Abstract
NF-κB influences the initiation, progression, and maintenance of diverse cancer types. Despite current therapeutic efforts to block hyperactive NF-κB in cancer cells, the in vivo effects of a drug upon this complex pathway are unclear. We monitored NF-κB activity and a fast-expressing reporter level simultaneously in head and neck squamous carcinoma cells by quantitative live microscopy. The real time single cell assay revealed the TNF-α induced oscillation of NF-κB was echoed by equally dynamic reporter expression rate. Bortezomib is a proteasome inhibitor whose anti-cancer action is partly mediated through inhibition of NF-κB. When administered to pre-activated cells, the drug gave rise to distinct inhibition dynamics, with discrete pulses of reporter induction remaining for hours. These findings suggest that, contrary to a simplistic presumption for a pathway ‘blockade’, the network dynamics and the intracellular pharmacokinetics of the inhibitor must be critically evaluated in developing strategies for optimal intervention of oncogenic pathways.
Introduction
Recent trends in clinical investigations clearly tend towards molecularly targeted approaches that are based on mechanisms underlying the pathophysiology of the disease. Often the goal is to block the activity of a specific pathway that has been implicated in the disease process, by targeting a key component with a small molecule or an antibody, for example. It is seldom known whether the desired ‘blockade’ is achieved in the relevant tissue and why paradoxical outcomes occur in certain cases. Here we show that NF-κB activity in tumor cells is altered by the action of a proteasome inhibitor in a complex way that cannot sufficiently be explained by the simplistic notion of ‘pathway blockade’.
NF-κB/Rel is a master regulator of inflammatory processes and has a growing list of cancers and other common diseases that require its aberrant activity (Aggarwal, 2004; Chang and Van Waes, 2005; Greten et al., 2004; Jackson-Bernitsas et al., 2006; Karin, 2006; Li et al., 2005; Ondrey et al., 1999; Pikarsky et al., 2004; Rosenwald and Staudt, 2003). The activation of this latent transcription factor is controlled by IκB proteins that keep them mostly in the cytoplasm. Upstream signaling activates the IκB kinase complex (IKK) that causes the release of NF-κB from IκB and its subsequent nuclear import and transcription of its target genes (Pahl, 1999). One of the target genes is its own inhibitor IκBα, giving rise to a classical negative feedback loop. Interestingly, it has recently been demonstrated that NF-κB can be activated with complex oscillatory dynamics, largely driven by this negative feedback (Friedrichsen et al., 2006; Ganguli et al., 2005; Hoffmann et al., 2002; Nelson et al., 2004). Other pathways that share similar negative feedback loops have also been shown to exhibit oscillations, suggestive of a rather broad phenomenon (Hirata et al., 2002; Lahav et al., 2004).
It is then important to ask if and how this previously unappreciated dynamics manifests in the therapeutic setting. However, dynamic patterns are largely lost in assays using snapshot measurements because cell-to-cell variability and time-dependent changes are indistinguishable. Even in single cell measurements taken from cells conditioned for differing duration of time, meaningful temporal changes are difficult to ascertain due to the inherent cell-to-cell heterogeneity. Therefore it is necessary to obtain continuous single cell data in real time for an accurate investigation of cellular dynamics.
In the present study, we monitored the activity of NF-κB and the expression of an NF-κB inducible IL-8 promoter-driven reporter, in living cells under conditions that mimic the real time active process of NF-κB inhibition by a small molecule. We chose Bortezomib (also known as Velcade or PS-341) in light of its well-documented anti-cancer effects, partly mediated by inhibition of NF-κB (Van Waes, 2007). Bortezomib is a proteasome inhibitor that stabilizes the crucial endogenous inhibitor IκBα that sequesters NF-κB in the cytoplasm (Richardson et al., 2005; Zavrski et al., 2005). Its therapeutic use had been approved for multiple myeloma. Current clinical trials are examining its efficacy for other types of cancer, including head and neck squamous cell carcinoma (HNSCC) which we focused in our study. Given the transient nature of NF-κB activity, NF-κB and reporter kinetics were obtained by live microscopy observations of single cells. We analyzed the real time data and interpreted the dynamic patterns using a simple mathematical model that captures the essence of the canonical NF-κB network and the inhibitor interaction. Finally, based on the general agreement between our time course data and model simulations, we were able to infer a plausible intracellular kinetics of drug activity.
Materials and Methods
Cell culture and treatments
The human head and neck squamous cell carcinoma cell line UM-SCC9 is from T. E. Carey at the University of Michigan. The cells were maintained in Eagle’s Minimal Essential Medium supplemented with 10% fetal bovine serum and penicillin/streptomycin. Serum level was reduced to 1%, 24 hours before 50 ng/ml TNF-α treatment. hTNF-α was purchased from R&D Systems. Bortezomib was provided by Millennium Pharmaceuticals under a materials-cooperative research and development agreement.
Plasmids, transfection, and cell sorting
We transfected UM-SCC9 cells with EGFP-tagged p65 expression vector under a CMV promoter (a gift from M.R.H. White, University of Liverpool) (Nelson et al., 2004) and generated a stable cell line by cell sorting based on the fluorescence signal. Specifically, UM-SCC9 cells were plated on a 24-well culture plate to reach 50~70 % confluency. The p65:EGFP plasmid (0.3 µg/well) was transfected into the cells according to the Effectene Transfection Reagent (Qiagen) protocol. Cells were passaged and grown to reach 90% confluence in T-75 flasks before cell sorting. Transfected UM-SCC9 cells were sorted using a FACSAria (Becton Dickinson, San Jose, CA) equipped with Coherent Sapphire 488 blue and FACSDiva software. EGFP fluorescence was measured with a 530/30 emission filter. Sorted cells from the flow cytometry were maintained in culture with supplemented phenol red-free EMEM (Gibco). The reporter gene plasmid pIL-8:mCherry was constructed by replacing the luciferase gene within an IL-8 promoter-luciferase reporter (described in (Mukaida et al., 1994; Wolf et al., 2001)) with mCherry (from R. Tsien, UC San Diego). More specifically, the plasmid containing the complete coding sequence (pRSET-B mCherry) was kindly provided by Dr. R. Tsien (UC San Diego). The mCherry coding sequence was digested by restriction enzymes BamHI (5’) and HindIII (3’), purified, and inserted into pXP2 IL-8-133-luc plasmid (containing −133 to+44 bp portion of human IL-8 promoter sequence, under the permission by Dr. N. Mukaida, Kanazawa, Japan. (Mukaida et al., 1994; Mukaida et al., 1989; Ondrey et al., 1999)) in the HindIII site of multiple cloning sites between IL-8 promoter and luciferase reporter gene. This construct thereby drives the expression of mCherry controlled by the IL-8 promoter and was transfected into our p65:EGFP-stable cells 48 hours before imaging (Lipofectamine). The reported maturation half life of mCherry is 15 minutes which is far superior to other reporter tags which take several hours to emit signals (Shaner et al., 2004).
