Abstract
Intron removal during pre-messenger RNA (pre-mRNA) splicing involves arrangement of snRNAs into conformations that promote the two catalytic steps. The Prp19 complex [nineteen complex (NTC)] can specify U5 and U6 snRNA interactions with pre-mRNA during spliceosome activation. A candidate for linking the NTC to the snRNAs is the NTC protein Cwc2, which contains motifs known to bind RNA, a zinc finger and RNA recognition motif (RRM). In yeast cells mutation of either the zinc finger or RRM destabilize Cwc2 and are lethal. Yeast cells depleted of Cwc2 accumulate pre-mRNA and display reduced levels of U1, U4, U5 and U6 snRNAs. Cwc2 depletion also reduces U4/U6 snRNA complex levels, as found with depletion of other NTC proteins, but without increase in free U4. Purified Cwc2 displays general RNA binding properties and can bind both snRNAs and pre-mRNA in vitro. A Cwc2 RRM fragment alone can bind RNA but with reduced efficiency. Under splicing conditions Cwc2 can associate with U2, U5 and U6 snRNAs, but can only be crosslinked directly to the U6 snRNA. Cwc2 associates with U6 both before and after the first step of splicing. We propose that Cwc2 links the NTC to the spliceosome during pre-mRNA splicing through the U6 snRNA.
INTRODUCTION
The removal of intron regions from pre-messenger RNA (pre-mRNA) by the process of pre-mRNA splicing is an essential step during gene expression. Intron regions must be removed accurately and efficiently to produce mRNA that retains the original protein coding potential of the gene. Pre-mRNA splicing is carried out by a large RNA/protein complex called the spliceosome. The spliceosome contains five small nuclear ribonucleoprotein particles (snRNPs) that are RNA/protein complexes named after the single small nuclear RNA (U1, U2, U4, U5 or U6 snRNA) that each contains. In addition to the snRNPs, there are also numerous non-snRNP protein factors that associate with the spliceosome that are essential for pre-mRNA splicing (1).
A number of interactions take place to form the active spliceosome which is competent for intron removal (2,3). First, the U1 snRNP interacts with the 5′-splice site region. Next, the U2 snRNP interacts with the branchpoint sequence. Following interaction of the U1 and U2 snRNPs with the pre-mRNA, the U4/U6.U5 tri-snRNP interacts with the pre-mRNA and the bound U1 and U2 snRNPs. After joining of the tri-snRNP, structural rearrangements follow that destabilize the association of the U1 and U4 snRNPs with the spliceosome. These structural rearrangements include changes in RNA/RNA interactions that set up the spliceosome for catalysis. Specifically the U1 snRNA base pairing at the 5′-splice site is replaced by a U5 snRNA interaction with the 5′-exon and a U6 snRNA interaction with the 5′-end of the intron. In addition, U4/U6 base pairing is replaced by U6 base pairing with the U2 snRNA. These RNA/RNA interactions form the active site of the spliceosome for the first step of splicing. The second step of splicing occurs rapidly after the first step producing the mRNA molecule. The spliceosome then disassembles and the snRNPs are recycled for use in another round of splicing.
Protein factors have been implicated in mediating the RNA rearrangements required for spliceosome activation. In yeast, Prp28 is involved in the displacement of U1 from the 5′-splice site (4). Brr2 catalyzes U4/U6 snRNA unwinding during spliceosome activation (5) and U2/U6 unwinding after splicing, with Brr2 activity being regulated by Snu114 (6). Following U1 and U4 destabilization from the spliceosome, a protein complex associated with the Prp19 protein termed the NTC (nineteen complex) is required for the stable association of the U5 and U6 snRNAs with the 5′-splice site region (7,8). The NTC specifies and stabilizes dynamic interactions of U5 and U6 with the 5′-splice site (7). The NTC is also required for dissociation of the Lsm complex from the 3′-end of U6 allowing the 3′-end to contact the intron (7). The NTC then remains associated with the spliceosome during the two catalytic steps of splicing. The NTC consists of at least 10 protein components in the yeast Saccharomyces cerevisiae (8,9). In addition to their role in activation of the spliceosome, proteins of the NTC have also been implicated in 3′-splice site surveillance as a number of NTC proteins interact with second step splicing factors (9–12) and the human NTC complex is required for the second step of splicing (13). The NTC has recently been found to have an indirect role in spliceosome recycling by influencing U4/U6 stability/biogenesis (14).
The function of the NTC during activation of the spliceosome undoubtedly involves interactions with the snRNAs, but no NTC component has been found to bind RNA. Cwc2/Ntc40 is the only protein of the NTC complex that contains known motifs for RNA binding, namely a zinc finger and an RNA recognition motif (RRM) (9). Cwc2 is required for splicing and has been demonstrated to interact with Prp19 by far western, in vitro binding and two-hybrid analysis (9,15). Two-hybrid analysis has further defined the Cwc2/Prp19 interaction domains to the C-terminal portion of Cwc2 (downstream from the zinc finger and RRM) and the C-terminal two-thirds of Prp19 including the WD40 domain (9). Cwc2 also interacts with Ntc30/Isy1, another protein of the NTC (9). To date, there is no information regarding whether Cwc2 interacts with the RNA components of the spliceosome.
To further define the function of the NTC in the activation of the spliceosome, we set out to investigate if Cwc2 interacts with the RNAs of the spliceosome. Depletion of Cwc2 in vivo results in the destablization of U1, U4, U5 and U6 snRNA, leading to reduced levels of the U4/U6 complex. Purified Cwc2 can bind directly to all five spliceosomal snRNAs and three different pre-mRNAs in vitro. In splicing extracts Cwc2 is not snRNP associated, but during splicing Cwc2 interacts with spliceosomes containing the U2, U5 and U6 snRNAs. Mutation of Cwc2 revealed that the zinc finger motif and the RRM are essential for Cwc2 stability. A purified RRM fragment of Cwc2, lacking the zinc finger, binds U1, U2, U4 and U6 snRNAs in vitro, but not U5 snRNA. We propose that Cwc2 displays general RNA binding properties that are mediated at least in part by the RRM and possibly the zinc finger motif. Within splicing extracts Cwc2 crosslinks directly to the U6 snRNA and we suggest that the general RNA binding properties of Cwc2 may be regulated by other NTC/spliceosomal components. Overall, we propose that Cwc2 provides the link between the NTC and the spliceosome through the U6 snRNA.
MATERIALS AND METHODS
Plasmid construction and mutagenesis
The CWC2 (YDL209C) gene was cloned by PCR amplification of S. cerevisiae genomic DNA (Promega) with primers 209FG-5′-GGTCTAGATTTCTAATATGGTTAAGAAGCG and 209BG-5′-GGGGATCCTTGCAGGTATCTACTACAAAG containing restriction sites for XbaI and BamHI, respectively. The PCR product was digested with XbaI and BamHI, then ligated into pRS416 (16) digested with the same enzymes to produce pRS416-Cwc2. The identity of the insert was confirmed by sequencing. The XbaI/BamHI fragment containing the CWC2 gene was then excised from pRS416-Cwc2 and ligated into pRS413 to produce pRS413-Cwc2. A 3FLAG tagged version of Cwc2 in pRS413 was produced by overlap PCR of pRS413-Cwc2 with primer pairs 209FG/Cwc2FLAGB (5′-CTTTGTCGTCATCGTCCTTGTAATCTT TGTCATCATCGTCTTTGTAATCC TCATCAGAGGAGAGGT) and 209BG/Cwc2FLAGF (5′-TTACAAGGACGATGACGACAAAGATTACAAGGATGATGATGATAAGTAGAGAAGAAAATCGTTTCC) then ligation into pRS413 to produce pRS413-Cwc2-3FLAG. Mutagenesis of pRS413-Cwc2-3FLAG was carried out according to the method of Kunkel (17) with primers: C73Y-5′-TGCAAAAAATAGATAGAAGAACAACTG; T136V-5′-ACCTACATACAAAACCTTGTTTTTCTT; Y34A-5′-CCCTTGGGACCATTTATTAGCCCATATATTAAACTGCAATCC; W37A-5′-CGCAAACCCTTGGGAAGCTTTATTATACCATATATTAAACG; C73Y-5′-TGCAAAAAATAGATAGAAGAACAACTG; C86H-5′-GGTATATGATGCAGATATTCATGTTTAGGGCCAAGGCAAC; Y138A-5′-AATACCACCTACCGCCAATGTCTTGTT; F183D-5′-GCATTAGCTTGATATTTAAACTTCACATCTCCACAGTTTTTGCTCTCAAC; F186D-5′-CAAATTCTGCATTAGCTTGATATTTATCCTTCACAAATCCACAGTTTTTG. The ACT1 pre-mRNA was excised from plasmid p283 (18) with SacI and XbaI and inserted into SacI/XbaI digested pBluescript II KS+ (Stratagene) in the T7 orientation to produce plasmid pBS-Actin. Mutagenesis of pBS-Actin to produce pBS-ActinACAC for production of ACAC pre-mRNA was carried out according to the method of Kunkel (17) with primer ActACAC-5′-GCTATATTATATGTTT ACACGTTGCTGCTTTGG. All mutagenesis was confirmed by sequencing.
