Abstract
Currently, almost all FDA approved therapeutic antibodies (except ReoPro, Lucentis and Cimzia which are Fabs), and the vast majority of those in clinical trials are full-size antibodies mostly in IgG1 format of about 150 kDa size. A fundamental problem for such large molecules is their poor penetration into tissues (e.g., solid tumors) and poor or absent binding to regions on the surface of some molecules (e.g., on the HIV envelope glycoprotein) which are fully accessible only by molecules of smaller size. Therefore, much work especially during the last decade has been aimed at developing novel scaffolds of much smaller size and high stability. Here I briefly describe a proposition to use the immunoglobulin (Ig) constant CH2 domain (CH3 for IgE and IgM) as a scaffold. CH2 is critical for the Ig effector functions. Isolated CH2 is stable monomer in contrast to all other constant domains and most of the variable domains. CH2 and engineered CH2 domains with improved stability can be used as scaffolds for construction of libraries containing diverse binders to various antigens. Such binders based on a CH2 scaffold could also confer some effector functions. Because the CH2 domains are the smallest independently folded antibody domains that can be engineered to contain simultaneously antigen-binding sites and binding sites mediating effector and stability functions, and to distinguish them from domain antibodies which are used to denote engineered VH or VL domains or nanobodies which are used to denote camelid VHH, I termed them nanoantibodies (nAbs).
Key words: CH2, nanoantibodies, scaffold, Fabs, antibodies
Antibody-based therapeutics currently enjoy unprecedented success, growth and recognition of their potential; 22 monoclonal antibodies (mAbs) are approved in the United States for clinical use, hundreds are in clinical trials for treatment of various diseases including cancers, immune disorders and infections, and second and third generations of antibodies are under development, mostly to improve already existing antibody specificities. However, this success mostly reflects discoveries made one-two decades ago. During the last decade the basic concepts and methodologies for antibody generation have not changed significantly but have been applied to develop antibodies to numerous new targets. Is it possible to produce conceptually new antibodies able to resolve long-standing problems including efficient oral delivery, efficient penetration into solid tumors and low cost of production which are major drawbacks of antibodies in comparison to small molecules?
Although I do not expect fundamental changes in the current paradigm of research and development, one area where I could forsee conceptually novel antibody-based candidate therapeutics although within the current paradigm is going beyond traditional full size antibody structures. Currently, almost all FDA approved therapeutic antibodies (except ReoPro, Lucentis, and Cimzia which are Fabs), and the vast majority of those in clinical trials are full-size antibodies mostly in IgG1 format of about 150 kDa size. A fundamental problem for such large molecules is their poor penetration into tissues (e.g., solid tumors) and poor or absent binding to regions on the surface of some molecules (e.g., on the HIV envelope glycoprotein) which are accessible by molecules of smaller size. Therefore, a large amount of work especially during the last decade has been aimed at developing novel scaffolds of much smaller size and high stability. Such scaffolds are based on various human and non-human molecules of high stability.1–8 Of those the domain antibodies (dAbs) are one of the most promising as candidate therapeutics.7,8
Firstly, the size (12–15 kDa) of the dAbs is about an order of magnitude smaller than the size of an IgG1 (about 150 kDa). The small size leads to relatively good penetration into tissues and the ability to bind into cavities or active sites of protein targets which may not be accessible to full size antibodies. This could be particularly important for the development of therapeutics against rapidly mutating viruses, e.g., HIV. Because these viruses have evolved in humans to escape naturally occurring antibodies of large size, some of their surface regions which are critical for the viral life cycle may be vulnerable for targeting by molecules of smaller size including dAbs. Secondly, dAbs may be more stable than full size antibodies in the circulation and can be relatively easily engineered to further increase their stability. For example, some dAbs with increased stability could be taken orally or delivered via the pulmonary route or may even penetrate the blood-brain barrier, and retain activity even after being subjected to harsh conditions, such as freeze-drying or heat denaturation. In addition, dAbs are typically monomeric, of high solubility and do not significantly aggregate or can be engineered to reduce aggregation. Their half-life in the circulation can be relatively easily adjusted from minutes or hours to weeks. The dAbs are human molecules which decreases the likelihood of undesirable immune responses. In contrast to conventional antibodies, domain antibodies are well expressed in bacterial, yeast and mammalian cell systems. Finally, the small size of dAbs allows for higher molar quantities per gram of product, which should provide a significant increase in potency per dose and reduction in overall manufacturing cost (www.domantis.com, www.ablynx.com).
