Abstract
Microbes evolved to produce natural products that inhibit growth of competing soil microorganisms. In many cases, these compounds act on fungi, which are eukaryotes closely related to metazoans, including humans. The calcineurin inhibitors CsA and FK506, the Tor inhibitor rapamycin, and the Hsp90 inhibitor geldanamycin all act via targets conserved from yeast to humans. This allows use of genetically tractable fungi as models to elucidate how these drugs and their targets function in yeast and human cells. They also enable studies to harness their intrinsic antimicrobial activities to develop novel antifungal therapies. Extensive studies have revealed a globally conserved role for Tor in regulating growth and proliferation in response to nutrients, and targeting its essential functions results in robust antifungal action. Similarly, a conserved and essential role for calcineurin in fungal virulence has been discovered that could be targeted by inhibitors in therapeutic use in a variety of clinical settings. Finally, the discovery that inhibitors of calcineurin or Hsp90 result in dramatic synergism with either azoles or glucan synthase inhibitors (candins) provides another therapeutic vantage point. Taken together, these fungal targets and their inhibitors provide a robust platform from which to develop novel antimicrobial therapies.
Keywords: Tor, Calcineurin, Hsp90, Rapamycin, FK506, Cyclosporin A, Geldanamycin
Introduction
The limited drug armamentarium and increasing drug resistance to some current antifungal therapies based on 5-flucytosine, polyenes, azoles and candins, has created a need for novel molecular targets and drugs to combat fungal infections. The central roles of the Tor kinase and the Ca2+/calmodulin-dependent protein phosphatase calcineurin and its modulator Hsp90 in regulating cell growth and responses to stress in fungi have raised interest in the use of their inhibitors: rapamycin (Tor), FK506 and cyclosporin A (CsA) (calcineurin), and geldanamycin (GD) (Hsp90), as antifungal drugs. The potent immunosuppressive effects of these inhibitors have fueled development of novel, less immunosuppressive analogs, several of which are now under preclinical and clinical study. Recent findings reveal an interesting interplay between Tor, calcineurin and Hsp90 in regulating polarized growth and stress responses, which suggests that combination therapies with inhibitor analogs could confer synergistic antifungal effects (Figure 1). Here we present an overview of the Tor, calcineurin and Hsp90 signaling pathways in the model yeast S. cerevisiae and pathogenic fungi and discuss recent developments in targeting these pathways for antifungal therapy.
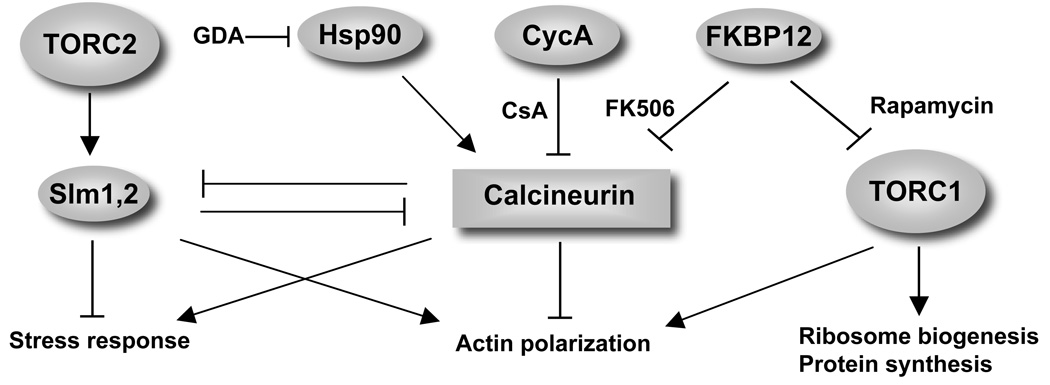
Figure 1. The antifungal drugs cyclosporin A, FK506, rapamycin and geldanamycin target conserved signaling pathways with inter-related functions in regulating responses to nutrients and stress.
CsA and FK506 form stable complexes with the prolyl isomerases cyclophilin A (CycA) and FKBP12 respectively, which subsequently bind to and inhibit calcineurin function. The Ca2+/calmodulin-dependent protein phosphatase calcineurin is a complex, composed of a regulatory and two catalytic subunits hypothesized to be stabilized (or maintained in a signaling competent form) by Hsp90, and this effect can be blocked by Hsp90 inhibitors, including geldanamycin (GD). Calcineurin plays important roles in responding to stress-induced Ca2+ signals and acts by controlling transcription of stress responsive genes while counteracting actin cytoskeleton polarization. Stress activates calcineurin to dephosphorylate the Slm PH domain proteins, thereby antagonizing TORC2-Slm signaling. Under optimal growth conditions these calcineurin functions are opposed by the rapamycin insensitive TORC2 kinase via direct phosphorylation of Slm1 and Slm2. In complex with FKBP12, rapamycin binds TORC1 and thereby blocks its functions in ribosome biogenesis, protein synthesis and actin polarization.
Tor in Saccharomyces cerevisiae
The Tor kinases were first identified in S. cerevisiae as the targets of the antifungal and immunosuppressive drug rapamycin, a cyclic macrolide produced by the soil bacterium Streptomyces hygroscopicus [1]. When rapamycin diffuses into the cell it forms a complex with the FKBP12 prolyl isomerase, which subsequently binds to the Tor kinase and blocks its functions. Both FKBP12 and Tor are ubiquitously conserved in eukaryotic organisms from yeasts to humans.
Aside from the protein kinase domain, the Tor proteins have putative domains for protein-protein interactions, including multiple N-terminal HEAT repeats and the FAT domain [2–4]. Overexpression of the FAT domain is toxic in S. cerevisiae and mutations in this domain in Schizosaccharomyces pombe tor1+ or mammalian Tor renders Tor constitutively active [2,5]. In addition, the Tor proteins feature the highly conserved FRB domain, which serves as the binding site for FKBP12-rapamycin [6].
The Tor proteins populate two multi-protein complexes in S. cerevisiae: TOR complex 1 (TORC1) formed by either Tor1 or Tor2 in association with Kog1, Lst8, and Tco89, and Tor complex 2 (TORC2) consisting of Tor2, Lst8, Avo1, Avo2, Avo3, Bit2 and Bit61 [7–9]. Recently, novel components of these complexes have been identified in the fission yeast S. pombe and several TORC components have putative homologs in the fungal pathogens C. albicans and Cryptococcus neoformans [10,11](Table 1). Importantly, except for S. cerevisiae and S. pombe that possess two Tor homologs, other eukaryotic organisms ranging from fungi to mammals have only one Tor protein. However, where examined, this single Tor protein is capable of forming two distinct protein complexes equivalent to TORC1 and TORC2.
Table 1.
