mutations, respectively, and the sequences of their TgHDAC3 gene were determined. The sequences from the R20D9 and R01E11 clones contained the T99A- or T99I-encoding mutations, respectively, confirming that the transfected fragments had replaced the endogenous TgHDAC3 in the clones (Fig. 2 B). Note that FR235222 treatment had no effect on DNA replication and IMC1-delineated daughter cells in the TgHDAC3T99A and TgHDAC3T99I reconstructed mutants when compared with the WT parasites (Fig. 1 E and not depicted). We conclude that the T99A and T99I mutations in TgHDAC3 are each sufficient to confer resistance to FR235222. Moreover, this provides additional support for the conclusion that the growth phenotype is not caused by the effect of FR235222 on the host cell.
Next, we compared resistance to FR235222 of the NEU-mutagenized M190D4 and M3135C3 clones and of the recombinant R20D9 and R01E11 clones (Fig. 2 C). The M190D4 and the R20D9 clones that both harbor the T99A mutation displayed similar levels of resistance to FR235222 (50 nM), although M190D4 was slightly more resistant than R20D9 in the presence of 60 nM FR235222. Of the clones carrying the T99I mutation, M3135C3 was significantly more resistant than R01E11, and both were slightly more resistant than the clones carrying the T99A mutation (Fig. 2 C). These data suggested that the NEU-mutagenized clones (M190D4 and M3135C3) might contain additional FR235222 resistance mutations besides the TgHDAC3T99A and TgHDAC3T99I reconstructed mutants, and that the latter might confer higher levels of resistance to FR235222 than TgHDAC3T99A. Thus, it is possible that FR235222 has a minor secondary mode of action or that alternative mechanisms of resistance exist. Of note, all mutated parasites grew less well than the WT in the absence of drug (Table S1), which could mean that there is a fitness cost for the parasite escaping the drug.
FR235222 inhibits HDAC3 activity in T. gondii
We next examined histone H4 acetylation in the resistant lines before and after exposure to FR235222 by immunoblot analysis (Fig. 1 F). In the absence of drug, basal AcH4 signals were somewhat lower in the R20D9 TgHDAC3T99A line, but higher in the R01E11 TgHDAC3T99I line, than in WT parasites. Immunoblotting also showed that similar levels of TgHDAC3 were produced by the WT and resistant lines, indicating that the mutations did not affect HDAC3 expression or stability (Fig. 1 F). Therefore, the T99A and T99I mutations in TgHDAC3 likely affect the enzymatic activity on the histone H4 substrate. Additionally, under 20-nM FR235222 treatment, the levels of AcH4 signals were increased approximately eightfold in the WT but were only increased approximately threefold and remained unchanged in the TgHDAC3T99A and TgHDAC3T99I resistant lines, respectively. These data are consistent with the hypothesis that the TgHDAC3 mutations confer resistance to the FR2352222-induced histone H4 hyperacetylation. Because these data indicate that AcH4s are native substrates for TgHDAC3, we used hyperacetylated chromatin and partially purified TgHDAC3-HA-FLAG enzyme to test its sensitivity to FR235222 in an in vitro HDAC assay. Fig. 2 D shows that FR235222 directly inhibits the enzymatic activity of TgHDAC3. Thus, these data are consistent with TgHDAC3 inhibition responsible for growth defects. Note that the pan-HDACi TSA and FR235222 have similar inhibition activities on purified TgHDAC3, which contrasts with their respective in vivo activities (Fig. 1 A). A simple explanation could be that FR235222 has a higher ability to cross the host cell and parasitic membranes than TSA.
FR235222 affects histone H4 acetylation at bradyzoite- or stage-specific genes
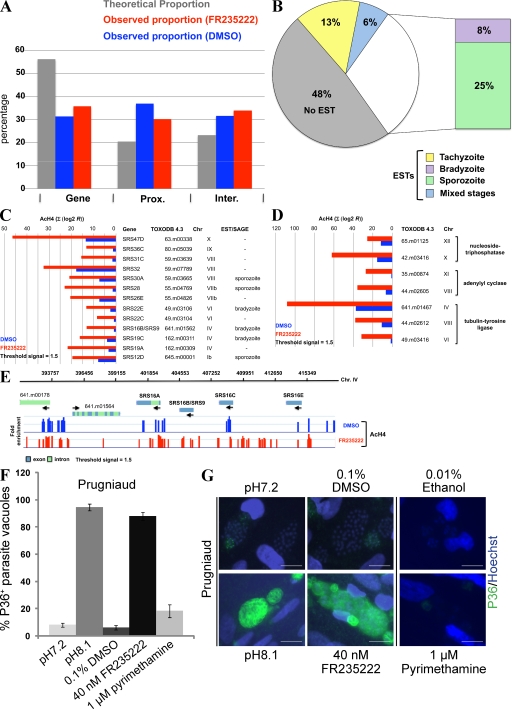
Because FR235222 inhibits TgHDAC3 activity, we reasoned that the anti-TgHDAC3 property of FR235222 could be used to identify the genes that are controlled by TgHDAC3. To this end, we performed genome-wide AcH4 measurements after FR235222 treatment (Fig. 3 and Table S2). Using ChIP and microarray hybridization (ChIP-on-chip assay), we analyzed the patterns of DNA-bound AcH4 after treatment of freshly lysed WT RH T. gondii with 40 nM FR235222 for 4 h. Upon treatment with FR235222, H4 acetylation distribution was significantly altered across all 14 chromosomes, indicative of a large-scale nuclear reorganization event. Approximately 5% of the ∼7,817 predicted genes (available from ToxoDB 4.3) had increased H4 acetylation levels, defining the Up group (369 genes; Table S2). The majority of AcH4 enrichment sites localized within 1 kb upstream of the gene start codon, with maximal enrichment immediately downstream of the transcription start site (TSS; Fig. 3 A and not depicted), possibly reflecting in some downstream effects at the transcriptional level of the genes affected.
Figure 3.
Genome-wide analysis of the effect of FR235222 treatment on histone H4 acetylation in T. gondii tachyzoites. (A) Statistical analyses of the AcH4 ChIP signal repartition across the T. gondii genome (see Materials and methods). The acetylated mark is more frequently enriched in both proximal and intergenic regions than within an internal gene region. (B) Diagram of the distribution of 369 FR235222 target genes (the up group) over Toxoplasma life-cycle stages, according to EST expression (tachyzoite, bradyzoite, and sporozoite). Numbers represent the percentage of genes that are either shared between two or three stages, or that are detected in a single stage. (C and D) The graphs indicate the fold enrichment of AcH4 over several FR235222 target genes. The signal from multiple closely spaced probes will often form a peak at or near the binding site. For each peak, the signals (selected cutoff = 1.5) were summed (Σratio [log2]) over the distance of the peak (1,000 bp upstream the start codon). (E) A representative view of Toxoplasma ChIP-on-chip analysis. High resolution mapping of AcH4 location on chromosome IV (SRS9 locus) after FR235222 treatment versus mock DMSO. Each vertical bar represents the log2 signal ratio of the test sample signal divided by the input control signal. The x axis denotes the genomic position of each probe. A threshold of 1.5 was applied to each experiment. Green (intron) and blue (exon) boxes indicate ToxoDB 4.3 gene annotation. The data are visualized by Genobrowser software. (F and G) FR235222 treatment induces T. gondii Prugniaud strain differentiation. Intracellular parasites (12h after infection) were incubated in the presence of either alkaline growth media to induce in vitro differentiation or 40 nM FR235222 for 2–3 d. As a control, parasites were treated with 1 µM pyrimethamine. Bradyzoite-differentiated parasites were identified by IFA using an anti-P36 antibody. Percentages of SRS9/P36-positive parasitophorous vacuoles in each condition are shown. Means ± SD of P36+ vacuoles are shown (n = 2 experiments). P36 (SRS9) expression was analyzed by IFAs in intracellular tachyzoites (pH 7.2) and in bradyzoite-differentiated parasites in vitro (pH 8.2). Tachyzoites were grown in the presence of 0.1% DMSO or 0.01% ethanol for 24 h (pH 7.2) or for 2–3 d (pH 8.2), 40 nM FR235222 for 3 d, or 1 µM pyrimethamine for 2–3 d. Bars, 10 µm.
