Abstract
CUGBP2 (ETR-3/NAPOR/BRUNOL3) promotes inclusion of cardiac troponin T (cTNT) exon 5 via binding between positions 21 and 74 of the downstream intron. The molecular mechanism by which CUGBP2 activates cTNT exon 5 inclusion is unknown. Our results suggest that CUGBP2 promotes exon inclusion by a novel mechanism in which CUGBP2 directly interacts with components of the activated U2 snRNP and enhances binding of U2 snRNP to the branch site located upstream of the exon. Using an in vitro splicing assay, we show that recombinant CUGBP2 enhances complex A formation of a cTNT pre-mRNA. Enhanced complex A assembly requires both the upstream and downstream introns consistent with dual requirements for the downstream CUGBP2-binding site and an upstream branch site for U2 snRNP binding. We also show that CUGBP2 enhances binding of U2 snRNA to the cTNT pre-mRNA consistent with enhanced complex A assembly. Purification of CUGBP2-interacting proteins using tandem affinity purification leads to the demonstration that the core 17S U2 snRNP components, SF3b145 and SF3b49 bind directly to CUGBP2. We conclude that CUGBP2 activates exon inclusion by forming direct interactions with components of the 17S snRNP complex and recruits and/or stabilizes binding of U2 snRNP.
INTRODUCTION
Pre-mRNA splicing removes introns and joins exons together to generate mature mRNA. Splicing requires highly conserved 5′ and 3′ splice sites at the exon–intron junctions and is catalyzed by the spliceosome which is composed of five small nuclear ribonucleoproteins (snRNPs) and >150 proteins (1). Recognition of the splice site sequences by base-pairing of the snRNA components of the snRNPs not only identifies exons within pre-mRNAs but also directs cleavage and rejoining (transesterification) at the correct nucleotides (2). Characterization of spliceosome function in vitro has established that the spliceosome assembles by a stepwise assembly of complexes (H->E->A->B->C) representing a dynamic addition and exchange of its components (3). First the U1 snRNP binds to the 5′ splice site in an ATP-independent manner to form complex E followed by ATP-dependent binding of U2 snRNP to the branch site sequence, typically located 18–40 nt upstream of the exon, which forms complex A. Incorporation of U4/U6.U5 tri-snRNP into complex A generates complex B. Dissociation of U1 and U4 snRNPs and U6 base-pairing with 5′ splice site creates complex C. SnRNP and other components that bind to the splice site sequences upstream and downstream of the exon communicate across the exon to ‘define’ and exon for removal (4,5).
Recent estimates indicate that at least 89% of human genes undergo alternative splicing, generating multiple mRNAs from each gene (6,7). A large fraction of alternative splicing events are regulated to meet the functional demands of the cell or tissue (8,9). Alternative splicing is commonly regulated by RNA-binding proteins that bind to sequence motifs located within the alternative exon or within the flanking intron sequences (10). Only a few studies have revealed how these RNA-binding proteins communicate with the basal splicing machinery to promote or repress splicing (11–15). With a few exceptions (16), most splicing regulators characterized to date modulate the early stages of spliceosome assembly including formation of complex E and A. For example, binding of Fox1/Fox2 to the consensus sequence, UGCAUG, in the upstream intron of calcitonin exon 4 inhibits complex E′ formation by blocking binding of SF1 to the branch site (17). Binding of Nova to a cluster of binding sites within an alternative exon blocks inclusion by inhibiting U1 snRNP binding (13). Binding of TIA1 to U-rich stretches downstream of an exon facilitates recognition of U1 snRNP to the 5′ splice site (18). Binding of RBM25 to CGGGCA in exon 2 of Bcl-x enhances U1 snRNP recruitment to the weak 5′ splice site (14).
