Abstract
The proximal promoter consists of binding sites for transcription regulators and a core promoter. We identified an overrepresented motif in the proximal promoter of human genes with an Initiator (INR) positional bias. The core of the motif fits the INR consensus but its sequence is more strict and flanked by additional conserved sequences. This strict INR (sINR) is enriched in TATA-less genes that belong to specific functional categories. Analysis of the sINR-containing DHX9 and ATP5F1 genes showed that the entire sINR sequence, including the strict core and the conserved flanking sequences, is important for transcription. A conventional INR sequence could not substitute for DHX9 sINR whereas, sINR could replace a conventional INR. The minimal region required to create the major TSS of the DHX9 promoter includes the sINR and an upstream Sp1 site. In a heterologous context, sINR substituted for the TATA box when positioned downstream to several Sp1 sites. Consistent with that the majority of sINR promoters contain at least one Sp1 site. Thus, sINR is a TATA-less-specific INR that functions in cooperation with Sp1. These findings support the idea that the INR is a family of related core promoter motifs.
INTRODUCTION
The promoter of RNA polymerase II genes consists of two types of DNA regulatory sequences, enhancers and core promoters. Enhancer elements, which are gene specific, serve as the binding sites of transcription regulatory factors and can be divided into two classes: those that function independently of their position relative to the transcription start site (TSS) and those that can activate transcription only when located proximal to the TSS. The core promoter is situated around the TSS and is the site on which RNA polymerase II and general transcription factors (GTFs) assemble into a preinitiation complex [for review see (1)]. Each gene has a unique transcriptional control program that is determined by a specific combination of regulatory elements that vary between individual genes. Among these sequences are features common to many genes, in particular proximal elements and core promoter motifs, which contribute to the overall expression of the gene.
The best-characterized core promoter elements are the TATA box and the Initiator (INR), which are regarded as universal elements (1,2). However, recent bioinformatics studies revealed that the TATA box is present in a smaller fraction of pol II genes than initially estimated: between 20% to 46% in yeast, depending on the definition of the TATA box sequence (3,4), ∼30% in Drosophila genes (5) and 10–24% in human genes (6–8). The TATA box has a strict location at −35 to −25 relative to the TSS and is recognized by the TATA binding protein (TBP) subunit of the GTF TFIID. The INR is located around the TSS (9) and is recognized by the TAF1 and TAF2 subunits of TFIID (10–12). Additional documented core promoter elements are the DPE that is located at +28 relative to the TSS (13,14), two TFIIB recognition elements (BREs) (15,16) and a TAF1 recognition element DCE (17). The two BREs and the DCE function only in conjunction with a TATA element.
In this study, we combined bioinformatics with molecular analysis to investigate the core promoter region of mammalian genes. We focused on an INR-like element that is present in 1.5% of human genes and is characterized by a strict sequence compared with the more diverged INR, and is enriched in TATA-less promoters of genes in specific functional categories. Detailed molecular analysis indicates that this strict INR (sINR) cooperates with Sp1 to direct accurate transcription initiation of TATA-less promoters. Our findings suggest that the INR is a family of core promoter motifs that share a common basis and have in addition specific distinguishing features.
MATERIALS AND METHODS
Bioinformatics analysis of the human proximal promoter
Human proximal promoter regions from −60 to +40 relative to the TSS were retrieved from the EPD (http://www.epd.isb-sib.ch/), HPD (http://zlab.bu.edu/∼mfrith/HPD.html) and the DBTSS (http://dbtss.hgc.jp/), and analyzed by the MEME (Multiple EM for Motif Elicitation) program (18), using the default parameters, inquiring for the most significant motifs of 6–12 nt. For the gene functional annotation clustering, the Database for Annotation, Visualization and Integrated Discovery (DAVID), fifth version (http://david.abcc.ncifcrf.gov/gene2gene.jsp) was used, with default parameters at medium classification stringency.
Plasmid construction
The promoter regions of the DHX9 and ATP5F1 genes (from −150 to +50 and −155 to +60, respectively) were cloned by genomic PCR into pGL2-Basic (Promega) via SmaI and HindIII sites. Mutation of sINR and the DHX9 promoter deletions were carried out using PCR technique. To construct sINR in a heterologous context, the SV40 early core promoter in the pGL2-promoter plasmid (Promega) was replaced by sINR, the TATA box or a random sequence by digesting the plasmid with NcoI and StuI and inserting oligonucleotides with appropriate restriction sites. Construction of the luciferase reporter gene under the Pel98 promoter and its INR mutant are described in K.Gazit et al. (submitted for publication). Pel 98 sINR mutant was generated by PCR. Primer sequences used for plasmid construction are shown in Supplementary Data 1. All plasmids constructed in this study were verified by sequencing.
Transient transfection assays and RNA analysis
The 293T and ts13 cells were maintained and transfected as described (19). Twenty-four hours after transfection total RNA was prepared using Tri-reagent (MRC Inc.). Primer extension was performed as previously described (20) using 20 μg of total RNA for the luciferase primer and 2 μg RNA for the puro-GFP primer. Primer extension of endogenous genes was performed using 20 μg total RNA prepared from nontransfected cells. The sequencing reaction was carried out with the Sequenase Version 2.0 kit (USB Corporation). Results were visualized with a Phosphoimager (Fuji, BAS 2500). For determining the effect of Ying Yang 1 (YY1) depletion on the activity of the DHX9 promoter, 293T cells were grown on six-well plate and transfected with 500 ng YY1 siRNA expression plasmid, generously provided by Yang Shi (21), YY2 siRNA or with the pSuper parental plasmid as a control. Forty-eight hours later RNA was extracted using the RNeasy kit (Qiagen) and was quantified by real time PCR. To analyze the effect of YY1 depletion on the reporter gene, 293T cells grown in six-well plates were transfected with 500 ng YY1 siRNA expression plasmid and 48 h later were transfected again with either 1.5 μg of the YY1 siRNA expression plasmid or a control, and with 100 ng reporter plasmid containing DHX9 luciferase reporter plasmid together with EGFP-N1. After an additional 24 h, RNA was extracted using the RNeasy kit (Qiagen) and was quantified by real time PCR. The neomycin resistance gene under the control of the SV40 early promoter within the EGFP-N1, was used to normalize transfection efficiency.
Electrophoretic mobility shift assay
Fluorescently labeled oligonucleotides of the DHX9 sINR sequence were annealed and used as probes in binding reactions containing 2 μg of poly(dI-dC) and 2 μg of HeLa nuclear extract prepared as described previously (22), in binding buffer consisting of 25 mM HEPES (pH 7.9), 50 mM KCl, 1 mM DTT and 10% glycerol. The reaction mix was incubated on ice for 10 min after which 50 femtomole probe was added for an additional 20 min. Competitor DNAs were added prior to the addition of the probe. In the super shift reactions, 400 ng of YY1 antibody (SantaCruz, C20) was added to the primary mix and incubated for 15 min at RT. Then the probe was added and the mix was incubated on ice for an additional 20 min. The reactions were separated by native electrophoresis at 4°C in a 4.87% polyacryamide gel with 1× Tris–Glycine buffer at 185 V. The gel visualized with the Typhoon 9400 instrument (Amersham Biosciencs).