Analysis of the genome-wide hyperacetylation pattern induced by FR235222 led to three main observations. First, based on the abundance of stage-specific expressed sequence tags (ESTs), roughly one third of the genes in the Up group were found to be expressed exclusively in the bradyzoite and/or sporozoite stages (Fig. 3, B and C; and Table S2). Thus, when T. gondii type II Prugniaud strain was cultivated in the presence of 40 nM FR235222, growth was inhibited and ∼80% of the parasitic vacuoles expressed the bradyzoite-specific antigen surface antigen glycoprotein–related sequence (SRS) 9/P36 at levels comparable to alkaline-stress parasites, which is known to induce stage conversion (Fig. 3, F and G). Very similar effects were observed by staining with the cyst-specific CC2 mAb (unpublished data) (19, 20). A very weak induction of SRS9/P36 was observed in parasites treated with pyrimethamine (a non-HDACi compound) when compared with FR235222-treated parasites; this indicates that the effect of FR235222 on SRS9/P36 expression is not solely caused by growth inhibition.
Then, we tested whether FR235222 was able to induce SRS9/P36 expression in the type I RH strain, which is characterized by a poor ability to differentiate in bradyzoites in vitro. Fig. S6 A shows that RH strain intracellular tachyzoites grown in the presence of FR235222 expressed SRS9/P36 at higher levels than untreated parasites. Consistent with SRS9/P36 up-regulation, nucleosomes at the level of the SRS9 locus were more acetylated in drug-treated parasites than in DMSO control (Fig. 3, C and E). However, SRS9 mRNA levels were not increased in response to HDACi treatment (unpublished data), suggesting that FR235222 might affect SRS9 expression in a more complicated fashion than solely through chromatin structure.
Given that mutations within TgHDAC3 confer resistance to FR235222, one could expect that induction of bradyzoite genes by HDACi's would be abolished in the drug-resistant lines. To test this hypothesis, SRS9/P36 levels were monitored by immunofluorescence analyses (IFAs) in the WT and TgHDAC3T99A mutant strains. Fig. S6 (A and B) shows that in the absence of HDACi's, P36 expression became constitutively expressed in the TgHDAC3T99A (R20D9) mutant strain compared with WT parasites. A very similar phenotype was observed in the TgHDAC3T99I (R01E11) mutant strain (unpublished data). Thus, these data indicate that mutations T99A and T99I must affect the regulatory role of TgHDAC3 on SRS9 expression.
However, many of the well-known bradyzoite-specific genes such as BAG1, LDH2, and ENO1 were not affected upon FR235222 treatment, showing that bradyzoite-specific genes are not controlled exclusively via histone H4 acetylation/deacetylation, as previously suggested (21). Second, almost half of the genes hyperacetylated by FR235222 have not been reported to be expressed at any of the parasite stages examined (Fig. 3 B), suggesting that they may be genes expressed at low levels or at an intermediate stage not covered by ESTs. Third, FR235222-mediated histone hyperacetylation affected functionally and structurally related genes (Fig. 3, C and D). For example, the 13 genes encoding for the SRS antigen family, which are scattered over seven different chromosomes, were all hyperacetylated in FR235222-treated parasites. This indicates that histone acetylation induced by FR235222 can target physically unlinked but functionally related genes in the parasite genome.
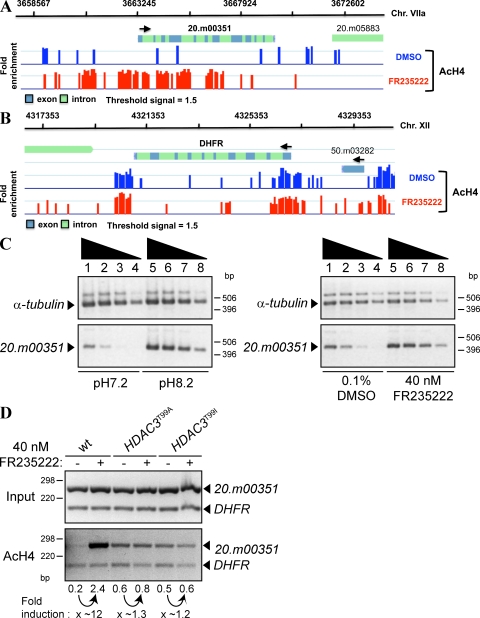
Next, we tested whether FR235222 was capable of inducing expression of the genes that have hyperacetylated histone H4. For this, we used 20.m00351 as a target gene, which encodes a bradyzoite-specific heat shock-related protein (Fig. 4 A) (22). Indeed, the 20.m00351 mRNA levels accumulated in FR235222-treated parasites, indicating that FR235222 can influence the expression of bradyzoite-specific genes at the transcriptional level (Fig. 4 C). Induction of 20.m00351 transcripts by HDACi treatment was similar to those observed in alkaline-induced bradyzoites (Fig. 4 C). In agreement with this, ChIP-on-chip assays showed that histone H4 was hyperacetylated at the 20.m00351 promoter region upon FR235222 treatment (Fig. 4 A), whereas the AcH4 levels remained unaffected by the drug in the neighborhood of the DHFR encoding gene that is steadily expressed in tachyzoite and bradyzoite parasites (Fig. 4 B). ChIP assays followed by semiquantitative PCR (scanning ChIP) confirmed that nucleosomes upstream of 20.m00351 were hyperacetylated in FR235222-treated parasites (∼12-fold induction; Fig. 4 D), whereas AcH4 signals upstream of the control DHFR gene were not modified in treated parasites (Fig. 4 D). Therefore, it is likely that histone H4 acetylation is the primary control mechanism of 20.m00351 transcription in response to FR235222 treatment.
Figure 4.
TgHDAC3 is a regulator of T. gondii differentiation. (A) High resolution ChIP-on-chip mapping of AcH4 location on chromosome VIIa (20.m00351 locus) after FR235222 treatment versus mock DMSO, as described in Fig. 3 E. (B) High resolution ChIP-on-chip mapping of AcH4 location on chromosome XII (DHFR locus) after FR235222 treatment versus mock DMSO, as described in Fig. 3 E. (C) FR235222 treatment induced the expression of the gene 20.m00351. mRNA levels of 20.m00351 were analyzed by RT-PCR in intracellular tachyzoites (pH 7.2) and in bradyzoite-differentiated parasites in vitro (pH 8.2). Tachyzoites were grown in the presence of 0.1% DMSO for 24 h (pH 7.2) or for 2–3 d (pH 8.2), or 40 nM FR235222 for 3 d. Lanes 1–4 and lanes 5–8 represent fourfold serial dilutions of the first-strand cDNA from parasites treated with 0.1% DMSO (pH 7.2 and 8.2) and 40 nM FR235222. α-Tubulin mRNA are expressed equally in each set of experiments and are presented as a control (n = 3 experiments). (D) Scanning ChIP experiments showing the effects of FR235222 on AcH4 levels in the presence of the TgHDAC3 WT, TgHDAC3T99A, and TgHDAC3T99I alleles at the promoter region of the bradyzoite-specific gene 20.m00351. ChIP analysis and quantification were performed as described in Materials and methods. The ratios of 20.m00351 and control DHFR signals present in input samples were used to calculate the relative precipitated fold enrichment shown below each lane (n = 3 experiments). A representative set of data is shown.