Tor pathway signaling components and putative homologs in model and pathogenic fungi
| Tor signaling components | S. cerevisiae homologs | S. pombe homologs | C. albicans homologs | C. neoformans var. neoformans homologs | C. neoformans var. grubii homologs |
|---|---|---|---|---|---|
| Upstream regulators | |||||
| Tsc1 | - | tsc1+ | orf19.4110 | - | - |
| Tsc2 | - | tsc2+ | orf19.1798 | CNC05620 | CNAG_02953.1 |
| Rheb | RHB1 1 | rhb1+ | orf19.5994 (RHB1) | CNB03660 (RHEB) | CNAG_03876.1 |
| TORC1 | |||||
| Tor1 | TOR1 | tor2+ | orf19.2290 (TOR1) | CNF03740 (TOR1) | CNAG_06642.1 |
| Tor2 | TOR2 | - | - | - | - |
| Kog1 | KOG1 | mip1+ | orf19.418 (KOG1) | CND01940 | CNAG_01063.1 |
| Tco89 | TCO89 | tco89+ | orf19.761 (TCO89) | - | - |
| Lst8 | LST8 | wat1+ | orf19.3862 | CNM01130 | CNAG_06107.1 |
| - | toc1+ | - | - | - | |
| YKL033W 3 | tti1+ 2 | orf19.3062 | CNB00940 | CNAG_06709.1 | |
| TEL2> 3 | tel2+ 2 | orf19.7101 | CNI02310 | CNAG_04318.1 | |
| CKA1 3 | orb5+ 2 | orf19.3530 (CKA2) | CNF02800 (CK2) | CNAG_05694.1 | |
| TORC2 | |||||
| Tor2 | TOR2 | tor1+ | - | - | - |
| Lst8 | LST8 | wat1+ | orf19.3862 | CNM01130 | CNAG_06107.1 |
| Avo1 | AVO1 | sin1+ | orf19.5221 | CNJ01240 | CNAG_04693.1 |
| Avo2 | AVO2 | - | orf19.215 | - | CNAG_02727.1 |
| Avo3 | AVO3/TSC11 | ste20+ | orf19.728 (TSC11) | CNM01660 (STE16) | CNAG_06165.1 |
| Bit61 | BIT61 | bit61+ | - | - | - |
| Bit2 | BIT2 | - | - | - | - |
| YKL033W 3 | tti1+ 2 | orf19.3062 | CNB00940 | CNAG_06709.1 | |
| TEL2 3 | tel2+ 2 | orf19.7101 | CNI02310 | CNAG_04318.1 | |
| CKA1 3 | orb5+ 2 | orf19.3530 (CKA2) | CNF02800 (CK2) | CNAG_05694.1 | |
| Downstream effector targets | |||||
| Sit4 | SIT4 | ppe1+ | orf19.5200 (SIT4) | CND05500 | CNAG_01436.1 |
| Tap42 | TAP42 | SPCC63.05 | orf19.4626 (SCH9) | CNA03170 | CNAG_00335.1 |
| Sch9 | SCH9 | sck1+ | orf19.829 | CNN00360 (SCH9) | CNAG_06301.1 (SCH9) |
| Ypk2 | YPK2 | gad8+ | orf19.399 | CNJ01100 | CNAG_04678.1 |
Uncoupled from S. cerevisiae TORC1 signaling.
TORC1 and TORC2 interacting proteins in S. pombe
Unknown if interacting with S.cerevisiae TORC1 and TORC2 complexes.
Tor pathway signaling homologs in pathogenic fungi were identified by comparing previously characterized S. cerevisiae and S. pombe components through reciprocal best-hit BLASTp searches.
TORC1 is rapamycin-sensitive and essential to promote growth by regulating transcription and translation whereas TORC2 is insensitive to rapamycin and regulates actin cytoskeleton polarization and responses to stress [7]. However, recent studies have shown that mutations in TORC1 components and rapamycin treatment also result in actin depolarization [12–14]. This functional overlap between these two complexes was previously unnoticed and underscores the complexity embedded within this signaling network.
Nutrient sensing and Tor
Treatment of yeast cells with rapamycin triggers events that mimic the effects of nutrient starvation, including inhibition of ribosome biogenesis and protein translation, and inducing autophagy and G0 entry (reviewed in [15]). These and other observations support the model that the Tor pathway responds to nutrient cues to regulate cell growth.
Several lines of evidence link amino acid sensing and Tor signaling to membranes of the vesicular trafficking system in S. cerevisiae (reviewed in [16]). First, Tor1 has been proposed to sense glutamine [17]. Second, Tor proteins and TORC components localize to internal membranes, including the vacuole, which is the major cellular amino acid reservoir [8,13,18]. Third, TORC1 and the vacuolar EGO/Gse complex, which in response to amino acids regulates sorting of the general amino acid permease Gap1, orchestrate microautophagy [19,20]. Fourth, a role for the class C Vps complex, which functions in vesicle trafficking between endosomes and the vacuole, has been proposed in mediating intracellular amino acid homeostasis for efficient Tor signaling [21].
The link between Tor and amino acid sensing is not limited to S. cerevisiae and has also been documented in S. pombe where tor1+ regulates amino acid uptake in a rapamycin-sensitive fashion controlled by Tsc/Rheb signaling [22,23]. Interestingly, homologs of the TORC1 upstream regulators Tsc1/Tsc2 and Rheb are also conserved in S. pombe, C. albicans and C. neoformans (Table 1). These findings suggest that amino acid sensing is a conserved regulatory mechanism among several pathogenic fungi and further understanding will lead to new discoveries in Tor regulation that can be exploited to control the pathogenesis associated with these particular organisms.
TORC1 effectors of transcription and translation
The transcriptional response to inhibition of TORC1 with rapamycin revealed global roles in positively regulating ribosome biogenesis, while blocking the expression of nitrogen catabolite regulated (NCR), retrograde response (RTG), and stress responsive (STRE) genes [24–28].
The Tap42-Sit4 PP2A-like phosphatase mediates both TORC1 inhibition of transcription and activation of translation and cell growth [25,29]. The prevailing model is that, under ample nutrient conditions, TORC1 phosphorylates Tap42, thereby favoring Tap42-Sit4 complex formation [30]. Nutrient deprivation or rapamycin treatment results in dissociation of this complex and targeting of Sit4 towards specific substrates. An alternative model posits that inactive Tap42-Sit4 complex is tethered to membranes via TORC1, and rapamyicn treatment or nitrogen starvation releases activated Tap42-Sit4 complex into the cytosol [31]. However, this model is not supported by TORC1 characterization studies and awaits further confirmation. The NCR genes, regulated by the transactivators Gln3 and Gat1 and the repressor Ure2, are among the best-studied examples of TORC1 transcriptional regulation. TORC1 activity prevents Sit4-mediated dephosphorylation of Gln3 and Gat1 and thereby blocks nuclear translocation [25]. However, recent findings indicate this pathway is even more complex. Gln3 nuclear localization in response to nitrogen source quality requires Golgi to endosome trafficking, and regulation of Gat1 is not strongly Ure2- or Sit4-dependent [32–34]. In general, control of nuclear translocation has emerged as a common mechanism by which Tor regulates gene expression.
In addition, Tap42-Sit4 and Sit4 interactions with its associated proteins, Sap185 and Sap190, regulate the phosphorylation levels of Gcn2 and eIF2α to control the rate of translation and in particular Gcn4 translation [35,36].
Both the TORC1 and the cAMP-PKA pathways govern ribosome biogenesis in response to nutrients. This process entails the coordinated expression of ribosomal protein, Ribi, rRNA, and tRNA genes, and therefore involves the activity of Pol I, Pol II, and Pol III, respectively [37]. Recent studies have identified Sch9 kinase as an important TORC1 effector of ribosome biogenesis [38,39]. Moreover, Sch9 partially mediates TORC1 effects on the Rim15 kinase and control over G0 entry [40].
TORC2 effectors of actin polarization and stress responses
A requirement for TORC2 in actin polarization (via control of the Rho1/Pkc1/MAPK cell integrity pathway) was first revealed by genetic studies indicating that TORC2 activates Rom2, the guanine nucleotide-exchange factor for Rho1 [41]. Recently, the AGC kinase Ypk2 and the PH domain proteins Slm1 and Slm2, all of which drive actin polarization, were shown to be direct TORC2 substrates [42–44]. How these TORC2 effector branches coordinately regulate cell integrity and actin polarization remains to be determined. In addition, the activities of both Ypk2 and Slm1,2 are influenced by phytosphingolipids and required to regulate ceramide synthesis, a process important in stress response [43]. Interestingly, TORC2 mutants show reduced ceramide syntheses and this defect and the inability of slm1 and slm2 cells to cope with oxidative and heat stresses, are both alleviated by calcineurin defects [45–47]. Moreover, upon stress conditions Slm1 and Slm2 are dephosphorylated by calcineurin, and this event is required to activate stress responses [46–48].