Yeast manipulation
For analysis of CWC2 mutations, the diploid strain Y23907 (BY4743; Mat a/α; his3Δ1/his3Δ1; leu2Δ0/leu2Δ0; lys2Δ0/LYS2; MET15/met15Δ0; ura3Δ0/ura3Δ0; YDL209c::kanMX4/YDL209c) was obtained from EUROSCARF and was transformed with pRS416-Cwc2 by lithium acetate transformation (19). Viable G418-resistant and Ura+ transformants were sporulated and tetrads dissected. The haploid progeny were analyzed to identify the strain YCWC2KO (BY4743; Mat a; his3Δ1; leu2Δ0; lys2Δ0; MET15; ura3Δ0; YDL209c::kanMX4; pRS416-Cwc2). This strain was transformed with pRS413-Cwc2-3FLAG plasmids containing different CWC2 mutations and cells were tested for viability by plasmid shuffle on 5-fluoro-orotic acid (5-FOA) (20). A yeast strain with the CWC2 gene under control of the GAL1 promoter was constructed by PCR of plasmid pFA6a-kanMX6-PGAL1 (21) with primers 209GALF-5′-ATTATGTATTGAAGGCTGGATCATAATAA AGACTACTGGCGATGGGAATTCGAGCTCGTTTAAAC and 209GALB-5′- TGACTCCTTCACTTGCACTTTCGCGGATTTATCCCTCCA AGATGTCATTTTGAGATCCGGGTTTT. The resulting PCR product was transformed into yeast strain BY4743 (Mat a/α; his3Δ1/his3Δ1; leu2Δ0/leu2Δ0; lys2Δ0/LYS2; MET15/met15Δ0; ura3Δ0/ura3Δ0) and G418-resistant colonies were selected. Integration of the GAL1 promoter was confirmed by PCR and these strains were sporulated and tetrads dissected. Haploid progeny were analyzed to identify the strain YGALCWC2 (BY4743; Mat a; his3Δ1; leu2Δ0; lys2Δ0; MET15; ura3Δ0; kanMX6:PGAL1-CWC2). TAP tagged CWC2, PRP21 and PRP11 were produced by PCR of plasmid pYM13 (22) with primers Cwc2TAPF-5′-GGTAAGCTTGGTGGTCCCTTGCTCGATTA CCTCTCCTCTGATGAGGATCGTACGCTGCAGGTCGAC and Cwc2TAPB-5′-TGTTAATGGCAGATACTTAGAATTTGGTGGAAA CGATTTTCTTCTCTAATCGATGAATTCGAGCTCG, Prp21TAPF-5′-CGTAAAATT AGAGCTGTGGGAGAGACAAGGTTAAAGAAAA GTAAAAAACGTACGCTGCAGGTCGAC and Prp21TAPB-5′-CATCAACCTTTACATACTTCTA GCTAACTTTGATTTTCATGAGGCTTAATC GATGAATTCGAGCTCG or Prp11TAPF-5′-TACGTTCAATTTTTCTTTAAAC AAGCCGAACAAGAACAAGCCGATGTACGTAC GCTGCAGGTCGAC and Prp11TAPB-5′-AGAGTTTTGTATGCCGA TTATTAACGTTACCCAAAAATA ATACTGTCAATCGATGAATTCGAGCTCG. The resulting PCR products were transformed into yeast strain BJ2168 (Mat a; prc1-407; prb1-122; pep4-3; leu2; trp1; ura3-52; gal2) and G418-resistant colonies were selected. Integration of the TAP tag was confirmed by PCR and the strains called YCWC2TAP, YPRP21TAP and YPRP11TAP. Growth curves in liquid culture were obtained as previously described (23).
Analysis of Cwc2 expression
Yeast strain YCWC2KO was transformed with the CEN HIS3 pRS413-Cwc2-3FLAG plasmid containing either wild-type or mutant Cwc2-3FLAG. Overnight cultures grown under selection for the CEN URA3 plasmid, containing the wild-type CWC2 gene, and the CEN HIS3 plasmid were used to seed 25 ml cultures at an OD600 of 0.5. These were grown for 4 h at 30°C, harvested at 2000g for 5 min, washed, repelleted and resuspendend in 200 µl SDS sample buffer (60 mM Tris–HCl pH 6.8, 10% glycerol, 2% SDS, 5% 2-mercaptoethanol, 0.5% Bromophenol blue). Acid-washed glass beads (Sigma, 425–600 µ) were added to the liquid level before addition of a further 200 µl SDS sample buffer. Samples were then boiled for 5 min, vortexed for 3 min and 30 µl was used for SDS–PAGE and western blotting. Anti-FLAG M2 (Sigma) was used at a concentration of 1:20 000 and anti-Zwf1 (G6PDH, Sigma) antibody at 1:50 000.
Primer extension and immunoprecipitation
RNA isolation and primer extension to determine U3 snoRNA splicing and snRNA levels in the YGALCWC2 strain were as previously described (23) using primers U3snoRNA-RT, U1H, U5RT, U2RT2, U6B or U4S2. For immunoprecipitation of snRNAs from the YCWC2TAP and YPRP21TAP strains, yeast whole-cell extract was produced as described (24). Extract (50 µl) was either diluted with 350 µl IPP150 (10 mM Tris–HCl pH 8, 150 mM NaCl, 0.5% Igepal-630) or used in a 125 µl splicing reaction for 30 min at 23°C, then diluted with 275 µl IPP150. Splicing reactions were carried out according to published procedures (25) except that 4 nM Ac/Cla or ACAC pre-mRNA was utilized to accumulate spliceosomes. Extract diluted with IPP150 was mixed with 50 µl rabbit IgG agarose beads (Sigma) and incubated at 4°C for 45 min. Beads were then washed four times with IPP150, the last wash removed, then beads suspended in splicing diluent (0.3 M NaOAc pH 3, 1 mM EDTA, 0.1% SDS, 25 µg/ml E.coli tRNA). The beads were phenol extracted three times and RNA precipitated from the aqueous phase. Precipitated RNA and RNA isolated directly from the extract were subjected to primer extension as previously described (23) with primers U1RT136-5′-GACCAAGGAGTTTGCATCAATGAC, U2RTAll-5′-GCCAAAAAATGTGTATTGT AAC, U4RTAll-5′-GGTATTCCAAAAATTCCCTACATAGTC, U5RT-5′-AAAAATATGGCAGG CCTACAGTAACGG and U6RTAll-5′-TCATCCTTATGCAGGG. Extension reactions were separated on 40 cm 6% acrylamide/7 M urea gels.
Crosslinking
YCWC2TAP and YPRP11TAP whole-cell extract (100 µl) was used in 250 µl splicing reactions for 30 min at 23°C. Splicing reactions were carried out according to published procedures (25) except that 4 nM Ac/Cla or ACAC pre-mRNA was utilized to accumulate spliceosomes. Reactions were irradiated three times at 411 mJ/cm2 in a Stratalinker (Stratagene model 2400) on ice before addition of 250 µl 2× binding buffer (20 mM Tris–HCl pH 7.5, 1 M NaCl, 1% SDS, 0.2 mM EDTA). Streptavidin coated Dynabeads (Dynal m270) were prepared by washing 200 µl slurry three times with 400 µl 1× binding buffer, incubation at room temperature for 15 min with 1 µM biotinylated oligonucleotide complementary to U5, U2 or U6 (U5BioTEG 5′-AAGTTCCAAAAAATATGGCAAGC, U2BioTEG 5′-TTGGGTGCCAAAAAATGTG, U6BioTEG 5′-AAAACGAAATAAATCTCTTTG) in 1× binding buffer and washing a further three times with 400 µl 1× binding buffer. Beads were then added to the crosslinked splicing reactions and incubated at 30°C for 1 h, mixing once during this time. After washing three times with 1 ml 1× binding buffer proteins were eluted in 400 µl elution buffer (10m Tris–HCl pH 7.5, 1 mM EDTA) with 4 µl RNase cocktail (Ambion) at 37°C for 30 min. Proteins were then precipitated by addition of 100 µl 100% TCA, 0.4% sodium deoxycholate on ice for 20 min and pelleted at 16 100g at 4°C for 15 min. Pellets were washed with cold acetone and resuspended in SDS sample buffer. Western blotting was then carried out using anti-TAP antibody (Open Biosystems) at a dilution of 1:2000.