FR23522-resistance mutations in TgHDAC3 decrease the enzyme activity
Finally, we examined by scanning ChIP assays the AcH4 levels in the 5′ regulatory region of the 20.m00351 locus in the WT and in the TgHDAC3T99A and TgHDAC3T99I resistant lines in the presence or absence of FR235222. As expected, in the WT, AcH4 levels at the 20.m00351 locus were increased 12-fold in the presence of the drug, whereas in the TgHDAC3T99A resistant line no difference in the AcH4 levels were noticed in the presence or absence of the drug (Fig. 4 D). However, in the absence of the drug, the AcH4 levels of the resistant lines were approximately threefold higher than those of the WT (Fig. 4 B). This showed that the T99A and T99I mutations also affect the basal activity of TgHDAC3 at the 20.m00351 locus. This is in agreement with our earlier finding that resistant parasites constitutively express SRS9 in the absence of drug treatment (Fig. S6).
DISCUSSION
Compared with the widespread use of HDACi's in the cancer field, much less is known about the effects of these compounds on Apicomplexan parasites. In this study, we provide new insight into the effects of HDACi's on Plasmodium species, T. gondii, and N. caninum, with the characterization of the mode of action of a novel compound, FR235222, that efficiently inhibits parasite growth in vitro. We found that point mutations within TgHDAC3 were sufficient to decrease the sensitivity of T. gondii parasites to FR235222 or apicidin (Fig. 1 A and not depicted), thus providing genetic evidence that TgHDAC3 is the drug-targeted enzyme.
The basis for selective inhibition of HDACi's is one of the key unsolved questions concerning these compounds. The active site of class I and II HDAC is characterized by a structurally conserved catalytic core containing a divalent zinc cation. Crystallographic structures of the human HDAC7 (23) and HDAC8 (24, 25), along with the bacterial HDAC-like protein (HDLP) (26), showed that the mechanism of inhibition by HDACi's relies on the delivery of a zinc binding group to the bottom of a narrow active site pocket formed by loops L1 to L7 (Fig. S4) (26). Based on sequence homology, mutations in TgHDAC3 (T99A and T99I) conferring resistance to FR235222 localize to the L2 loop of HDLP, where the residue Y91 localized at the rim of the active site contacts the cognate HDACi (TSA; Fig. 2 A and Fig. S4) (26). Structural data of HsHDAC8 pointed out the role of the residue D101 in both substrate and HDACi recognition; HsHDAC8D101A mutated enzyme was inactive on protein substrates and binding efficiency to hydroxamate inhibitor was decreased (25). Given that D101 is localized in the vicinity of T99 of TgHDAC3, these data further strengthen the hypothesis of a direct inhibition of TgHDAC3 by FR235222 and are consistent with a role of T99 in the interactions with cyclopeptide inhibitors. T99A and T99I change amino acid polarity; it is therefore tempting to speculate that polar interactions at the rim of the active site support the binding to HDACi's, as proposed by Vannini et al. (25). We predict that the binding efficiency of HDACi's to TgHDAC3 would be diminished in the T99A and T99I mutated versions of TgHDAC3. However, an effect of these mutations on the regulation and/or activity of TgHDAC3 compensating the decrease in HDAC activity caused by drug inhibition cannot be excluded. In summary, the data presented in this study show the unexpected role of the Apicomplexa-conserved T99 residue for the resistance to cyclopeptide HDACi's.
Gene expression in Apicomplexan parasites is considerably dynamic, with large numbers of mRNAs exclusively expressed in a single developmental stage. In P. falciparum, microarray studies revealed a remarkably tight regulatory process that generates a continuous cascade of gene expression. Most genes are induced maximally at a time when they are presumably required for the parasite, after which the genes are down-regulated, leading to the hypothesis of the “just-in-time” manufacturing process (27). Similar observations have been made in T. gondii, where the primary developmental transitions are also accompanied by temporal changes at the level mRNAs from genes that are dispersed across all chromosomes (5). The details of how the expression of genes is controlled in these parasites have not been determined yet, although alterations in chromatin structure have been associated with changes in expression (2, 28). Indeed, it is now becoming increasingly clear that acetylation balance is greatly altered during parasite differentiation. How these changes are regulated at the molecular level remains unknown. Our data indicate that FR235222-mediated histone hyperacetylation affects also functionally and structurally related genes (Fig. 3, C and D). Moreover, the long-range histone acetylation pattern induced by FR235222 expands over linked families of genes, suggesting once more a common transcriptional regulation through chromatin structure (Fig. 3 E and Fig. S5 C). It is possible, then, that the loci containing clustered homologous genes are generally regulated by enhancer regions that impose stricter control of initiation than typically observed at other genes. Interestingly, when ChIP-on-chip data of AcH4 were compared with ChIP-on-chip data obtained using an anti-Myc–SET8 antibody (29), we noticed broad DNA regions with low levels of histone acetylation embedded into heterochromatic regions. The histone methylase SET8 is known to contribute to marking silent heterochromatic domains in Apicomplexan genomes by methylating H4K20 (29). It is noteworthy that FR235222 is unable to spread the hyperacetylation over the constitutive heterochromatin domains marked by SET8 (Fig. S5 D). Thus, the regions affected by FR235222 seem to correspond to facultative heterochromatin; constitutive heterochromatin cannot be hyperacetylated, probably because of methylation of histone H4 at lysine 20.
In this study, we provide direct evidence that the HDAC3 family plays a role in gene expression, differentiation, and cell-cycle control. Drug inhibition of TgHDAC3 prevents the formation of the daughter cells. Moreover, parasites treated with low doses of FR235222 are committed to differentiate into bradyzoites. This is consistent with the observations that during bradyzoite conversion a transient slowing of S phase leads to mature bradyzoites, which possess a uniform genome content (1N DNA) consistent with cell-cycle arrest in G1/G0 (30, 31). Moreover, drug inhibition of TgHDAC3 and parasites mutated at TgHDAC3 (TgHDAC3T99A and TgHDAC3T99I) lead to the expression of the bradyzoite marker SRS9 (Fig. S6). These data support previous observations that TgHDAC3 plays a role in the regulatory pathway driving parasite conversion (11).
A recent study indicated that a strong inducer of bradyzoite differentiation known as compound 1 up-regulated the transcription of bradyzoite-specific genes (21). By comparing the effect of FR235222 on nucleosome acetylation levels with the effect of compound 1 on transcript levels, we could determine that a core set of genes is common to bradyzoite differentiation induced by compound 1 and FR235222, as well as sets of genes that are specifically affected by each molecule (Fig. S5 A and Table S2). This further indicates that FR235222 affects the regulation of bradyzoite-specific genes at an epigenetic level (Table S2). However, with the exception of the B-NTPase gene (42.m03416), none of the well-known bradyzoite-specific genes (e.g., BAG1, LDH2, and ENO1, available from ToxoDB under accession nos. 55.m00009, 80.m00010, and 59.m03411, respectively), which are strongly induced by compound 1, were affected upon FR235222 treatment. One explanation could be that this group of genes is regulated by a different mechanism than through histone H4 acetylation or deacetylation, as previously suggested (21). However, whether these reflect differences in the effect of compound 1 and FR235222 or discrepancies between histone H4 acetylation levels and transcriptional activity has not been investigated further.