This illustrates that Ypk2 and the Slm proteins integrate nutrient (nitrogen) and lipid signals and TORC2-Slm and calcineurin signaling antagonistically govern stress survival (Figure 1).
Rapamycin effectors in human fungal pathogens
Currently, systemic mycoses are treated with an armamentarium of antifungal drugs consisting of nucleic acid inhibitors (5-flucytosine), polyenes (amphotericin B and nystatin), ergosterol biosynthesis inhibitors (azoles) and echinocandins (caspofungin, micafungin) (reviewed in [49]). A separate group of antifungal compounds are the immunophilin-targeting drugs CsA, FK506 (tacrolimus), and rapamycin (sirolimus), which due to their immunosuppressive activity have been less appreciated as potential antifungal agents [50]. Recently, there is renewed interest in the antifungal activity of rapamycin fostered by the development of less immunosuppressive rapamycin analogs and findings that lipid-formulated rapamycin, amphotericin B, and 5-flucytosine act synergistically in vitro [50,51].
A single Tor homolog (Tor1) has been identified in the human fungal pathogens C. neoformans and C. albicans and the fungicidal activity of rapamycin in these two species is exerted via conserved FKBP12-rapamycin complexes that bind Tor1 and thereby inhibit its activity [52,53].
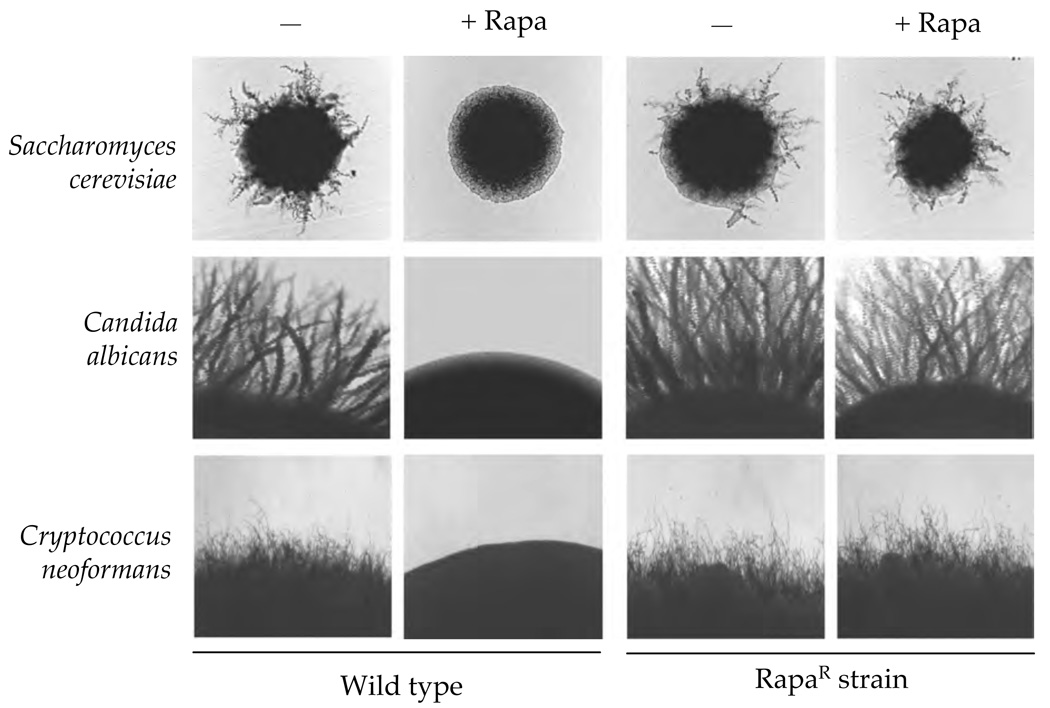
Interestingly, at sublethal concentrations, rapamycin blocks filamentous differentiation in S. cerevisiae, C. albicans, and C. neoformans [54,55] (Figure 2). In C. albicans filamentous growth is essential for virulence and consequently is a potential target for antifungal therapy [56–58]. In this pathogen, evidence-linking Tor1 to filamentous growth continues to mount with several Tor1 signaling components being associated with this developmental transition. A clear example is the link between nitrogen availability and filamentous growth in C. albicans [59]. As discussed above, in S. cerevisiae Tor1 signaling mediates the cellular response to nitrogen limitation via the Sit4 protein phosphatase and the GATA transactivators Gln3 and Gat1, which regulate the expression of nitrogen utilization genes, including the ammonium permease Mep2 [25–28]. In C. albicans, mep2/mep2 and gln3/gln3 loss of function mutations block filamentous growth under limiting nitrogen conditions and gln3/gln3 mutants are avirulent in murine models of disseminated disease [59–61]. Additionally, the protein phosphatase Sit4 also plays an important role during filamentous growth and virulence of C. albicans [29,62].
Figure 2. Rapamycin inhibits hyphal growth of model and pathogenic fungi.
Hyphal growth of model and pathogenic fungi is inhibited by rapamycin. S. cerevisiae wild type (MLY61a/ α) and rapamycin resistant strain TOR1-4/TOR1[54] were grown for 3 days at 30°C on SLAD medium with or without 10 nM rapamycin. C. albicans wild type (strain SC5314) and TOR1-1/TOR1 [53] strains were grown on Spider medium with or without 20 nM rapamycin for seven days at 37°C. C. neoformans teleomorphic forms (Filobasidiella neoformans) were generated from JEC20 (MAT a) and JEC21 (MATα) matings and JEC20 and JEC21 TOR1-1 [52] matings on V8 media (pH 7). Matings were performed in the presence or absence of 150 nM rapamycin at room temperature for 3 days.
It is increasingly evident that the Tor1 nutrient sensing pathway regulates important virulence traits in C. albicans. Furthermore, components of this pathway including TORC1 and TORC2, upstream regulators (Tsc1,2 and Rheb) and effectors (Tap42, Sit4, Sch9, and Ypk2) appear to be conserved among several fungal organisms, including pathogenic fungi (Table 1). This conservation will ultimately allow the use of model and pathogenic fungi for further characterization of Tor signaling and identification of fungal-specific Tor effectors that can be harnessed as potential targets for antifungal therapy.
Calcineurin
The calcineurin inhibitors FK506 and CsA were initially isolated as potent immunosuppressive drugs [63,64] and subsequently became cornerstones of therapy in solid organ and bone marrow transplantation. More recent evidence has suggested a role for these drugs, and non-immunosuppressive analogs, as novel antifungal therapeutics [50,65]. Calcineurin is conserved from yeasts to humans and is crucial for mediating cellular stress responses. Functional calcineurin consists of two subunits, a catalytic A and a regulatory B subunit, both of which are essential for function [66]. When Ca2+ fluxes into the cytosol from either intracellular stores or extracellular sources, calcineurin is bound by Ca2+-calmodulin causing a conformational change that relieves repression of the catalytic site by an autoinhibitory domain [66]. FK506 and CsA form intracellular complexes with FKBP12 and cyclophilin, respectively [67], and these complexes then bind to and block calcineurin function [68–72](Figure 1). FK506-FKBP12 and CsA-cyclophilinA also inhibit calcineurin in pathogenic fungal species including C. neoformans, C. albicans, and Aspergillus fumigatus [70,73–79].
In pathogenic fungi, calcineurin plays a crucial role in virulence. In C. neoformans calcineurin mutants are attenuated for virulence in animal models of infection due to their inability to grow at body temperature (37°C) [75,80]. Similarly, C. albicans calcineurin mutants are attenuated in a murine systemic infection model [81–83]; in this case calcineurin mutants are not temperature sensitive, but serum and cation sensitive and thus unable to survive calcium stress imposed by serum [84]. Interestingly, the role of calcineurin in C. albicans virulence appears to be host niche specific, as strains lacking calcineurin are attenuated for virulence in systemic and ocular infection models [81–83,85], but fully virulent in murine pulmonary, vaginal, and oropharyngeal models [86](Reedy, Filler, and Heitman unpublished data). In the pulmonary pathogen A. fumigatus, calcineurin is required for morphogenesis. Strains lacking calcineurin form short, blunted filaments and are thus significantly attenuated for virulence through a third distinct mechanism of action [87].