Analysis of the U4/U6 complex
The U4/U6 complex was analyzed by solution hybridization (26). Briefly, a 5 ml overnight culture of the YGALCWC2 strain in YPGal was used to start 25 ml 4 and 8 h cultures in YPD and YPGal at an OD600 of 0.5 and 0.2, respectively. These were grown for 4 or 8 h to an OD600 between 1 and 2. Cells were then diluted with 25 ml ice-cold water and pelleted at 2000g for 5 min. Pellets were resuspended in 1 ml cold RNA buffer (0.5 M NaCl, 200 mM Tris–HCl pH 7.5, 10 mM EDTA), transferred to a 1.5 ml tube and centrifuged at 4.5g for 5 s. Pellets were then resuspended in 300 µl RNA buffer, vortexed with ∼200 µl acid washed glass beads for 2 min at 4°C and centrifuged at 15600g for 1 min. Each sample was extracted twice with an equal volume of sodium acetate-buffered phenol/chloroform/iso-amyl alcohol (25:24:1) before ethanol precipitation. A total of 2.3 µg of RNA was annealed to 0.2 pmol 5′ 32P-labeled primer U6D: 5′-AAAACGAAATAAATCTCTTTG or U4B: 5′-AGGTATTCCAAAAATTCCC in annealing buffer (150 mM NaCl, 50 mM Tris–HCl pH 7.5, 1 mM EDTA) at 37°C for 15 min. Reactions were then electrophoresed on 15 cm, 0.4 mm thick, 9% nondenaturing polyacrylamide gels (30:1 acrylamide:bisacrylamide) at 150 V for 4 h at 4°C.
Expression and purification of recombinant Cwc2 and Cwc2 RRM fragment
The Cwc2 open reading frame was PCR amplified from pRS416-Cwc2 with primers 209PETF 5′-CTAGTACATATGACATCTTGGAGGGATAAATCCGCG and 209PETB 5′-CAGCTACTCGAGATCCTCATCAGAGGAGAGGTAATCGAGC containing restriction sites for NdeI and XhoI. The Cwc2 RRM fragment was PCR amplified with primers Cwc2PET-Frag6 5′-CATATGGGCGGGATCGGTTCATTTAGGAAG and 209PETB. The PCR products were digested with XhoI then cloned into XhoI/EcoRV digested pBluescript II KS+ to produce pBS-Cwc2 and pBS-Cwc2RRM, which were then sequenced. The Cwc2 full-length and RRM fragment open reading frame were excised from pBS-Cwc2 and pBS-Cwc2RRM with NdeI and XhoI, then cloned into similarly digested pET30b (Novagen) to produce pET30b-Cwc2 and pET30b-Cwc2RRM, respectively. The plasmids were transformed into Escherichia coli strain Rosetta 2 (DE3) (Novagen). C-terminally 6His-tagged Cwc2 and RRM fragments were expressed in 500 ml Overnight Express medium (Novagen) for 24 h at 19°C and 37°C, respectively. 6His-tagged Cwc2 and the RRM fragment were purified according to a previously published purification protocol (23).
RNA transcription and electrophoretic mobility shift assay
The U1, U2, U3, U4, U5, U6, U14, ACT1, CYH2 and RPS10B RNAs were transcribed from PCR-generated template primed with UpG to allow end labeling according to published procedures (23,27). All RNAs were full length except for U2, which lacked ∼1 kb of yeast-specific sequence, and U3, which lacked stem loops 2, 3 and 4. The U6 snRNA 3′-end contained a cyclic phosphate produced by ribozyme cleavage (28). Plasmid pBS-Actin was digested with EcoRV or ClaI for run-off transcription of Actin or Actin Ac/Cla pre-mRNA, respectively. Plasmid pBS-ActinACAC was digested with EcoRV for run-off transcription of Actin ACAC pre-mRNA. Electrophoretic mobility shift assay (EMSA) reactions were performed with a range of protein concentrations in 20 mM HEPES–KOH pH 7.9, 300 mM NaCl, 10 µM ZnCl2, 10% glycerol, 10 mM DTT, 0.45 mg/ml E. coli tRNA and 0.8 mg/ml BSA. 15 000 dpm of end-labeled RNA was added and reactions were incubated at room temperature for 10 min before addition of 0.4 volume glycerol loading dye (0.05% bromophenol blue, 10% glycerol). Reactions were then resolved on 15 cm, native 5% acrylamide gels (37.5:1 acrylamide:bisacrylamide) in 1× Tris–borate buffer containing 6% glycerol for 3.5 h at 150 V.
RESULTS
Cwc2 contains a zinc finger and a nonconsensus RRM
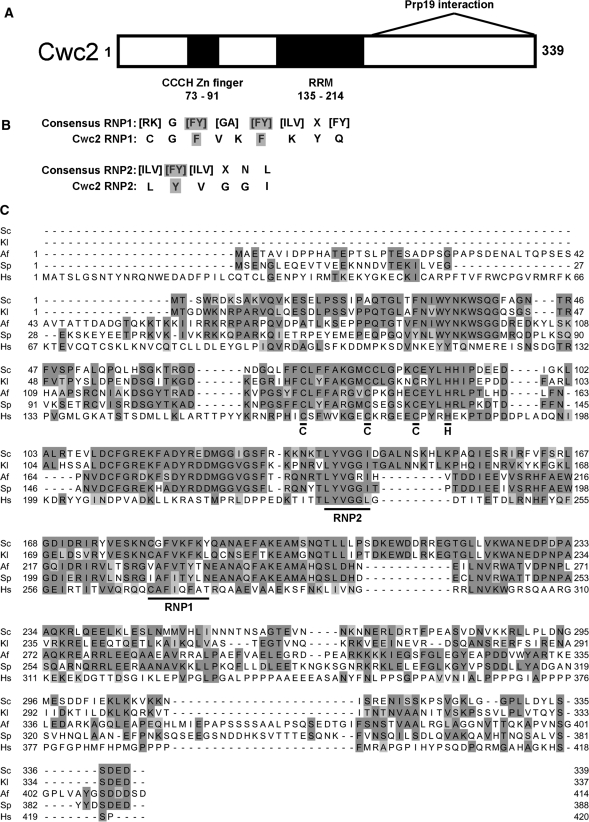
Analysis of the Cwc2 protein sequence reveals two specific motifs that may be involved in nucleic acid and/or protein interactions (Figure 1). The first motif, amino acids 73–91, is a CCCH zinc finger of the Cx7Cx5Cx3H type. The second motif, amino acids 135–214, is an RRM. The RRM contains a well-conserved RNP2 motif, however, the RNP1 motif is less conserved compared with the consensus for that motif (Figure 1B). The RNP1 of Cwc2 has an extra amino acid separating the two aromatic residues that are important for RNA binding. The RNP1 of Cwc2 also does not fit the consensus for the U2AF homology motif (UHM), a protein interaction variant of the RRM, even though Cwc2 has a lower pI (7.97) characteristic of other UHMs (29). Alignment of Cwc2 with its orthologs in other organisms reveals that it has 54%, 43% and 40% amino acid identity with other fungal Cwc2 genes from Kluyveromyces lactis, Aspergillus fumigatus and Schizosaccharomyces pombe, respectively (Figure 1C). The closest human ortholog is RBM22. However, amino acid identity is only 27% with similarities between Cwc2 and RBM22 only in the central region containing the zinc finger and RRM with the N- and C-terminus of Cwc2 and RBM22 being quite divergent. RBM22 has recently been found to be one of the core proteins of a purified human spliceosomal C complex able to carry out the second step of splicing (30). The presence of the zinc finger and RRM indicates that these regions may be important for RNA and/or protein binding of Cwc2.