CUGBP2 (ETR-3, BRUNOL3, NAPOR) is one of six members of the CELF (CUGBP1 and ETR-3 like factor) family. CUGBP2 contains three RNA recognition motif (RRM) domains and directly binds to intronic sequences to regulate splicing of chicken and human cardiac troponin T exon 5 (cTNT), insulin receptor exon 11, Tau exon 2 and 3, and NMDA R1 exons 5 and 21 (19–22). Analysis of natural binding sites as well as results from SELEX have demonstrated that CUGBP2 binds to UG rich sequence motifs (21). One of the best characterized splicing events regulated by CUGBP2 is the activation of chicken cTNT exon 5. Troponin T is one of three subunits of the troponin complex that regulates the actin–myosin interactions that mediate muscle contraction (23). Exon 5 inclusion is regulated during heart development such that >90% of mRNAs include the exon in embryonic heart and >95% of mRNAs exclude the exon in the adult (24). The embryonic exon inclusion pattern was also favored in differentiated skeletal muscle cultures and the cis acting elements required for enhanced inclusion in differentiated skeletal muscle were mapped to the intron downstream of cTNT exon 5 (19). Using transient transfection of cTNT minigenes, we subsequently showed that binding of CELF proteins (including CUGBP2) to U/G-rich elements within these cis elements was required to activate exon inclusion in vivo (21,25). We also demonstrated that bacterially expressed recombinant CUGBP2 activates cTNT exon 5 in an in vitro splicing assay and that activation required two CUGBP2-binding sites located 21 and 74 nt downstream of the exon (19). However, it remains unknown how CUGBP2 communicates with basal splicing machinery to activate exon 5 inclusion after binding to intron 5.
In this study, we show that CUGBP2 enhances complex A formation on an RNA containing only exon 5 flanked by segments of the upstream and downstream introns. CUGBP2-enhanced complex A assembly requires the upstream intron and psoralen cross-linking assays were used to demonstrate that CUGBP2 increases binding of U2 snRNA to the pre-mRNA. We also demonstrate that CUGBP2 directly binds to SF3b145 and SF3b49 that are core components of the 17S U2 snRNP complex. These results strongly support a model in which CUGBP2 binds downstream of exon 5 and recruits or stabilizes binding of U2 snRNP to the branch site upstream of the exon consistent with a mechanism of enhanced exon definition.
MATERIALS AND METHODS
Reagents
Rabbit polyclonal anti-SF3b 145 was kindly provided by Dr Robin Reed or purchased (Proteintech Group, Inc; PGI). Rabbit polyclonal anti-SF3b49 was purchased from PGI. Anti-SF3b125 was kindly provided by Dr Reinhard Lührmann. Rabbit anti-TAP antibody was from Jackson Immuno Research laboratories. 35S protein labeling mix was purchased from Perkin Elmer. Glutathione sepharose was purchased from Novagen. The TNT coupled recticulocyte lysate system was from Promega. IgG-sepharose 6 fast flow was purchased from Amersham. DSP (dithiobis [succinimidylpropionate]) was from Pierce. HeLa nuclear extract was prepared as described (19,26) or purchased from Protein One. AMT (4′-Aminomethyltrioxalen hydrochloride)-Psoralen was from Sigma. Digoxigenin-labeled (5′ end) oligonucleotide was synthesized by IDT. Anti-Digoxigenin-HRP, DIG wash buffer, DIG block buffer set and DIG easy hybridization buffer were purchased from Roche. Pre-mRNAs were in vitro transcribed with or without 32P-UTP and gel isolated as described (19).