Research on novel antibody-derived scaffold continues. We have identified a VH based scaffold which is stable and highly soluble.9 It was used for construction of a large-size (20 billion clone) dAb phage library by grafting CDR3s and CDR2s from five of our other Fab libraries and randomly mutagenizing CDR1. Panning of this library with an HIV Env complexed with CD4 resulted in the identification of a very potent broadly cross-reactive dAb against HIV, m36, which neutralized primary HIV isolates from different clades with IC50s and IC90s in the low µg/ml range.10
I have hypothesized that CH2 domain (CH2 of IgG, IgA and IgD, and CH3 of IgE and IgM) as a scaffold could offer additional advantages compared to those listed above for dAbs because it contains binding sites or portions of binding sites conferring effector and stability functions. It is the only constant domain which is appropriate monomeric scaffold for binders because in the immunoglobulin Fc the CH2s from the two chains are only weakly interacting through carbohydrates (Figs. 1 and 2), and an isolated human CH2 domain would be soluble and stable even in the absence of glycosylation, and monomeric especially in the absence of glycosylation. Because of their small size and the CH2 role in antibody effector functions, I termed them nanoantibodies, i.e., the smallest antibody fragments that could be engineered to exhibit simultaneously antigen binding, and effector and stability functions. We found that isolated human γ1 CH2 is highly soluble, monomeric and relatively stable (Vu, Gong, Dimitrov, et al., in preparation); its melting temperature (TM) was significantly higher than a previously reported Tm for a murine isolated unglycosylated CH2 of 41°C.11 We also found that the crystal structure of an isolated human CH2 is almost identical to that of CH2 in Fc (Fig. 2);12 its dynamics is being studied by NMR.13 Several large libraries based on CH2 (up to 50 billion clones) were constructed by mutagenesis of the loops and antigen-specific binders identified (Xiao, Feng, Vu and Dimitrov, submitted). We improved further the stability of the wild type CH2 by identifying a mutant with almost 20°C higher Tm (Gong, Dimitrov, et al., in preparation). Libraries have been also made by grafting CDRs from our phage-displayed antibody libraries onto this improved CH2 scaffold, and CH2-based binders are being tested for binding to complement (CH2 contains binding site for C1q) and Fc receptors especially to the neonatal Fc receptor (FcRn) (Gong, Vu, Xiao and Dimitrov, unpublished work). We have hypothesized that binding to the FcRn could increase the antibody stability in vivo, and binding to complement could lead to lysis of the target be a virion, bacteria or cell. I believe that further development of these and other CH2-based nAbs could provide new opportunities for identification of potentially useful therapeutics.
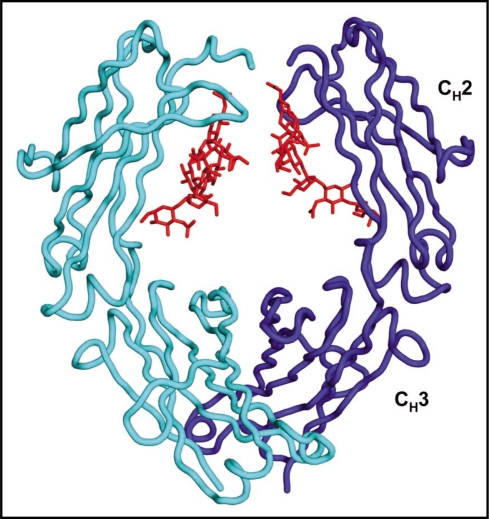
Figure 1.
Crystal structure of Fc (PDB code 1IIS)14 (the two heavy chains are in light and dark blue and the N-linked oligosaccharides attached to Asn297 is in red) indicating weak carbohydrate-mediated inter-chain CH2 interactions in contrast to CH3.
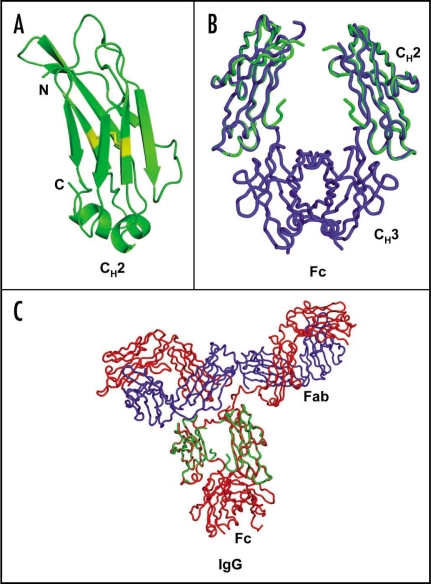
Figure 2.