Figure 1.
Domain structure and alignment of Cwc2. (A) Depiction of the protein motifs found in Cwc2. The location of the zinc finger and RRM are shown. The region of Cwc2 (amino acids 228–339) found to interact with Prp19 by two-hybrid and pull-down analysis is also shown (9). (B) Comparison of consensus RNP1 and RNP2 sequences with Cwc2 RNP1 and RNP2 sequences (X is for any amino acid). The aromatic residues, which are important for RNA binding, are highlighted. (C) The amino acid sequence of Cwc2 from S. cerevisiae (Sc, NCBI-GI: 6319992) was aligned with orthologs from K. lactis (Kl, NCBI-GI: 50305857), A. fumigatus (Af, NCBI-GI: 70998470), S. pombe (Sp, NCBI-GI: 19114249) and Homo sapiens (Hs, NCBI-GI: 8922328). Orthologs were identified using KEGG and the alignment was made with ClustalW and Jalview.
Cwc2 is essential for splicing in vivo and for the levels of the U1, U4, U5 and U6 snRNAs
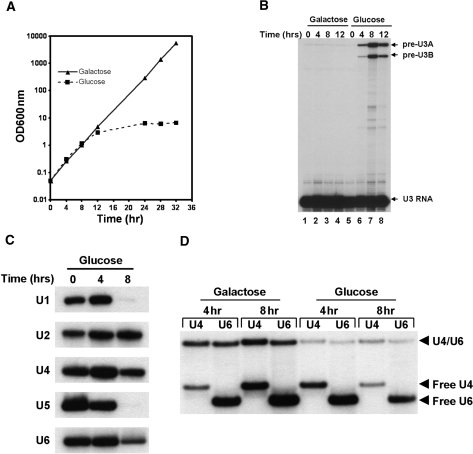
Recent work has implicated the NTC in spliceosome recycling and U4/U6 snRNP stability/biogenesis (14). Specifically, when expression of essential NTC genes PRP19, NTC90 or NTC77 are switched off there is an accumulation of the free form of U4 snRNP (14). A more detailed study of strains deleted for the nonessential NTC25, or NTC25 and NTC30 together, revealed a decrease in levels of free U6 snRNA resulting in reduced levels of the U4/U6 di-snRNP and increased levels of free U4 snRNP (14). It was suggested that the deficiency in U4/U6 stability/biogenesis resulted from inhibition of spliceosome recycling and may be a general effect of deletion or depletion of NTC proteins. We produced a GAL1 regulated CWC2 gene to investigate the levels of snRNAs and the U4/U6 snRNA complex when expression of CWC2 is turned off. Growth of the GAL1 regulated CWC2 in glucose to shut off expression of CWC2 results in the inhibition of cell growth (Figure 2A). Switching off the expression of CWC2 also results in the accumulation of unspliced precursor RNA, typical of inhibition of splicing (Figure 2B). Primer extension analysis of total RNA isolated at 0, 4 and 8 h following the switch to glucose reveals that levels of U6 are decreased when expression of CWC2 is turned off (Figure 2C), similar to that described for other NTC proteins (14). Interestingly, when expression of CWC2 is switched off, we also observe a significant decrease in levels of the U1 and U5 snRNAs and a small decrease in U4 levels that has not been observed for other NTC proteins (Figure 2C). Levels of U2 snRNA do not decrease. In addition, native gel electrophoresis revealed a dramatic decrease in the level of base-paired U4/U6 as well as decreased levels of free U4 and U6 (Figure 2D). This agrees with the primer extension results, which demonstrate a reduction in total levels of U4 rather than just base-paired U4. Surprisingly, this is in contrast to depletion of other NTC proteins where the reduced levels of base-paired U4/U6 resulted in increased levels of free U4 (14). This suggests that CWC2 is required for maintaining U1, U4 and U5, as well as U6, snRNA levels. Decreased levels of U4 and U6, in turn, results in a reduction of the U4/U6 snRNA complex.
Figure 2.
Cwc2 is required for cell growth, pre-mRNA splicing, the levels of the U1, U4, U5 and U6 snRNAs and maintenance of the U4/U6 complex. (A) The CWC2 gene was put under control of the GAL1 promoter. Growth over time of the GAL1-CWC2 yeast in the presence of galactose, which allows expression of Cwc2, or glucose, which represses Cwc2 expression. (B) Primer extension analysis of splicing of the U3 intron-containing genes U3A and U3B. When CWC2 is switched off by growth in glucose there is an accumulation of U3 precursor RNA indicating a defect in pre-mRNA splicing. (C) Depletion of Cwc2 influences the levels of the U1, U4, U5 and U6 snRNAs. Primer extension analysis of snRNA levels before and following the switch of the GAL1-CWC2 strain to glucose. The RNA utilized for snRNA primer extension is identical to that used in (B). (D) Depletion of Cwc2 reduces the levels of U4/U6 complex and free U4 and U6. RNA was isolated under nondenaturing conditions from the GAL1-CWC2 strain grown in galactose or glucose, hybridized to radiolabeled probes complementary to U4 or U6 and separated on a 9% polyacrylamide gel.
Cwc2 interacts directly with snRNAs and different pre-mRNAs in vitro
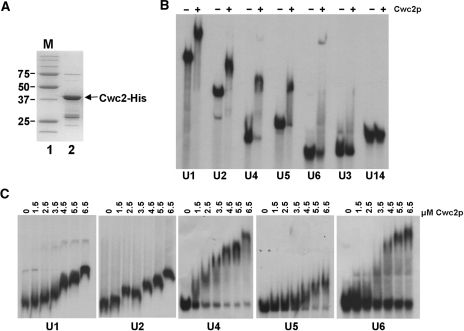
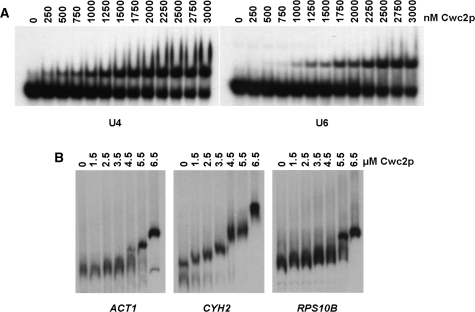
As Cwc2 depletion influences the levels of U1, U4, U5 and U6 snRNAs, it was of interest to determine whether Cwc2 is associated with any of the snRNAs of the spliceosome. Therefore, we expressed and purified C-terminally 6His-tagged Cwc2 (Cwc2-His) from bacteria (Figure 3A) to determine if it interacted with snRNAs in vitro. Incubation of purified Cwc2-His with U1, U2, U4, U5 or U6 snRNAs and analysis by EMSA indicate that the Cwc2-His is able to bind to all five snRNAs (Figure 3B). However, under the same conditions Cwc2-His did not bind U3 or U14 snoRNAs, demonstrating some specificity for snRNAs (Figure 3B). Titration of the concentration of Cwc2-His between 1.5 µM and 6.5 µM followed by analysis of snRNA binding by EMSA reveals that snRNA binding is variable, with Cwc2-His binding to U4 and U6 occurring at lowest concentrations of the protein followed by U2, U5 and U1 (Figure 3C). Using a range of lower concentrations of Cwc2-His revealed that with the U4 and U6 snRNAs, binding is apparent at concentrations as low as 250 and 1000 nm, respectively (Figure 4A). Cwc2-His, therefore, appears to have the ability to bind to all the snRNAs in vitro with varying efficiency.
Figure 3.
Binding of Cwc2 to snRNAs in vitro. (A) SDS–PAGE analysis of purified, His-tagged, Cwc2 (lane 2). Molecular weight markers (lane 1) are indicated to the left of the panel in KiloDaltons. (B) EMSA using 32P end-labeled RNAs that are indicated below the panel. RNA was incubated without (−) and with (+) 4.5 µM Cwc2-His, then complexes were resolved on 5% nondenaturing polyacrylamide gels. (C) Increasing concentrations of Cwc2-His protein were incubated with fixed amounts of the individual snRNAs indicated below the panels. Concentrations of Cwc2-His are indicated above each panel. Complexes were resolved on 5% nondenaturing polyacrylamide gels.
Figure 4.