Transient transfection of IL-8:mCherry
The stable cell line expressing p65:EGFP was plated in 3×105 cells/ml onto a 35 mm glass bottom dish (MatTek, Ashland, MA) in antibiotic- and phenol red-free medium. 48 hours before microscopy, cells were transfected with 0.3 µg/ml plasmid DNA and 1 µl/ml Lipofectamine Transfection Agent (Invitrogen) for 4 hours using Opti-MEM (Invitrogen) in a total of 200 µl transfection volume. 3 ml of phenol red-free EMEM was added for incubation overnight. 1 % serum, phenol red-free EMEM was then applied to the transfected cells 24 hours before microscropy experiments.
Immunofluorescence
Appropriately treated cells in LabTek II 6-well chambers with coverslip bottom were fixed with 1.5% formaldehyde. Standard immunostaining protocol was performed with anti-p65 (sc-372, Santa Cruz Biotechnology, inc. Santa Cruz, CA) and donkey anti-rabbit IgG conjugated to Texas Red dye (Jackson ImmunoResearch Laboratories, inc. West Grove, PA) as the secondary Ab. Vectashield with DAPI was used as the mounting medium. Cells were imaged on an inverted epifluorescence microscope system controlled by MetaMorph (Molecular Devices), which consisted of inverted Nikon TE300 microscope with a 60X 1.4 N.A. objective (Nikon), Lambda 10–2 filter changer, and Cool Snap ES CCD camera (Roper Scientific/Photometrics). Excitation light was attenuated with a ND filter with 32% light transmission. Filters were from 86000 Sedat Quadruple Filter Set for FITC (Ex 490/20; Em 528/38; Polychroic mirror), RD-TR-PE ( Ex 555/28; Em 617/73; Polycroic mirror), and DAPI (Ex 360/40; Em 457/50, Polychroic mirror) (Chroma Technology Corp.). The DAPI signal was used to automatically detect nuclear boundaries which were then used to quantify the nuclear mean intensity of each channel. Cell boundaries were drawn manually. All quantification was done in MATLAB.
Live cell microscopy
Time lapse images were collected every 15 minutes on a Zeiss LSM510 confocal microscope with a 40x oil-immersion objective (1.3 N.A.). Focus was adjusted with the Autofocus procedure in the LSM510 software before each time point by xz line scanning (z range = 60 µm) of the reflection from the coverslip bottom, using a 633 nm laser line outside of the excitation spectra for EGFP (488 nm) and mCherry (543 nm). This autofocus setting based on coverslip reflection with no fluorophore excitation minimized the additional photobleaching and phototoxicity on the cells. Typically 1 or 2 z-sections were acquired for each time point with low laser power. Incubation condition was maintained onstage throughout the time lapse microscopy using an environmental chamber BC-500W (20/20 Technology, Inc.) with humidified 5% CO2 and 37 °C. Treatment solution (either TNF-α or Bortezomib) was added over the cell medium through an injection port and the time lapse image acquisition was started immediately. LSM files were exported as 12-bit grayscale tiff image files for further analyses.
The live cell imaging experiment in Movie S3 was generated on Nikon Biostation IM, compact incubation box with a built-in wide-field optical system. 40x air objective was used. The dish of cells was placed in an internal sealed chamber with humidified 5% CO2 and 37 °C. The 10 minute-time lapse image acquisition started 15 minutes after TNF-α treatment, because focus had to be re-adjusted manually after the perturbation introduced by opening the chamber and pipetting TNF-α into the dish (No autofocus function was available in this microscope). Nikon format files were exported to 12-bit grayscale tiff files for further analyses.
Image analysis
Background constant was subtracted from the time course images and a median filter was applied to handle speckle-type noise pixels. To quantify the nuclear and total fluorescent intensities, regions of interest were drawn manually around individual nuclear and cell boundaries. Cells that moved outside the field of imaging or those that significantly overlapped with other cells for any time point were excluded from the analysis, as well as dividing or dying cells. Region measurements for the area and the integrated intensity were extracted and minimally smoothed using locally weighted regression to remove high-frequency noise. Nuclear concentration of p65 was approximated as the mean intensity of nuclear p65:EGFP normalized by the total cellular mean intensity. The expression level of pIL-8:mCherry was represented by the cellular mean fluorescent intensity for each cell. The rate of change in mCherry expression was estimated by the following procedure (modified from a similar method in (Friedman et al., 2005)) to avoid the photo-toxicity inherent in repeated whole-cell photobleaching: To estimate the time derivative of mCherry expression, we smoothed the time course data for the mCherry intensity and the cell volume using locally weighted regression. The reporter expression rate was then defined as the difference of smoothed mean mCherry intensities between the current and previous time points. All analysis methods were implemented using our custom-written programs in MetaMorph (Universal Imaging Corporation, Molecular Devices), R (http://www.r-project.org), and MATLAB (The MathWorks).
Mathematical modeling and computational simulations
The basic model of NF-κB regulation described in (Sung and Simon, 2004) was used for simulations of various kinetic profiles of intracellular bortezomib. Dynamical models in such a form of differential equations encode the interaction kinetics between essential pathway components. Briefly, the model includes reaction kinetics representing: activation of IKK, reversible binding of NF-κB to IκBα and to IKK, IKK-regulated degradation of IκBα by the ubiquitin-proteasome pathway, constitutive degradation of IκBα, nuclear import/export of NF-κB and IκBα, and the delayed synthesis of IκBα induced by NF-κB. Even though our model is based on a simplified description of the pathway with numerous omissions, it can produce simulations that match the observed single cell behavior reasonably well (Nelson et al., 2005). Sensitivity analysis of a more complicated NF-κB model through extensive variation of parameter values indicated that the characteristics of the system dynamics are captured by a few key pathway components (A.E.C. Ihekwaba, 2004), supporting the utility of a simple model. The simulation setting for approximating the inhibition dynamics from bortezomib was specified to match our inhibition experiment. The inhibitor was introduced after 10 hours of constitutive activation of NF-κB by IKK. Time delay between nuclear NF-κB activation and IκBα protein synthesis was set to be 60 min as it produced a good agreement between the simulated and the observed time course of p65. Various pharmacodynamic profiles were simulated including variants from (Sung and Simon, 2004). All computations were carried out using MATLAB.