The attenuated virulence of fungal calcineurin mutants suggests that inhibition of calcineurin alone could have therapeutic potential; moreover, calcineurin inhibitors can also be utilized in combination therapy with current antifungal agents. In in vitro studies with C. albicans, A. fumigatus, C. neoformans, and the dermatophyte Trichophyton mentagrophytes calcineurin inhibitors convert normally fungistatic azoles, as well as other clinically available antifungals, into fungicidal compounds. This synergistic action extends to azole resistant fungal strains [73,77–79,88–91]. An in vivo proof-of principle study demonstrated that a CsA-fluconazole combination was more effective than either drug alone at treating candidal endocarditis infection in rats [92,93]. Subsequent studies document improved survival and disease resolution with combination rather than monotherapy in the treatment of C. albicans murine keratitis [85], catheter biofilms [94], and treatment of T. mentagrophytes model skin infections [90]. The clinical use of calcineurin inhibitors is limited by their immunosuppressive activity, however non-immunosuppressive analogs (FK506: L-685,818 from Merck and CsA: 211–810 and 209–825 from Novartis) are available that still inhibit fungal calcineurin and spare host calcineurin [73,74,88]. Research aimed at identifying additional components of the calcineurin signaling that could bypass the immunosuppressive activity of calcineurin inhibition is currently underway [65,95–100].
Hsp90
Recent studies reveal that Hsp90, a component of a chaperone complex induced by heat stress, governs the trajectory of drug resistance in fungi [96]. Using genetically engineered yeast strains in which Hsp90 expression can be reduced, or small molecule inhibitors of Hsp90 (geldanamycin), Hsp90 was shown to be required for both the rapid emergence of azole drug resistance, and for its maintenance. Hsp90-dependent drug resistance involves alterations in the ergosterol biosynthetic pathway targeted by azole drugs, whereas azole resistance conferred by over-expression of pumps that extrude drugs is Hsp90-independent. Potent drug synergism was observed between Hsp90 inhibitors and azoles in C. albicans, and with candins in A. fumigatus [96]. These findings parallel previous studies in which calcineurin inhibitors exhibited synergistic antifungal activity with azoles against C. albicans [76,83]. A model has been advanced suggesting calcineurin might therefore be a direct client protein of Hsp90 [101], and both genetic and protein interaction data suggests the two proteins physically and functionally interact in S. cerevisiae [98,102].
An attractive feature is that several calcineurin inhibitors are already FDA approved for clinical use and phase II and III clinical trials are ongoing for geldanamycin and its analogs (17-AAG ,17-DMAG) for a variety of oncological indications based on their chemotherapeutic potential [101]. Given limited success of current antifungal regimens for many systemic and topical fungal infections, the emergence of drug resistance, and increasing numbers of susceptible patients, combinatorial drug approaches hold considerable appeal. These combinations potentiate activity of azoles or candins, in some cases rendering them fungicidal rather than merely fungistatic, extend their therapeutic range, and concomitantly block emergence of some classes of drug resistant mutants. It will be a challenge to combine drugs inhibiting targets highly conserved between fungi and humans, and which in some cases have immunosuppressive or toxic side effects. This is particularly relevant for the Hsp90 inhibitors geldenamycin and radicicol that target Hsp90′s highly conserved ATP binding pocket to block ATP-dependent chaperone activity [103]. Nevertheless, even minor structural differences in conserved drug binding pockets can be exploited to develop target specific inhibitors. A champion example is the Cox2 specific inhibitors that exploit a single amino acid difference in the active site of Cox1 and Cox2 [104–106]. Additionally, the Hsp90 chaperone complex includes not only Hsp90 but also many additional co-factors, which might be targets for fungal specific inhibitors. Fungal specific calcineurin inhibitor analogs have also been identified [74], and the wealth of structural and enzymatic data for these targets (FKBP12, cyclophilin A, calcineurin, and Hsp90) renders these pathways attractive from a medicinal chemistry perspective [107]. The challenge ahead is to translate these in vitro findings to studies in heterologous host and animal models as a prelude to clinical testing as novel antimicrobial approaches in humans.
Conclusions and outlook
Small bioactive compounds produced by soil microorganisms have potent and specific activities against conserved cellular pathways providing molecular tools for dissection of cellular functions. The immunosuppressive drugs cyclosporin A, FK506, rapamycin, and geldanamycin are all microbial products that inhibit targets conserved from unicellular yeasts and pathogenic fungi, such as S. cerevisiae, S. pombe, C. albicans and C. neoformans to complex organisms including humans. The molecular targets of these drugs (cyclophilin A, FKBP12, calcineurin, Tor, and Hsp90) function in conserved signaling cascades that couple environmental stimuli to cell growth and proliferation (from yeasts to humans). Thus, studies of natural product action in model genetic systems are contributing to our understanding of therapeutic action in humans. Moreover, these agents have broad spectrum, potent antimicrobial activities, both alone and in combination with established antifungal drugs including the azoles and candins, and therefore represent novel, lead strategies for antifungal therapeutic development.
Figure 3. The structure of 17-AAG.
Acknowledgements
This work was supported by R01 CA114107 from the National Cancer Institute (to Maria E. Cardenas) and AI 50438 from the National Institute of Allergy and Infectious Diseases (to Joseph Heitman and Maria E. Cardenas).
References and recommended reading
Papers of special interest have been highlighted as:
* of especial interest
** of outstanding interest
- 1. Heitman J, Movva NR, Hall MN. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253(5022):905–909. doi: 10.1126/science.1715094. **This study presents a genetic screen that identified for the first time Tor1 and Tor2 as the targets of rapamycin in S. cerevisiae.