Binding of Cwc2 to the U4 and U6 snRNAs and different pre-mRNAs in vitro. (A) EMSA using 32P end-labeled U4 and U6 snRNAs. Increasing concentrations of Cwc2-His protein were incubated with a fixed amount of a labeled RNA. The identity of the RNA is indicated below the panels. Concentrations of Cwc2-His are indicated above each panel. Complexes were resolved on 5% nondenaturing polyacrylamide gels. (B) Binding of Cwc2 to the ACT1, CYH2 and RPS10B pre-mRNAs in vitro. EMSA using 32P end-labeled pre-mRNAs. Increasing concentrations of Cwc2-His protein were incubated with a fixed amount of a labeled RNA. The identity of the RNA is indicated below the panels. Concentrations of Cwc2-His are indicated above each panel. Complexes were resolved on 5% nondenaturing polyacrylamide gels.
As Cwc2-His can bind differentially to more than one snRNA, its ability to bind pre-mRNA was also tested by EMSA. Three different intron-containing pre-mRNAs were tested, ACT1, CYH2 and RPS10B (Figure 4B). Cwc2-His bound to all three pre-mRNAs in vitro, although requiring higher concentrations of protein than the snRNAs. Cwc2-His, therefore, binds both snRNAs and different pre-mRNAs in vitro. During spliceosome activation, NTC association specifies snRNA/pre-mRNA interactions (7,8). Thus if Cwc2-RNA binding occurs within spliceosomes, Cwc2 could be involved in specifying these RNA/RNA interactions.
Cwc2 associates predominantly with the U2 and U6 snRNAs in assembled spliceosomes
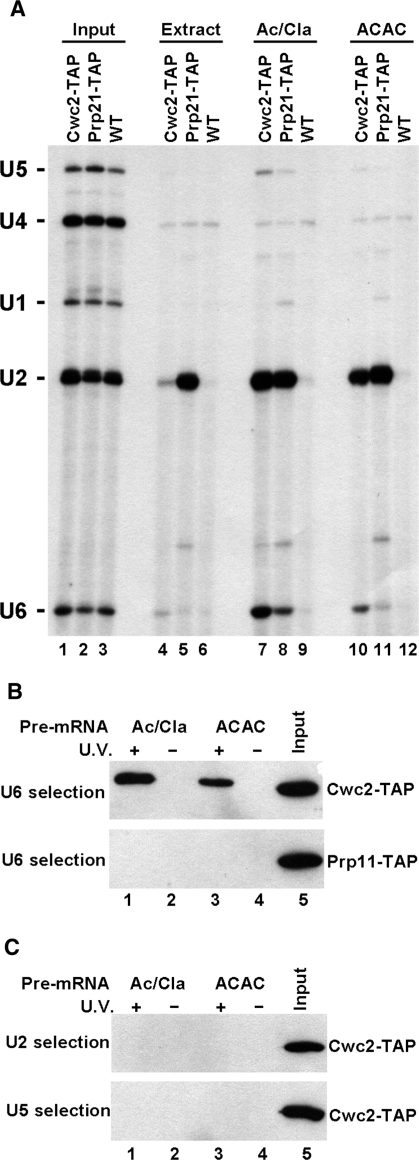
To determine whether any of the snRNA interactions of Cwc2-His found in vitro occur in assembled snRNPs or spliceosomes, a yeast strain was constructed where the chromosomal CWC2 gene was fused to the TAP tag (31). As a control for snRNA association the U2 snRNP protein Prp21 was also TAP-tagged. Yeast whole-cell splicing extract was then produced from these two strains and a wild-type, untagged, strain. Primer extension of RNA isolated from extracts indicated that each strain contains equivalent levels of snRNAs (Figure 5A, lanes 1–3). Cwc2-TAP or Prp21-TAP was then precipitated from extracts with IgG-coated beads, and the RNA associated with Cwc2-TAP or Prp21-TAP was extracted from the beads. The presence of all five snRNAs was then analyzed by primer extension. In extract alone no snRNAs were associated with Cwc2-TAP, whereas Prp21-TAP could precipitate U2 snRNA (Figure 5A, lanes 4 and 5). This indicates that Cwc2 does not associate with any of the snRNAs in yeast whole-cell splicing extracts. This agrees with similar experiments carried out with Prp19 demonstrating that it is also not associated with snRNPs in extracts (32). Next, extracts were incubated under splicing conditions with an ACT1 pre-mRNA (Ac/Cla) which does not contain sequences after the branch point so cannot carry out the first step of splicing, but allows spliceosome assembly and NTC binding (33). Under these conditions the U2 and U6 snRNAs and a small amount of U5 snRNA associated with Cwc2-TAP, whereas U2 and to a lesser extent U6, associated with the Prp21-TAP (Figure 5A, lanes 7 and 8). An ACT1 pre-mRNA with a 3′-splice site, ACAC mutation was utilized to accumulate spliceosomes that have completed the first step of splicing but are blocked for the second step of splicing (8,34). Extracts incubated under splicing conditions with the ACAC pre-mRNA reveal that after the first step of splicing the U2 and U6 snRNAs remain associated with Cwc2-TAP and U2 snRNA with Prp21-TAP (Figure 5A, lanes 10 and 11). There is a slight reduction in U6 snRNA association with the Cwc2-TAP and very little U5 snRNA association (Figure 5A, lane 10). In each condition tested, the wild-type untagged extract did not precipitate any snRNAs (Figure 5A, lanes 6, 9 and 12). This indicates that the U2 and U6 snRNAs predominantly associate with Cwc2 in assembled spliceosomes under native conditions.
Figure 5.
Cwc2 is only snRNA-associated within spliceosomes and contacts the U6 snRNA directly in first and second step spliceosomes. (A) Yeast whole-cell extract was made from yeast strains containing TAP-tagged Cwc2 (Cwc2-TAP), TAP-tagged Prp21 (Prp21-TAP) or a non-TAP-tagged wild-type (WT) strain. Total RNA was isolated from 1.25% of extract used in the precipitations from extracts and splicing reactions, then subjected to primer extension with primers complementary to the U1, U2, U4, U5 and U6 snRNAs (lanes 1–3, Input). Extract alone (lanes 4–6, Extract), extract incubated under splicing conditions with an ACT1 pre-mRNA truncated after the branchpoint sequence (lanes 7–9, Ac/Cla) or extract incubated under splicing conditions with an ACT1 pre-mRNA with a 3′-splice site mutation (lanes 10–12, ACAC) were precipitated with IgG-coated beads. RNA extracted from the beads was subjected to primer extension with primers complementary to the U1, U2, U4, U5 and U6 snRNAs. The positions of each snRNA are shown on the left. (B) Yeast whole-cell extract was made from yeast strains containing TAP-tagged Cwc2 (Cwc2-TAP) (top panel) and TAP-tagged Prp11 (Prp11-TAP) (bottom panel). Splicing reactions with Ac/Cla (lanes 1 and 2) or ACAC (lanes 3 and 4) pre-mRNA were UV irradiated (lanes 1 and 3) before selection with a biotinylated oligonucleotide complementary to U6 snRNA, under denaturing conditions. Cwc2-TAP or Prp11-TAP were detected with the anti-TAP antibody by immunoblotting. As a control, identical splicing reactions were subjected to selection of U6 under the same denaturing conditions without previous UV irradiation (lanes 2 and 4). Lane 5 shows 1% of the TAP-tagged extract used in splicing reactions. (C) As in (B) except selection from Cwc2-TAP extract splicing reactions after crosslinking utilized biotinylated oligonucleotides complementary to U2 snRNA (top panel) or U5 snRNA (bottom panel).
Cwc2 contacts U6 snRNA directly in active spliceosomes
As the NTC stabilizes U6 association within active spliceosomes, we next wanted to determine if the Cwc2-U6 association in spliceosomes detected under native conditions reflected a direct RNA–protein interaction. Spliceosomes blocked before the first and second steps of splicing, using the Ac/Cla and ACAC pre-mRNAs, respectively, were assembled in Cwc2-TAP extract and UV crosslinked. A biotinylated oligonucleotide complementary to U6 snRNA was then used to purify U6 snRNA and associated crosslinked proteins using streptavidin-coated magnetic beads under stringent denaturing conditions. Crosslinking of Cwc2-TAP to U6 snRNA was then analyzed by immunoblotting with anti-TAP antibody. Cwc2-TAP was found to crosslink to U6 snRNA in both first and second step spliceosomes (Figure 5B, top panel, lanes 1 and 3). Interestingly, direct crosslinking of Cwc2 to U6 decreased following the first step of splicing similar to that observed under native conditions. Without UV irradiation this Cwc2–U6 interaction was dissociated under the selection conditions used (Figure 5B, top panel, lanes 2 and 4). The requirement for UV for detection of the Cwc2–U6 interaction demonstrates a direct RNA–protein interaction in the first and second step spliceosomes. The U2 snRNP protein, Prp11, was not found to crosslink to U6 snRNA in an identical experiment carried out in Prp11-TAP extract (Figure 5B, bottom panel). Selection of crosslinked first and second step spliceosomes with biotinylated oligonucleotides complementary to the U2 or U5 snRNAs did not co-purify any crosslinked Cwc2-TAP, implying that Cwc2 does not contact these snRNAs directly (Figure 5C). The direct contact with U6 snRNA suggests that Cwc2 may be involved in the role of the NTC in anchoring U6 to the spliceosome after U4 dissociation and specifying its interactions with the pre-mRNA.
