Abstract
A tissue preparation protocol for MALDI (Matrix-Assisted Laser Desorption/Ionization) imaging mass spectrometry of an integral membrane protein was developed using ocular lens and retina tissues as model samples. Frozen bovine and human lenses were cryosectioned equatorially or axially at −20 °C into 20 μm-thick tissue sections. Lens sections were mounted onto gold-coated MALDI targets by methanol soft-landing to maintain tissue integrity. Tissue sections underwent extensive water washing to deplete the samples of highly abundant water-soluble proteins. Automated matrix deposition was achieved using an acoustic reagent multispotter, with sinapinic acid as matrix and high percentage acetonitrile as solvent, with a center-to-center spot spacing of 200–300 μm. Molecular images of full-length Aquaporin-0 (AQP0) and its most abundant truncation products were obtained from mass spectral data acquired across whole bovine and human lens sections. In equatorial and axial sections of bovine lenses, full-length AQP0 was detected throughout the lens. A truncation product corresponding to AQP0 (1–260) was detected in the bovine lens core at low abundance. In axial lens sections, no antero-posterior variation was detected. In 11 year-old human lens sections, full-length AQP0 was most abundant in the lens periphery, but was detected throughout the lens. The major truncation product, consisting of AQP0 residues 1–246, was absent from the lens periphery and increased in abundance in the lens core. This tissue preparation protocol was then applied to image the distribution of the G-protein coupled receptor, opsin, in the rabbit retina. This protocol has expanded the variety of target analytes which can be detected by MALDI imaging mass spectrometry to include integral membrane proteins.
Keywords: MALDI imaging mass spectrometry, AQP0, membrane protein, lens
INTRODUCTION
Matrix-assisted laser desorption/ionization (MALDI) imaging mass spectrometry is a relatively new analytical technique that combines the sensitivity of mass spectrometry with positional information to map the distribution of multiple analytes simultaneously from thin tissue sections.1 There are two modes of direct tissue analysis by MALDI mass spectrometry; profiling and imaging. In the profiling experiment, matrix solution is applied to distinct anatomical regions of thin tissue sections mounted on a MALDI target. Mass spectra are acquired and compared from each distinct region. For the imaging experiment, matrix solution is applied either by a spray of fine droplets, which results in a thin, homogenous layer of matrix across the entire tissue, or in a high density microdroplet array using an automated deposition instrument. The MALDI laser then performs a raster across the entire tissue section and the intensity of any m/z signal observed plotted as a function of position to construct a molecular image.
Typically, target analytes include abundant soluble proteins and peptides within a given tissue,1, 2 although lipids3 and drug compounds4, 5 have also been successfully analysed. Integral membrane proteins are critical components of biological systems, which can act as transmembrane transporters, channels, receptors and cell adhesion molecules. As with more traditional proteomics approaches, integral membrane proteins remain under-represented due to inherent solubility difficulties for mass spectral analysis. Traditional detergent solubilization techniques used in biochemistry to analyse hydrophobic proteins lead to ion suppression effects and are not, for the most part, compatible with mass spectrometric analysis. The ability to detect this class of proteins using the MALDI imaging mass spectrometry experiment would be a significant step toward analysis of tissue distributions of these biologically important molecules.