- 2.Alarcon CM, Heitman J, Cardenas ME. Protein kinase activity and identification of a toxic effector domain of the target of rapamycin TOR proteins in yeast. Mol Biol Cell. 1999;10(8):2531–2546. doi: 10.1091/mbc.10.8.2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bosotti R, Isacchi A, Sonnhammer EL. FAT: a novel domain in PIK-related kinases. Trends Biochem Sci. 2000;25(5):225–227. doi: 10.1016/s0968-0004(00)01563-2. [DOI] [PubMed] [Google Scholar]
- 4.Perry J, Kleckner N. The ATRs, ATMs, and TORs are giant HEAT repeat proteins. Cell. 2003;112(2):151–155. doi: 10.1016/s0092-8674(03)00033-3. [DOI] [PubMed] [Google Scholar]
- 5.Urano J, Sato T, Matsuo T, Otsubo Y, Yamamoto M, Tamanoi F. Point mutations in TOR confer Rheb-independent growth in fission yeast and nutrient-independent mammalian TOR signaling in mammalian cells. Proc Natl Acad Sci U S A. 2007;104(9):3514–3519. doi: 10.1073/pnas.0608510104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen J, Zheng XF, Brown EJ, Schreiber SL. Identification of an 11-kDa FKBP12-rapamycin-binding domain within the 289-kDa FKBP12-rapamycin-associated protein and characterization of a critical serine residue. Proc Natl Acad Sci U S A. 1995;92(11):4947–4951. doi: 10.1073/pnas.92.11.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo JL, Bonenfant D, Oppliger W, Jenoe P, Hall MN. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell. 2002;10(3):457–468. doi: 10.1016/s1097-2765(02)00636-6. **This study together with references 8–11, show the characterization of the rapamycin-sensitive TORC1 and the rapamycin-insensitive TORC2 in S. cerevisiae (ref 1–9) and S. pombe(ref 10–11)
- 8.Wedaman KP, Reinke A, Anderson S, Yates J, 3rd, McCaffery JM, Powers T. Tor kinases are in distinct membrane-associated protein complexes in Saccharomyces cerevisiae. Mol Biol Cell. 2003;14(3):1204–1220. doi: 10.1091/mbc.E02-09-0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reinke A, Anderson S, McCaffery JM, Yates J, 3rd, Aronova S, Chu S, Fairclough S, Iverson C, Wedaman KP, Powers T. TOR complex 1 includes a novel component, Tco89p (YPL180w), and cooperates with Ssd1p to maintain cellular integrity in Saccharomyces cerevisiae. J Biol Chem. 2004;279(15):14752–14762. doi: 10.1074/jbc.M313062200. [DOI] [PubMed] [Google Scholar]
- 10.Hayashi T, Hatanaka M, Nagao K, Nakaseko Y, Kanoh J, Kokubu A, Ebe M, Yanagida M. Rapamycin sensitivity of the Schizosaccharomyces pombe tor2 mutant and organization of two highly phosphorylated TOR complexes by specific and common subunits. Genes Cells. 2007;12(12):1357–1370. doi: 10.1111/j.1365-2443.2007.01141.x. [DOI] [PubMed] [Google Scholar]
- 11.Matsuo T, Otsubo Y, Urano J, Tamanoi F, Yamamoto M. Loss of the TOR kinase Tor2 mimics nitrogen starvation and activates the sexual development pathway in fission yeast. Mol Cell Biol. 2007;27(8):3154–3164. doi: 10.1128/MCB.01039-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang H, Jiang Y. The Tap42-protein phosphatase type 2A catalytic subunit complex is required for cell cycle-dependent distribution of actin in yeast. Mol Cell Biol. 2003;23(9):3116–3125. doi: 10.1128/MCB.23.9.3116-3125.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Araki T, Uesono Y, Oguchi T, Toh EA. LAS24/KOG1, a component of the TOR complex 1 (TORC1), is needed for resistance to local anesthetic tetracaine and normal distribution of actin cytoskeleton in yeast. Genes Genet Syst. 2005;80(5):325–343. doi: 10.1266/ggs.80.325. [DOI] [PubMed] [Google Scholar]
- 14.Aronova S, Wedaman K, Anderson S, Yates J, 3rd, Powers T. Probing the membrane environment of the TOR kinases reveals functional interactions between TORC1, actin, and membrane trafficking in Saccharomyces cerevisiae. Mol Biol Cell. 2007;18(8):2779–2794. doi: 10.1091/mbc.E07-03-0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Virgilio C, Loewith R. Cell growth control: little eukaryotes make big contributions. Oncogene. 2006;25(48):6392–6415. doi: 10.1038/sj.onc.1209884. [DOI] [PubMed] [Google Scholar]
- 16.Rohde JR, Bastidas R, Puria R, Cardenas ME. Nutritional control via Tor signaling in Saccharomyces cerevisiae. Curr Opin Microbiol. 2008;11(2):153–160. doi: 10.1016/j.mib.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crespo JL, Powers T, Fowler B, Hall MN. The TOR-controlled transcription activators GLN3, RTG1, and RTG3 are regulated in response to intracellular levels of glutamine. PNAS. 2002;99(10):6784–6789. doi: 10.1073/pnas.102687599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cardenas ME, Heitman J. FKBP12-rapamycin target TOR2 is a vacuolar protein with an associated phosphatidylinositol-4 kinase activity. EMBO J. 1995;14(23):5892–5907. doi: 10.1002/j.1460-2075.1995.tb00277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dubouloz F, Deloche O, Wanke V, Cameroni E, De Virgilio C. The TOR and EGO Protein Complexes Orchestrate Microautophagy in Yeast. Molecular Cell. 2005;19(1):15–26. doi: 10.1016/j.molcel.2005.05.020. [DOI] [PubMed] [Google Scholar]
- 20.Gao M, Kaiser CA. A conserved GTPase-containing complex is required for intracellular sorting of the general amino-acid permease in yeast. Nat Cell Biol. 2006;8(7):657–667. doi: 10.1038/ncb1419. [DOI] [PubMed] [Google Scholar]
- 21.Zurita-Martinez SA, Puria R, Pan X, Boeke JD, Cardenas ME. Efficient Tor signaling requires a functional class C Vps protein complex in Saccharomyces cerevisiae. Genetics. 2007;176(4):2139–2150. doi: 10.1534/genetics.107.072835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weisman R, Roitburg I, Nahari T, Kupiec M. Regulation of leucine uptake by tor1+ in Schizosaccharomyces pombe is sensitive to rapamycin. Genetics. 2005;169(2):539–550. doi: 10.1534/genetics.104.034983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Slegtenhorst M, Carr E, Stoyanova R, Kruger WD, Henske EP. tsc1+ and tsc2+ regulate arginine uptake and metabolism in Schizosaccharomyces pombe. J Biol Chem. 2004;279(13):12706–12713. doi: 10.1074/jbc.M313874200. [DOI] [PubMed] [Google Scholar]
- 24.Powers T, Walter P. Regulation of ribosome biogenesis by the rapamycin-sensitive TOR-signaling pathway in Saccharomyces cerevisiae. Mol Biol Cell. 1999;10(4):987–1000. doi: 10.1091/mbc.10.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beck T, Hall MN. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature. 1999;402(6762):689–692. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- 26.Cardenas ME, Cutler NS, Lorenz MC, Di Como CJ, Heitman J. The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 1999;13(24):3271–3279. doi: 10.1101/gad.13.24.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hardwick JS, Kuruvilla FG, Tong JK, Shamji AF, Schreiber SL. Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc Natl Acad Sci U S A. 1999;96(26):14866–14870. doi: 10.1073/pnas.96.26.14866. **In this work and that presented in reference 26, genome-wide gene expression analysis was employed to reveal that the transcriptional profile induced by rapamycin closely resembles that induced by nutrient limitation. Results presented in references 25 and 28 demonstrated that Tor signaling regulates gene expression by controlling the nuclear translocation of the target transcription factors.