Oligonucleotides and probes
The following DNA oligonucleotides and 5′ DIG-labeled probes were used: R5S (5′-TAGATCAGACGAGATA-3′ (27), U1 snRNA (1–12nt): 5′-CCAGGTAAGTAT-3′ (27), U2 snRNA (26–43nt): 5′-GAACAGATACTACACTTG-3′. DIG-E5: 5′-CTCAAGCCATTCTTCCTCTT-3′, DIG-U2 snRNA: 5′-GAGGTACTGCAATACCAGGTC-3′
Expression and purification of recombinant proteins
GST-CUGBP2 and GST-CUGBP1 were induced in BL21 by 0.2 mM IPTG for 4 h at 30°C. Cells were harvested by centrifugation at 6000× g for 10 min and resuspended in extraction buffer [25 mM HEPES (pH 7.5), 20% glycerol, 1 mM DTT, 20 mM NaCl, 0.2 mM EDTA, 0.05% SDS, 1% TritonX-100 and proteinase inhibitor cocktail buffer]. Cells were lysed by sonicating three times for 10 s each. Supernatants were collected after centrifugation at 14 800× g for 10 min.
His-tagged CUGBP2 was induced like a GST proteins and purified using the Novagen kit according to the manufacture's protocol.
GST pull down and cross-linking
GST proteins were incubated with 30 μl of glutathione sepharose resin in extraction buffer containing 150 mM NaCl and incubated on a rotating platform for 2 h at 4°C then washed with the same buffer three times. [35S] Methionine/Cysteine labeled SF3b145 and SF3b49 were prepared using TNT coupled reticulocyte lysate systems (Promega). 35S -labeled SF3b145 and SF3b49 were incubated with GST, GST-CUGBP1, or GST-CUGBP2 in binding buffer [20 mM Tris (pH 7.4), 50 mM NaCl, 0.05% NP40] with rotation at 4°C. After 2 h, unbound proteins were removed by washing with binding buffer five times. Bound proteins were released with 1× SDS PAGE loading buffer and loaded on 10% SDS–PAGE gel. The gel was dried and the bands visualized using a cyclone phosphoimager.
For protein cross-linking, DSP (final concentration, 2 mM) was added in cross-linking buffer (0.5 M HEPES in 0.5× PBS) and incubated for 30 min at 25°C. Cross-linking was stopped with 50 mM Tris (pH 7.4) and incubation at 25°C for 10 min. For RNase A treatment, RNase A (final concentration 50 μg/ml) was added and incubated for 20 min at 25°C. The unbound proteins were washed with cross-linking buffer three to four times. Cross-linking was reversed by the thiol groups in SDS–PAGE sample buffer.
Tandem affinity purification (TAP)
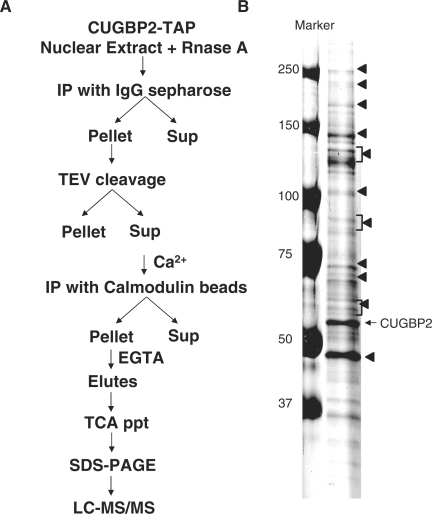
TAP and CUGBP-2 TAP proteins were purified according to (28) with modifications. Stably over-expressed tet-inducible TAP (CA1) and CUGBP2-TAP (DE5) HeLa cell lines were maintained in DMEM containing 200 μg/ml hygromycin. TAP and CUGBP2-TAP were induced by 10 μg/ml (final concentration) doxycycline. After 48 h, cells were scraped in 1× PBS and spun down by centrifugation at 6000 × g for 10 min. Nuclear extracts were prepared and dialyzed against buffer D (26). For the first step affinity purification, NP40 (0.05%) and 50 μg/ml RNase A were added to nuclear extracts and incubated with 50 μl of IgG Sepharose (50% slurry) for 16 h at 4°C. Bound protein was washed three times with washing buffer [0.1% NP40, 20 mM Tris (pH 7.4), 25% glycerol, 1.5 mM MgCl2, 0.2 mM EDTA, 120 mM KCl, 10 mM 2-β-mercaptoethanol and 0.2 mM PMSF]. The Sepharose pellet was washed with TEV cleavage buffer [10 mM Tris (pH 8.0), 120 mM KCl, 0.1% NP40, 0.5 mM EDTA, 1 mM DTT]. The tagged proteins were cleaved by adding 25 