Structure of the CH2 antibody domain and structural comparison with the corresponding region in the Fc and IgG structures.12 (A) Ribbon diagram of the isolated, unglycosylated CH2 domain from IgG g1 is shown. (B) The isolated CH2 domain (green) was superimposed by a least-squares algorithm using the Cα traces of the CH2 domains of Fc structure (blue; PDB code 2DTQ). (C) Superposition of the isolated CH2 structure (green) with that of CH2 portions of an intact IgG (PDB code 1HZH) using the Cα trace alignment. Heavy and light chains of IgG are shown in red and blue respectively. Carbohydrate moieties in between the CH2 domains of Fc and IgG structures in (B) and (C) are omitted for clarity.
Acknowledgements
This work was supported by the NIH NCI CCR intramural program, the NIH intramural AIDS program (IATAP) and the NIH intramural biodefense program. I would like to thank to Dr. Prabakaran Ponraj for helpful discussions and help with the figures.
Footnotes
Previously published online as a mAbs E-publication: http://www.landesbioscience.com/journals/mabs/article/7480
References
- 1.Kolmar H, Skerra A. Alternative binding proteins get mature: rivalling antibodies. FEBS J. 2008;275:2667. doi: 10.1111/j.1742-4658.2008.06437.x. [DOI] [PubMed] [Google Scholar]
- 2.Skerra A. Alternative non-antibody scaffolds for molecular recognition. Curr Opin Biotechnol. 2007;18:295–304. doi: 10.1016/j.copbio.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 3.Nygren PA, Skerra A. Binding proteins from alternative scaffolds. J Immunol Methods. 2004;290:3–28. doi: 10.1016/j.jim.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 4.Binz HK, Amstutz P, Pluckthun A. Engineering novel binding proteins from nonimmunoglobulin domains. Nat Biotechnol. 2005;23:1257–1268. doi: 10.1038/nbt1127. [DOI] [PubMed] [Google Scholar]
- 5.Hey T, Fiedler E, Rudolph R, Fiedler M. Artificial, non-antibody binding proteins for pharmaceutical and industrial applications. Trends Biotechnol. 2005;23:514–522. doi: 10.1016/j.tibtech.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 6.Holliger P, Hudson PJ. Engineered antibody fragments and the rise of single domains. Nat Biotechnol. 2005;23:1126–1136. doi: 10.1038/nbt1142. [DOI] [PubMed] [Google Scholar]
- 7.Holt LJ, Herring C, Jespers LS, Woolven BP, Tomlinson IM. Domain antibodies: proteins for therapy. Trends Biotechnol. 2003;21:484–490. doi: 10.1016/j.tibtech.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 8.Saerens D, Ghassabeh GH, Muyldermans S. Single-domain antibodies as building blocks for novel therapeutics. Curr Opin Pharmacol. 2008;8:600–608. doi: 10.1016/j.coph.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 9.Chen W, Zhu Z, Feng Y, Xiao X, Dimitrov DS. Construction of a large phage-displayed human antibody domain library with a scaffold based on a newly identified highly soluble, stable heavy chain variable domain. J Mol Biol. 2008;382:779–789. doi: 10.1016/j.jmb.2008.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen W, Zhu Z, Feng Y, Dimitrov DS. Human domain antibodies to conserved sterically restricted regions on gp120 as exceptionally potent cross-reactive HIV-1 neutralizers. Proc Natl Acad Sci USA. 2008;105:17121–17126. doi: 10.1073/pnas.0805297105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feige MJ, Walter S, Buchner J. Folding mechanism of the CH2 antibody domain. J Mol Biol. 2004;344:107–118. doi: 10.1016/j.jmb.2004.09.033. [DOI] [PubMed] [Google Scholar]
- 12.Prabakaran P, Vu BK, Gan J, Feng Y, Dimitrov DS, Ji X. Crystal structure of an isolated, unglycosylated antibody CH2 domain. Acta Crystallogr B. 2008;64:1062–1067. doi: 10.1107/S0907444908025274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vu BK, Walsh J, Dimitrov DS, Ishima R. Dynamics of antibody domains studied by solution NMR. Meth Mol Biol. 2008 doi: 10.1007/978-1-59745-554-1_29. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin WL, West AP, Jr, Gan L, Bjorkman PJ. Crystal structure at 2.8 A of an FcRn/heterodimeric Fc complex: mechanism of pH-dependent binding. Mol Cell. 2001;7:867–877. doi: 10.1016/s1097-2765(01)00230-1. [DOI] [PubMed] [Google Scholar]