Recently, the distributions of ocular integral membrane proteins were analysed using the profiling approach.6 Porcine rhodopsin was detected in retinal preparations, while Aquaporin-0 (AQP0) and its major truncation products were mapped throughout the ocular lens in a variety of species. AQP0 is the most abundant integral lens membrane protein, consists of 263 amino acids, and has a mass of approximately 28 kDa. It is predicted to span the membrane six times7 and is thought to act as a transmembrane water channel8–11 and in the formation of intercellular junctions.12, 13 Knockout and mutation studies indicate that AQP0 is critical for lens structure and function.14–16 An extensive literature exists on age-related post-translational processing of AQP0. Truncation of AQP0 in whole human lens homogenates has been studied with a variety of techniques, including gel electrophoresis,17–19 and atomic force microscopy.20 However, these studies lack spatial information across the whole tissue. Limited spatial information on post-translational processing of AQP0 has been acquired through manual tissue dissection and subsequent MALDI mass spectrometry analysis,21, 22 which shows that AQP0 is truncated as a function of location in the lens; young cells in the lens periphery contain full-length AQP0, while older cells in the center of the lens contain truncated AQP0. These approaches are spatially limited to the resolution of the dissected lens regions, typically 4–5 regions per lens. Furthermore, tissue mass spectrometric profiling of AQP0 is limited to a lateral resolution of approximately 1mm due to the manual deposition of matrix solution and does not produce an image of protein distribution.6
This study presents a sample preparation protocol that allowed the acquisition of the first molecular images of the tissue distribution of integral membrane proteins using MALDI imaging mass spectrometry. Extensive water washing to deplete tissue sections of abundant soluble proteins and automated application of high percentage organic solvent matrix solution allowed detection of a 28 kDa integral lens membrane protein; AQP0 and its major truncation products in the bovine and human ocular lens. These results expand the range of analytes that MALDI imaging mass spectrometry can detect. This technique may be applicable to other tissue systems to enhance the growing field of spatially-resolved proteomics.
EXPERIMENTAL
Reagents
High-performance liquid chromatography (HPLC)-grade water, acetonitrile and methanol were purchased from Thermo Fisher Scientific Inc. (Hampton, NH). Sinapinic acid (SA) was purchased from Sigma-Aldrich (St Louis, MO). Tissue freezing medium (TFM) was obtained from Triangle Biomedical Sciences, Inc. (Durham, NC). Frozen bovine lenses and fresh whole rabbit eyes were obtained from Pel-Freeze Biologicals (Rogers, AR). Frozen human lenses were obtained from the National Disease Research Interchange (Philadelphia, PA), and stored at −80 °C until further use. Unless otherwise stated, all other reagents were purchased from Sigma-Aldrich.
Tissue preparation for MALDI analysis
Frozen bovine and human lenses were attached to cold specimen chucks with application of a small amount of TFM embedding medium at the base of the tissue only. Lenses were sectioned equatorially or axially at −20 °C into 20 μm-thick tissue sections using a disposable blade stage-equipped cryostat (Leica CM3050 S, Wetzlar, Germany). Lens polarity was determined using standard histological staining of a sister axial section and identifying the epithelial cell monolayer which covers the lens anterior pole (data not shown). For retinal studies, neural retina was dissected in phosphate buffered saline from fresh whole rabbit eyes, and frozen at −80 °C until further use. Frozen retinal tissue was orientated to collect sections in the plane of the retinal cell layers, encased in TFM embedding medium, and 16 μm-thick tissue sections collected. To collect frozen sections, a thin, uniform layer of methanol (room temperature, r.t.) was applied to a gold-coated MALDI target plate (r.t.) (lens tissue) or conductive glass slide (r.t.) (retinal tissue), and cryosections thaw-mounted by touching the MALDI target to the tissue section prior to evaporation of methanol. After air drying, tissue sections were washed 10 × 1 minute by manually pipetting water on to the sections. After each 1 minute wash, water was removed by pipette, and remaining water was removed by vacuum dessication.
Matrix deposition
Automated matrix deposition was carried out using a Portrait 630 acoustic reagent multispotter (Labcyte Inc., Sunnyvale, CA). Forty passes of one spot (170 pL) per pass of 20 mg/mL sinapinic acid in 90%ACN/0.2%TFA (lens tissue) or 40 mg/mL 2,5-dihydroxybenzoic acid in 90%ACN/0.2%TFA (retinal tissue) were applied with a center-to-center spot distance of 300 μm for bovine lenses and retinal tissue. For human lenses, four arrays with center-to-center spot distance of 400 μm were applied successively, shifting each array 200 μm in x and/or y to create an array of matrix spots with center-to-center spot distance of 200 μm. This array printing method ensured no contamination of adjacent matrix spots.