The potential antiparasitic activity of HDACi's has been reported before by others (13, 32–34), but obtaining drugs selective of Apicomplexan parasites is the major challenge. We observed that FR235222 inhibits parasite proliferation without affecting dramatically normal human host cells; parasites are ∼10 times more sensitive to FR235222 than HFF cells in our in vitro conditions, thus revealing a certain specificity of action of FR235222 toward parasites (Fig. 1 A and Fig. S7). Though FR235222 is relatively nontoxic to primary host human cells (Fig. S7) (16), an increase in AcH4 levels in human cells was observed at high concentrations (1 µM) of FR235222, indicating that this compound is somehow active on one or multiple mammalian HDACs (17). Possibly, the mammalian enzymes inhibited are not involved in essential biological functions, at least in our conditions. Given that no comparative analyses of mammalian versus Apicomplexan HDACs are currently available, no clear-cut conclusion about the isoenzyme selectivity of FR235222 can be drawn. Mutations in TgHDAC3 were at least sufficient to confer resistance to FR235222, and the mutated residue T99 of TgHDAC3 is conserved exclusively in the homologous enzymes present in all Apicomplexan parasites; therefore, an attractive model is that sensitivity to cyclopeptide HDACi's is carried by specific residues present only on parasitic HDACs but absent in mammalian homologues. Thus, a selective inhibition of Apicomplexan HDAC provides an elegant and novel way to interfere with parasite proliferation. Although it has been shown that FR235222 is bioactive in animal models (35, 36), a direct demonstration of its effectiveness in vivo against T. gondii remains to be performed.
MATERIALS AND METHODS
Parasite strains, cell culture, and in vitro bradyzoite differentiation.
The parasite strains used in this study are the following: the T. gondii type I RH strain that has lost the ability to complete the two-host life cycle, the T. gondii type II Prugniaud strain that is capable of robust bradyzoite differentiation, and the T. gondii type III CTG strain. All T. gondii and N. caninum strains were maintained by serial passage in HFF monolayer under tachyzoite conditions in DMEM (Invitrogen) supplemented with 10% (vol/vol) FBS (Invitrogen). T. gondii type II Prugniaud strain was maintained under tachyzoite conditions in DMEM supplemented with 10% (vol/vol) FBS and 25 mM Hepes buffer, pH 7.2. To induce in vitro bradyzoite differentiation, extracellular tachyzoites were allowed to invade HFF cells for 16 h, and the culture medium was removed and replaced by RPMI1640 supplemented with 1% FBS and 50 mM Hepes buffer, pH 8.2. After 2–3 d of culture in alkaline medium, bradyzoite induction was assessed for P36 expression by IFA.
Mutagenesis.
Parasites were chemically mutagenized according to a modification of a previously published protocol (37). 107 tachyzoites (freshly emerged from their host cells) were incubated at 37°C for 2 h in serum-deficient minimal essential medium containing 150 µg/ml NEU (diluted from a 1% stock solution in PBS). Parasites were washed two times and were allowed to recover in a fresh T25 flask of HFF monolayer in the absence of drug for 3 d. Released tachyzoites were then inoculated into fresh cell monolayers in medium containing 90 nM FR235222 and incubated until viable extracellular tachyzoites emerged 20 d later. Surviving parasites were passaged once more under continued FR235222 treatment and cloned by limiting dilution. Three cloned mutants (M190D4, M3135C3, and M3135D8) were isolated. TgHDAC3 open reading frame (orf) was amplified by RT-PCR using primers PstI-HDAC1-PCR3 (5′-AACTGCAGGATCGGAACCTTTTGGTCTCTG-3′) and HDAC1-PCR5-BamHI (5′-CGGGATCCGCGCTCAGTGCGCTGCGCAAGC-3′). The resulting PCR products were cloned into pCRII-TOPO cloning vector using the TOPO TA cloning kit (Invitrogen) and sequenced.
Transformation and selection of FR235222-resistant transformants.
DNA fragments containing the DNA sequences flanking the codon coding for the residue T99, and the mutated residues T99A and T99I of TgHDAC3 were generated by PCR amplification of the genomic DNA of the WT RH strain, M190D4, and M3135C3 resistant lines, respectively. Primers HD1-oligo2-PCR5 (5′-GACCTCTACAAACATATGGTGAGTG-3′) and HD1-oligo3-PCR3 (5′-GATACACGTCCACGCACTTGGTGATG-3′) amplify the TgHDAC3 sequence from nucleotides 133 to 1930 of the TgHDAC3 orf, producing a PCR product of 1,798 bp (Fig. S4).
Approximately 107 tachyzoites (RH WT strain) were transfected with ∼6 µg of linear DNA fragments by using electroporation parameters established previously (38). After electroporation, parasites were allowed to recover overnight in HFF monolayer in the absence of drug, and parasites were selected in 90 nM FR235222; resistant parasites were single-cell cloned and the TgHDAC3 DNA sequence was determined as described. Note that transfection of the type II Prugniaud strain was unfruitful in providing resistant parasites.
Proliferation assays.
The extent of the growth of T. gondii in monolayer cultures was determined by measuring the incorporation of [3H]uracil into acid-precipitable material. Human fibroblasts were grown to confluence in 24-well plates and infected with 103 T. gondii tachyzoites per well in the absence or the presence of FR235222. After 72 h, the culture was washed with PBS to remove noninvading parasites and replenished with fresh medium containing 5 µCi [3H]uracil/well. After 4 h, the monolayers were washed with PBS to remove nonincorporated [3H]uracil, and the remaining materials were solubilized in 500 µl of lysis/scintillation solution (Optiphase Super Mix; Perkin Elmer). The radioactivity was quantified by liquid scintillation spectrometry using a multidetector liquid scintillation counter (MicroBeta TriLux; Perking Elmer). EC50 has been determined according to nonlinear regression analysis using Prism 5 software (GraphPad Software, Inc.).
Quantitative assessment of the antimalarial activity of FR235222 was performed as previously described (39) on P. falciparum clones 3D7 and Dd2, except that the parasites were in contact with the drug for 48 and 20 h for the asynchronous 3D7 and Dd2 cultures and plasmion-enriched 3D7 schizonts, respectively. EC50 has been determined according to nonlinear regression analysis using HN-NonLin V1.1 software (available at http://malaria.farch.net).
Synchronized cultures of P. berghei GFP@hsp70 ANKA strain were performed as previously described (40). Cultures were grown in the presence of DMSO alone or with FR235222 added at 5, 11, and 22 h. Cultures were examined at 5, 11, 22, and 24 h after Giemsa staining and by FACs after DNA staining with Hoechst 33258 (Invitrogen).
The effect of HDACi's on mammalian host cell proliferation was assayed on nonconfluent HFF monolayers treated with FR235222 or TSA. Cell proliferation was quantified using CellTiter Aquous Phase kit (Promega). EC50 has been determined according to nonlinear regression analysis using Prism 5 software.
Immunofluorescence microscopy.