Results
TNF-α induces oscillations of the level of nuclear NF-κB and the dynamics are detectable only by live microscopy
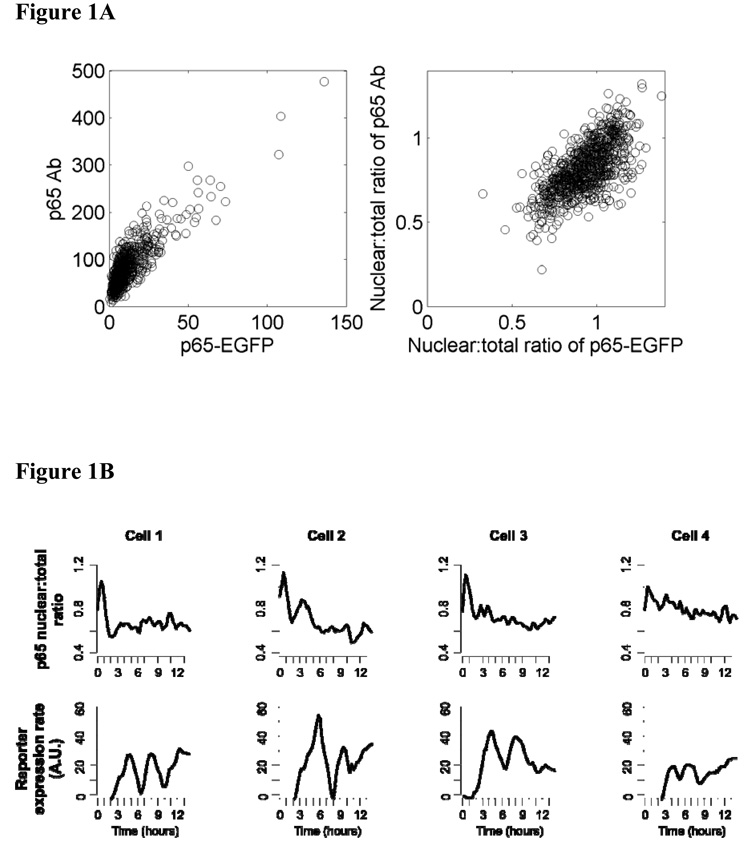
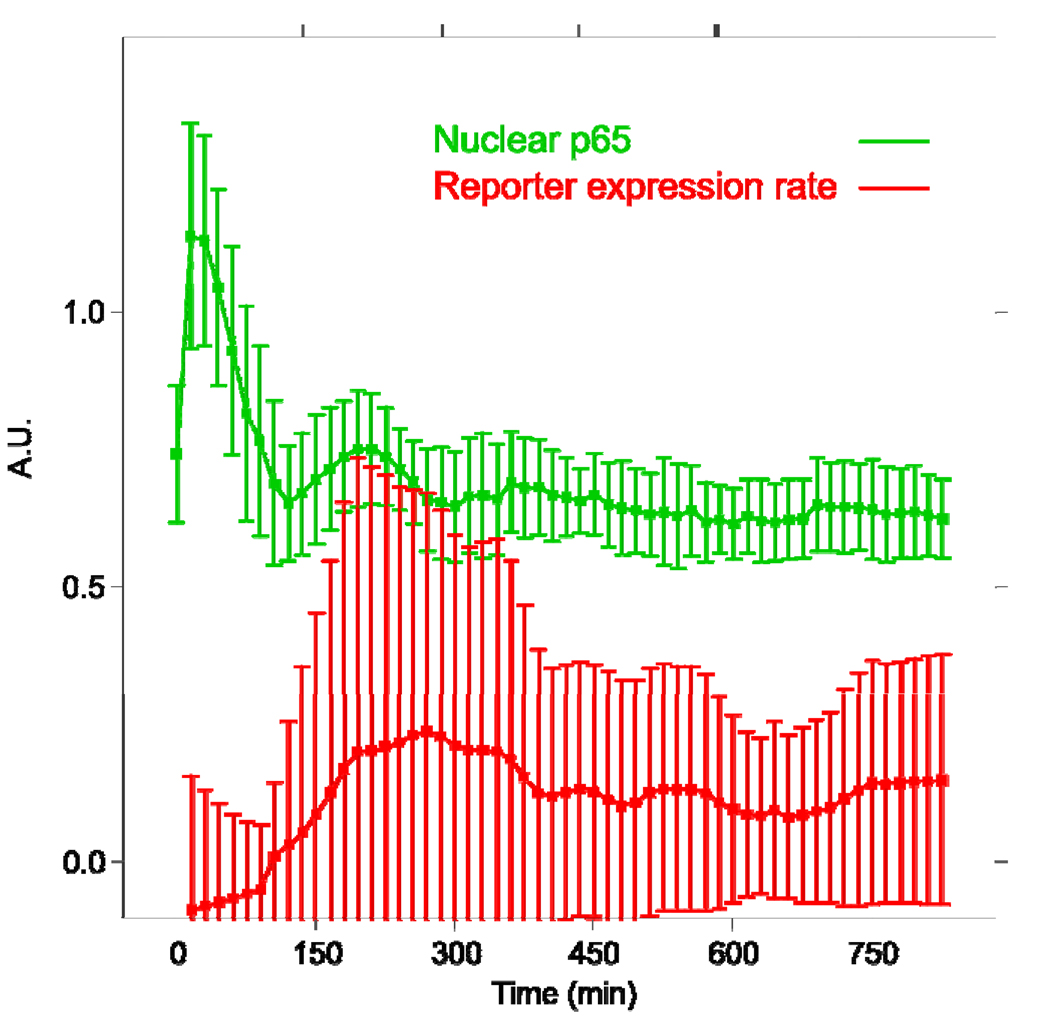
We examined real time kinetics of NF-κB in HNSCC cells UM-SCC9 that stably express p65:EGFP using time lapse fluorescence microscopy. p65 is a transactivation subunit of NF-κB that forms the classical heterodimer p65/p50. We attempted to derive stable lines from other cell lines in the UM-SCC series as well, but were successful only with UM-SCC9, whose endogenous p65 expression was the lowest among them. Perhaps the cell lines expressing a constitutively high level of p65 could not tolerate the ectopic expression of p65:GFP. We then first verified that the level and the nuclear translocation of the measured p65:EGFP closely approximate those of the endogenous p65 in individual cells (Figure 1A; see Supplemental Data Figure S4). Next we confirmed that the nuclear level of p65 oscillates after TNF-α stimulation in the cell line UM-SCC9 as reported for other cell systems (Hoffmann et al., 2002; Nelson et al., 2004). There was an observable damped oscillation of NF-κB in most cells (Figure 1B, upper panel; Figure S2, upper panel; Movie S1, Movie S3). The initial peak nuclear translocation was the strongest and it started before imaging of some cells (Figure 1B, cells 1, 2), making the first trough appear slightly lower than the p65 nuclear:total ratio at the first time point. The cyclic peaks of the oscillations in individual cells were not synchronized, resulting in a typical one-peak activity curve in the population average plot (Figure 1C). Large standard deviations of the cells from the population average value demonstrate that this averaged time course is not representative of the constituent cells at any given time. This confirms that temporal dynamics of living cells cannot be appreciated in assays based on measurements from cell populations.
Figure 1. Both nuclear NF-κB and target protein expression are oscillatory in single living cells and this dynamics is not detectable in measurements from cell populations.