MALDI Imaging
Mass spectrometric analyses were performed in the linear, positive mode at +20 kV accelerating potential on a time-of-flight mass spectrometer (Bruker Autoflex II Linear; Bruker Daltonik, Bremen, Germany), which was equipped with a Smartbeam laser capable of operating at a repetition rate of 200 Hz. Delayed extraction parameters were optimized for signal intensity and mass resolution at a focus mass of 28 kDa. A linear, external calibration was applied to the instrument prior to data collection using a protein mixture of insulin (M+H+ = 5,734), cytochrome C (M+H+ = 12,361), myoglobin (M+H+ = 16,952), and trypsinogen (M+H+ = 23,982). Mass spectral data sets were acquired over whole bovine lens sections using flexImaging™ software (Bruker Daltonik, Bremen, Germany) in the mass range of m/z 21,000 to 35,000, with a raster step size of 300 μm and 300 laser shots per spectrum. For human lenses, data sets were acquired over whole lens sections in the mass range of m/z 10,000 to 35,000, with a raster step size of 200 μm and 300 laser shots per spectrum. For retinal tissue, data sets were acquired in the mass range of m/z 10,000 to 55,000, with a raster step size of 300 μm and 300 laser shots per spectrum. After data acquisition, molecular images were reconstituted using the flexImaging™ software. Data was normalized to total ion current using flexImaging™ software, and each m/z signal plotted ± 0.15% mass-to-charge units. For display purposes, data was interpolated, and pixel intensities were rescaled in flexImaging™ for each individual signal to utilize the entire dynamic range (lens tissue), or plotted on the same monochromatic intensity scale (retinal tissue). Assignments of protein identifications to m/z signals were made based on previous ocular membrane protein profiling work6, 21 by matching observed m/z values to predicted values of AQP0 and its prominent truncation products.
RESULTS AND DISCUSSION
We have developed a tissue preparation protocol which maps the distribution of a 28 kDa integral membrane protein by MALDI imaging mass spectrometry. Extensive water washing of the tissue prior to automated application of sinapinic acid in a high percentage organic matrix solvent was required to produce strong signals for full-length and truncated forms of ocular lens AQP0 in both bovine and human lens sections.
Standardized methods for MALDI imaging of proteins and peptides have been reported.23 For example, frozen tissue sections are typically washed in ethanol to remove phospholipids, physiological salts, and fix proteins, which increases ion yield in a variety of tissues. For protein imaging, sinapinic acid and 50%ACN/0.1%TFA is the most common matrix/solvent combination and has been shown to give high quality mass spectral information.24 However, for membrane protein imaging both the tissue washing procedure and matrix/solvent conditions were altered in order to detect strong membrane protein ion signals. Firstly, extensive water washing was used to deplete the tissue of abundant water-soluble proteins which are the most readily detected class of proteins using standard MALDI imaging mass spectrometry tissue preparation protocols. Subsequent ethanol washing was initially trialled, as it was hypothesized that delipidation may enhance membrane protein ion signals; however, application of matrix solution to water/ethanol-washed tissue sections led to matrix solution spreading, poor matrix crystalization, and poor membrane protein ion signals. Rehydration of the tissue following water/ethanol washing was also attempted, but no improvement in ion intensity was observed. Therefore, tissue sections were washed with water only prior to matrix application.
Membrane proteins have previously been detected directly from ocular tissue by manual application of matrix using solvent conditions involving, first, application of a mixture of 7:3 formic acid:hexafluoroisopropanol, followed by repeated application of sinapinic acid in 90%ACN/0.1%TFA.6 Since the mixture of 7:3 formic acid:hexafluoroisopropanol tended to spread on tissue, matrix application procedures utilizing matrix in 50–90% organic solvent only were tested. No membrane protein signals were detected when matrix was applied using 50%ACN/0.1%TFA; however, strong membrane protein signals were detected when matrix was applied in 90%ACN/0.2%TFA. In addition, spreading of the matrix solution upon application to tissue was limited such that 200 μm spot spacing was achieved using an automated acoustic reagent multispotter. Matrix application using the acoustic reagent multispotter was considered crucial for detection of strong membrane protein signals since matrix application using a gas-powered sprayer gave inconsistent results.