Primers
Sequences of the primers used throughout the study are shown in Supplementary Data 1.
RESULTS
Identification of a strict INR element
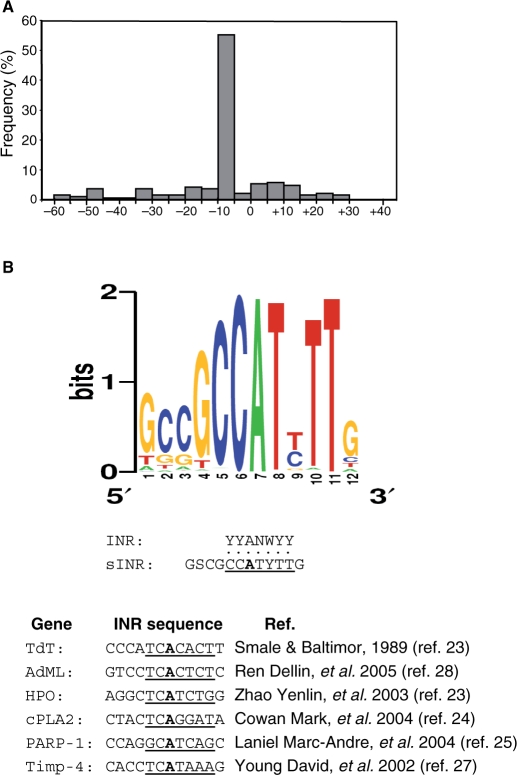
To characterize the proximal promoter of human genes, we retrieved promoter sequences with verified TSSs from the EPD (1871 promoters), HPD (2004 promoters) and the DBTSS (14 681 promoters), and, using the MEME program (18), searched for motifs overrepresented in the −60 to +40 region relative to the TSS. This program looks for conserved un-gapped blocks in a set of query sequences, and returns motifs of 6–12 nt. A motif with the sequence GSCGCCATYTTG (Table 1) appeared in the three databases with a frequency of ∼1.5% in the proximal promoter region of human genes. The distribution of this motif relative to the TSS (Figure 1A) was determined and found to be strictly localized around the TSS, from position −10 up to +5, similar to the INR element. Indeed, the core sequence of this motif (CCATYTT) shares 7 nt with the INR sequence YYANA/TYY. However, the consensus of this motif is less divergent and it has additional conserved flanking sequences that are missing from the INR (Figure 1B). Comparing the sequence of this motif to several experimentally verified INR elements (23–28) confirms that the consensus of this motif differs from the INR sequence of the tested genes in the stringency of the INR core and the flanking sequences (Figure 1C). We have therefore designated this motif sINR for strict INR.
Table 1.
Features of sINR
| Database | Consensus | No. of sites | Frequency (%) | E-value |
|---|---|---|---|---|
| EPD | GSCGCCATYTTG | 55 | 2.9 | 7.7e-32 |
| HPD | GSMGCCATYTTG | 31 | 1.5 | 7e-10 |
| DBTSS | GSCGCCATYTTG | 191 | 1.3 | 4.8e-18 |
The consensus of the sINR element identified by analyzing 1847, 2004 and 14 628 human proximal promoter sequences from −60 to +40 relative to the TSS (from EPD, HPD and DBTSS, respectively), by the MEME program. The frequency of the motif and the E-value are shown.
Figure 1.
(A) sINR is located mainly around the TSS. The distribution of sINR at 5 nt intervals throughout the proximal promoter region (−60 to +40 relative to the TSS) as determined by the DBTSS. (B) The sINR is a strict consensus and differs from other known INRs in its flanking sequences. The upper panel shows a graphical representation of multiple sequence alignment (http://weblogo.berkeley.edu/logo.cgi) of sINR from 112 genes out of 191 that contain the element around their TSSs. Alignment of all 191 genes resulted in the same consensus (data not shown). Comparison of the sINR consensus to the broad INR consensus (middle panel) and to INR sequences of subset of genes that contain functional INR in their promoter (lower panel).
Functional classification of sINR-containing genes revealed statistically significant enrichment in specific biological activities associated with various aspects of RNA metabolism such as RNA processing and synthesis, regulation of transcription, nucleic acid metabolism and chromosome organization and biogenesis (Table 2). These categories differ from those found in TATA-containing genes, which are enriched, for example, in development, response to wounding, response to external stimulus and inflammatory response categories (29), all absent from sINR genes. These findings support the notion that core promoter type is linked to specific gene function.
Table 2.
Functional classification of genes bearing sINR element
| Cluster | Enrichment | Term | P-value |
|---|---|---|---|
| 1 | 9.52 | RNA processing | 6.3e-23 |
| 1 | 9.52 | RNA metabolism | 1.6e-21 |
| 2 | 8.42 | Regulation of transcription | 4.6e-27 |
| 2 | 8.42 | Nucleic acid metabolism | 4.7e-22 |
| 3 | 5.54 | Chromosome organization and biogenesis | 1.4e-7 |
The table describes clusters with the highest enrichment score within the gene list and the most significant terms within each cluster with their P-value.
sINR is an important transcriptional regulatory element
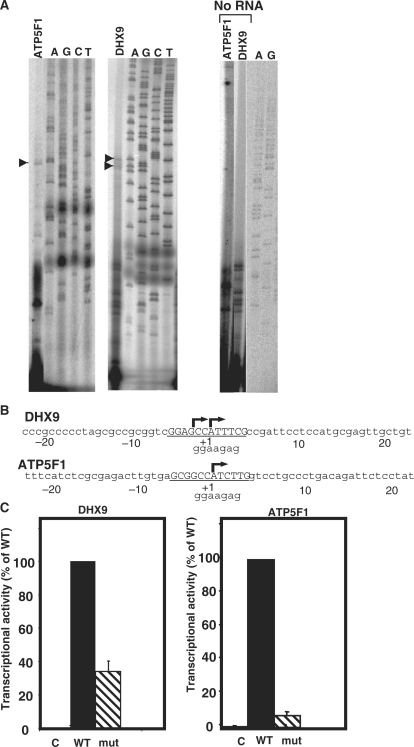
To assess the role of sINR in transcription, two sINR-containing genes, DHX9 and ATP5F1, were selected. First, we verified, by primer extension assay, that their TSS is located within their sINR. We used primers corresponding to +70 and +73 of DHX9 and ATP5F1, respectively, relative to the TSS assigned by the databases. In both genes, we found TSSs located within the sINR motif (Figure 2A and B). The short products seen in the analysis of DHX9 and ATP5F1 genes are RNA-independent primer extension products (right panel, no RNA lanes) and therefore are nonspecific. In the DHX9 gene an additional TSS, 2 nt upstream to the TSS specified in the database, was also observed.