- 28.Bertram PG, Choi JH, Carvalho J, Ai W, Zeng C, Chan TF, Zheng XF. Tripartite regulation of Gln3p by TOR, Ure2p, and phosphatases. J Biol Chem. 2000;275(46):35727–35733. doi: 10.1074/jbc.M004235200. [DOI] [PubMed] [Google Scholar]
- 29.Di Como CJ, Arndt KT. Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev. 1996;10(15):1904–1916. doi: 10.1101/gad.10.15.1904. [DOI] [PubMed] [Google Scholar]
- 30.Jiang Y, Broach JR. Tor proteins and protein phosphatase 2A reciprocally regulate Tap42 in controlling cell growth in yeast. EMBO J. 1999;18(10):2782–2792. doi: 10.1093/emboj/18.10.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan G, Shen X, Jiang Y. Rapamycin activates Tap42-associated phosphatases by abrogating their association with Tor complex 1. EMBO J. 2006;25(15):3546–3555. doi: 10.1038/sj.emboj.7601239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cox KH, Kulkarni A, Tate JJ, Cooper TG. Gln3 phosphorylation and intracellular localization in nutrient limitation and starvation differ from those generated by rapamycin inhibition of Tor1/2 in Saccharomyces cerevisiae. J Biol Chem. 2004;279(11):10270–10278. doi: 10.1074/jbc.M312023200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Puria R, Zurita-Martinez SA, Cardenas ME. Nuclear translocation of Gln3 in response to nutrient signals requires Golgi-to-endosome trafficking in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2008;105(20):794–7199. doi: 10.1073/pnas.0801087105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Georis I, Tate JJ, Cooper TG, Dubois E. Tor pathway control of the nitrogen-responsive DAL5 gene bifurcates at the level of Gln3 and Gat1 regulation in Saccharomyces cerevisiae. J Biol Chem. 2008;283(14):8919–8929. doi: 10.1074/jbc.M708811200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cherkasova VA, Hinnebusch AG. Translational control by TOR and TAP42 through dephosphorylation of eIF2alpha kinase GCN2. Genes Dev. 2003;17(7):859–872. doi: 10.1101/gad.1069003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rohde JR, Campbell S, Zurita-Martinez SA, Cutler NS, Ashe M, Cardenas ME. TOR controls transcriptional and translational programs via Sap-Sit4 protein phosphatase signaling effectors. Mol Cell Biol. 2004;24(19):8332–8341. doi: 10.1128/MCB.24.19.8332-8341.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Warner JR, Vilardell J, Sohn JH. Economics of ribosome biosynthesis. Cold Spring Harb Symp Quant Biol. 2001;66:567–574. doi: 10.1101/sqb.2001.66.567. [DOI] [PubMed] [Google Scholar]
- 38.Jorgensen P, Rupes I, Sharom JR, Schneper L, Broach JR, Tyers M. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 2004;18(20):2491–2505. doi: 10.1101/gad.1228804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Urban J, Soulard A, Huber A, Lippman S, Mukhopadhyay D, Deloche O, Wanke V, Anrather D, Ammerer G, Riezman H, Broach JR, et al. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol Cell. 2007;26(5):663–674. doi: 10.1016/j.molcel.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 40.Swinnen E, Wanke V, Roosen J, Smets B, Dubouloz F, Pedruzzi I, Cameroni E, De Virgilio C, Winderickx J. Rim15 and the crossroads of nutrient signalling pathways in Saccharomyces cerevisiae. Cell Div. 2006;1:3. doi: 10.1186/1747-1028-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmidt A, Bickle M, Beck T, Hall MN. The yeast phosphatidylinositol kinase homolog TOR2 activates RHO1 and RHO2 via the exchange factor ROM2. Cell. 1997;88(4):531–542. doi: 10.1016/s0092-8674(00)81893-0. [DOI] [PubMed] [Google Scholar]
- 42.Kamada Y, Fujioka Y, Suzuki NN, Inagaki F, Wullschleger S, Loewith R, Hall MN, Ohsumi Y. Tor2 directly phosphorylates the AGC kinase Ypk2 to regulate actin polarization. Mol Cell Biol. 2005;25(16):7239–7248. doi: 10.1128/MCB.25.16.7239-7248.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Audhya A, Loewith R, Parsons AB, Gao L, Tabuchi M, Zhou H, Boone C, Hall MN, Emr SD. Genome-wide lethality screen identifies new PI4,5P2 effectors that regulate the actin cytoskeleton. EMBO J. 2004;23(19):3747–3757. doi: 10.1038/sj.emboj.7600384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fadri M, Daquinag A, Wang S, Xue T, Kunz J. The pleckstrin homology domain proteins Slm1 and Slm2 are required for actin cytoskeleton organization in yeast and bind phosphatidylinositol-4,5-bisphosphate and TORC2. Mol Biol Cell. 2005;16(4):1883–1900. doi: 10.1091/mbc.E04-07-0564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aronova S, Wedaman K, Aronov PA, Fontes K, Ramos K, Hammock BD, Powers T. Regulation of ceramide biosynthesis by TOR complex 2. Cell Metab. 2008;7(2):148–158. doi: 10.1016/j.cmet.2007.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mulet JM, Martin DE, Loewith R, Hall MN. Mutual antagonism of target of rapamycin and calcineurin signaling. J Biol Chem. 2006;281(44):33000–33007. doi: 10.1074/jbc.M604244200. [DOI] [PubMed] [Google Scholar]
- 47.Daquinag A, Fadri M, Jung SY, Qin J, Kunz J. The yeast PH domain proteins Slm1 and Slm2 are targets of sphingolipid signaling during the response to heat stress. Mol Cell Biol. 2007;27(2):633–650. doi: 10.1128/MCB.00461-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bultynck G, Heath VL, Majeed AP, Galan JM, Haguenauer-Tsapis R, Cyert MS. Slm1 and Slm2 are novel substrates of the calcineurin phosphatase required for heat stress-induced endocytosis of the yeast uracil permease. Mol Cell Biol. 2006;26(12):4729–4745. doi: 10.1128/MCB.01973-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sanglard D, Odds FC. Resistance of Candida species to antifungal agents: molecular mechanisms and clinical consequences. Lancet Infect Dis. 2002;2(2):73–85. doi: 10.1016/s1473-3099(02)00181-0. [DOI] [PubMed] [Google Scholar]
- 50.Blankenship JR, Steinbach WJ, Perfect JR, Heitman J. Teaching old drugs new tricks: reincarnating immunosuppressants as antifungal drugs. Curr Opin Investig Drugs. 2003;4(2):192–199. [PubMed] [Google Scholar]
- 51. Vakil R, Knilans K, Andes D, Kwon GS. Combination antifungal therapy involving Amphotericin B, rapamycin and 5-Fluorocytosine using PEG-phospholipid micelles. Pharm Res in press. 2008 doi: 10.1007/s11095-008-9588-1. *First study showing the fungicidal synergistic effects of lipid-formulated rapamycin, amphotericin B and 5-flucytosine on C. albicans viability.
- 52.Cruz MC, Cavallo LM, Gorlach JM, Cox G, Perfect JR, Cardenas ME, Heitman J. Rapamycin antifungal action is mediated via conserved complexes with FKBP12 and TOR kinase homologs in Cryptococcus neoformans. Mol Cell Biol. 1999;19(6):4101–4112. doi: 10.1128/mcb.19.6.4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cruz MC, Goldstein AL, Blankenship J, Del Poeta M, Perfect JR, McCusker JH, Bennani YL, Cardenas ME, Heitman J. Rapamycin and less immunosuppressive analogs are toxic to Candida albicans and Cryptococcus neoformans via FKBP12-dependent inhibition of TOR. Antimicrob Agents Chemother. 2001;45(11):3162–3170. doi: 10.1128/AAC.45.11.3162-3170.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cutler NS, Pan X, Heitman J, Cardenas ME. The TOR signal transduction cascade controls cellular differentiation in response to nutrients. Mol Biol Cell. 2001;12(12):4103–4113. doi: 10.1091/mbc.12.12.4103. **Results in this study and those presented in references 51 and 52, demonstrate that sublethal concentrations of rapamycin block filamentous differentiation via conserved FKBP12 and Tor homologs in S. cerevisiae and in the human fungal pathogens C. albicans and C. neoformans.