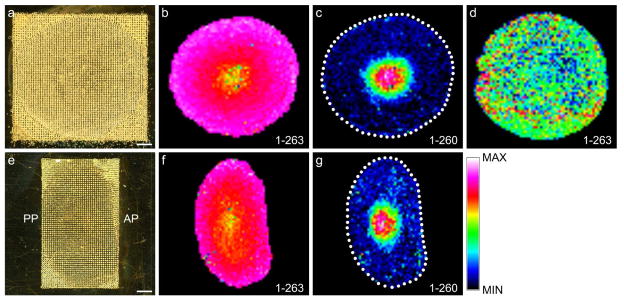
Figure 1 shows the distribution of bovine AQP0 in equatorial and axial tissue sections. Equatorial lens sections possess young, newly differentiating lens cells at the tissue edge, and older, terminally differentiated lens cells in the center of the tissue, while axial lens sections add antero-posterior information to the lens cell age gradient. Water-washed tissue sections were spotted with matrix solution at a spot spacing of 300 μm (Figure 1a, e). Full-length AQP0 (residues 1–263) was detected throughout the bovine lens in the summary mass spectrum at the predicted m/z 28,224 ± 0.07%, with highest intensity in young, peripheral lens cells, and with decreased intensity in the older lens cells (Figure 1b, f). Loss of intact AQP0 signal has also been observed by immunohistochemistry (IHC) methods in whole rat lens sections;25 however, detection of specific truncation products (as shown below) is not possibly by IHC. Truncated AQP0 (residues 1–260) was detected in the summary mass spectrum at the predicted m/z 27,912 ± 0.06% in the lens core at low abundance (Figure 1c, g). No antero-posterior variability was detected for either form of AQP0 in axial sections. In contrast, tissue sections not washed with water gave poor signal for full-length AQP0, and the corresponding molecular image contained no meaningful spatial information (Figure 1d), indicating the importance of the water washing steps. The observed distributions of AQP0 and truncated AQP0 are in good agreement with previous on-tissue MALDI profiling of AQP0, where very little truncation of AQP0 was detected in the bovine lens,6 and with laser capture microdissected samples, where truncation at Thr260 was reported as the main truncation product in the bovine lens core.26 In addition, the consistency of the tissue distributions of each form of AQP0 in equatorial and axial sections indicates that results obtained using this tissue preparation technique are highly reproducible.
Figure 1. MALDI Imaging of AQP0 in bovine lenses.
Lenses were sectioned in both equatorial (a–d) and axial (e–g) orientation. (a, e) Optical scan of bovine lens sections prepared for integral membrane protein imaging with high-density matrix microdroplet array applied at 300 μm spot spacing. Distribution of the mass spectral signal for full-length AQP0 (1–263) (plotted m/z 28,203 equatorial and 28,221 axial) in water-washed (b, f), and unwashed tissue sections (d). Only in the core of the lens is the truncation product AQP0 (1–260) (plotted m/z 27,895 equatorial and 27,911 axial) detected (c, g). AP = anterior pole, PP = posterior pole, scale bars = 2 mm. The dotted lines indicate the edge of the tissue.