Figure 2.
sINR is essential for transcription directed by DHX9 and ATP5F1 promoters. (A) Determination, by primer extension, of the TSSs of the endogenous DHX9 and ATP5F1 genes using gene-specific primers as probes and total RNA prepared from 293T cells. The primer-extension products were run together with sequencing ladders (marked A, C, G and T). The TSSs are indicated by arrowheads and their positions are shown in (B). (B) The DNA sequences and the positions of TSSs of the DHX9 and ATP5F1 promoters. The TSSs are indicated by arrows and correspond to the TSS bands shown in (A). The sINR motif is underlined and the lower case letters underneath indicate the sequence of the mutation as shown in (C). (C) The effect of sINR mutation on transcription. The promoters of the DHX9 and ATP5F1 genes (from −150 to +50 and −55 to +60 of DHX9 and ATP5F1, respectively) were cloned in front of a firefly luciferase reporter gene and then subjected to site-directed mutagenesis to create sINR mutants. The wild type (WT) or mutated (mut) promoter or the promoter-less parental plasmid (pGL2-basic, C) was co-transfected into 293T cells with RSV-renilla luciferase that serves as a reference for transfection efficiency. Twenty-four hours post transfection firefly and renilla luciferase activities were measured. The results shown are the average ± SD of four independent experiments, the activity of the wild type promoters being 100%. Primer extension analysis of the wild type or mutated DHX9 promoter is shown in Supplementary Data 2.
Next, the promoters of these genes (from −150 to +50 and −155 to +60 of DHX9 and ATP5F1, respectively) were cloned in front of a firefly luciferase reporter gene, and then site-directed mutagenesis was used to generate sINR mutants. The constructs were co-transfected into 293T cells together with a RSV-renilla luciferase reporter plasmid that serves as a reference for transfection efficiency. Twenty-four hours post transfection, firefly and renilla luciferase activities were measured. As shown in Figure 2C, both promoters (WT columns) displayed significant promoter activity compared with the promoter-less control construct pGL2-basic (C columns). The mutation in the sINR caused a dramatic decrease in the activity of both promoters, indicating that the sINR motif is important for transcription.
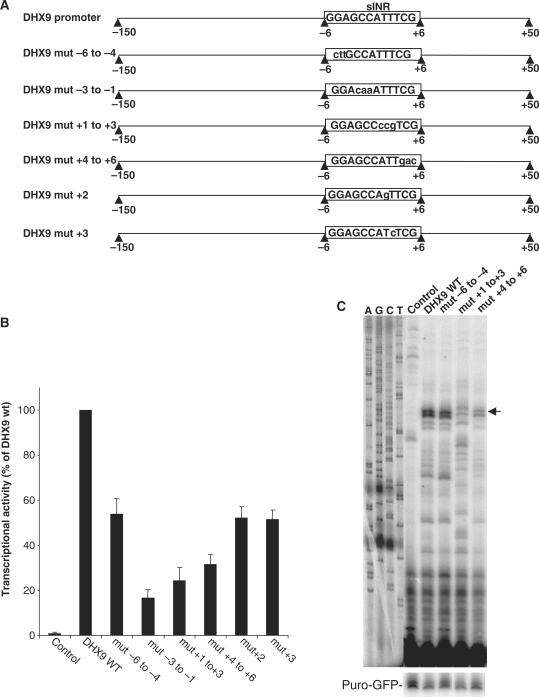
The sequence requirements for sINR to act as a transcriptional element were examined in more detail by mutating several successive blocks within the motif in the DHX9 promoter. For substitution, nucleotides were selected that occur least frequently in each position as determined by the bioinformatics analysis. In addition, two point mutations were generated in which the thymidine at position +2 was replaced by a guanosine residue and the thymidine at position +3 was changed to a cytosine residue (Figure 3A). The wild type and mutated constructs were transfected into 293T cells and their luciferase activity was measured. All the mutations within the motif, including the nucleotides flanking the core INR and single substitutions of the thymidine residues in positions +2 and +3, decreased transcription (Figure 3B). The mutations in positions −3 to +6 of the core INR had the most severe effect on transcription. Figure 3C shows the effect of some of the mutations on the TSS by primer extension. The mutation in position +1 to +3 changed the TSS and reduced transcription; whereas, mutations in other positions affected the promoter strength. These results show that the entire sequence of the sINR is important for transcription. The point mutation in position +2 (T to G) is of particular interest as this position is dispensable in the broad INR (YYANWYY); whereas, its mutation in sINR significantly decreased DHX9 promoter activity.
Figure 3.
The sequence requirements for sINR function. (A) A scheme of sINR mutants. The mutated sequences within sINR of the DHX9 promoter are shown in lower case letters. (B) The wild type and mutated promoters (fused to firefly luciferase reporter gene) were transfected into 293T cells together with RSV-renilla that serves as internal control. Twenty-four hours post transfection firefly and renilla luciferase activities were measured. The normalized results are the mean of at least four independent experiments (±SD), wild type DHX9 promoter activity being 100%. (C) Position of the TSS of wild type (WT) and the indicated representative mutants were determined by primer extension. The primer-extension products were run together with sequencing ladders (A, C, G and T).
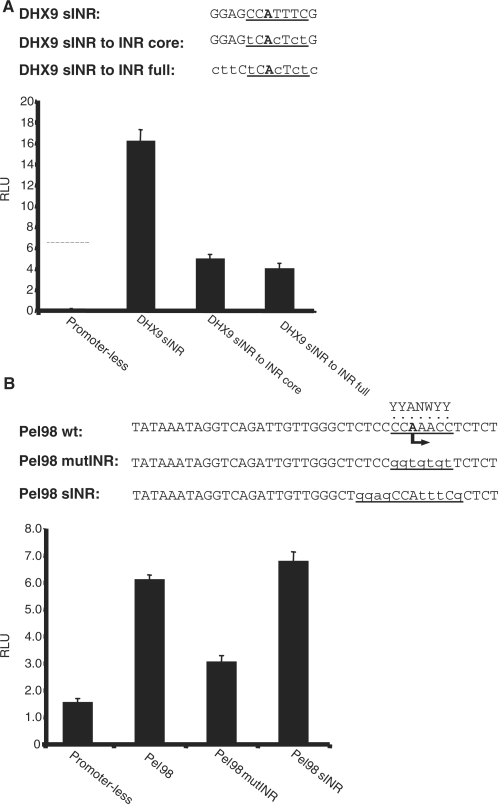
DHX9 sINR was not efficiently substituted by a conventional INR
To examine in more detail the functional relationship between the general INR to sINR, we constructed a DHX9 promoter in which sINR was mutated to a conventional INR sequence derived from the AdML promoter (TCACTCT), with or without the flanking sequences of sINR (Figure 4A). These constructs were transfected into 293T cells and their luciferase activity was compared with that of the wild type DHX9 promoter. Remarkably, a DHX9 promoter bearing the AdML INR had very low activity (Figure 4A), comparable to the activities of sINR mutants that do not match the INR consensus (Figures 2 and 3). These findings suggest that a conventional INR could not efficiently substitute for sINR function, whether or not sINR flanking sequences were present. We also constructed an AdML promoter bearing sINR instead of its INR, but neither the wild type nor sINR-containing promoter had significant activity relative to the promoter-less plasmid in transfected cells. We therefore replaced the INR of another gene, Pel98, that we showed to have a functional INR (K.Gazit et al., submitted for publication), with the DHX9 sINR (Figure 4B). Wild type, INR mutant and sINR replacement mutant were transfected into MEFs, where the Pel98 promoter is active, and their luciferase activity was measured. The results confirm that the Pel98 INR is functional, as when mutated luciferase activity is decreased 2-fold (Figure 4B). sINR fully compensated for the Pel98 INR activity (Figure 4B). Thus, sINR can effectively replace a conventional INR.