Infected HFFs grown on coverslips were washed in PBS and fixed/permeabilized for 20 min at room temperature with PBS containing 3% (vol/vol) formaldehyde and 0.2% Triton X-100 (vol/vol). Blocking was performed with PBS containing 5% FBS and 5% goat serum for 1 h at room temperature. Samples were incubated in PBS containing 1% FBS with the primary antibodies indicated in the figures (1:1,000 dilutions of anti-AcH4 antiserum [Millipore], anti-P36 monoclonal mouse antibodies, and anti-IMC1 monoclonal mouse antibodies), followed by the secondary antibodies goat anti–mouse IgG coupled with Alexa Fluor 488 and goat anti–rabbit IgG coupled with Alexa Fluor 568 (Invitrogen) at a 1:1,000 dilution each in PBS–1% FBS. Nuclei of host cells and parasites were stained for 10 min at room temperature with Hoechst 33258 at 2 µg/ml in PBS. After four washes in PBS, coverslips were mounted on a glass slide with Mowiol mounting medium (48 mM Tris-HCl [pH 8.5], 4.8% Mowiol 4-88 [wt/vol], 12% glycerol [vol/vol]), and images were acquired with a fluorescence microscope (Axioplan 2; Carl Zeiss, Inc.) equipped with 40 and 100× objectives and a charge-coupled device camera (Axiocam MRm; Carl Zeiss, Inc.) coupled to Axiovision 4.5 software (Carl Zeiss, Inc.). U. Gross (University of Göttingen, Göttingen, Germany), W. Bohne (University of Göttingen, Göttingen, Germany), G.E. Ward (University of Vermont, Burlington, VT), and J.F. Dubremetz (Centre National de la Recherche Scientifique, Montpellier, France) provided antibodies.
Isolation of T. gondii [3H]acetyl-labeled histones.
T. gondii (type I RH strain) extracellular tachyzoites (∼3 × 108 parasites) were resuspended in fresh medium containing 2.5 mCi sodium [3H]acetate (0.1 Ci/mmol; PerkinElmer) and 10 mM sodium butyrate. Parasites were incubated at 37°C for 1 h. Parasites were then washed in cold PBS and resuspended in 700 µl RSB buffer (10 mM Hepes [pH 8], 10 mM KCl, 1.5 mM MgCl2, 0.5% NP-40 [vol/vol], 0.5 mM dithiothreitol [DTT], and 1 mM PMSF). Parasites were lysed using a dounce (tight plunger) and lysate was centrifugated at 10,000 g for 10 min at 4°C. To acid extract the histones, the nuclear pellet was resuspended in 0.2 ml 5 M MgCl2 and 0.2 ml 0.8 M HCl was added. The mixture was incubated overnight at 4°C. Insoluble material was removed by centrifugation at 16,000 g for 10 min at 4°C, and the soluble fraction containing radiolabed histones was precipitated with 33% TCA final concentration. Precipitated histones were washed twice with ice-cold acetone and resuspended in 100 µl of water. Protein concentration was determined by Bradford assay using BSA as a standard, and specific activity was determined as 154.5 cpm/µg.
HDAC assay.
TgHDAC3-HA-FLAG was partially purified from transgenic extracellular tachyzoites expressing TgHDAC3-HA-FLAG ectopically, as previously described (11). Parasite pellets (∼3 × 109 tachyzoites) containing TgHDAC3-HA-FLAG recombinant proteins were resuspended in 15 ml of lysis buffer (20 mM Tris-HCl [pH 8], 500 mM KCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 20% glycerol [vol/vol], 0.5% NP-40 [vol/vol],1 mM PMSF, and protease inhibitor cocktail [Roche]). Parasites were lysed using a dounce (tight plunger), and lysate was centrifugated at 20,000 g for 30 min at 4°C. The soluble fraction was applied to an anti-FLAG M2 affinity gel column (500 µl; Sigma-Aldrich) equilibrated with lysis buffer. The column was washed with a 40-column volume of lysis buffer and the beads–anti-FLAG–TgHDAC3–HA–FLAG matrix was resuspended in 500 µl HDAC buffer (50 mM Tris-HCl [pH 8], 137 mM NaCl, 2.7 mM KCl, 1 mM MgCl2). 50-µl aliquots of this matrix were used as a source of TgHDAC3-HA-FLAG in the HDAC assays. Reaction mixtures were assembled in a 100-µl final volume of HDAC buffer containing 1 mg/ml BSA, a 50-µl aliquot of the HDAC-bead matrix described, and 0–1,000-nM final concentrations of HDACi's (TSA and FR235222). The mixtures were incubated at room temperature for 15 min to allow HDACi–HDAC complex formation. HDAC reactions were initiated by the addition of [3H]acetyl-labeled histones (3,708 cpm) as the substrate. After 1 h of incubation at 37°C, the reactions were stopped by the addition of 10 µl of 11 M HCl. The released [3H]acetic acid was extracted with 1 ml ethyl acetate. 900 µl of the solvent layer was then mixed with 5 ml of toluene scintillation solution, quantitated by scintillation counting, and used as a measure of HDAC activity.
Electrophoresis and immunoblot analysis of proteins.
Whole-cell extracts were prepared as follows: ∼107 extracellular parasites (needle passed, and filtered using a 3-µm nucleopore membrane) were lysed in 50 µl of lysis buffer (10 mM Tris-HCl [pH 6.8], 0.5% SDS [vol/vol], 10% glycerol [vol/vol], 1 mM EDTA) and sonicated. Protein concentration was determined by using the Bradford protein assay kit (Bio-Rad Laboratories) and BSA as a standard. A total of 50 µg of protein per sample was analyzed by using NuPAGE 4–12% Bis-Tris polyacrylamide gels with MES-SDS running buffer (Invitrogen), transferred onto nitrocellulose membrane (Hybond-ECL; GE Healthcare) or polyvinylidene fluoride membrane (Immobilon-P; Millipore), and probed with a 1:1,000 dilution of an anti-AcH4 antiserum (AcH4K5K8K12K16; Millipore), a 1:500 dilution of an anti–histone H4 antiserum (H4; Millipore), a 1:1,000 dilution of an antiacetyl–histone H2B antiserum (AcH2BK12K15; Abcam), a 1:1,000 dilution of an anti-H3 antiserum (H3; Abcam), a 1:1,000 dilution of an antiacetyl–histone H3K9 antiserum (AcH3K9; Abcam), a 1:2,000 dilution of a monoclonal anti–α-tubulin antibody (Sigma-Aldrich), and a 1:1,000 dilution of an anti-TgHDAC3 antibody (see Materials and methods). The blots were developed with the SuperSignal West Pico Chemiluminescent Substrate kit (Thermo Fisher Scientific).
Anti-TgHDAC3 antibodies.
Antiserum against TgHDAC3 was produced by Eurogentec using the Super Speedy immunization protocol and peptide #10 (C+DFEDMADRDQKVPI-COOH). Specific antibodies were affinity purified against peptide #10. For immunoblot analysis and IFAs, purified antibodies were used at 1:500 dilutions. IFAs confirmed that the majority of TgHDAC3 signals are localized to the nucleus of the intracellular tachyzoites (Fig. S1 A). Immunoblotting using 30 µg of proteins/lane revealed a signal at the expected size (∼50 kD) for TgHDAC3 (Fig. S1 B). Immunoblot analysis of the RH strain transfected with Pgra1-TgHDAC3-HA-FLAG (11) revealed the presence of two bands: the first one corresponding to the signal at ∼50 kD (endogenous TgHDAC3) was also detected in the WT parasites, and a second signal with a higher molecular mass (∼3 kD) that corresponds to the addition of the HA-FLAG tags in the C-terminal position of the protein.