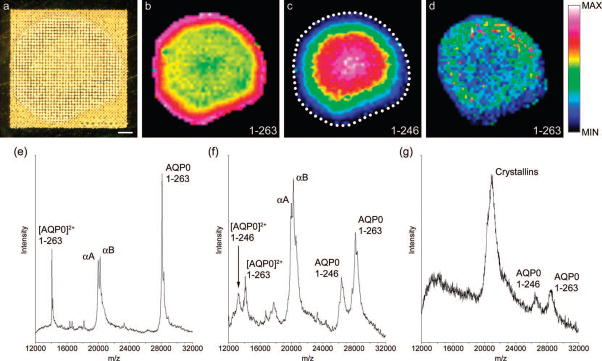
The distributions of full-length AQP0 and its truncation products were different in the 11 year-old human lens (Figure 2). Due to the smaller size of the human lens section, matrix microdroplets were deposited to a final center-to-center spot distance of 200 μm, which was the highest spatial resolution possible while avoiding cross-contamination of adjacent matrix spots (Figure 2a). Full-length AQP0 was again detected throughout the lens in the summary mass spectrum at the predicted m/z 28,123 ± 0.02%, but was most intense in lens cells up to 600 μm from the lens edge (Figure 2b). At this location in the lens, signal for full-length AQP0 decreased and remained fairly constant throughout the deeper-lying lens cells. In contrast, signal for the main truncation product AQP0 (1–246) at the predicted m/z 26,359 ± 0.07% was not detected in peripheral cells, and increased in intensity in the lens core (Figure 2c). Similar distributions of each AQP0 form were observed when MALDI images of the doubly-charged AQP0 ions were generated (data not shown). When a molecular image of full-length AQP0 was plotted from human lens tissue not washed with water, minimal spatial information was obtained (Figure 2d). Full-length AQP0 appeared most abundant in the mid-cortex, probably due to suppression of AQP0 signal in the lens periphery by abundant crystallin signal, and poor matrix crystal formation in the lens core. Example mass spectra extracted from individual sampling locations in the peripheral (Figure 2e) and core (Figure 2f) regions of the MALDI tissue imaging data set of water-washed lens sections indicate the strong signals detected for each AQP0 form, as well as for the abundant soluble crystallin proteins αA- and αB- crystallin (19 & 20 kDa), which were not completely washed away during water washing. However, the water washing greatly enhanced signal for each detected form of AQP0 as evidenced by the mass spectrum extracted from the cortex of unwashed human lens tissue, which is characterized by broad, noisy protein signals and abundant crystallin signals (Figure 2g). The detected AQP0 truncation products are consistent with previous studies of human AQP0 post-translational modification, which identified Asn259 and Asn246 as major sites of truncation,27 and that these forms of AQP0 increase in the lens core.21
Figure 2. MALDI Imaging of AQP0 in a human lens.
(a) Optical scan of an equatorial section from an 11 year-old human lens prepared for integral membrane protein imaging with high-density matrix microdroplet array applied at 200 μm spot spacing. (b) Signal for full-length AQP0 (1–263) (plotted m/z 28,129) is most intense around the edge of the lens, and persists in the lens core. (c) The major truncation product in the human lens AQP0 (1–246) (plotted m/z 26,378) increases in intensity in the core of the lens. (d) Signal for full-length AQP0 (1–263) in unwashed tissue sections, indicating the importance of the washing step. (e) Extracted spectrum from the lens periphery showing singly- and doubly-charged AQP0 signals and signals for the abundant soluble proteins α A- and αB-crystallin. (f) Extracted spectrum from the lens core, showing singly- and doubly-charged full-length AQP0, AQP0 (1–259) and AQP0 (1–246). Signals for crystallin proteins are also abundant. (g) Extracted spectrum from the lens cortex of unwashed tissue, showing noisy, poorly resolved mass spectral signals for AQP0 1–263 and 1–246, and abundant crystallin signal. Scale bar = 1 mm. The dotted line indicates the edge of the tissue.