- 55.Martins LF, Montero-Lomeli M, Masuda CA, Fortes FS, Previato JO, Mendonca-Previato L. Lithium-mediated suppression of morphogenesis and growth in Candida albicans. FEMS Yeast Res. 2008;8(4):615–621. doi: 10.1111/j.1567-1364.2008.00376.x. [DOI] [PubMed] [Google Scholar]
- 56.Lo HJ, Kohler JR, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink GR. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90(5):939–949. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- 57.Saville SP, Lazzell AL, Monteagudo C, Lopez-Ribot JL. Engineered control of cell morphology in vivo reveals distinct roles for yeast and filamentous forms of Candida albicans during infection. Eukaryot Cell. 2003;2(5):1053–1060. doi: 10.1128/EC.2.5.1053-1060.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zheng X, Wang Y, Wang Y. Hgc1, a novel hypha-specific G1 cyclin-related protein regulates Candida albicans hyphal morphogenesis. EMBO J. 2004;23(8):1845–1856. doi: 10.1038/sj.emboj.7600195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Biswas K, Morschhauser J. The Mep2p ammonium permease controls nitrogen starvation-induced filamentous growth in Candida albicans. Mol Microbiol. 2005;56(3):649–669. doi: 10.1111/j.1365-2958.2005.04576.x. [DOI] [PubMed] [Google Scholar]
- 60.Dabas N, Morschhauser J. Control of ammonium permease expression and filamentous growth by the GATA transcription factors Gln3 and Gat1 in Candida albicans. Eukaryot Cell. 2007;6(5):875–888. doi: 10.1128/EC.00307-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liao WL, Ramon AM, Fonzi WA. GLN3 encodes a global regulator of nitrogen metabolism and virulence of C. albicans. Fungal Genet Biol. 2008;45(4):514–526. doi: 10.1016/j.fgb.2007.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee CM, Nantel A, Jiang L, Whiteway M, Shen SH. The serine/threonine protein phosphatase Sit4 modulates yeast-to-hypha morphogenesis and virulence in Candida albicans. Mol Microbiol. 2004;51(3):691–709. doi: 10.1111/j.1365-2958.2003.03879.x. [DOI] [PubMed] [Google Scholar]
- 63.Borel JF. Comparative study of in vitro and in vivo drug effects on cell-mediated cytotoxicity. Immunology. 1976;31(4):631–641. [PMC free article] [PubMed] [Google Scholar]
- 64.Goto T, Kino T, Hatanaka H, Okuhara M, Kohsaka M, Aoki H, Imanaka H. FK 506: historical perspectives. Transplant Proc. 1991;23(6):2713–2717. [PubMed] [Google Scholar]
- 65. Steinbach WJ, Reedy JL, Cramer RA, Jr, Perfect JR, Heitman J. Harnessing calcineurin as a novel anti-infective agent against invasive fungal infections. Nat Rev Microbiol. 2007;5(6):418–430. doi: 10.1038/nrmicro1680. *An excellent review of calcineurin biology in C. neoformans, C. albicans and A. fumigatus and its potential use for antifungal therapy.
- 66.Hemenway CS, Heitman J. Calcineurin. Structure, function, and inhibition. Cell Biochem Biophys. 1999;30(1):115–151. doi: 10.1007/BF02737887. [DOI] [PubMed] [Google Scholar]
- 67.Ho S, Clipstone N, Timmermann L, Northrop J, Graef I, Fiorentino D, Nourse J, Crabtree GR. The mechanism of action of cyclosporin A and FK506. Clin Immunol Immunopathol. 1996;80(3 Pt 2):S40–S45. doi: 10.1006/clin.1996.0140. [DOI] [PubMed] [Google Scholar]
- 68.Fruman DA, Klee CB, Bierer BE, Burakoff SJ. Calcineurin phosphatase activity in T lymphocytes is inhibited by FK 506 and cyclosporin A. Proc Natl Acad Sci U S A. 1992;89(9):3686–3690. doi: 10.1073/pnas.89.9.3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Clipstone NA, Fiorentino DF, Crabtree GR. Molecular analysis of the interaction of calcineurin with drug-immunophilin complexes. J Biol Chem. 1994;269(42):26431–26437. [PubMed] [Google Scholar]
- 70.Heitman J, Cardenas ME, Breuder T, Hemenway C, Muir RS, Lim E, Goetz L, Zhu D, Lorenz M, Dolinski K. Antifungal effects of cyclosporine and FK 506 are mediated via immunophilin-dependent calcineurin inhibition. Transplant Proc. 1994;26(5):2833–2834. [PubMed] [Google Scholar]
- 71.Cardenas ME, Muir RS, Breuder T, Heitman J. Targets of immunophilin-immunosuppressant complexes are distinct highly conserved regions of calcineurin A. EMBO J. 1995;14(12):2772–2783. doi: 10.1002/j.1460-2075.1995.tb07277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cardenas ME, Zhu D, Heitman J. Molecular mechanisms of immunosuppression by cyclosporine, FK506, and rapamycin. Curr Opin Nephrol Hypertens. 1995;4(6):472–477. doi: 10.1097/00041552-199511000-00002. [DOI] [PubMed] [Google Scholar]
- 73.Odom A, Del Poeta M, Perfect J, Heitman J. The immunosuppressant FK506 and its nonimmunosuppressive analog L-685,818 are toxic to Cryptococcus neoformans by inhibition of a common target protein. Antimicrob Agents Chemother. 1997;41(1):156–161. doi: 10.1128/aac.41.1.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cruz MC, Del Poeta M, Wang P, Wenger R, Zenke G, Quesniaux VF, Movva NR, Perfect JR, Cardenas ME, Heitman J. Immunosuppressive and nonimmunosuppressive cyclosporine analogs are toxic to the opportunistic fungal pathogen Cryptococcus neoformans via cyclophilin-dependent inhibition of calcineurin. Antimicrob Agents Chemother. 2000;44(1):143–149. doi: 10.1128/aac.44.1.143-149.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fox DS, Cruz MC, Sia RA, Ke H, Cox GM, Cardenas ME, Heitman J. Calcineurin regulatory subunit is essential for virulence and mediates interactions with FKBP12-FK506 in Cryptococcus neoformans. Mol Microbiol. 2001;39(4):835–849. doi: 10.1046/j.1365-2958.2001.02295.x. [DOI] [PubMed] [Google Scholar]
- 76. Cruz MC, Goldstein AL, Blankenship JR, Del Poeta M, Davis D, Cardenas ME, Perfect JR, McCusker JH, Heitman J. Calcineurin is essential for survival during membrane stress in Candida albicans. EMBO J. 2002;21(4):546–559. doi: 10.1093/emboj/21.4.546. **This study features the mechanism by which calcineurin inhibitors cyclosporine A and FK506 are toxic in combination with the ergosterol biosynthesis inhibitor fluconazole on C. albicans viability. In addition, this synergism was observed with all azole antifungals tested, even among azole resistant strains and several pathogenic fungi. Non-immunosuppressive cyclosporine analogs were also found to exert synergistic fungicidal activities against C. albicans.