Semiquantitative RT-PCR.
mRNA levels of 42.m03416, 20.m00351, and α-tubulin (583.m00022) were analyzed by RT-PCR in FR235222-treated parasites and in bradyzoites induced in vitro. T. gondii type II Prugniaud strain was cultured under tachyzoite conditions in DMEM supplemented with 10% (vol/vol) FBS and 25 mM Hepes buffer, pH 7.2. To induce in vitro bradyzoite differentiation, extracellular tachyzoites were allowed to invade HFF cells for 16 h, and the culture medium was removed and replaced by RPMI 1640 supplemented with 1% FBS and 50 mM Hepes buffer, pH 8.2. Parasites were allowed to differentiate for 2 d of culture in alkaline medium. To monitor the effect of FR235222 on mRNA levels, extracellular tachyzoites were allowed to invade HFF cells for 16 h under tachyzoite growth conditions (pH 7.2), and 40 nM FR235222 was added to the medium. After 3 d of growth in the presence of 40 nM FR235222 or 24 h of growth in the presence of 0.1% DMSO (mock experiment), infected HFF monolayers containing intracellular parasites were scraped from culture flasks, needle passed, and filtered using a 3-µm nucleopore membrane to separate parasites from host-cell debris. Parasites were resuspended in 1 ml TRIzol reagent (Invitrogen). All subsequent purification steps were performed according to the manufacturer's instructions. Concentrations of RNA samples were determined by measuring the OD value at wavelength 260. RT reactions were performed using 5 µg of total RNA, 100 µM of an oligo-(dT)25-(N)7 primer, and 400 U SuperScript III RT (Invitrogen). RT-PCR was performed according to the manufacturer's instructions. cDNA of 42.m03416, 20.m00351, and α-tubulin were amplified by PCR using DyNAzyme EXT DNA Polymerase (Finnzymes) and 1 µl of different dilutions of the cDNA products as DNA template. 20.m00351 was amplified using primers 20.m00351 F1 (5′-TGTATGAAGGCGATTCGTGC-3′) and 20.m00351 R1 (5′-ACCTTCTGGATGTCAGTCTC-3′); α-tubulin was amplified using primers α-TUB-F (5′-GTTATCAGCATCCACGTCGG-3′) and α-TUB-R (5′-CTTCTTGCCGTAGTCAACAGAC-3′). PCR products were resolved via 1% agarose gel electrophoresis and visualized by ethidium bromide staining.
ChIP assay.
Freshly released tachyzoites were needle passed and filtered using a 3-µm nucleopore membrane. Parasites were resuspended into fresh DMEM supplemented with 10% (vol/vol) FBS and 25 mM Hepes buffer, pH 7.2. Parasites were incubated in the presence of 40 nM FR235222 and 0.1% DMSO for 4 h at 37°C with 5% CO2. For ChIP-chip experiments, freshly released tachyzoites (∼5 × 109 at ∼12 × 107 parasites/ml) were fixed for 15 min in 1% formaldehyde. An increase in AcH4 signals was verified by immunoblotting to verify that FR235222 treatment was effective. To prepare chromatin samples, fixed parasites were lysed in MNase buffer (0.32 M sucrose, 50 mM Tris-HCl [pH 7.8], 4 mM MgCl2, 3 mM CaCl2, 100 mM NaCl, 0.25% NP-40 [vol/vol], 5% glycerol [vol/vol], protease inhibitor EDTA-free cocktail [Roche]), and DNA was digested for 4 min at 37°C by 2 U/ml MNase. Digestion was stopped with 20 mM EDTA, and chromatin was recovered in the soluble fraction after centrifugation at 10,000 g at 4°C; this constituted the S1 fractions. Pelleted materials were resuspended in dialysis buffer (1 mM Tris-HCl [pH 7.8], 0.2 mM EDTA) containing 1 mM PMSF and protease inhibitor cocktail and were dialyzed overnight at 4°C against the same solution. Dialyzed materials were then centrifuged and supernatants were harvested; this constituted the S2 fractions. For ChIPs, fractions S1 and S2 were pooled and DNA quality was verified by electrophoresis on 2% agarose gels; oligonucleosome ladders of 100–1,000 bp were obtained. The histone–DNA complexes were immunoprecipitated with anti-AcH4 antibodies according to NimbleGen's protocol (available at http://www.vmrf.org/researchcenters/gene-chip/chromatin_immunoprecipitation.pdf). In brief, sheep anti–rabbit IgG Dynabeads (1 ml per reaction; Invitrogen) were incubated with 200 µl anti-AcH4. Dynabeads (beads–IgG–anti–AcH4 complexes) were then washed with 5 mg/ml BSA in PBS and incubated with ∼500 µg chromatin. Dynabead complexes were washed twice in RIPA buffer (50 mM Hepes [pH 8], 1 mM EDTA, 1% NP-40 [vol/vol], 0.7% deoxycholate [wt/vol], 0.5 M LiCl) containing protease inhibitor cocktail, followed by two washes in 1× TE buffer (10 mM Tris-HCl [pH 8], 1 mM EDTA). Bound chromatin was eluted in 250 µl of elution buffer (10 mM Tris-HCl [pH 8], 1 mM EDTA, 1% SDS [vol/vol]). Histone–DNA cross-links were reversed by incubation at 65°C overnight in the presence of 150 mM NaCl and was phenol-chloroform purified. Isolated DNA samples (input and immunopurified DNA) were arrayed on custom synthetic oligo arrays (387,000 features per array, 46–50mer oligos; NimbleGen) according to the manufacturer's protocol, and data analysis was performed as previously described (29). The data are visualized by Genobrowser software (available at http://www.genostar.com/en/genostar-software/genomic-analysis-tools/genobrowser.html). The raw data discussed in this paper have been deposited in the Gene Expression Omnibus under accession no. GSE15241. The genome-wide data have been integrated into the Toxoplasma database (ToxoDB).
For scanning ChIP assays, freshly released tachyzoites (∼2 × 108 at ∼2 × 107 parasites/ml) were fixed as described above. Immunoprecipitated DNA was subjected to PCR analysis. 20.m00351 was amplified by PCR using primers 20.m00351_B F (5′-AGCTGTGCCGTTTGCGTGCCG-3′) and 20.m00351_B R (5′-TCCTACACCACGGGCGATTC-3′). DHFR was amplified using primers DHFR_B F (5′-CAAACCCGAGAAAAAGAAACG-3′) and DHFR_B R (5′-TGCTGTTCGCAGTCCCAGTAG-3′). All PCR reactions were performed in a 50-µl final volume, starting with 4 min at 94°C and followed by 30–35 cycles (depending on the region being amplified) of 94°C, 55°C, and 72°C (30 s each), with a final elongation of 10 min at 72°C. PCR products were separated on 2% agarose gels and analyzed using ImageJ software (available at http://rsbweb.nih.gov/ij/). Note that no PCR products were detected when DNA samples were assayed by ChIP using beads coupled with anti-GST antibodies instead of anti-AcH4 antibodies, indicating that our immunoprecipitated DNA fragments were specific to anti-AcH4 antibodies.
Statistical analyses of AcH4 ChIP-chip signal repartition across the genome.