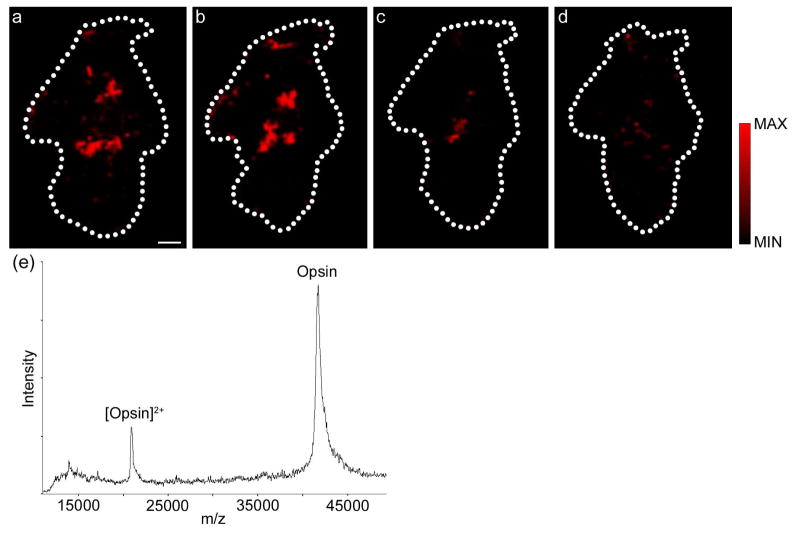
To demonstrate the applicability of the method to other tissue, rabbit retinal sections were targeted to map the distribution of the 42 kDa G protein-coupled receptor opsin. Opsins are enriched in the photoreceptor cells of the retina and are responsible for the initiation of visual perception. The retinal photoreceptor cell layer is approximately 50 μm thick,28 and cannot be resolved in cross-section using the current method. Hence, retinal sections in the plane of the photoreceptor cell layer were collected and prepared for membrane protein imaging. Figure 3 shows the distribution of opsin in 16 μm-thick serial sections of rabbit central retina, which was detected at the predicted m/z 41,706 ± 0.1%. For enhanced opsin signal, 2,5-dihydroxybenzoic acid was substituted as matrix, which is consistent with previous MALDI profiling of opsin.6 The localization of opsin becomes more central (Figure 3a–c), before disappearing in retinal sections not containing the photoreceptor cell layer (Figure 3d) due to each tissue section not originating entirely from this retinal cell layer and the initial spheroid shape of the eye cup. Nevertheless, application of this tissue preparation procedure to retinal tissue has allowed the detection of a 42 kDa integral membrane protein (Figure 3e), the mass of which lies outside the mass range of most analytes that are detected by imaging mass spectrometry.23
Figure 3. MALDI Imaging of Opsin in the rabbit retina.
Serial sections of rabbit central retina showing the localization of opsin. Collected sections were not entirely within the photoreceptor cell layer, hence opsin localization shifts towards the center of adjacent tissue sections (a–c, plotted m/z 41,677, 41,655, and 41,735, respectively), before disappearing in a retinal tissue section not containing photoreceptor cells (d, plotted m/z 41,670). (e) Extracted mass spectrum from central rabbit retina section (from panel a) showing singly- and doubly-charged ions for opsin. Scale bar = 2mm. The dotted lines indicate the edge of the tissue sections.
In conclusion, extensive water washing of frozen tissue sections, and automated application of matrix solution using a high percentage organic solvent have allowed MALDI images of integral membrane proteins to be generated for the first time. Application of this new tissue preparation procedure to other tissues may allow the spatial distributions of other integral membrane proteins to be generated thereby expanding the field of accessible analytes to this functionally important class of biomolecules.