A. Cells treated with TNF-α for 0, 30, 90, 150, 240 min were fixed and stained with antibody to p65. The GFP signal and immunofluorescence channels were imaged and quantified separately to estimate the p65-EGFP and total (endogenous + fusion) levels, respectively, in single cells. The correlation between GFP and immunofluorescence was 0.85 and 0.68 for mean cellular intensities and nuclear:total ratios, respectively. All data are pooled in the plots (n = 873 cells) since individual time point data showed similar correlations. B. Nuclear p65 level exhibits damped oscillations and the reporter gene has discrete expression pulses in individual UM-SCC9 cells treated with 50 ng/ml TNF-α. The upper panel shows the nuclear to total ratio of p65:EGFP mean fluorescence intensities. The lower panel presents the rate of expression change extracted from the mCherry time course (see Methods). The quantifications are from the cells shown in Supplementary Figure S1 and the time lapse images are presented in the Supplementary Movie S1. C. Population average of nuclear p65 level and IL-8 reporter induction rate from all cells in view. The green curve is the average of nuclear mean intensities for p65:EGFP. The average of the mCherry expression rates is shown by the red curve. Bars represent standard deviations.
Peaks in NF-κB oscillations are correlated with reporter expression pulses
The expression time course of an NF-κB inducible reporter, pIL-8:mCherry, was monitored in parallel as a means of assessing whether the later nuclear p65 peaks were functional and correlated with gene regulation. Interleukin-8 (IL-8) is a cytokine induced by NF-κB with physiological effects ranging from angiogenesis to inflammation. We have previously shown that NF-κB is constitutively activated and is a critical regulator of IL-8 gene expression in this and other UM-SCC cell lines as well as primary tumor tissues for HNSCC (Chen et al., 1999; Duffey et al., 1999; Wolf et al., 2001). A monomer red fluorescent protein mCherry was chosen for its photostability and quick maturation time, making it an excellent reporter for monitoring expression kinetics (Shaner et al., 2004). Therefore, the rate of change in the mCherry fluorescence intensity closely approximates the induction or repression of the IL-8 promoter. Calculating this rate of change involves subtracting the signal from the previous time point and therefore this method is akin to removing the previous signal by photobleaching techniques. Our method of distinguishing the signal corresponding to the newly produced reporter molecules, however, is non-invasive because it teases out the de novo synthesis population at the image analysis step (see Methods). On the other hand, parallel monitoring of the reporter and p65:EGFP was challenging for several reasons. First, our attempts to generate a stable cell line expressing the reporter out of the p65:EGFP cells were unsuccessful. As a consequence, it was difficult to efficiently generate time lapse imaging data due to a low transient transfection efficiency of the reporter construct, coupled with a lack of marker to identify the cells transfected with the inducible reporter (before TNF-α induction) at the start of imaging.
The IL-8 promoter-driven reporter exhibited multiple production surges in cells treated with TNF-α, reflected in the discretely timed pulses in the reporter expression rate (Figure 1B, lower panel). The expression rate quantification procedure applied to cells that were not transfected with the reporter construct did not produce any expression bursts (Figure S2). The asynchronous reporter expression pulses were completely undetectable in the population-averaged measurements (Figure 1C), again highlighting that single cell time course data are necessary to capture real time dynamics. Instead the population average showed a typical kinetic profile that reaches a plateau level after activation.
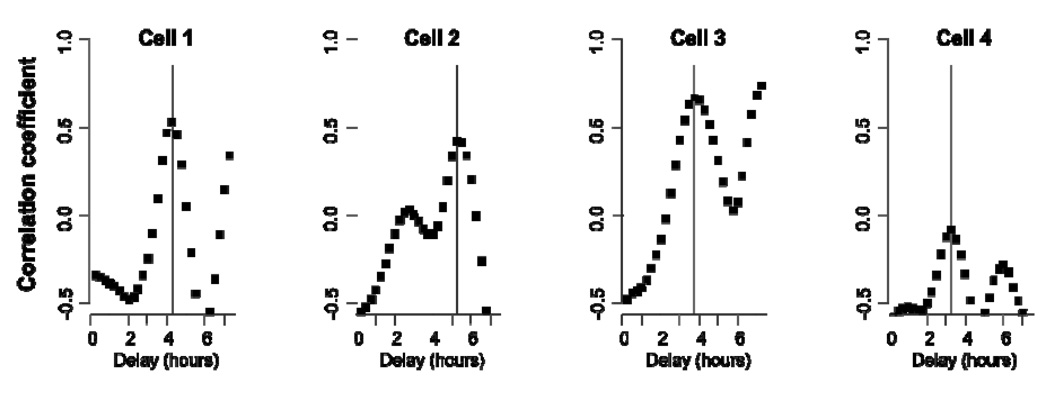
The expression rate had an oscillatory time course with roughly the same period as that of p65 oscillation, apparently echoing the oscillation of p65. We computed the correlation coefficients between p65 and reporter time course data, using varying time delays. Figure 2 shows that the correlation between the two processes is maximal at a distinct cell-dependent delay value. Our finding supports that oscillatory gene expression dynamics is detectable and correlated with dynamic NF-κB activity, even at the level of protein. The tight relationship also highlights the sensitive control of IL-8 gene expression by NF-κB over other factors, as the promoter may contain sites for other factors such as AP-1 (Mukaida et al., 1994).
Figure 2. Correlation between nuclear NF-κB peaks and pulses of target reporter expression.
The plot shows the correlation coefficient between the nuclear p65 time course and the pIL-8:mCherry reporter expression rate data shifted by the delay time labeled on the x-axis. The vertical line marks the time delay that gives a maximal correlation. By nature of periodicity, integer multiples of this delay time also produce significant correlations. The time course data and cell labeling correspond to those used in Figure 1B.