Figure 4.
Functional relationship between sINR and INR. (A) The core or the full sINR of DHX9 promoter was mutated to fit the AdML INR sequence as shown in the upper panel. The wild type and mutated promoters were analyzed as in Figure 3. The results are average of four independent experiments (±SD). (B) The INR of the Pel98 promoter was either mutated or replaced by sINR as shown in the upper panel. The Pel98 promoter derivatives (fused to firefly luciferase reporter gene) were transfected into MEFs together with RSV-renilla that serves as internal control. Twenty-four hours post transfection firefly and renilla luciferase activities were measured. The results are average of four independent experiments (±SD).
Upstream Sp1 site and a unique inverted repeat sequence cooperate with sINR to activate the DHX9 promoter
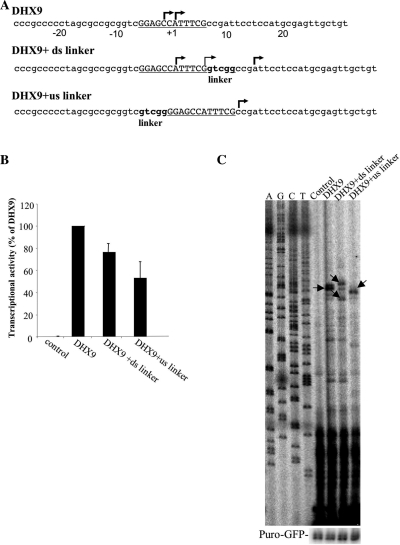
The INR core promoter element functions in combination with additional core promoter elements such as TATA or DPE. Both DHX9 and ATP5F1 promoters, analyzed above, lack these elements. To determine whether there is an element(s) that cooperates with sINR and is located at a specific position and distance from it, we inserted a 5-nt linker either downstream or upstream to the sINR motif in the DHX9 gene promoter as illustrated in Figure 5A. Assuming the existence of an additional element, insertion of the linker would determine whether it is located at a fixed distance from the sINR, like the DPE and the INR elements that are co-localized on the same core promoter at a fixed distance (13). The 5-nt length of the linker is expected to change the spatial proximity of sINR with upstream or downstream elements, by a half helical turn. Wild type and modified DHX9 promoters were co-transfected into 293T cells and 24 h post transfection firefly and renilla luciferase activities were measured. Both the downstream and the upstream linker in the DHX9 promoter caused reduction in the activity of the reporter gene (Figure 5B), but the effect of the upstream linker was more severe (∼2-fold P = 3.95 × 10−8) than the reduction with the downstream linker (∼1.25 fold P = 3.34 × 10−7).
Figure 5.
(A) The sequence of DHX9 wild type and linker mutated constructs. (B) Firefly luciferase reporter gene driven by the DHX9 promoter and the linker mutant derivatives and the promoter-less reporter were transfected into 293T cells together with RSV-renilla luciferase that served as control for transfection efficiency. Twenty-four hours later firefly and renilla luciferase activities were measured and the relative activity is presented as ratio of DHX9 wild type promoter. Control indicates the activity of the promoter-less reporter, ‘ds’ and ‘us’ denote downstream and upstream, respectively. (C) The effect of DHX9 promoter linker mutants on TSS selection. Wild type and mutated constructs were transfected into 293T cells together with puro-GFP and their mRNA levels were monitored by primer extension and normalized with puro-GFP mRNA. Control indicates the activity of the promoter-less reporter, ‘ds’ and ‘us’ denote downstream and upstream, respectively. The positions of the TSSs are shown in (A) by arrows.
We next analyzed the effect of these linkers on TSS location using primer extension (Figure 5C). We observed that the main TSS (at the A residue position +1) of the wild type construct appears in the construct with the downstream linker that also has two more TSSs (the TSS positions are shown in Figure 5A). In the presence of the upstream linker, however, the main TSS at position +1 disappears and there is a shift of the TSS outside of the sINR motif at position +7. These results confirm the presence of a cooperating element upstream to the sINR element in the DHX9 promoter, which might be dependent on the distance or the phasing of the sINR.
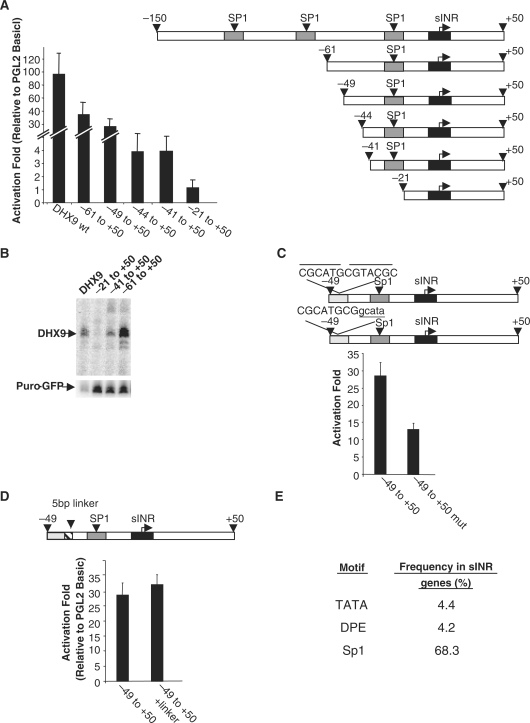
The DHX9 promoter contains three Sp1 sites upstream to the sINR element. To define the minimal upstream promoter sequence important for accurate transcription and to determine whether the Sp1 sites cooperate with the sINR element, constructs of the DHX9 promoter were created through progressive dissections as illustrated in Figure 6A (right panel) and the activity of these constructs was measured (Figure 6A, left panel). The −21 to +50 construct containing the sINR element but without Sp1 sites had no promoter activity as the observed level of activity was no different from the promoter-less pGL2-Basic. However, when the first upstream Sp1 site was included in the −41 to +50 and −44 to + 50 constructs, there was a 4-fold activation of the promoter over the pGL2 Basic. Addition of 5-bp upstream up to position −49 caused a dramatic increase in activity, up to 30-fold over the basic activity. These results show that the Sp1 site around position −21 and an additional element between positions −44 and −49 are critical for the DHX9 promoter activity. Sequences upstream to −49 also contribute to DHX9 promoter activity (Figure 6A).