Acknowledgments
Grant #s: NIH grants EY13462 (KLS) & GM58008 (RMC)
References
- 1.Caprioli RM, Farmer TB, Gile J. Molecular Imaging of Biological Samples: Localization of Peptides and Proteins Using MALDI-TOF MS. Analytical Chemistry. 1997;69(23):4751–60. doi: 10.1021/ac970888i. [DOI] [PubMed] [Google Scholar]
- 2.Stoeckli M, Chaurand P, Hallahan DE, Caprioli RM. Imaging Mass Spectrometry: A New Technology for the Analysis of Protein Expression in Mammalian Tissues. Nature Medicine. 2001;7:493–496. doi: 10.1038/86573. [DOI] [PubMed] [Google Scholar]
- 3.Jackson SN, Wang HYJ, Woods AS. Direct Profiling of Lipid Distribution in Brain Tissue Using MALDI-TOF MS. Analytical Chemistry. 2005;77:4523–4527. doi: 10.1021/ac050276v. [DOI] [PubMed] [Google Scholar]
- 4.Reyzer M, Hsieh Y, Ng K, Korfmacher WA, Caprioli RM. Direct Analysis of Drug Candidates in Tissue by Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry. Journal of Mass Spectrometry. 2003;38:1081–1092. doi: 10.1002/jms.525. [DOI] [PubMed] [Google Scholar]
- 5.Troendle FJ, Reddick CD, Yost RA. Detection of Pharmaceutical Compounds in Tissue by Matrix-Assisted Laser Desorption/Ionization and Laser Desorption/Chemical Ionization Tandem Mass Spectrometry with a Quadrupole Ion Trap. Journal of the American Society for Mass Spectrometry. 1999;10:1315–1321. [Google Scholar]
- 6.Thibault DB, Gillam CJ, Grey AC, Han J, Schey KL. MALDI Tissue Profiling of Integral Membrane Proteins from Ocular Tissues. Journal of the American Society for Mass Spectrometry. 2008;19(6):814–22. doi: 10.1016/j.jasms.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jung JS, Preston GM, Smith BL, Guggino WB, Agre P. Molecular Structure of the Water Channel Through Aquaporin CHIP. The Hourglass Model. Journal of Biological Chemistry. 1994;269(20):14648–14654. [PubMed] [Google Scholar]
- 8.Németh-Cahalan KL, Hall JE. pH and Calcium Regulate the Water Permeability of Aquaporin 0. Journal of Biological Chemistry. 2000;275(10):6777–82. doi: 10.1074/jbc.275.10.6777. [DOI] [PubMed] [Google Scholar]
- 9.Németh-Cahalan KL, Kalman K, Hall JE. Molecular Basis of pH and Ca2+ Regulation of Aquaporin Water Permeability. The Journal of General Physiology. 2004;123:573–580. doi: 10.1085/jgp.200308990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Varadaraj K, Kumari S, Shiels A, Mathias RT. Regulation of Aquaporin Water Permeability in the Lens. Investigative Ophthalmology and Visual Science. 2005;46(4):1393–1402. doi: 10.1167/iovs.04-1217. [DOI] [PubMed] [Google Scholar]
- 11.Varadaraj K, Kushmerick C, Baldo GJ, Bassnett S, Shiels A, Mathias RT. The Role of MIP in Lens Fiber Cell Membrane Transport. Journal of Membrane Biology. 1999;170:191–203. doi: 10.1007/s002329900549. [DOI] [PubMed] [Google Scholar]
- 12.Zampighi GA, Eskandari S, Hall JE, Zampighi L, Kreman M. Micro-Domains of AQP0 in Lens Equatorial Fibers. Experimental Eye Research. 2002;75:505–519. doi: 10.1006/exer.2002.2041. [DOI] [PubMed] [Google Scholar]
- 13.Zampighi GA, Hall JE, Ehring GR, Simon SA. The Structural Organization and Protein Composition of Lens Fiber Junctions. Journal of Cell Biology. 1989;108:2255–2275. doi: 10.1083/jcb.108.6.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Al-Ghoul KJ, Kirk T, Kuszak AJ, Zoltoski RK, Shiels A, Kuszak JR. Lens Structure in MIP-deficient Mice. The Anatomical Record. 2003;273(2):714–30. doi: 10.1002/ar.a.10080. [DOI] [PubMed] [Google Scholar]