Bortezomib induces distinct reporter expression pulses while it attenuates the overall level of NF-κB activity
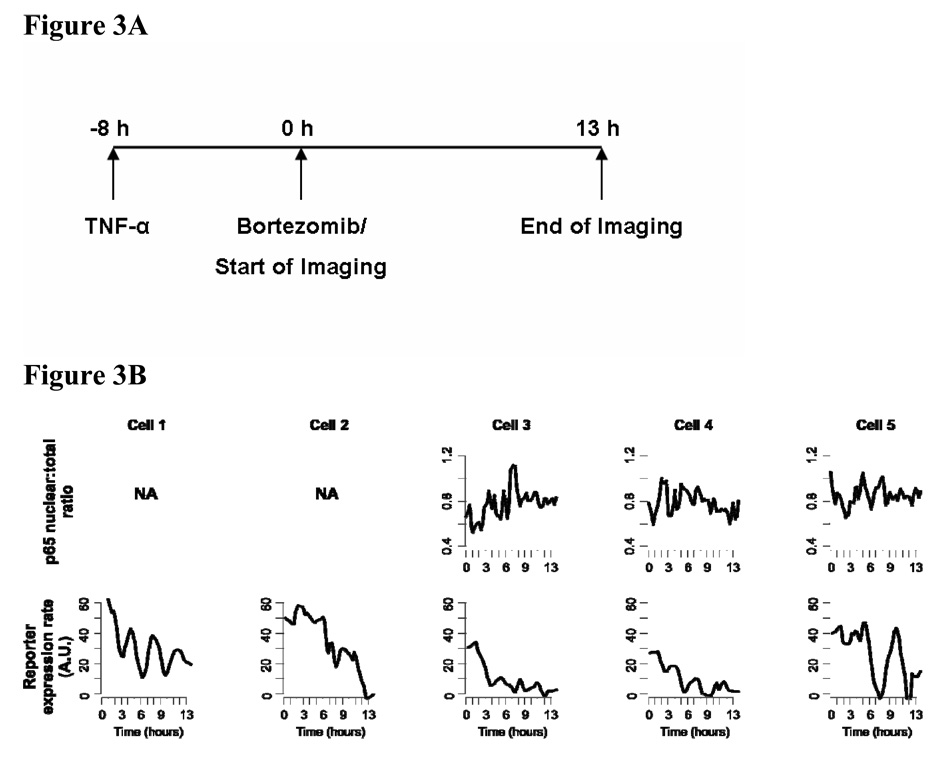
Next we investigated how the ongoing dynamic activities of NF-κB can be disrupted by an inhibitor of NF-κB. Among its reported mechanisms of action, NF-κB is thought to be a major target pathway for Bortezomib as it inhibits the degradation of IκBα proteins in the canonical activation pathway for NF-κB. Bortezomib was previously found to inhibit NF-kB activation in UM-SCC9 and tumor specimens from patients (Sunwoo et al., 2001; Van Waes et al., 2005). We designed our inhibition experiment to test the effect of the drug treatment on cancer cells with pre-existing activity of NF-κB. Therefore drug treatment and live cell microscopy were started in cells pretreated with TNF-α (Figure 3A). We used a low drug concentration that has minimal cytotoxicity to assess its specific effect on gene expression and also to emulate a sub-optimal drug concentration at the target cells due to pharmacokinetic constraints in vivo.
Figure 3. NF-κB activity maintains its natural oscillatory dynamics while being down-modulated by an inhibitor.
A. The protocol timeline for NF-κB activation by TNF-α (50 ng/ml), bortezomib treatment, and live cell imaging. Bortezomib concentration in all inhibitor experiments was 20 ng/ml. B. Quantified microscopy data in the same format as in Figure 1B. Some cells do not show robust signal from both fluorescence channels above the detection limit. The data is from a time course imaging that is representative of five separate experiments.
The induction kinetics of IL-8 showed decreased overall expression rate in most cells after Bortezomib treatment (Figure 3B, lower panel), whereas, in control experiments without the inhibitor, reporter induction continued for 2–3 days (data not shown). Importantly, during this overall modulation the reporter expression pulses persisted in many cells, as predicted by our theoretical model (Sung and Simon, 2004). Cells without detectable signal from the transiently transfected reporter did not show such a time course profile (Movie S2). Cells 1 through 5 had little or no p65:EGFP signals, complicating the interpretation of the quantified nuclear p65 level (Figure 3B, upper panel; Movie S2).
Nonetheless, the reporter expression consistently demonstrated that the transcriptional activity of NF-κB was clearly affected by the inhibitor. The inhibition dynamics for all cells could be summarized as a ‘down-trend oscillation’, as seen by the reporter kinetics (Figure 3B, lower panel). This pattern applied to cells with seemingly differing efficiency of transcription/translation machineries (Note the different absolute levels of expression rates in cells 1, 2, 5 versus cells 3, 4). While Bortezomib may also influence other cellular factors, it did not significantly inhibit ERK or AP-1 activation, or STAT3, when HNSCC tumor samples were assayed 24 hours post-treatment (Allen et al., 2008). Therefore, we focused here on its effect upon NF-κB and attempted to minimize secondary effects by a low drug concentration and an instant, not pre-, treatment. Unlike typical inhibitor experiments where the cells are pre-conditioned on the drug for sufficiently long, we assessed a physiological perturbation of NF-κB activity by an inhibitor. This interactive assay showed that the oscillatory mode of NF-κB action is still maintained while the overall level is being modulated by Bortezomib.
Estimation of the intracellular drug activity through quantitative in silico systems modeling of inhibition dynamics
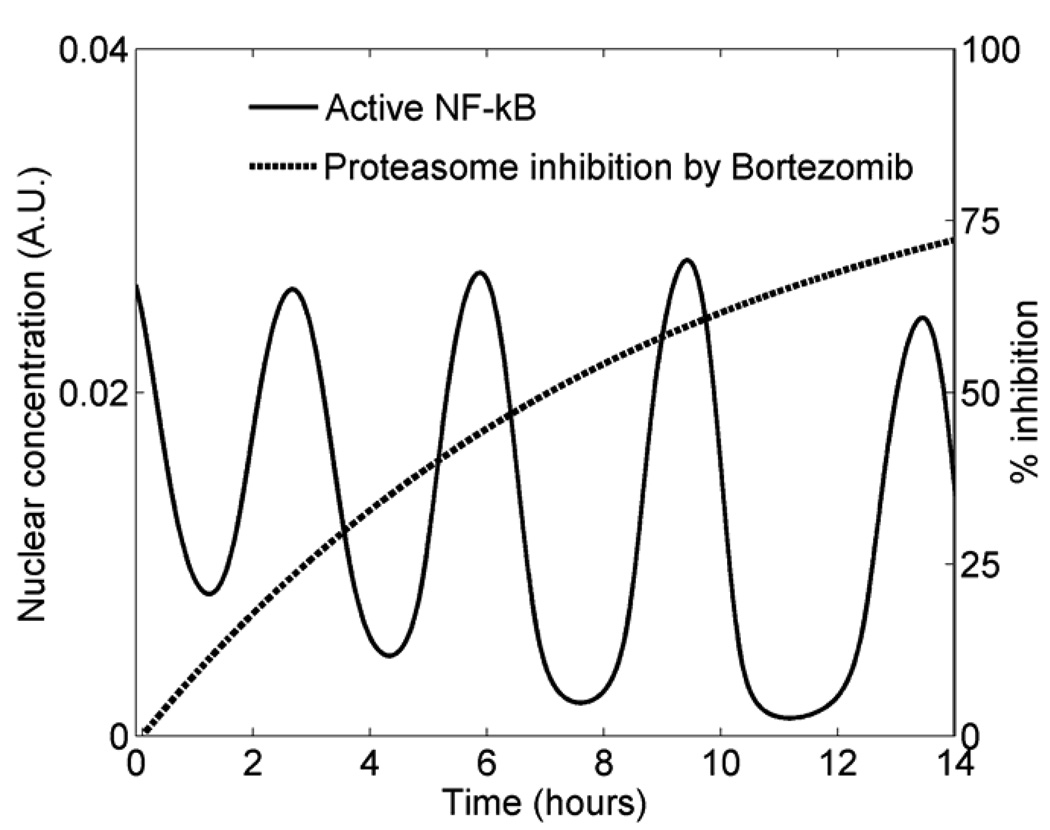
The inhibition dynamics after drug treatment also enabled us to estimate the time course of the intracellular drug effect in individual cells, a rarely measured quantity. Through simulations of various ‘intracellular pharmacodynamic profiles’ within the previously developed modeling framework for NF-κB (Sung and Simon, 2004), we found that the observed inhibition dynamics is consistent with a saturation kinetics during this time window (Figure 4). Given the cell-cell variability, experimental noise, and other unknowns, we opted for a qualitative comparison of the simulations and the microscopy data. The maximum inhibition level and the half-time (time to reach 50% of the maximum level) used to produce the simulation shown in Figure 4 were 90% and 6 hours, respectively. This inferred kinetic profile fits within the available pharmacodynamic data of bortezomib that indicates detectable proteasome inhibition in the tumor up to 48 hours after drug treatment (Van Waes et al., 2005).