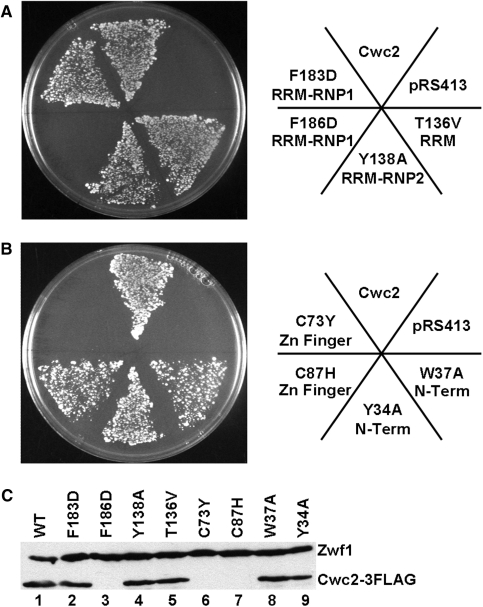
Mutation of the Cwc2 zinc finger and RRM destabilize Cwc2 and inhibit growth in vivo
The presence of a CCCH zinc finger and an RRM motif in Cwc2 suggests that one, or both, of these motifs may be important for Cwc2–RNA binding in pre-mRNA splicing. Each motif is known to bind RNA although RRMs are also known to bind protein. To determine whether specific amino acids within and around these motifs were important for Cwc2 function in vivo a yeast strain was constructed (YCWC2KO) in which the CWC2 coding sequence was deleted and complemented by a CEN URA3 plasmid containing the wild-type CWC2 gene. This strain was transformed with CEN HIS3 plasmids expressing either wild-type Cwc2 with three C-terminal FLAG tags (Cwc2-3FLAG) or Cwc2-3FLAG containing mutations in Cwc2. Transformants were transferred to media containing 5-FOA, which selects against the wild-type CEN URA3 plasmid and tests the ability of CWC2 mutants to support growth of yeast cells as the sole source of CWC2. RRM motifs contain three aromatic residues that are important for RNA binding (35). Two of these aromatic residues are in the RNP1 and the third in the RNP2 consensus sequences (Figure 1B). Mutation in the Cwc2 RRM of the conserved residue adjacent to the RNP2 motif (T136V) and the aromatic residue within RNP2 (Y138A) supported growth of yeast cells as the sole source of Cwc2 (Figure 6A). Mutation of one of the aromatic residues within RNP1 (F183D) also supported growth of yeast cells (Figure 6A). In contrast, mutation of the second aromatic residue in RNP1 F186D did not support growth (Figure 6A). Mutation of a cysteine in the zinc finger domain of Cwc2 to a histidine (C87H), which still has the potential to coordinate zinc, allows yeast growth (Figure 6B). However, mutation of a cysteine residue in the zinc finger to an amino acid that does not coordinate zinc (C73Y) does not support growth of yeast cells as the sole source of Cwc2 (Figure 6B). Finally, mutation of two highly conserved amino acids in Cwc2 (Y34A and W37A) were each able to support growth of yeast cells (Figure 6B). Expression of the mutant proteins, in the presence of the CEN URA3 plasmid containing the wild-type CWC2 gene, was analyzed by immunoblotting using anti-FLAG antibody (Figure 6C). The two lethal Cwc2 mutant proteins (F186D and C73Y) were not stably expressed in vivo (Figure 6C, lanes 3 and 6). The viable mutant, C87H, was also not stably expressed in vivo (Figure 6C, lane 7); however, longer exposure revealed very low levels of the F186D, C73Y and C87H mutant proteins, with slightly higher levels of C87H maybe explaining why it produces a viable phenotype (data not shown). The significant reduction in mutant protein explains the effect these mutations have on viability as the sole source of Cwc2 protein.
Figure 6.
Mutations in the Cwc2 zinc finger and RRM are lethal in vivo due to reduced expression of the mutant protein. A haploid yeast strain with the chromosomal CWC2 gene knocked-out and CWC2 on a URA3 marked plasmid (pRS416-Cwc2) was created. This strain was transformed with either a HIS3 marked plasmid alone (pRS413) or pRS413 with wild-type Cwc2 with three C-terminal FLAG tags (Cwc2-3FLAG) or Cwc2-3FLAG with the indicated Cwc2 mutations in the RRM domain (A) or mutations of conserved N-terminal residues including the zinc finger domain (B). Single colonies from these transformations were streaked onto 5-FOA to select against the wild-type pRS416-Cwc2 plasmid. The plates were photographed after incubation at 30°C for 3 days. (C) Immunoblot with the anti-FLAG antibody and the G6PDH antibody (against Zwf1 protein, as a loading control) of extracts from strains containing the pRS416-Cwc2 wild-type plasmid and the pRS413-Cwc2-3FLAG wild-type or mutant plasmid.
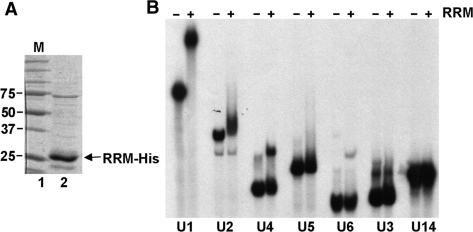
Cwc2 RRM interacts with U1, U2, U4 and U6 but not U5 snRNA in vitro
Mutagenesis of the RRM and zinc finger motifs destabilizes Cwc2 in vivo, probably by preventing proper folding of these domains, consequently we decided to analyze the RNA binding of these domains in vitro. We attempted to express and purify from bacteria His-tagged fragments of Cwc2 containing these two domains individually for use in in vitro binding assays. We were unable to express a soluble zinc finger-containing fragment (data not shown), but an RRM-containing fragment was expressed and purified successfully and used for EMSAs (Figure 7A). The RRM fragment interacts with the U1, U2, U4 and U6 snRNAs in vitro but did not appear to interact with the U5 snRNA (Figure 7B). Like full-length Cwc2, the RRM fragment did not interact with the U3 or U14 snoRNAs (Figure 7B). Titration of the concentration of the RRM fragment revealed that it interacted with U1 and U6 more efficiently, and with U4 snRNA less efficiently, than the full-length Cwc2 (data not shown). The Cwc2 RRM may, therefore, be responsible for the binding of U6 snRNA in spliceosomes. The rest of the protein also appears to have a role in binding some RNAs, especially U5.
Figure 7.
Binding of Cwc2 RRM fragment to snRNAs in vitro. (A) SDS–PAGE analysis of purified, His-tagged, Cwc2 RRM fragment (lane 2). Molecular weight markers (lane 1) are indicated to the left of the panel in KiloDaltons. (B) EMSA using 32P end-labeled RNAs. The identity of the RNA is indicated below the panel. RNA was incubated without (−) and with (+) 4.5 µM Cwc2-His RRM fragment, then complexes were resolved on 5% nondenaturing polyacrylamide gels.
DISCUSSION
The NTC plays an important role in the catalytic activation of the spliceosome. The NTC is known to promote the stable association of the U5 and U6 snRNAs with the spliceosome (7,8). To determine how the NTC associates with the snRNAs of the spliceosome, we have investigated the RNA interactions of Cwc2, the only putative RNA binding protein of the NTC. Here, we demonstrate that Cwc2 has the ability to bind all five snRNAs in vitro. Cwc2 can also bind three different pre-mRNAs in vitro, although with less efficiency than snRNA binding. Binding to U4, followed by U6, occurred most efficiently at the lowest concentrations of Cwc2, however, in assembled spliceosomes we find that Cwc2 interacts predominantly with U2 and U6 snRNAs and crosslinks directly to the U6 snRNA. Finally, Cwc2 depletion appears to have a more dramatic effect on snRNA levels than other NTC proteins, reducing the levels of U1, U4 and U5 as well as U6. Taken together, we propose that Cwc2 functionally links the NTC with the pre-mRNA and snRNAs of the spliceosome.