Statistical analyses of the ChIP signal repartition were performed with the R package software, as previously described (29). The chromosomal regions were clustered into three main DNA sequences: the coding sequence (CDS; specified here as “gene”), the proximal region (1,000 bp upstream and downstream of each CDS), and the intergenic region. The enrichment within these three regions (“observed proportion”) were compared with what should be expected if signals have a uniform repartition (“theoretical proportion”). The test is a classical proportion test. For each chromosome, we computed the following formula: C is the chromosome size, L is the region size, NC is the number of signals in the chromosome, NL is the number of signals in the region, PC is the theoretical proportion, and P is the observed proportion. The computed value is assumed to follow the standard normal distribution when the two conditions CV1 = NC × PC and CV2 = NC × (1 − PC) are at least equal to 5. When this is the case, the value |z| = 1.96 has a probability of 0.05 to be exceeded.
Online supplemental material.
Fig. S1 provides a control experiment showing that growth arrest does not increase histone H4 acetylation in parasites. Fig. S2 provides data showing that FR235222 specifically affects AcH4 levels but not acetylation levels of H3K9 and H2BK12K15. Fig. S3 provides data showing the characterization of anti-TgHDAC3 antibodies. Fig. S4 shows a detailed multiple sequence alignment of selected deacetylases combined with structural data. Fig. S5 provides a comparison of compound 1–induced genes with genes that harbor proximal FR235222-induced histone H4 hyperacetylation. Fig. S6 shows the effect of TgHDAC3T99A mutation on P36 (SRS9) expression. Fig. S7 shows the effect of FR235222 and TSA on human host cell proliferation. Table S1 provides the properties of the different FR235222-resistant lines isolated. Table S2 provides the list of the genes containing increased AcH4 signals upon FR235222 treatment as determined by ChIP-on-chip assays. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20082826/DC1.
Acknowledgments
A. Bougdour performed the mutagenesis experiments, the in vitro HDAC assays, the IFAs, the ChIP scanning assays, the immunoblots, and the RT-PCR experiments. D. Maubon performed the proliferation assays on the T. gondii strains, N. caninum, and HFF host cells. A. Bouillon and J.-C. Barale determined drug sensitivity on P. falciparum. P. Baldacci performed the experiments on P. berghei. M.-A. Hakimi performed the ChIP-on-chip assay. P. Ortet, O. Bastien, and M.-A. Hakimi performed ChIP-on-chip analysis. A. Bougdour, R. Ménard, and M.-A. Hakimi conceived and designed the experiments. A. Bougdour, H. Pelloux, R. Ménard, and M.-A. Hakimi wrote the manuscript. We thank Astellas Pharma Inc. for their kind gift of FR235222 compound. We thank A. Curt for her technical help.
M.-A. Hakimi is supported by grants from Centre National de la Recherche Scientifique (ATIP+), the Agence National de la Recherche (ANR; MIME program), Lyon-Biopôle, and the Institut National de la Santé et de la Recherche Médicale (Contrat d'Interface). A. Bougdour is supported by an ANR-MIME grant. Genomic data were provided by the Institute for Genomic Research (supported by National Institutes of Health grant AI05093) and by the Sanger Center (Wellcome Trust). Expressed sequence tags were generated by Washington University (National Institutes of Health grant 1R01AI045806-01A1).
The authors have no conflicting financial interests.
Footnotes
Abbreviations used: AcH4, acetylated histone H4; ChIP, chromatin immunoprecipitation; EST, expressed sequence tag; HDAC, histone deacetylase; HDACi, HDAC inhibitor; HDLP, HDAC-like protein; HFF, human foreskin fibroblast; IFA, immunofluorescence analysis; IMC1, inner membrane complex protein 1; NEU, N-nitroso-N-ethylurea; SRS, surface antigen glycoprotein–related sequence; TgHDAC3, T. gondii HDAC3; TSA, trichostatin A; TSS, transcription start site.
References
- Dubey J.P., Lindsay D.S., Speer C.A. 1998. Structures of Toxoplasma gondii tachyzoites, bradyzoites, and sporozoites and biology and development of tissue cysts.Clin. Microbiol. Rev. 11:267–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakimi M.A., Deitsch K.W. 2007. Epigenetics in Apicomplexa: control of gene expression during cell cycle progression, differentiation and antigenic variation.Curr. Opin. Microbiol. 10:357–362 [DOI] [PubMed] [Google Scholar]
- Maubon D., Ajzenberg D., Brenier-Pinchart M.P., Darde M.L., Pelloux H. 2008. What are the respective host and parasite contributions to toxoplasmosis? Trends Parasitol. 24:299–303 [DOI] [PubMed] [Google Scholar]
- Cleary M.D., Singh U., Blader I.J., Brewer J.L., Boothroyd J.C. 2002. Toxoplasma gondii asexual development: identification of developmentally regulated genes and distinct patterns of gene expression.Eukaryot. Cell. 1:329–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radke J.R., Behnke M.S., Mackey A.J., Radke J.B., Roos D.S., White M.W. 2005. The transcriptome of Toxoplasma gondii.BMC Biol. 3:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh U., Brewer J.L., Boothroyd J.C. 2002. Genetic analysis of tachyzoite to bradyzoite differentiation mutants in Toxoplasma gondii reveals a hierarchy of gene induction.Mol. Microbiol. 44:721–733 [DOI] [PubMed] [Google Scholar]
- Anantharaman V., Iyer L.M., Aravind L. 2007. Comparative genomics of protists: new insights into the evolution of eukaryotic signal transduction and gene regulation.Annu. Rev. Microbiol. 61:453–475 [DOI] [PubMed] [Google Scholar]
- Sullivan W.J., Jr., Hakimi M.A. 2006. Histone mediated gene activation in Toxoplasma gondii.Mol. Biochem. Parasitol. 148:109–116 [DOI] [PubMed] [Google Scholar]
- Shahbazian M.D., Grunstein M. 2007. Functions of site-specific histone acetylation and deacetylation.Annu. Rev. Biochem. 76:75–100 [DOI] [PubMed] [Google Scholar]
- Gissot M., Kelly K.A., Ajioka J.W., Greally J.M., Kim K. 2007. Epigenomic modifications predict active promoters and gene structure in Toxoplasma gondii.PLoS Pathog. 3:e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saksouk N., Bhatti M.M., Kieffer S., Smith A.T., Musset K., Garin J., Sullivan W.J., Jr., Cesbron-Delauw M.F., Hakimi M.A. 2005. Histone-modifying complexes regulate gene expression pertinent to the differentiation of the protozoan parasite Toxoplasma gondii.Mol. Cell. Biol. 25:10301–10314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle J.P., Rajasekar B., Saeij J.P., Ajioka J.W., Berriman M., Paulsen I., Roos D.S., Sibley L.D., White M.W., Boothroyd J.C. 2006. Just one cross appears capable of dramatically altering the population biology of a eukaryotic pathogen like Toxoplasma gondii.Proc. Natl. Acad. Sci. USA. 103:10514–10519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darkin-Rattray S.J., Gurnett A.M., Myers R.W., Dulski P.M., Crumley T.M., Allocco J.J., Cannova C., Meinke P.T., Colletti S.L., Bednarek M.A., et al. 1996. Apicidin: a novel antiprotozoal agent that inhibits parasite histone deacetylase.Proc. Natl. Acad. Sci. USA. 93:13143–13147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone R.W., Licht J.D. 2003. Histone deacetylase inhibitors in cancer therapy: is transcription the primary target? Cancer Cell. 4:13–18 [DOI] [PubMed] [Google Scholar]