Figure 4. Saturating intracellular pharmacodynamics is consistent with NF-κB inhibition dynamics via quantitative modeling.
The solid curve represents the concentration of nuclear NF-κB that is not bound by IκBα from simulating a dynamical model (Sung and Simon, 2004). The red curve shows the kinetics of % proteasome inhibition by bortezomib (introduced at t = 0) that was used to produce the NF-κB time course. The simulation setting is described in Methods.
Discussion
Our study demonstrates that the effect of a specific inhibitor that targets an oncogenic pathway can be complex in cancer cells. The ongoing NF-κB activity in HNSCC cells is highly dynamic and the interaction of Bortezomib with the pathway does not result in a simple ‘blockade’. Reporter expression kinetics reflect the dynamic oscillations of NF-κB as shown by real time microscopy of single cells. Instead of blocking the pathway activity, the drug treatment induces a distinct signaling effect, and expression of certain endogenous target genes may be transiently enhanced by the oscillatory activity of NF-κB after Bortezomib treatment. Computational modeling of the NF-κB regulatory network predicts similar non-intuitive effects for inhibitors targeting other steps of the pathway (Sung and Simon, 2004). These phenomena may constrain a ‘clean’ inhibition of NF-κB by specific inhibitors, including IKK inhibitors. Therefore, further studies will be necessary for other classes of targeted inhibitors to examine their in vivo effects. We note that there might be some differences between our cells and primary tumor tissues, such as microenvironment and/or cell-cell adhesion effects. But the lack of real time dynamic assays suitable for primary tumor cells merits the use of cell lines expressing GFP fusion proteins, especially because little is understood about the dynamic systems aspect of cell signaling, and knowledge gained from real time studies can potentially provide unique insights to numerous translational and therapeutic efforts.
We also illustrated that unsynchronized temporal phenomena are mostly missed by conventional assays. Even single cell assays based on time snapshots such as immunofluorescence are also significantly limited because reconstruction of individual real time dynamics is complicated by the natural cell-to-cell variability. Our live cell imaging techniques captured temporal patterns sensitively also by employing rapidly maturing fluorescent reporter in combination with analysis methods that minimized photo-damage on imaged cells. While there is an increasing use of genomic/proteomic approaches for identifying clinically relevant genes, results from such assays are complicated by the phenomenon highlighted here. The pre-/post-treatment gene activity may be temporally complex and missed in cell population average measurements (as illustrated in Fig. 1C), and yet the in vivo consequence may be rather significant.
Some features of evaluating gene regulation with reporter proteins are noteworthy. The stage of folded proteins is a physiological readout as it represents the final product of gene expression that is capable of its intended cellular function. However, protein dynamics is not as direct a measurement as mRNA abundance because RNA processing, translation, protein folding kinetics, and protein degradation all probably smooth out ‘rough edges’ in transcription kinetics (Raj et al., 2006). Therefore the extent of oscillation in our reporter induction rate is likely to be an underestimate of the dynamic nature of transcription controlled by NF-κB in living cells.
The oscillation in NF-κB transcriptional activity is largely a consequence of the delayed negative feedback loop provided by IκBα (Hoffmann et al., 2002; Sung, 2008). The transient peaks in NF-κB oscillations, although subtle, were capable of eliciting functional gene expression pulses from the pIL-8 reporter, even after inhibitor treatment. Discrete pulses of gene transcription in single cells has been reported previously (Chubb et al., 2006; Friedman et al., 2005; Shorte et al., 2002; Yu et al., 2006) and our results show how such expression dynamics can be linked to the periodic activity of a transcription factor. This is consistent with a multi-threshold switch relating stimulus duration to gene production, contrary to the view of monotonous signal accumulation. The oscillation of NF-κB activity may be a robust feature as it persists even under inhibitory perturbations. The distinct dynamical features of NF-κB signaling may allow diverse activation modes, thereby eliciting differential gene induction responses. This extends the role of a classic negative feedback mechanism from a signaling terminator to an active participant of distinct gene regulation.
Our findings question the notion of ‘pathway blockade’ that ignores the intricate regulatory network inherent for most inducible transcription factors. We propose the ‘inhibition dynamics’ on the in vivo effects of targeted inhibitors as a requisite issue for therapeutic strategies. The drug-induced signaling dynamics is functionally relevant as shown by its relationship to gene expression kinetics. With an increasing set of therapeutically relevant intracellular signaling pathways operating with similar temporal complexity (Dolmetsch et al., 1998; Dyachok et al., 2006; Hirata et al., 2002; Lahav et al., 2004; Violin et al., 2003), it will be important to re-evaluate more cellular processes using techniques suitable to detect real time dynamics and assess the signaling consequences of perturbing the complex oncogenic interaction networks in cancer cells.
Supplementary Material
Acknowledgments
We thank John Rendina (20/20 Technology, Inc.) for modifying the environmental chamber for our live cell microscopy experiments. We thank Ning T. Yeh, Padma Sheila Rajagopal, and Mindy Lin for the technical support. We also thank Tom Misteli for his comments on an early version of this manuscript.
This work was supported by NIDCD intramural project Z01-DC-00016 and by the NIH Intramural Research Program for Center for Cancer Research, National Cancer Institute. Dr. Van Waes has received Bortezomib and clinical trial support under an NIH approved Cooperative Research and Development Agreement with Millennium Pharmaceuticals.