units of TEV proteinase in 250 µl of cleavage buffer and incubating at 25°C with rotation for 2 h. For the second step affinity purification, three volumes of IPP 150 buffer [10 mM Tris (pH 8.0), 150 mM NaCl, 1 mM MgOAc, 1 mM imidazole, 2 mM CaCl2, 10 mM 2-β-mercaptoethanol] plus 3 μl of 1 M CaCl2 and 300 μl of calmodulin resin (50% slurry) were added to the eluant from the first step and incubated on rotating platform for 3 h at 4°C. Calmodulin resin was washed three times with IPP 150 buffer. Proteins bound to calmodulin resin were eluted five times with 200 ml of calmodulin elution buffer containing 10 mM Tris (pH 8.0), 150 mM Nacl, 0.02% NP40, 1 mM MgOAc, 1 mM imidazole, 20 mM EGTA (pH 8.0), 10 mM 2-β-mercaptoethanol. Eluted proteins were precipitated in 25% TCA, following 1 h on ice and centrifugation at 14 800 × g for 20 min at 4°C. TCA precipitates were washed with 300 μl of cold acetone and acetone was evaporated by incubating pellet on 65°C for 1 min. Pellets were dissolved in SDS–PAGE loading buffer and then analyzed by SDS–PAGE.
NanoLC–MS/MS
TCA-precipitated proteins eluted from the calmodulin resin were resolved on a 4–20% gradient gel and stained with Coomassie Blue 250 R. Bands were gel isolated and the proteins were identified at ProtTech, Inc (Norristown, PA) using the Nano LC–MS/MS peptide sequencing technology.
Western blotting
The samples from TAP were resolved by SDS–PAGE and transferred into the PVDF membrane. Anti-SF3b145, anti-SF3b49, anti-SF3b125 and anti-CUGBP2-HRP antibodies were used and pico substrate (Pierce) was used to visualize bands.
In vitro splicing & spliceosome assembly
In vitro splicing reactions were performed with 32P-GTP or 32P-UTP-labled RNA in splicing buffer (30% HeLa nuclear extract, 1 mM ATP, 20 mM creatine phosphate, 2 mM MgOAc, 1 mM DTT, 3% PVA) at 30°C for indicated times. Ninety percent of each reaction was treated with Proteinase K buffer [100 mM Tris–HCl (pH 7.5), 12.5 mM EDTA, 150 mM NaCl, 1% SDS and 0.2 mg/ml proteinase K] at 37°C for 30 min. RNA was extracted by phenol:chloroform:isoamyl alcohol followed by ethanol precipitation. RNA was dissolved in gel loading buffer (Ambion, Inc). RNA was run on 8 M urea 5% acrylamide gel, dried and visualized by autoradiography. Ten percent of each splicing reaction was run on native 0.4% agarose–4% acrylamide gel or 1.5–2% agarose gel. The gel was dried and bands were visualized by autoradiography. For ATP-depleted spliceosome assembly, HeLa nuclear extract was incubated at 30°C for 30 min prior to initiating the splicing reaction and the assembly reaction was done in the absence of ATP and creatine phosphate. The complexes were resolved on a 1.5–2% native agarose gels after adding loading buffer (12 mg/ml heparin and 25% glycerol). The gel was fixed in 10% methanol and 10% acetic acid at room temperature for 30 min, dried and visualized by autoradiography. For E complexes, heparin was removed from loading buffer.
snRNAs digestion by RNase H
DNA oligonucleotides complementary to U1 snRNA (1–12 nt) (27), U2 snRNA (26–43 nt), or non-specific olionucleotide (R5S) (27), were incubated with HeLa nuclear extract in the presence of RNase H (two unit) at 30°C for 30 min. The nuclear extract was used for in vitro splicing reactions as described above.