- Marks P., Rifkind R.A., Richon V.M., Breslow R., Miller T., Kelly W.K. 2001. Histone deacetylases and cancer: causes and therapies.Nat. Rev. Cancer. 1:194–202 [DOI] [PubMed] [Google Scholar]
- Mori H., Urano Y., Abe F., Furukawa S., Tsurumi Y., Sakamoto K., Hashimoto M., Takase S., Hino M., Fujii T. 2003. FR235222, a fungal metabolite, is a novel immunosuppressant that inhibits mammalian histone deacetylase (HDAC). I. Taxonomy, fermentation, isolation and biological activities.J. Antibiot. (Tokyo). 56:72–79 [PubMed] [Google Scholar]
- Gomez-Paloma L., Bruno I., Cini E., Khochbin S., Rodriquez M., Taddei M., Terracciano S., Sadoul K. 2007. Design and synthesis of cyclopeptide analogues of the potent histone deacetylase inhibitor FR235222.ChemMedChem. 2:1511–1519 [DOI] [PubMed] [Google Scholar]
- Mori H., Urano Y., Kinoshita T., Yoshimura S., Takase S., Hino M. 2003. FR235222, a fungal metabolite, is a novel immunosuppressant that inhibits mammalian histone deacetylase III. Structure determination.J. Antibiot. (Tokyo). 56:181–185 [DOI] [PubMed] [Google Scholar]
- Bohne W., Gross U., Ferguson D.J., Heesemann J. 1995. Cloning and characterization of a bradyzoite-specifically expressed gene (hsp30/bag1) of Toxoplasma gondii, related to genes encoding small heat-shock proteins of plants.Mol. Microbiol. 16:1221–1230 [DOI] [PubMed] [Google Scholar]
- Gross U., Bormuth H., Gaissmaier C., Dittrich C., Krenn V., Bohne W., Ferguson D.J. 1995. Monoclonal rat antibodies directed against Toxoplasma gondii suitable for studying tachyzoite-bradyzoite interconversion in vivo.Clin. Diagn. Lab. Immunol. 2:542–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnke M.S., Radke J.B., Smith A.T., Sullivan W.J., Jr., White M.W. 2008. The transcription of bradyzoite genes in Toxoplasma gondii is controlled by autonomous promoter elements.Mol. Microbiol. 68:1502–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen J., Fleige T., Gross U., Bohne W. 2008. Identification of novel bradyzoite-specific Toxoplasma gondii genes with domains for protein-protein interactions by suppression subtractive hybridization.Mol. Biochem. Parasitol. 157:228–232 [DOI] [PubMed] [Google Scholar]
- Schuetz A., Min J., Allali-Hassani A., Schapira M., Shuen M., Loppnau P., Mazitschek R., Kwiatkowski N.P., Lewis T.A., Maglathin R.L., et al. 2008. Human HDAC7 harbors a class IIa histone deacetylase-specific zinc binding motif and cryptic deacetylase activity.J. Biol. Chem. 283:11355–11363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannini A., Volpari C., Filocamo G., Casavola E.C., Brunetti M., Renzoni D., Chakravarty P., Paolini C., De Francesco R., Gallinari P., et al. 2004. Crystal structure of a eukaryotic zinc-dependent histone deacetylase, human HDAC8, complexed with a hydroxamic acid inhibitor.Proc. Natl. Acad. Sci. USA. 101:15064–15069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannini A., Volpari C., Gallinari P., Jones P., Mattu M., Carfi A., De Francesco R., Steinkuhler C., Di Marco S. 2007. Substrate binding to histone deacetylases as shown by the crystal structure of the HDAC8-substrate complex.EMBO Rep. 8:879–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnin M.S., Donigian J.R., Cohen A., Richon V.M., Rifkind R.A., Marks P.A., Breslow R., Pavletich N.P. 1999. Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors.Nature. 401:188–193 [DOI] [PubMed] [Google Scholar]
- Bozdech Z., Llinas M., Pulliam B.L., Wong E.D., Zhu J., DeRisi J.L. 2003. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum.PLoS Biol. 1:E5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherf A., Lopez-Rubio J.J., Riviere L. 2008. Antigenic variation in Plasmodium falciparum.Annu. Rev. Microbiol. 62:445–470 [DOI] [PubMed] [Google Scholar]
- Sautel C.F., Cannella D., Bastien O., Kieffer S., Aldebert D., Garin J., Tardieux I., Belrhali H., Hakimi M.A. 2007. SET8-mediated methylations of histone H4 lysine 20 mark silent heterochromatic domains in apicomplexan genomes.Mol. Cell. Biol. 27:5711–5724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerome M.E., Radke J.R., Bohne W., Roos D.S., White M.W. 1998. Toxoplasma gondii bradyzoites form spontaneously during sporozoite-initiated development.Infect. Immun. 66:4838–4844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radke J.R., Guerini M.N., Jerome M., White M.W. 2003. A change in the premitotic period of the cell cycle is associated with bradyzoite differentiation in Toxoplasma gondii.Mol. Biochem. Parasitol. 131:119–127 [DOI] [PubMed] [Google Scholar]
- Chen Y., Lopez-Sanchez M., Savoy D.N., Billadeau D.D., Dow G.S., Kozikowski A.P. 2008. A series of potent and selective, triazolylphenyl-based histone deacetylases inhibitors with activity against pancreatic cancer cells and Plasmodium falciparum.J. Med. Chem. 51:3437–3448 [DOI] [PubMed] [Google Scholar]
- Dow G.S., Chen Y., Andrews K.T., Caridha D., Gerena L., Gettayacamin M., Johnson J., Li Q., Melendez V., Obaldia N., III, et al. 2008. Antimalarial activity of phenylthiazolyl-bearing hydroxamate-based histone deacetylase inhibitors.Antimicrob. Agents Chemother. 52:3467–3477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobl J.S., Cassell M., Mitchell S.M., Reilly C.M., Lindsay D.S. 2007. Scriptaid and suberoylanilide hydroxamic acid are histone deacetylase inhibitors with potent anti-Toxoplasma gondii activity in vitro.J. Parasitol. 93:694–700 [DOI] [PubMed] [Google Scholar]
- Mori H., Abe F., Furukawa S., Sakai F., Hino M., Fujii T. 2003. FR235222, a fungal metabolite, is a novel immunosuppressant that inhibits mammalian histone deacetylase (HDAC) II. Biological activities in animal models.J. Antibiot. (Tokyo). 56:80–86 [PubMed] [Google Scholar]
- Marks P.A., Richon V.M., Rifkind R.A. 2000. Histone deacetylase inhibitors: inducers of differentiation or apoptosis of transformed cells.J. Natl. Cancer Inst. 92:1210–1216 [DOI] [PubMed] [Google Scholar]
- Morrissette N.S., Mitra A., Sept D., Sibley L.D. 2004. Dinitroanilines bind alpha-tubulin to disrupt microtubules.Mol. Biol. Cell. 15:1960–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messina M., Niesman I., Mercier C., Sibley L.D. 1995. Stable DNA transformation of Toxoplasma gondii using phleomycin selection.Gene. 165:213–217 [DOI] [PubMed] [Google Scholar]
- Desjardins R.E., Canfield C.J., Haynes J.D., Chulay J.D. 1979. Quantitative assessment of antimalarial activity in vitro by a semiautomated microdilution technique.Antimicrob. Agents Chemother. 16:710–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janse C.J., Van Vianen P.H. 1994. Flow cytometry in malaria detection.Methods Cell Biol. 42:295–318 [DOI] [PubMed] [Google Scholar]