Figure 6.
Defining the minimal sequence for DHX9 promoter with activity. (A and B) Schematic representation of promoter dissections (right panel). The positions of Sp1 and sINR are indicated. The reporter constructs were transfected into 293T cells and promoter activity was measured by luciferase assay (A) and primer extension (B) (left panel). (C) The inverted repeat sequence located at −49 to −37 of the DHX9 promoter (each repeat is indicated by an overhead line) and its mutant derivative. The promoter activity of these constructs after transfection to 293T cells was measured by the luciferase assay (lower panel). (D) The importance of the distance between sINR and the inverted repeat element. A 5-bp linker was inserted between sINR and the inverted repeat sequence (upper panel). Promoter activity, after transfection into 293T cells of the wt or the linker-bearing constructs, was measured by the luciferase assay. (E) The frequency of TATA, DPE and Sp1 in sINR-containing genes. These elements were searched for in sINR promoters using the minimal TATA box consensus TATAAA, the DPE consensus RGWYV and Sp1 concensus CCCCGCCCC allowing up to two mismatches, at their defined locations: TATA −35 to −25; DPE at +28, in addition to a G residue at position +14; Sp1 at position −300 to +100 in both strands relative to the TSS. The frequency of the motif in sINR promoters is presented as a percentage.
To determine which of the proximal elements contribute to TSS selection we performed primer extension assays (Figure 6B). The results show that the major TSS appears only when the first Sp1 is added to the promoter in the construct −41 to +50 and the intensity of this TSS is significantly enhanced in the −61 to +50 construct. These findings demonstrate the importance of the proximal Sp1 site for transcription initiation through sINR and the contribution of the −44 to −49 positions for promoter strength.
Inspection of the sequence of the enhancer element at position −44 to −49 and its surroundings revealed an inverted repeat between positions −37 to −49 (Figure 6C, right panel). When we mutated part of the repeat, at positions −37 to −41, promoter activity decreased significantly compared with the −49 to +50 construct (Figure 6C) suggesting that this inverted repeat is crucial for efficient transcription activity.
Since the Sp1 site at position −21 to −27 and the inverted repeat element at position −37 are both important for transcription from the DHX9 promoter, we next wanted to test which of these two upstream elements cooperates with the sINR element in a distance-dependent manner. We inserted a 5-nt linker just before the inverted repeat sequence at position −37, and it did not decrease the promoter activity (Figure 6D). In contrast, the linker between sINR and Sp1 did decrease transcriptional activity and also altered the TSS (Figure 5). These results suggest that sINR and Sp1 sites are responsible for accurate and efficient transcription initiation and that the distance between them is important. The inverted repeat element at position −37 to −49 acts as a strong enhancer element.
We checked the relevance of these findings to other sINR-containing genes by determining the tendency of sINR to occur with an upstream Sp1 or with a TATA box or DPE at their expected locations. The results (Figure 6E) revealed that while only a very small fraction of sINR genes bear TATA and DPE (4.4 and 4.2%, respectively), the vast majority (68.3%) of sINR-containing genes contain at least one Sp1 site.
sINR can substitute for a TATA element in a heterologous context containing Sp1 sites
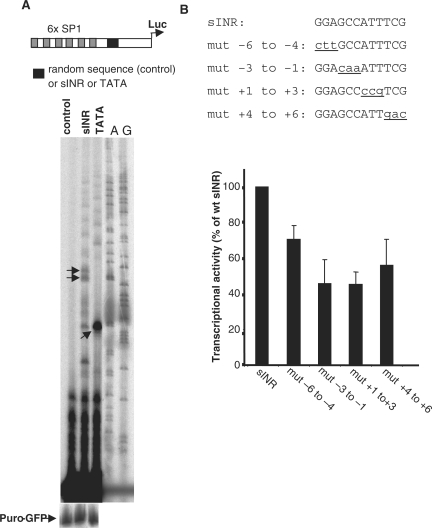
Investigating further the function of sINR as a core promoter element, we tested whether it can substitute for the TATA-like element of the SV40 early promoter, which contains six upstream tandem Sp1 binding sites. The core promoter region of the SV40 early promoter, from −45 to +42, was replaced with an oligonucleotide containing the sINR sequence. Oligonucleotides of the same size bearing a canonical TATA box consensus or an unrelated sequence were used as positive and negative controls, respectively. The constructs were transfected into 293T cells together with CMV-puro-GFP and mRNA levels of the transfected genes were analyzed by primer extension. As expected, the canonical TATA box replacement of the SV40 core promoter efficiently directed transcription from a single major initiation site located 29 nt downstream of the TATA box (Figure 7A). Likewise, sINR exhibits core promoter activity as evident from two visible initiation sites at the expected central A residue (+1) and at −2 position within sINR, as seen in the DHX9 promoter, whereas no activity was detected with the control sequence (Figure 7A). Together, these findings suggest that sINR, like the TATA box, is capable of directing efficient transcription initiation when positioned downstream to Sp1 sites. In this heterologous promoter context, we also examined the influence of several mutations in sINR (the mutations are illustrated at Figure 6B top panel) using the luciferase assay. The results show that all four mutations decreased promoter activity (Figure 7B). The most severe decrease in activity is observed in the mutations at position −3 to −1 and +1 to +3 of the sINR sequence, as occurred also with the DHX9 promoter (Figure 3B).
Figure 7.
sINR can substitute for the TATA-like element of the early SV40 promoter. (A) The TATA-like core promoter of the early SV40 promoter was replaced with either a random sequence (control), sINR sequence or canonical TATA box (upper panel). The SV40 promoter derivatives were transfected into 293T cells together with CMV puro-GFP and analyzed by primer extension 24 h later (lower panel). (B) The effect of mutations in sINR in the heterologous context of the SV40 promoter. Mutated sINR sequences (top panel) were analyzed by the luciferase assay. Normalized results (mean ± SD) of four independent experiments, the activity of the wild type sINR being 100%, are shown in the lower panel.