- 77.Kontoyiannis DP, Lewis RE, Osherov N, Albert ND, May GS. Combination of caspofungin with inhibitors of the calcineurin pathway attenuates growth in vitro in Aspergillus species. J Antimicrob Chemother. 2003;51(2):313–316. doi: 10.1093/jac/dkg090. [DOI] [PubMed] [Google Scholar]
- 78.Steinbach WJ, Schell WA, Blankenship JR, Onyewu C, Heitman J, Perfect JR. In vitro interactions between antifungals and immunosuppressants against Aspergillus fumigatus. Antimicrob Agents Chemother. 2004;48(5):1664–1669. doi: 10.1128/AAC.48.5.1664-1669.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Steinbach WJ, Cramer RA, Jr, Perfect BZ, Henn C, Nielsen K, Heitman J, Perfect JR. Calcineurin inhibition or mutation enhances cell wall inhibitors against Aspergillus fumigatus. Antimicrob Agents Chemother. 2007;51(8):2979–2981. doi: 10.1128/AAC.01394-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Odom A, Muir S, Lim E, Toffaletti DL, Perfect J, Heitman J. Calcineurin is required for virulence of Cryptococcus neoformans. EMBO J. 1997;16(10):2576–2589. doi: 10.1093/emboj/16.10.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bader T, Bodendorfer B, Schroppel K, Morschhauser J. Calcineurin is essential for virulence in Candida albicans. Infect Immun. 2003;71(9):5344–5354. doi: 10.1128/IAI.71.9.5344-5354.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Blankenship JR, Wormley FL, Boyce MK, Schell WA, Filler SG, Perfect JR, Heitman J. Calcineurin is essential for Candida albicans survival in serum and virulence. Eukaryot Cell. 2003;2(3):422–430. doi: 10.1128/EC.2.3.422-430.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sanglard D, Ischer F, Marchetti O, Entenza J, Bille J. Calcineurin A of Candida albicans: involvement in antifungal tolerance, cell morphogenesis and virulence. Mol Microbiol. 2003;48(4):959–976. doi: 10.1046/j.1365-2958.2003.03495.x. [DOI] [PubMed] [Google Scholar]
- 84.Blankenship JR, Heitman J. Calcineurin is required for Candida albicans to survive calcium stress in serum. Infect Immun. 2005;73(9):5767–5774. doi: 10.1128/IAI.73.9.5767-5774.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Onyewu C, Afshari NA, Heitman J. Calcineurin promotes infection of the cornea by Candida albicans and can be targeted to enhance fluconazole therapy. Antimicrob Agents Chemother. 2006;50(11):3963–3965. doi: 10.1128/AAC.00393-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bader T, Schroppel K, Bentink S, Agabian N, Kohler G, Morschhauser J. Role of calcineurin in stress resistance, morphogenesis, and virulence of a Candida albicans wild-type strain. Infect Immun. 2006;74(7):4366–4369. doi: 10.1128/IAI.00142-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Steinbach WJ, Cramer RA, Jr, Perfect BZ, Asfaw YG, Sauer TC, Najvar LK, Kirkpatrick WR, Patterson TF, Benjamin DK, Jr, Heitman J, Perfect JR. Calcineurin controls growth, morphology, and pathogenicity in Aspergillus fumigatus. Eukaryot Cell. 2006;5(7):1091–1103. doi: 10.1128/EC.00139-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Onyewu C, Blankenship JR, Del Poeta M, Heitman J. Ergosterol biosynthesis inhibitors become fungicidal when combined with calcineurin inhibitors against Candida albicans, Candida glabrata, and Candida krusei. Antimicrob Agents Chemother. 2003;47(3):956–964. doi: 10.1128/AAC.47.3.956-964.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Reedy JL, Husain S, Ison M, Pruett TL, Singh N, Heitman J. Immunotherapy with tacrolimus (FK506) does not select for resistance to calcineurin inhibitors in Candida albicans isolates from liver transplant patients. Antimicrob Agents Chemother. 2006;50(4):1573–1577. doi: 10.1128/AAC.50.4.1573-1577.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Onyewu C, Eads E, Schell WA, Perfect JR, Ullmann Y, Kaufman G, Horwitz BA, Berdicevsky I, Heitman J. Targeting the calcineurin pathway enhances ergosterol biosynthesis inhibitors against Trichophyton mentagrophytes in vitro and in a human skin infection model. Antimicrob Agents Chemother. 2007;51(10):3743–3746. doi: 10.1128/AAC.00492-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kontoyiannis DP, Lewis RE, Alexander BD, Lortholary O, Dromer F, Gupta KL, John GT, Del Busto R, Klintmalm GB, Somani J, Lyon GM, et al. Calcineurin inhibitor agents interact synergistically with antifungal agents in vitro against Cryptococcus neoformans isolates: correlation with outcome in solid organ transplant recipients with cryptococcosis. Antimicrob Agents Chemother. 2008;52(2):735–738. doi: 10.1128/AAC.00990-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Marchetti O, Entenza JM, Sanglard D, Bille J, Glauser MP, Moreillon P. Fluconazole plus cyclosporine: a fungicidal combination effective against experimental endocarditis due to Candida albicans. Antimicrob Agents Chemother. 2000;44(11):2932–2938. doi: 10.1128/aac.44.11.2932-2938.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Marchetti O, Moreillon P, Glauser MP, Bille J, Sanglard D. Potent synergism of the combination of fluconazole and cyclosporine in Candida albicans. Antimicrob Agents Chemother. 2000;44(9):2373–2381. doi: 10.1128/aac.44.9.2373-2381.2000. *This is the first report showing that the calcineurin inhibitor cyclosporine in conjunction with the ergosterol biosynthesis inhibitor fluconazole exhibits synergistic fungicidal activity against C. albicans.
- 94.Uppuluri P, Nett J, Heitman J, Andes D. Synergistic effect of calcineurin inhibitors and fluconazole against Candida albicans biofilms. Antimicrob Agents Chemother. 2008;52(3):1127–1132. doi: 10.1128/AAC.01397-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Onyewu C, Wormley FL, Jr, Perfect JR, Heitman J. The calcineurin target, Crz1, functions in azole tolerance but is not required for virulence of Candida albicans. Infect Immun. 2004;72(12):7330–7333. doi: 10.1128/IAI.72.12.7330-7333.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Cowen LE, Lindquist S. Hsp90 potentiates the rapid evolution of new traits: drug resistance in diverse fungi. Science. 2005;309(5744):2185–2189. doi: 10.1126/science.1118370. **A prescient study revealing a novel role for the molecular chaperone Hsp90 in the fixation and evolution of drug resistant traits among fungi.
- 97.Santos M, de Larrinoa IF. Functional characterization of the Candida albicans CRZ1 gene encoding a calcineurin-regulated transcription factor. Curr Genet. 2005;48(2):88–100. doi: 10.1007/s00294-005-0003-8. [DOI] [PubMed] [Google Scholar]
- 98.Cowen LE, Carpenter AE, Matangkasombut O, Fink GR, Lindquist S. Genetic architecture of Hsp90-dependent drug resistance. Eukaryot Cell. 2006;5(12):2184–2188. doi: 10.1128/EC.00274-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Karababa M, Valentino E, Pardini G, Coste AT, Bille J, Sanglard D. CRZ1, a target of the calcineurin pathway in Candida albicans. Mol Microbiol. 2006;59(5):1429–1451. doi: 10.1111/j.1365-2958.2005.05037.x. [DOI] [PubMed] [Google Scholar]
- 100.Pardini G, De Groot PW, Coste AT, Karababa M, Klis FM, de Koster CG, Sanglard D. The CRH family coding for cell wall glycosylphosphatidylinositol proteins with a predicted transglycosidase domain affects cell wall organization and virulence of Candida albicans. J Biol Chem. 2006;281(52):40399–40411. doi: 10.1074/jbc.M606361200. [DOI] [PubMed] [Google Scholar]
- 101.Cowen LE. The evolution of fungal drug resistance: modulating the trajectory from genotype to phenotype. Nat Rev Microbiol. 2008;6(3):187–198. doi: 10.1038/nrmicro1835. [DOI] [PubMed] [Google Scholar]
- 102.Imai J, Yahara I. Role of HSP90 in salt stress tolerance via stabilization and regulation of calcineurin. Mol Cell Biol. 2000;20(24):9262–9270. doi: 10.1128/mcb.20.24.9262-9270.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Roe SM, Prodromou C, O'Brien R, Ladbury JE, Piper PW, Pearl LH. Structural basis for inhibition of the Hsp90 molecular chaperone by the antitumor antibiotics radicicol and geldanamycin. J Med Chem. 1999;42(2):260–266. doi: 10.1021/jm980403y. [DOI] [PubMed] [Google Scholar]
- 104.FitzGerald GA, Patrono C. The coxibs, selective inhibitors of cyclooxygenase-2. N Engl J Med. 2001;345(6):433–442. doi: 10.1056/NEJM200108093450607. [DOI] [PubMed] [Google Scholar]
- 105.Flower RJ. The development of COX2 inhibitors. Nat Rev Drug Discov. 2003;2(3):179–191. doi: 10.1038/nrd1034. [DOI] [PubMed] [Google Scholar]
- 106.Mitchell JA, Warner TD. COX isoforms in the cardiovascular system: understanding the activities of non-steroidal anti-inflammatory drugs. Nat Rev Drug Discov. 2006;5(1):75–86. doi: 10.1038/nrd1929. [DOI] [PubMed] [Google Scholar]
- 107.Fox DS, Heitman J. Good fungi gone bad: the corruption of calcineurin. Bioessays. 2002;24(10):894–903. doi: 10.1002/bies.10157. [DOI] [PubMed] [Google Scholar]