Abbreviations
- NF-κB
nuclear factor kappa B
- TNF-α
tumor necrosis factor alpha
- IκB
inhibitor of kappa B
- IKK
IκB kinase
- HNSCC
head and neck squamous cell carcinoma
- EGFP
enhanced green fluorescent protein
Contributor Information
Myong-Hee Sung, Laboratory of Receptor Biology and Gene Expression, National Cancer Institute, National Institutes of Health, Bethesda, MD 20892.
Lorena Bagain, Head and Neck Surgery Branch, National Institute on Deafness and Communication Disorders, National Institutes of Health, Bethesda, MD 20892.
Zhong Chen, Head and Neck Surgery Branch, National Institute on Deafness and Communication Disorders, National Institutes of Health, Bethesda, MD 20892.
Tatiana Karpova, Laboratory of Receptor Biology and Gene Expression, National Cancer Institute, National Institutes of Health, Bethesda, MD 20892.
Xinping Yang, Head and Neck Surgery Branch, National Institute on Deafness and Communication Disorders, National Institutes of Health, Bethesda, MD 20892.
Christopher Silvin, Genetics and Molecular Biology Branch, National Human Genome Research Institute, National Institutes of Health, Bethesda, MD 20892.
Ty Voss, Laboratory of Receptor Biology and Gene Expression, National Cancer Institute, National Institutes of Health, Bethesda, MD 20892.
James McNally, Laboratory of Receptor Biology and Gene Expression, National Cancer Institute, National Institutes of Health, Bethesda, MD 20892.
Carter Van Waes, Head and Neck Surgery Branch, National Institute on Deafness and Communication Disorders, National Institutes of Health, Bethesda, MD 20892.
Gordon L. Hager, Laboratory of Receptor Biology and Gene Expression, National Cancer Institute, National Institutes of Health, Bethesda, MD 20892
References
- Ihekwaba DSB AEC, Grimley RL, Benson N, Kell DB. Sensitivity analysis of parameters controlling oscillatory signalling in the NF-kappaB pathway: the roles of IKK and IkappaBalpha. Systems Biology. 2004;1(1):93–103. doi: 10.1049/sb:20045009. [DOI] [PubMed] [Google Scholar]
- Aggarwal BB. Nuclear factor-kappaB: the enemy within. Cancer Cell. 2004;6(3):203–208. doi: 10.1016/j.ccr.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Allen C, Saigal K, Nottingham L, Arun P, Chen Z, Van Waes C. Bortezomib-induced apoptosis with limited clinical response is accompanied by inhibition of canonical but not alternative nuclear factor-{kappa}B subunits in head and neck cancer. Clin Cancer Res. 2008;14(13):4175–4185. doi: 10.1158/1078-0432.CCR-07-4470. [DOI] [PubMed] [Google Scholar]
- Chang AA, Van Waes C. Nuclear factor-KappaB as a common target and activator of oncogenes in head and neck squamous cell carcinoma. Adv Otorhinolaryngol. 2005;62:92–102. doi: 10.1159/000082476. [DOI] [PubMed] [Google Scholar]
- Chen Z, Malhotra PS, Thomas GR, Ondrey FG, Duffey DC, Smith CW, Enamorado I, Yeh NT, Kroog GS, Rudy S, McCullagh L, Mousa S, Quezado M, Herscher LL, Van Waes C. Expression of proinflammatory and proangiogenic cytokines in patients with head and neck cancer. Clin Cancer Res. 1999;5(6):1369–1379. [PubMed] [Google Scholar]
- Chubb JR, Trcek T, Shenoy SM, Singer RH. Transcriptional pulsing of a developmental gene. Curr Biol. 2006;16(10):1018–1025. doi: 10.1016/j.cub.2006.03.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolmetsch RE, Xu K, Lewis RS. Calcium oscillations increase the efficiency and specificity of gene expression. Nature. 1998;392(6679):933–936. doi: 10.1038/31960. [DOI] [PubMed] [Google Scholar]
- Duffey DC, Chen Z, Dong G, Ondrey FG, Wolf JS, Brown K, Siebenlist U, Van Waes C. Expression of a dominant-negative mutant inhibitor-kappaBalpha of nuclear factor-kappaB in human head and neck squamous cell carcinoma inhibits survival, proinflammatory cytokine expression, and tumor growth in vivo. Cancer Res. 1999;59(14):3468–3474. [PubMed] [Google Scholar]
- Dyachok O, Isakov Y, Sagetorp J, Tengholm A. Oscillations of cyclic AMP in hormone-stimulated insulin-secreting beta-cells. Nature. 2006;439(7074):349–352. doi: 10.1038/nature04410. [DOI] [PubMed] [Google Scholar]
- Friedman N, Vardi S, Ronen M, Alon U, Stavans J. Precise temporal modulation in the response of the SOS DNA repair network in individual bacteria. PLoS Biol. 2005;3(7):e238. doi: 10.1371/journal.pbio.0030238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrichsen S, Harper CV, Semprini S, Wilding M, Adamson AD, Spiller DG, Nelson G, Mullins JJ, White MR, Davis JR. Tumor necrosis factor-alpha activates the human prolactin gene promoter via nuclear factor-kappaB signaling. Endocrinology. 2006;147(2):773–781. doi: 10.1210/en.2005-0967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguli A, Persson L, Palmer IR, Evans I, Yang L, Smallwood R, Black R, Qwarnstrom EE. Distinct NF-kappaB regulation by shear stress through Ras-dependent IkappaBalpha oscillations: real-time analysis of flow-mediated activation in live cells. Circ Res. 2005;96(6):626–634. doi: 10.1161/01.RES.0000160435.83210.95. [DOI] [PubMed] [Google Scholar]
- Greten FR, Eckmann L, Greten TF, Park JM, Li ZW, Egan LJ, Kagnoff MF, Karin M. IKKbeta links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell. 2004;118(3):285–296. doi: 10.1016/j.cell.2004.07.013. [DOI] [PubMed] [Google Scholar]
- Hirata H, Yoshiura S, Ohtsuka T, Bessho Y, Harada T, Yoshikawa K, Kageyama R. Oscillatory expression of the bHLH factor Hes1 regulated by a negative feedback loop. Science. 2002;298(5594):840–843. doi: 10.1126/science.1074560. [DOI] [PubMed] [Google Scholar]
- Hoffmann A, Levchenko A, Scott ML, Baltimore D. The IkappaB-NF-kappaB signaling module: temporal control and selective gene activation. Science. 2002;298(5596):1241–1245. doi: 10.1126/science.1071914. [DOI] [PubMed] [Google Scholar]
- Jackson-Bernitsas DG, Ichikawa H, Takada Y, Myers JN, Lin XL, Darnay BG, Chaturvedi MM, Aggarwal BB. Evidence that TNF-TNFR1-TRADD-TRAF2-RIP-TAK1-IKK pathway mediates constitutive NF-kappaB activation and proliferation in human head and neck squamous cell carcinoma. Oncogene. 2006 doi: 10.1038/sj.onc.1209945. [DOI] [PubMed] [Google Scholar]
- Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441(7092):431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- Lahav G, Rosenfeld N, Sigal A, Geva-Zatorsky N, Levine AJ, Elowitz MB, Alon U. Dynamics of the p53-Mdm2 feedback loop in individual cells. Nat Genet. 2004;36(2):147–150. doi: 10.1038/ng1293. [DOI] [PubMed] [Google Scholar]
- Li Q, Withoff S, Verma IM. Inflammation-associated cancer: NF-kappaB is the lynchpin. Trends Immunol. 2005;26(6):318–325. doi: 10.1016/j.it.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Mukaida N, Okamoto S, Ishikawa Y, Matsushima K. Molecular mechanism of interleukin-8 gene expression. J Leukoc Biol. 1994;56(5):554–558. [PubMed] [Google Scholar]
- Mukaida N, Shiroo M, Matsushima K. Genomic structure of the human monocyte-derived neutrophil chemotactic factor IL-8. J Immunol. 1989;143(4):1366–1371. [PubMed] [Google Scholar]
- Nelson DE, Horton CA, See V, Johnson JR, Nelson G, Spiller DG, Kell DB, White MRH. Response to comment on ″Oscillations in NF-kappaB signaling control the dynamics of gene expression″. Science. 2005;308:52b. doi: 10.1126/science.1099962. [DOI] [PubMed] [Google Scholar]
- Nelson DE, Ihekwaba AE, Elliott M, Johnson JR, Gibney CA, Foreman BE, Nelson G, See V, Horton CA, Spiller DG, Edwards SW, McDowell HP, Unitt JF, Sullivan E, Grimley R, Benson N, Broomhead D, Kell DB, White MR. Oscillations in NF-kappaB signaling control the dynamics of gene expression. Science. 2004;306(5696):704–708. doi: 10.1126/science.1099962. [DOI] [PubMed] [Google Scholar]
- Ondrey FG, Dong G, Sunwoo J, Chen Z, Wolf JS, Crowl-Bancroft CV, Mukaida N, Van Waes C. Constitutive activation of transcription factors NF-(kappa)B, AP-1, and NF-IL6 in human head and neck squamous cell carcinoma cell lines that express pro-inflammatory and pro-angiogenic cytokines. Mol Carcinog. 1999;26(2):119–129. doi: 10.1002/(sici)1098-2744(199910)26:2<119::aid-mc6>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Pahl HL. Activators and target genes of Rel/NF-kappaB transcription factors. Oncogene. 1999;18(49):6853–6866. doi: 10.1038/sj.onc.1203239. [DOI] [PubMed] [Google Scholar]
- Pikarsky E, Porat RM, Stein I, Abramovitch R, Amit S, Kasem S, Gutkovich-Pyest E, Urieli-Shoval S, Galun E, Ben-Neriah Y. NF-kappaB functions as a tumour promoter in inflammation-associated cancer. Nature. 2004;431(7007):461–466. doi: 10.1038/nature02924. [DOI] [PubMed] [Google Scholar]
- Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S. Stochastic mRNA Synthesis in Mammalian Cells. PLoS Biol. 2006;4(10) doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson PG, Mitsiades C, Hideshima T, Anderson KC. Proteasome inhibition in the treatment of cancer. Cell Cycle. 2005;4(2):290–296. [PubMed] [Google Scholar]
- Rosenwald A, Staudt LM. Gene expression profiling of diffuse large B-cell lymphoma. Leuk Lymphoma. 2003;44(Suppl 3):S41–S47. doi: 10.1080/10428190310001623775. [DOI] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22(12):1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Shorte SL, Leclerc GM, Vazquez-Martinez R, Leaumont DC, Faught WJ, Frawley LS, Boockfor FR. PRL gene expression in individual living mammotropes displays distinct functional pulses that oscillate in a noncircadian temporal pattern. Endocrinology. 2002;143(3):1126–1133. doi: 10.1210/endo.143.3.8682. [DOI] [PubMed] [Google Scholar]
- Sung MH, De Lorenzi R, Salvatore L, Hendarwanto A, Hager GL, Bianchi ME, Pasparakis M, Agresti A. Manuscript in preparation. 2008 [Google Scholar]
- Sung MH, Simon R. In silico simulation of inhibitor drug effects on nuclear factor-kappaB pathway dynamics. Mol Pharmacol. 2004;66(1):70–75. doi: 10.1124/mol.66.1.70. [DOI] [PubMed] [Google Scholar]
- Sunwoo JB, Chen Z, Dong G, Yeh N, Crowl Bancroft C, Sausville E, Adams J, Elliott P, Van Waes C. Novel proteasome inhibitor PS-341 inhibits activation of nuclear factor-kappa B, cell survival, tumor growth, and angiogenesis in squamous cell carcinoma. Clin Cancer Res. 2001;7(5):1419–1428. [PubMed] [Google Scholar]
- Van Waes C. Nuclear factor-kappaB in development, prevention, and therapy of cancer. Clin Cancer Res. 2007;13(4):1076–1082. doi: 10.1158/1078-0432.CCR-06-2221. [DOI] [PubMed] [Google Scholar]
- Van Waes C, Chang AA, Lebowitz PF, Druzgal CH, Chen Z, Elsayed YA, Sunwoo JB, Rudy SF, Morris JC, Mitchell JB, Camphausen K, Gius D, Adams J, Sausville EA, Conley BA. Inhibition of nuclear factor-kappaB and target genes during combined therapy with proteasome inhibitor bortezomib and reirradiation in patients with recurrent head-and-neck squamous cell carcinoma. Int J Radiat Oncol Biol Phys. 2005;63(5):1400–1412. doi: 10.1016/j.ijrobp.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Violin JD, Zhang J, Tsien RY, Newton AC. A genetically encoded fluorescent reporter reveals oscillatory phosphorylation by protein kinase C. J Cell Biol. 2003;161(5):899–909. doi: 10.1083/jcb.200302125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf JS, Chen Z, Dong G, Sunwoo JB, Bancroft CC, Capo DE, Yeh NT, Mukaida N, Van Waes C. IL (interleukin)-1alpha promotes nuclear factor-kappaB and AP-1-induced IL-8 expression, cell survival, and proliferation in head and neck squamous cell carcinomas. Clin Cancer Res. 2001;7(6):1812–1820. [PubMed] [Google Scholar]
- Yu J, Xiao J, Ren X, Lao K, Xie XS. Probing gene expression in live cells, one protein molecule at a time. Science. 2006;311(5767):1600–1603. doi: 10.1126/science.1119623. [DOI] [PubMed] [Google Scholar]
- Zavrski I, Jakob C, Schmid P, Krebbel H, Kaiser M, Fleissner C, Rosche M, Possinger K, Sezer O. Proteasome: an emerging target for cancer therapy. Anticancer Drugs. 2005;16(5):475–481. doi: 10.1097/00001813-200506000-00002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.