Transcription factor YY1 binds to sINR, but is dispensable for sINR function
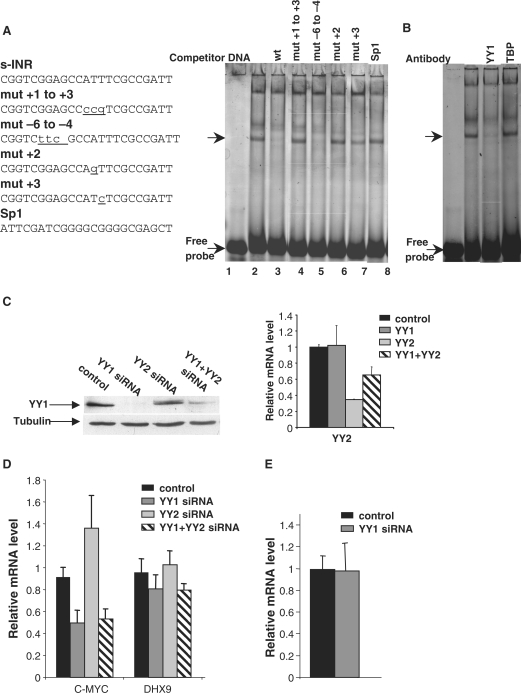
To determine which transcription factor binds to sINR, we employed the electrophoresis mobility shift assay (EMSA) using a fluorescently labeled oligonucleotide corresponding to the sINR sequence of DHX9 as a probe (Figure 8A, left panel) and nuclear extract prepared from HeLa cells. The results (Figure 8A, right panel) show that sINR formed two visible complexes with the extract (lane 2). The lower complex is specific to sINR since it is competed with by an excess of cold sINR but not with an oligo corresponding to Sp1 binding site (lanes 3 and 8). The complex was not competed with by an oligo bearing the +1 to +3 mutation or by substitution at position +2, but an oligo containing the mutation in the first 3 nt of the motif (−6 to −4 mutation) did compete efficiently with the probe. Moreover, the complex was slightly affected with by an oligo bearing the substitution at position +3. These findings are partially compatible with the functional analysis. The −6 to −4 mutation, which caused a decrease in transcription, retained binding activity; whereas, the other mutations (mut + 3 to +1, mut + 2 and mut + 3) that decreased transcription also failed to bind the complex. In light of these results, it is not clear whether the protein(s) that binds sINR in the nuclear extract also mediates its transcription regulatory function.
Figure 8.
Transcription factor YY1 binds sINR but is dispensable for sINR function. (A) EMSA using HeLa cell nuclear extract and a fluorescently labeled double stranded oligonucleotide containing DHX9 sINR. Lane 1 is the probe and lane 2 is the probe incubated with the nuclear extract. Competitor DNAs were added to the reactions in lanes 3–8 as indicated on the top. The specific protein–DNA complex and the free probes are indicated by arrows. The sequences of oligos used for binding and competition are shown in the left panel. (B) EMSA as in (A) with anti-YY1 and anti-TBP antibodies added to the reactions as indicated. (C) HeLa cells were transfected with YY1, YY2 or both (YY1 + YY2) siRNA expression plasmids or with the parental plasmid (control) along with 100 ng of puro-GFP. Twenty-four hours later puromycin was added for selection and after an additional 48 h the cells were harvested and subjected to RNA and protein extract preparation. YY1 depletion was monitored by western blot using YY1 and tubulin antibodies (left panel). YY2 depletion was monitored by real time PCR analysis using specific primers for the YY2 gene (right panel) and normalized to GAPDH mRNA. (D) DHX9, c-myc and GAPDH mRNA levels were quantified by real time PCR analysis using specific primers. DHX9 and c-myc levels were normalized to GAPDH and the mean ± SD of their relative levels from four independent transfection experiments are presented. (E) 293T cells were transfected with YY1 siRNA expression plasmid (siYY1) or with parental plasmid (control). Forty-eight hours later the cells were transfected again with either the YY1 siRNA expression plasmid or the control together with DHX9-luciferase reporter plasmid. Twenty hours later the cells were harvested and subjected to RNA and protein extract preparations. Luciferase mRNA level was quantified by real time PCR and the graph shows the mean ± SD of three independent transfection experiments. Normalization of transfection efficiency was done by measuring the mRNA level of the neomycin resistant gene under the SV40 early promoter that was co-transfected to the cells.
Previous studies have indicated that an INR sequence can be bound either by TFIID (10–12) or by the YY1 transcription factor (30). To test which of these proteins binds to sINR in nuclear extract, we added to the EMSA reactions antibodies specific to YY1 and the TFIID subunit TBP. As can be seen the YY1 antibodies supershifted the sINR complex, whereas the TBP antibody had no effect (Figure 8B). Thus, in vitro, YY1 appears to be the major sINR binding protein.
To examine further the involvement of YY1 in sINR function we used RNA interference (RNAi) to down-regulate the levels of YY1 or its paralog YY2. 293T cells were transfected with RNAi against YY1, YY2, both YY1 + YY2 or the parental expression vector pSuper. RNAi depletion of YY1 was confirmed by immunoblot (Figure 8C left panel). Since YY2-specific antibodies were not available to us, its depletion was verified by quantitative real time PCR using specific primers for YY2 (Figure 8C right panel). The effect of YY1 and YY2 depletion was determined on the mRNA levels of DHX9 and the c-myc (that serves as a control of a YY1 target gene), using quantitative real time PCR. As expected, the c-myc levels significantly decreased when YY1 was depleted (Figure 8D). In contrast, the DHX9 levels were unchanged by depletion of YY1, YY2 or both. We also determined the effect of YY1 knockdown on a luciferase reporter under the control of the DHX9 promoter. Forty-eight hours after transfection, total RNA was prepared and DHX9 mRNA quantitated using real-time RT PCR. We used the neomycin-resistant gene under the control of the SV40 promoter to normalize transfection efficiency. The results in Figure 8E show that down regulation of YY1 had no effect on the DHX9 promoter activity, as was observed for the endogenous DHX9. Thus, although YY1 binds the sINR in vitro and in vivo (data not shown), it seems to be dispensable for sINR function in transcription.
TAF1 is important for DHX9 promoter activity
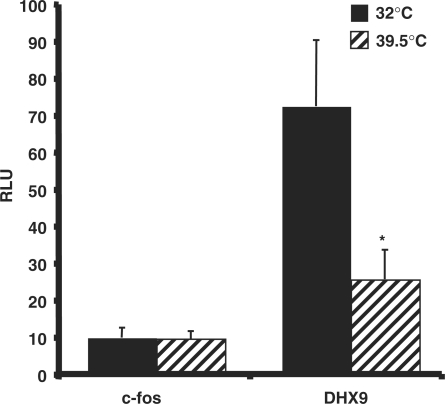
It is well established that TFIID can recognize and bind directly to the INR through the TAF1 and TAF2 subunits (10–12). To examine the involvement of TAF1 in transcription mediated by the sINR element, we used the ts13 temperature sensitive hamster cell line in which a point mutation in TAF1 renders TFIID inactive at 39.5°C. These cells were co-transfected with luciferase reporter plasmid driven by the DHX9 promoter or the TAF1-independent c-fos promoter. Twenty-four hours after the shift to 39.5°C luciferase activity was measured. The results in Figure 9A show that after the shift to the nonpermissive temperature, the DHX9 promoter activity is significantly decreased while c-fos promoter activity is being unchanged. This result implies that TAF1 and TFIID are important for transcription mediated by the DHX9 promoter.
Figure 9.
Dependency of the DHX9 promoter on TAF1. Hamster ts13 temperature sensitive cells were co-transfected with the indicated reporter plasmids. The cells were incubated at the permissive temperature (32°C) for 6 h, washed and then separated into two groups. One was grown at 32°C and the second at the nonpermissive temperature (39.5°C) for an additional 48 h, after which the cells were harvested and luciferase activity was measured. RSV promoter-driven renilla reporter luciferase plasmids served to normalize transfection efficiency. Results are the mean of three independent experiments, each with independent duplicates. The asterisk denotes that the difference of the DHX9 promoter activity at the different temperatures is significant, P < 0.01.