- 15.Shiels A, Bassnett S. Mutations in the Founder of the MIP Gene Family Underlie Cataract Development in the Mouse. Nature Genetics. 1996;12:212–5. doi: 10.1038/ng0296-212. [DOI] [PubMed] [Google Scholar]
- 16.Shiels A, Bassnett S, Varadaraj K, Mathias RT, Al-Ghoul K, Kuszak JR, Donoviel D, Lilleberg S, Friedrich G, Zambrowicz B. Optical Dysfunction of the Crystalline Lens in Aquaporin-0-Deficient Mice. Physiological Genomics. 2001;7:179–186. doi: 10.1152/physiolgenomics.00078.2001. [DOI] [PubMed] [Google Scholar]
- 17.Horwitz J, Robertson NP, Wong MM, Zigler JS, Kinoshita JH. Some Properties of Lens Plasma Membrane Polypeptides Isolated From Normal Human Lenses. Experimental Eye Research. 1979;28(3):359–65. doi: 10.1016/0014-4835(79)90098-8. [DOI] [PubMed] [Google Scholar]
- 18.Roy D, Spector A, Farnsworth PN. Human Lens Membrane: Comparison of Major Intrinsic Polypeptides from Young and Old Lenses Isolated by a New Methodology. Experimental Eye Research. 1979;28(3):353–8. doi: 10.1016/0014-4835(79)90097-6. [DOI] [PubMed] [Google Scholar]
- 19.Takemoto L, Takehana M. Major Intrinsic Polypeptide (MIP26K) from Human Lens Membrane: Characterization of Low-Molecular-Weight Forms in the Aging Human Lens. Experimental Eye Research. 1986;43:661–667. doi: 10.1016/s0014-4835(86)80032-x. [DOI] [PubMed] [Google Scholar]
- 20.Buzhynskyy N, Girmens JF, Faigle W, Scheuring S. Human cataract lens membrane at subnanometer resolution. Journal of Molecular Biology. 2007;374(1):162–9. doi: 10.1016/j.jmb.2007.09.022. [DOI] [PubMed] [Google Scholar]
- 21.Ball LE, Garland DL, Crouch RK, Schey KL. Post-translational Modifications of Aquaporin 0 (AQP0) in the Normal Human Lens: Spatial and Temporal Occurrence. Biochemistry. 2004;43:9856–9865. doi: 10.1021/bi0496034. [DOI] [PubMed] [Google Scholar]
- 22.Korlimbinis A, Berry Y, Thibault DB, Schey KL, Truscott RJ. Protein aging: Truncation of aquaporin 0 in human lens regions is a continuous age-dependent process. Experimental Eye Research. 2008 doi: 10.1016/j.exer.2008.12.008. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chaurand P, Norris JL, Cornett DS, Mobley JA, Caprioli RM. New Developments in Profiling and Imaging of Proteins From Tissue Sections by MALDI Mass Spectrometry. Journal of Proteome Research. 2006;5(11):2889–900. doi: 10.1021/pr060346u. [DOI] [PubMed] [Google Scholar]
- 24.Schwartz SA, Reyzer ML, Caprioli RM. Direct Tissue Analysis Using Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry: Practical Aspects of Sample Preparation. Journal of Mass Spectrometry. 2003;38(7):699–708. doi: 10.1002/jms.505. [DOI] [PubMed] [Google Scholar]
- 25.Grey AC, Li L, Jacobs MD, Schey KL, Donaldson PJ. Differentiation-dependent modification and subcellular distribution of aquaporin-0 suggests multiple functional roles in the rat lens. Differentiation. 2009;77(1):70–83. doi: 10.1016/j.diff.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Z, Han J, Schey KL. Spatial Differences in an Integral Membrane Proteome Detected in Laser Capture Microdissected Samples. Journal of Proteome Research. 2008;7(7):2696–2702. doi: 10.1021/pr700737h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schey KL, Little M, Fowler JG, Crouch RK. Characterization of Human Lens Major Intrinsic Protein Structure. Investigative Ophthalmology and Visual Science. 2000;41(1):175–82. [PubMed] [Google Scholar]
- 28.Ross MH, Pawlina W. Histology: A Text and Atlas With Correlated Cell and Molecular Biology. 5. Lippincott, Williams & Wilkins; Baltimore, MD: 2006. p. 906. [Google Scholar]