AMT-psoralen UV cross-linking and northern blot
Splicing complexes were assembled using the unlabeled chicken cTNT exon 5 substrate (MSE1-4, Figure 2A) at 30°C in HeLa nuclear extracts in splicing conditions described above. AMT-psoralen was added to a final concentration of 20 ng/ml and the reactions were UV (365 nm) irradiated on ice for 15 min. The cross-linked RNAs containing both exon 5 and U2 snRNA were detected by northern blot analysis using DIG-labeled probes. Briefly, RNA was extracted and resolved on 8 M urea 5% acrylamide gels, stained with ethidium bromide and transferred to nylon membrane. Blocking was done for 30 min at 42°C using Roche DIG Easy Hyb and DIG-probes were added for a 16 h incubation at 42°C. Membranes were washed two times for 15 min at 25°C with northern washing buffer (2× SSC, 0.5% SDS). Incubation with anti-DIG HRP antibody was done as manufacturer's protocol (Roche wash & block kit). Bands were detected using Pico substrate (Pierce).
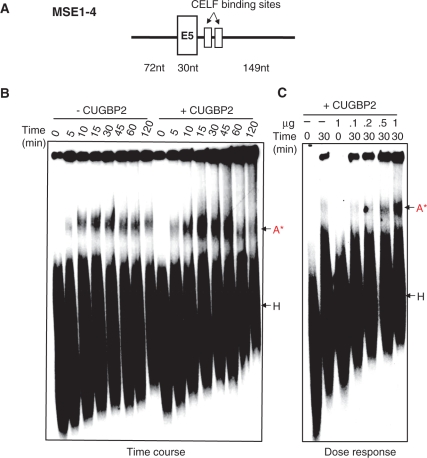
Figure 2.
CUGBP2 enhances formation of an A-like complex on an RNA containing only cTNT exon 5 and portions of its flanking introns. (A) Diagram of the MSE1-4 RNA. (B) Time course with and without 1 µg His-CUGBP2. (C) Dose-response to His-CUGBP2 in 30 min reactions. 32P-UTP labeled MSE1-4 RNA was incubated under splicing conditions and 10% of each reaction was displayed on a 0.4% agarose–4% acrylamide non-denaturing gel and detected by autoradiography. A-like complex indicated as A*.
Constructs and cell line
CUGBP2-TAP and TAP were inserted respectively into KpnI and Not I site of pBG2i vector (29). Tet-on inducible CUGBP2-TAP and TAP were stably expressed in HeLa. Cells were maintained in DMEM media containing hygromycin (200 μg/ml). SF3b145 and SF3b49 cDNAs for in vitro translation were provided by Dr Lührmann. E46NB and MSE 1–4 constructs have been described previously (19). The E45NA pre-mRNA contains (from 5′ to 3′) 76-nt vector, the last 14-nt cTNT exon 4, the 120-nt truncated intron 4 identical to that in E46NB (19), 30-nt exon 5 and the first 9 nt of intron 5. The E56BD pre-mRNA contains (from 5′ to 3′) 65 nt from the vector, the last 4 nt of intron 4, 30-nt exon 5, a 250-nt truncated intron identical to E46NB except for 6 nt missing 169 nt from exon 5, the complete 57-nt exon 6 and the first 15 nt of intron 6.