DISCUSSION
In this study, we have characterized sINR, a strict form of the mammalian INR core promoter element. sINR was found to occur in ∼1.5% of human promoters around the TSS. While the core sequence of sINR fits the consensus of the broad INR element consensus, it has several additional features that distinguish it from other initiators. The INR center of the motif is very strict and is flanked by additional conserved sequences. The sINR is specifically enriched in TATA-less genes which belong to specific functional categories including RNA processing and metabolism, regulation of transcription, nucleic acid metabolism and chromosome organization and biogenesis. Consistent with sINR being a TATA-less core promoter element, these functional groups are absent from those found with TATA-containing genes (29). Our findings provide support for the notion that core promoter type is linked to features beyond transcription initiation. For example, it has been reported that TATA box occurrence is correlated with increased divergence among yeast species, most likely due to higher rate of evolution (31) and that the TATA box is associated with a higher sensitivity of gene expression to mutations (32). A recent study from our lab demonstrated that in the NF-κB pathway, core promoter type is linked to differential regulation by transcription elongation factors (19). In addition, we found that the presence and the strength of a TATA box is tightly associated with gene length (29). Taking into consideration that the TATA box is linked to particular traits, it would be interesting to find the specific characteristics associated with sINR genes.
Detailed analysis of the DHX9 promoter sINR, which was used as a model, revealed that the entire sequence, including the conserved flanking sequences and the strict core, is important for transcription. Moreover, mutating sINR to a conventional INR sequence derived from the AdML promoter diminished DHX9 promoter activity to a level similar to sINR mutants that do not match INR consensus. In contrast, DHX9 sINR efficiently replaced a conventional INR of the Pel98 promoter. The minimal region of the DHX9 promoter required to drive promoter activity and accurate transcription initiation was found to include the sINR element and a proximal Sp1 site. In addition, the spatial arrangement of sINR and Sp1 appeared to be important as increasing the distance between the two elements decreased transcription and changed the TSS. The unique inverted repeat sequence at positions –37 to –49 is a strong enhancer of the DHX9 promoter. The cooperation between sINR and Sp1 is likely to be general as the sINR sequence could function as a core promoter element in a heterologous context when positioned downstream to several Sp1 sites. In addition the majority of native sINR-containing promoters contain Sp1. Thus sINR can be regarded as a TATA-less-specific INR that functions in cooperation with Sp1.
The transcriptional activity of Sp1 is manifested through several distinct activation domains (33,34), which are able to activate transcription through different core promoters (35). While full-length Sp1 activated transcription with equal efficiency through a TATA box or an INR core promoter, the isolated glutamine-rich activation domains preferentially activate an INR but not a TATA box containing core promoter suggesting that different mechanisms are used by Sp1 to communicate with the transcriptional machinery according to the core promoter type.
Our study suggests that two possible factors may be involved in sINR function. The YY1 protein was the major and specific sINR binding protein in nuclear extracts, and it also associates with the DHX9 promoter in vivo in chromatin immunoprecipitation assays (data not shown). However, YY1 and the related YY2 siRNA experiments did not provide evidence that YY1 is the major protein directing the activity of DHX9 sINR in transcription. This observation is also supported by the finding that the effect of certain mutations in sINR did not fully correlate with YY1 binding activity. YY1 has been implicated in INR function in the adeno-associated virus type 2 P5 promoter (30); however, its involvement in the function of the initiator of the human DNA polymerase β gene has been ruled out (36).
The second factor involved in sINR function is TFIID, a basal transcription factor that can recognize and bind directly to the INR through its TAF1 and TAF2 subunits (10–12). We found that the activity of the DHX9 promoter to be sensitive to TAF1 mutation suggesting that in this promoter TAF1 is important for sINR activity. However, considering the strict INR core and the conserved and functional flanking sequences it is possible that other TAFs or other factor(s) participate in the recognition of this element. The unique sequence and functional properties of sINR compared with other INR elements suggest that the INR core promoter element can be regarded as a family of related core promoter elements with common and distinct features.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Israel Science Foundation founded by the Israel Academy of Sciences and Humanities.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Yaara Azaria and Assaf Biran for constructing some of the plasmids used in this study and Dr. Sandra Moshonov for critical reading and editing of the manuscript.
REFERENCES
- 1.Smale ST, Kadonaga JT. The RNA polymerase II core promoter. Annu. Rev. Biochem. 2003;72:449–479. doi: 10.1146/annurev.biochem.72.121801.161520. [DOI] [PubMed] [Google Scholar]
- 2.Juven-Gershon T, Hsu JY, Kadonaga JT. Perspectives on the RNA polymerase II core promoter. Biochem. Soc. Trans. 2006;34:1047–1050. doi: 10.1042/BST0341047. [DOI] [PubMed] [Google Scholar]
- 3.Basehoar AD, Zanton SJ, Pugh BF. Identification and distinct regulation of yeast TATA box-containing genes. Cell. 2004;116:699–709. doi: 10.1016/s0092-8674(04)00205-3. [DOI] [PubMed] [Google Scholar]
- 4.Mencia M, Moqtaderi Z, Geisberg JV, Kuras L, Struhl K. Activator-specific recruitment of TFIID and regulation of ribosomal protein genes in yeast. Mol. Cell. 2002;9:823–833. doi: 10.1016/s1097-2765(02)00490-2. [DOI] [PubMed] [Google Scholar]
- 5.Ohler U, Liao GC, Niemann H, Rubin GM. Computational analysis of core promoters in the Drosophila genome. Genome Biol. 2002;3:RESEARCH0087. doi: 10.1186/gb-2002-3-12-research0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gershenzon NI, Ioshikhes IP. Synergy of human Pol II core promoter elements revealed by statistical sequence analysis. Bioinformatics. 2005;21:1295–1300. doi: 10.1093/bioinformatics/bti172. [DOI] [PubMed] [Google Scholar]
- 7.Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang C, Bolotin E, Jiang T, Sladek FM, Martinez E. Prevalence of the initiator over the TATA box in human and yeast genes and identification of DNA motifs enriched in human TATA-less core promoters. Gene. 2007;389:52–65. doi: 10.1016/j.gene.2006.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smale ST, Baltimore D. The “initiator” as a transcription control element. Cell. 1989;57:103–113. doi: 10.1016/0092-8674(89)90176-1. [DOI] [PubMed] [Google Scholar]