It has been shown that deletion or depletion of NTC proteins results in decreased U4/U6 stability/biogenesis as a result of inhibition of spliceosome recycling (14). Depletion of the essential NTC proteins Prp19, Ntc90 and Ntc77 or deletion of the nonessential Ntc25 and Ntc25/Ntc30 all resulted in decreased levels of U6 snRNA leading to reduced levels of the U4/U6 complex, and in turn increased levels of free U4 snRNA (14). It was proposed that free U4 accumulation is a general property of NTC protein depletion or deletion. We have found that depletion of Cwc2 also results in a decrease in U6 snRNA levels and reduced levels of the U4/U6 complex as observed with depletion or deletion of other NTC proteins. However, depletion of Cwc2 also results in reduced levels of U1, U5 and, to a lesser extent, U4 snRNA. There was also a reduction in free U4 snRNA despite the reduced levels of the U4/U6 complex. This reduction in snRNA levels occurs after 8 h of Cwc2 depletion and corresponds to a strong block in pre-mRNA splicing. Depletion of Cwc2 appears to exhibit more dramatic effects on the snRNAs than depletion of other NTC proteins. It will be interesting to determine exactly how Cwc2 depletion downregulates snRNAs and the relationship this has to snRNP biogenesis.
The influence of Cwc2 on snRNA levels is accompanied by its ability to bind all five snRNAs and three different pre-mRNAs in vitro. This suggests that Cwc2 binds RNA nonspecifically; however, it does not bind the U3 or U14 snoRNAs under similar conditions. The binding assays were carried out in the presence of excess unlabeled tRNA. In the absence of excess tRNA, Cwc2 can bind the U3 and U4 snoRNAs in vitro (data not shown), therefore, it appears that Cwc2 has general RNA binding properties but with higher specificity for snRNAs and pre-mRNAs. There may be a common structural or sequence element within the snRNAs that is recognized by Cwc2 that is not present within U3 or U14 snoRNAs. Full-length Cwc2 bound to U4, followed by U6, most efficiently at the lowest concentrations of Cwc2. A fragment of Cwc2 containing the RRM was able to bind all the snRNAs except U5. Like full-length Cwc2, the Cwc2 RRM did not bind the U3 or U14 snoRNAs under the conditions used. The RRM fragment of Cwc2 is, therefore, likely to be involved in the RNA binding activity but the rest of the protein must have an important role, especially in U5 binding in vitro. Cwc2 may be able to contact more than one snRNA at once through its two different motifs. Lethal mutations in the RRM and zinc finger affected expression of the protein, probably by causing incorrect folding of these domains. Further mutagenesis is required to assess the function of these domains in vivo.
The NTC has previously been shown to join the spliceosome after the dissociation of U1 and U4 (36). Prp19 was not found to be associated with the U4 containing spliceosome even in low ATP conditions, which inhibits U4 dissociation. Our immunoprecitation experiments agree with this timing, demonstrating that Cwc2 interacts with active spliceosomes containing U2, U6 and U5. Thus, even though Cwc2 can bind U1 and U4 by EMSA, it appears not to contact these snRNAs in snRNPs or in assembled spliceosomes. This is intriguing considering purified Cwc2 binds U4 with greatest efficiency at concentrations as low as 250 nM. However, before performing the immunoprecipitation, extracts were allowed to accumulate spliceosomes blocked just prior to the first step or after the first step of splicing, after U1 and U4 dissociation. Cwc2 immunoprecipitations contained a very small amount of U4 when spliceosomes were blocked earlier in assembly with low ATP concentrations (data not shown). Therefore, it is possible that Cwc2 does contact U4 and U1 in splicing extracts earlier in spliceosome assembly. Alternatively, while Cwc2 can bind all the snRNAs in vitro, Cwc2 RNA binding may be specified or regulated in splicing extracts by other components of the NTC/spliceosome.
Possible candidates for regulating the RNA binding activity of Cwc2 are Prp19 and Ntc30. Both Prp19 and Ntc30 interact with Cwc2 in the yeast two-hybrid assay (9). In addition, Prp19 interacts with Cwc2 directly in vitro (9). These protein–protein interactions could regulate the RNA binding ability of Cwc2 within the NTC. As Prp19 is in the U-box family of E3 ubiquitin ligases (37), and can interact directly with Cwc2 in vitro, Prp19 may also target Cwc2 for ubiquitination. Ubiquitination can result in a number of different consequences for a protein. For example, ubiquitination can influence the stability, conformation, activity or localization of a protein (38). Prp19 has been demonstrated to have ubiquitin ligase activity in vitro and interacts with subunits of the proteosome (39,40). However, no substrates for the ubiquitin ligase activity of Prp19 are known. An attractive hypothesis is that Cwc2 is the target for the Prp19 ubiquitin ligase activity and that ubiquitination of Cwc2 may regulate its RNA binding activity in the NTC. If Cwc2 RNA binding is regulated by components of the NTC, by protein–protein interactions or modification, Cwc2 would not be able to bind snRNAs in splicing extracts until the correct time in spliceosome assembly.
Cwc2 was found to contact the U6 snRNA directly in spliceosomes blocked both prior to the first and second steps of splicing. It has been demonstrated previously that the NTC is required for stable association of the U5 and U6 snRNAs with the active spliceosome and for specifying their interactions with the pre-mRNA before the first step of splicing (7,8). In order for this function to be carried out, it is expected that component(s) of the NTC will contact these snRNAs directly within spliceosomes. Until now, however, no proteins of the NTC have been demonstrated to be able to bind RNA. The fact that Cwc2 directly contacts U6 snRNA within spliceosomes suggests that it functions in this role of the NTC, stabilizing U6 snRNA association with the spliceosome. The NTC was found to specify the U6 interaction at the 5′-splice site and to bring about Lsm dissociation from the 3′-end of U6 enabling it to interact with the pre-mRNA intron (7,8). It will be interesting in the future to map the Cwc2 crosslink within the U6 snRNA to see if it is involved with one or both of these interactions.
The NTC remains associated with the spliceosome during the two catalytic steps of splicing. Cwc2 was also found to crosslink to U6 snRNA in spliceosomes blocked after the first but before the second step of splicing. Therefore, the NTC may be important for specifying U6 snRNA interactions at this stage, for example, during the proposed disruption of the U6-5′-splice site interaction (41). There appears to be a reduction in U6-Cwc2 association in both the crosslinking and immunoprecipitation experiments. This apparent reduction may represent remodeling of the Cwc2-U6 interaction, allowing U6 to form specific interactions required for the second step of splicing.
Although Cwc2 can interact with U2 and U5 in vitro and associates with active spliceosomes containing these snRNAs, we were not able to detect any crosslinks between Cwc2 and U2 or U5. This is intriguing as the NTC also stabilizes U5 association with the spliceosome (7,8). It may be that crosslinks were not selected by our method, although the U2 and U5 biotin oligonucleotides were able to hybridize to these snRNAs by native gel electrophoresis (data not shown). Alternatively, another protein of the NTC may bind U5 snRNA in spliceosomes. The ability of Cwc2 to bind to U6 snRNA in the spliceosome suggests that it may, at least in part, form the link between the NTC and the RNAs of the spliceosome. This link would allow the NTC to regulate important RNA/RNA dynamics during pre-mRNA splicing.
FUNDING
Wellcome Trust (082992 to R.T.O.). Funding for open access charge: Wellcome Trust.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank Heather Coyne for construction of some of the CWC2 mutations and members of the O’Keefe lab for advice on the manuscript.