RESULTS
CUGBP2 enhances complex A formation of the chicken cTNT pre-mRNA
We used a three-exon cTNT pre-mRNA substrate to determine which step of spliceosome assembly was enhanced by CUGBP2. This substrate was previously used to show activation of exon 5 inclusion by bacterially expressed recombinant His-tagged CUGBP2 (19). The cTNT substrate (E46NB, Figure 1A) was incubated in splicing conditions during a time course and 10% of the reaction from each time point was loaded on a native gel to assay spliceosome assembly (Figure 1B, left). RNA was extracted from the remaining sample and run on a denaturing gel to follow spliced products and intermediates (Figure 1B, right). Complex A formed within 15 min; this complex was demonstrated to be ATP- and U1- dependent (see below). Larger complexes, designated as B and C, formed subsequently, and their appearance at 45 min corresponded with appearance of spliced mRNAs containing and lacking exon 5 and lariat intermediates (Figure 1B, right).
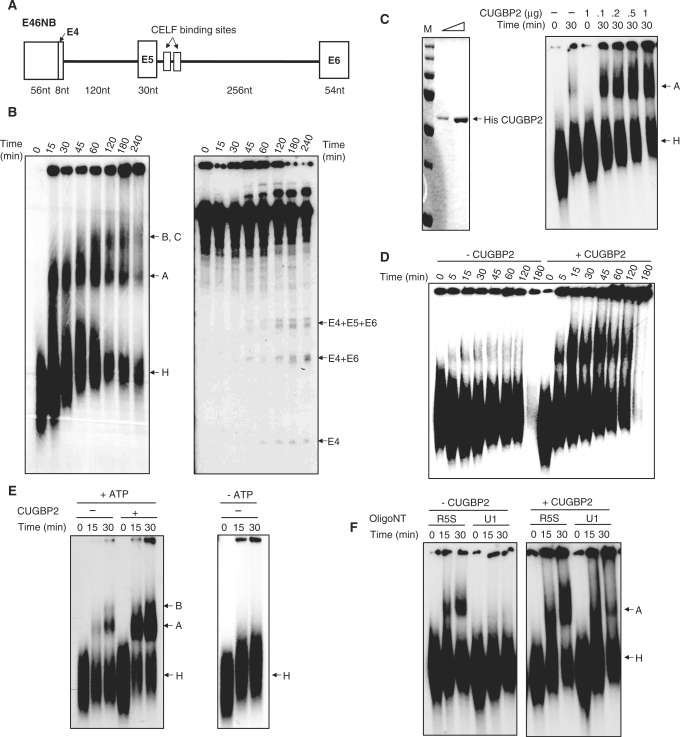
Figure 1.
CUGBP2 enhances complex A formation of a chicken cTNT pre-mRNA. (A) Diagram of chicken cTNT pre-mRNA substrate E46NB. (B) Spliceosome assembly during a time course for E46NB. 32P-UTP labeled E46NB was incubated under splicing conditions for the times indicated. Ten percent of each splicing reaction was loaded on 0.4% agarose–4% acrylamide non-denaturing gel (left). RNA was purified from what remained and was separated on an 8 M urea, 5% acrylamide gel (right). For both assays, RNA was visualized by autoradiography. (C) CUGBP2 increases complex A formation (dose response). Purified recombinant His-CUGBP2 (145 ng and 725 ng) is shown to be >95% pure by Coomassie staining (left). The indicated amounts of CUGBP2 were added to the splicing reactions, which were incubated for 30 min (right). (D) CUGBP2 (1 µg) increases complex A formation from the earliest time point tested. The complex enhanced by CUGBP2 is complex A based on (E) ATP-dependence and (F) U1 dependence. In F, complex assembly reactions were performed using HeLa nuclear extracts in which the first 12 nt of U1 snRNA were digested using a complementary oligo and RNase H. A non-specific DNA nucleotide (R5S) was used as a control (39).