- 10.Chalkley GE, Verrijzer CP. DNA binding site selection by RNA polymerase II TAFs: a TAF(II)250-TAF(II)150 complex recognizes the initiator. EMBO J. 1999;18:4835–4845. doi: 10.1093/emboj/18.17.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaufmann J, Smale ST. Direct recognition of initiator elements by a component of the transcription factor IID complex. Genes Dev. 1994;8:821–829. doi: 10.1101/gad.8.7.821. [DOI] [PubMed] [Google Scholar]
- 12.Verrijzer CP, Chen JL, Yokomori K, Tjian R. Binding of TAFs to core elements directs promoter selectivity by RNA polymerase II. Cell. 1995;81:1115–1125. doi: 10.1016/s0092-8674(05)80016-9. [DOI] [PubMed] [Google Scholar]
- 13.Burke TW, Kadonaga JT. The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes Dev. 1997;11:3020–3031. doi: 10.1101/gad.11.22.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burke TW, Willy PJ, Kutach AK, Butler JE, Kadonaga JT. The DPE, a conserved downstream core promoter element that is functionally analogous to the TATA box. Cold Spring Harb. Symp. Quant. Biol. 1998;63:75–82. doi: 10.1101/sqb.1998.63.75. [DOI] [PubMed] [Google Scholar]
- 15.Deng W, Roberts SG. A core promoter element downstream of the TATA box that is recognized by TFIIB. Genes Dev. 2005;19:2418–2423. doi: 10.1101/gad.342405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lagrange T, Kapanidis AN, Tang H, Reinberg D, Ebright RH. New core promoter element in RNA polymerase II-dependent transcription: sequence-specific DNA binding by transcription factor IIB. Genes Dev. 1998;12:34–44. doi: 10.1101/gad.12.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee DH, Gershenzon N, Gupta M, Ioshikhes IP, Reinberg D, Lewis BA. Functional characterization of core promoter elements: the downstream core element is recognized by TAF1. Mol. Cell Biol. 2005;25:9674–9686. doi: 10.1128/MCB.25.21.9674-9686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elkan TLBaC. Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers; pp. 28–36. [PubMed] [Google Scholar]
- 19.Amir-Zilberstein L, Ainbinder E, Toube L, Yamaguchi Y, Handa H, Dikstein R. Differential regulation of NF-kappaB by elongation factors is determined by core promoter type. Mol. Cell Biol. 2007;27:5246–5259. doi: 10.1128/MCB.00586-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ainbinder E, Amir-Zilberstein L, Yamaguchi Y, Handa H, Dikstein R. Elongation inhibition by DRB sensitivity-inducing factor is regulated by the A20 promoter via a novel negative element and NF-kappaB. Mol. Cell Biol. 2004;24:2444–2454. doi: 10.1128/MCB.24.6.2444-2454.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sui G, Affar El B, Shi Y, Brignone C, Wall NR, Yin P, Donohoe M, Luke MP, Calvo D, Grossman SR, et al. Yin Yang 1 is a negative regulator of p53. Cell. 2004;117:859–872. doi: 10.1016/j.cell.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 22.Amir-Zilberstein L, Dikstein R. Interplay between E-box and NF-kappaB in regulation of A20 gene by DRB sensitivity-inducing factor (DSIF) J. Biol. Chem. 2008;283:1317–1323. doi: 10.1074/jbc.M706767200. [DOI] [PubMed] [Google Scholar]
- 23.Cherrier M, D’Andon MF, Rougeon F, Doyen N. Identification of a new cis-regulatory element of the terminal deoxynucleotidyl transferase gene in the 5′ region of the murine locus. Mol. Immunol. 2008;45:1009–1017. doi: 10.1016/j.molimm.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 24.Cowan MJ, Yao XL, Pawliczak R, Huang X, Logun C, Madara P, Alsaaty S, Wu T, Shelhamer JH. The role of TFIID, the initiator element and a novel 5’ TFIID binding site in the transcriptional control of the TATA-less human cytosolic phospholipase A2-alpha promoter. Biochim. Biophys. Acta. 2004;1680:145–157. doi: 10.1016/j.bbaexp.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 25.Laniel MA, Poirier GG, Guerin SL. A conserved initiator element on the mammalian poly(ADP-ribose) polymerase-1 promoters, in combination with flanking core elements, is necessary to obtain high transcriptional activity. Biochim. Biophys. Acta. 2004;1679:37–46. doi: 10.1016/j.bbaexp.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 26.Ren D, Nedialkov YA, Li F, Xu D, Reimers S, Finkelstein A, Burton ZF. Spacing requirements for simultaneous recognition of the adenovirus major late promoter TATAAAAG box and initiator element. Arch. Biochem. Biophys. 2005;435:347–362. doi: 10.1016/j.abb.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 27.Young DA, Phillips BW, Lundy C, Nuttall RK, Hogan A, Schultz GA, Leco KJ, Clark IM, Edwards DR. Identification of an initiator-like element essential for the expression of the tissue inhibitor of metalloproteinases-4 (Timp-4) gene. Biochem. J. 2002;364:89–99. doi: 10.1042/bj3640089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao Y, Tang F, Cheng J, Li L, Xing G, Zhu Y, Zhang L, Wei H, He F. An initiator and its flanking elements function as a core promoter driving transcription of the Hepatopoietin gene. FEBS Lett. 2003;540:58–64. doi: 10.1016/s0014-5793(03)00158-3. [DOI] [PubMed] [Google Scholar]
- 29.Moshonov S, Elfakess R, Golan-Mashiach M, Sinvani H, Dikstein R. Links between core promoter and basic gene features influence gene expression. BMC Genomics. 2008;9:92. doi: 10.1186/1471-2164-9-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seto E, Shi Y, Shenk T. YY1 is an initiator sequence-binding protein that directs and activates transcription in vitro. Nature. 1991;354:241–245. doi: 10.1038/354241a0. [DOI] [PubMed] [Google Scholar]
- 31.Tirosh I, Weinberger A, Carmi M, Barkai N. A genetic signature of interspecies variations in gene expression. Nat. Genet. 2006;38:830–834. doi: 10.1038/ng1819. [DOI] [PubMed] [Google Scholar]
- 32.Landry CR, Lemos B, Rifkin SA, Dickinson WJ, Hartl DL. Genetic properties influencing the evolvability of gene expression. Science. 2007;317:118–121. doi: 10.1126/science.1140247. [DOI] [PubMed] [Google Scholar]
- 33.Courey AJ, Tjian R. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell. 1988;55:887–898. doi: 10.1016/0092-8674(88)90144-4. [DOI] [PubMed] [Google Scholar]
- 34.Kadonaga JT, Courey AJ, Ladika J, Tjian R. Distinct regions of Sp1 modulate DNA binding and transcriptional activation. Science. 1988;242:1566–1570. doi: 10.1126/science.3059495. [DOI] [PubMed] [Google Scholar]
- 35.Emami KH, Navarre WW, Smale ST. Core promoter specificities of the Sp1 and VP16 transcriptional activation domains. Mol. Cell Biol. 1995;15:5906–5916. doi: 10.1128/mcb.15.11.5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weis L, Reinberg D. Accurate positioning of RNA polymerase II on a natural TATA-less promoter is independent of TATA-binding-protein-associated factors and initiator-binding proteins. Mol. Cell Biol. 1997;17:2973–2984. doi: 10.1128/mcb.17.6.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.