REFERENCES
- 1.Will CL, Lührmann R. Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol. 2001;13:290–301. doi: 10.1016/s0955-0674(00)00211-8. [DOI] [PubMed] [Google Scholar]
- 2.Moore MJ, Query CC, Sharp PA. Splicing of precursors to mRNA by the spliceosome. In: Gesteland RF, Atkins JF, editors. The RNA World. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1993. pp. 303–357. [Google Scholar]
- 3.Nilsen TW. RNA-RNA interactions in nuclear pre-mRNA splicing. In: Simons RW, Grunberg-Manago M, editors. RNA Structure and Function. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1998. pp. 279–307. [Google Scholar]
- 4.Staley JP, Guthrie C. An RNA switch at the 5′ splice site requires ATP and the DEAD box protein Prp28p. Mol. Cell. 1999;3:55–64. doi: 10.1016/s1097-2765(00)80174-4. [DOI] [PubMed] [Google Scholar]
- 5.Raghunathan PL, Guthrie C. RNA unwinding in U4/U6 snRNPs requires ATP hydrolysis and the DEIH-box splicing factor Brr2. Curr. Biol. 1998;8:847–855. doi: 10.1016/s0960-9822(07)00345-4. [DOI] [PubMed] [Google Scholar]
- 6.Small EC, Leggett SR, Winans AA, Staley JP. The EF-G-like GTPase Snu114p regulates spliceosome dynamics mediated by Brr2p, a DExD/H box ATPase. Mol. Cell. 2006;23:389–399. doi: 10.1016/j.molcel.2006.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan SP, Cheng SC. The Prp19-associated complex is required for specifying interactions of U5 and U6 with pre-mRNA during spliceosome activation. J. Biol. Chem. 2005;280:31190–31199. doi: 10.1074/jbc.M505060200. [DOI] [PubMed] [Google Scholar]
- 8.Chan SP, Kao DI, Tsai WY, Cheng SC. The Prp19p-associated complex in spliceosome activation. Science. 2003;302:279–282. doi: 10.1126/science.1086602. [DOI] [PubMed] [Google Scholar]
- 9.Ohi MD, Gould KL. Characterization of interactions among the Cef1p-Prp19p-associated splicing complex. RNA. 2002;8:798–815. doi: 10.1017/s1355838202025050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ben-Yehuda S, Dix I, Russell CS, McGarvey M, Beggs JD, Kupiec M. Genetic and physical interactions between factors involved in both cell cycle progression and pre-mRNA splicing in Saccharomyces cerevisiae. Genetics. 2000;156:1503–1517. doi: 10.1093/genetics/156.4.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Makarov EM, Makarova OV, Urlaub H, Gentzel M, Will CL, Wilm M, Lührmann R. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science. 2002;298:2205–2208. doi: 10.1126/science.1077783. [DOI] [PubMed] [Google Scholar]
- 12.Villa T, Guthrie C. The Isy1p component of the NineTeen complex interacts with the ATPase Prp16p to regulate the fidelity of pre-mRNA splicing. Genes Dev. 2005;19:1894–1904. doi: 10.1101/gad.1336305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ajuh P, Kuster B, Panov K, Zomerdijk JC, Mann M, Lamond AI. Functional analysis of the human CDC5L complex and identification of its components by mass spectrometry. EMBO J. 2000;19:6569–6581. doi: 10.1093/emboj/19.23.6569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen CH, Kao DI, Chan SP, Kao TC, Lin JY, Cheng SC. Functional links between the Prp19-associated complex, U4/U6 biogenesis, and spliceosome recycling. RNA. 2006;12:765–774. doi: 10.1261/rna.2292106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tarn WY, Hsu CH, Huang KT, Chen HR, Kao HY, Lee KR, Cheng SC. Functional association of essential splicing factor(s) with PRP19 in a protein complex. EMBO J. 1994;13:2421–2431. doi: 10.1002/j.1460-2075.1994.tb06527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kunkel TA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl Acad. Sci. USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Keefe RT, Norman C, Newman AJ. The invariant U5 snRNA loop 1 sequence is dispensable for the first catalytic step of pre-mRNA splicing in yeast. Cell. 1996;86:679–689. doi: 10.1016/s0092-8674(00)80140-3. [DOI] [PubMed] [Google Scholar]
- 19.Gietz RD, Woods RA. Yeast transformation by the LiAc/SS Carrier DNA/PEG method. Methods Mol. Biol. 2006;313:107–120. doi: 10.1385/1-59259-958-3:107. [DOI] [PubMed] [Google Scholar]
- 20.Boeke JD, LaCroute F, Fink GR. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 21.Longtine MS, McKenzie A, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 22.Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- 23.Dobbyn HC, O’Keefe RT. Analysis of Snu13p mutations reveals differential interactions with the U4 snRNA and U3 snoRNA. RNA. 2004;10:308–320. doi: 10.1261/rna.5970404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ansari A, Schwer B. SLU7 and a novel activity, SSF1, act during the PRP16-dependent step of yeast pre-mRNA splicing. EMBO J. 1995;14:4001–4009. doi: 10.1002/j.1460-2075.1995.tb00071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin RJ, Newman AJ, Cheng SC, Abelson J. Yeast mRNA splicing in vitro. J. Biol. Chem. 1985;260:14780–14792. [PubMed] [Google Scholar]
- 26.Li Z, Brow DA. A rapid assay for quantitative detection of specific RNAs. Nucleic Acids Res. 1993;21:4645–4646. doi: 10.1093/nar/21.19.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McGrail JC, O’Keefe RT. The U1, U2 and U5 snRNAs crosslink to the 5′ exon during yeast pre-mRNA splicing. Nucleic Acids Res. 2008;36:814–825. doi: 10.1093/nar/gkm1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McGrail JC, Tatum EM, O’Keefe RT. Mutation in the U2 snRNA influences exon interactions of U5 snRNA loop 1 during pre-mRNA splicing. EMBO J. 2006;25:3813–3822. doi: 10.1038/sj.emboj.7601258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kielkopf CL, Lucke S, Green MR. U2AF homology motifs: protein recognition in the RRM world. Genes Dev. 2004;18:1513–1526. doi: 10.1101/gad.1206204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bessonov S, Anokhina M, Will CL, Urlaub H, Lührmann R. Isolation of an active step I spliceosome and composition of its RNP core. Nature. 2008;452:846–850. doi: 10.1038/nature06842. [DOI] [PubMed] [Google Scholar]
- 31.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 32.Tarn WY, Lee KR, Cheng SC. The yeast PRP19 protein is not tightly associated with small nuclear RNAs, but appears to associate with the spliceosome after binding of U2 to the pre-mRNA and prior to formation of the functional spliceosome. Mol. Cell Biol. 1993;13:1883–1891. doi: 10.1128/mcb.13.3.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng SC. Formation of the yeast splicing complex A1 and association of the splicing factor PRP19 with the pre-mRNA are independent of the 3′ region of the intron. Nucleic Acids Res. 1994;22:1548–1554. doi: 10.1093/nar/22.9.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vijayraghavan U, Parker R, Tamm J, Iimura Y, Rossi J, Abelson J, Guthrie C. Mutations in conserved intron sequences affect multiple steps in the yeast splicing pathway, particularly assembly of the spliceosome. EMBO J. 1986;5:1683–1695. doi: 10.1002/j.1460-2075.1986.tb04412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clery A, Blatter M, Allain FH. RNA recognition motifs: boring? Not quite. Curr. Opin. Struct. Biol. 2008;18:290–298. doi: 10.1016/j.sbi.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 36.Tarn WY, Lee KR, Cheng SC. Yeast precursor mRNA processing protein PRP19 associates with the spliceosome concomitant with or just after dissociation of U4 small nuclear RNA. Proc. Natl Acad. Sci. USA. 1993;90:10821–10825. doi: 10.1073/pnas.90.22.10821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aravind L, Koonin EV. The U box is a modified RING finger – a common domain in ubiquitination. Curr. Biol. 2000;10:R132–R134. doi: 10.1016/s0960-9822(00)00398-5. [DOI] [PubMed] [Google Scholar]
- 38.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 39.Loscher M, Fortschegger K, Ritter G, Wostry M, Voglauer R, Schmid JA, Watters S, Rivett AJ, Ajuh P, Lamond AI, et al. Interaction of U-box E3 ligase SNEV with PSMB4, the beta7 subunit of the 20 S proteasome. Biochem. J. 2005;388:593–603. doi: 10.1042/BJ20041517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohi MD, Vander Kooi CW, Rosenberg JA, Chazin WJ, Gould KL. Structural insights into the U-box, a domain associated with multi-ubiquitination. Nat. Struct. Biol. 2003;10:250–255. doi: 10.1038/nsb906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Konarska MM, Vilardell J, Query CC. Repositioning of the reaction intermediate within the catalytic center of the spliceosome. Mol. Cell. 2006;21:543–553. doi: 10.1016/j.molcel.2006.01.017. [DOI] [PubMed] [Google Scholar]