We next tested the effect of CUGBP2 on formation of E46NB spliceosome complexes. CUGBP2 strongly enhanced complex A formation in a dose-dependent manner beginning with the earliest time point tested (Figure 1C and D). To confirm that the enhanced complex was complex A and rule out CUGBP2-RNA complexes unrelated to the spliceosome, we demonstrated that this complex was not observed in the absence of ATP (Figure 1E, right). Formation of this complex was also blocked by RNase H-mediated digestion of the first 12 nt of the U1 snRNA by a complementary DNA oligonucleotide while a non-specific oligonucleotide (R5S) had no effect (Figure 1F). Efficient U1 snRNA digestion by RNase H was confirmed with ethidium bromide staining (Supplementary Figure 1). Only minimal assembly of complex A was observed when CUGBP2 was added to U1-digested nuclear extracts, probably reflecting enhanced assembly by residual U1 snRNP. From these results, we conclude that CUGBP2 strongly enhances complex A formation during E46NB pre-mRNA splicing.
Because the E46NB substrate contains two introns, the specific nature of the complexes formed on this RNA is unclear. We therefore examined the effect of CUGBP2 on A-like complex assembly on an RNA substrate containing a single exon and portions of the flanking introns: cTNT exon 5, the last 72 nt of intron 4 and the first 149 nt of intron 5 (MSE1-4, Figure 2A). This RNA contains the CUGBP2-binding sites in intron 5 and was previously shown to bind CUGBP2 (25). Isolated exons flanked by an upstream 3′ splice site and downstream 5′ splice site assemble an A-like complex (30). While it is unclear whether complex A and A-like complexes are identical, their clear similarities are consistent with the exon definition model in which exons are ‘defined’ by binding of the basal spliceosome components to the flanking splice sites prior to intron removal (4). The MSE1-4 substrate assembled an A-like complex that was ATP-dependent (Figure 2B and C and data not shown) and CUGBP2 enhanced A-like complex formation in dose- and time-dependent manner (Figure 2B and C). We conclude that CUGBP2 enhances assembly of an A-like complex on the isolated cTNT exon 5.
Enhanced complex A assembly by CUGBP2 requires both upstream and downstream introns
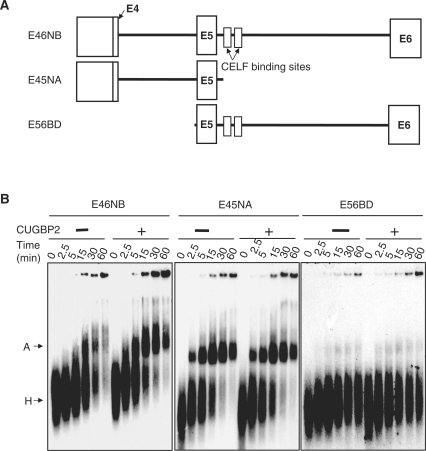
The results from Figure 2 showing that CUGBP2 enhanced assembly of an A-like complex on exon 5 alone suggested that CUGBP2 enhances definition of exon 5. If this was the case, the effect of CUGBP2 would require both the upstream and downstream introns. We generated two substrates from E46NB containing either exons 4 and 5 (E45NA) or exons 5 and 6 (E56BD) (Figure 3A). Spliceosome assembly reactions were performed over a time course in the presence or absence of CUGBP2 for E45NB, E45NA and E56BD substrates. All three RNA substrates assemble complex A in the absence of CUGBP2, as expected. CUGBP2 enhanced complex A formation of E46NB as demonstrated above, but the same CUGBP2 protein preparation did not enhance complex A assembly of either E45NA or E56BD (Figure 3B). E45NA lacks a CUGBP2-binding site, so the absence of an effect of CUGBP2 was expected. The results indicate that both two-exon substrates are competent to assemble A complexes in the absence of CUGBP2 but do not respond to a preparation of recombinant CUGBP2 that stimulated complex A assembly of E46NB and MSE1-4. In addition, these results demonstrate that recombinant CUGBP2 does not non-specifically stimulate complex A formation but rather exhibits specificity for exons flanked by its binding site downstream and an upstream 3′ splice site. Finally, these results strongly suggest that binding of CUGBP2 to its binding sites within intron 5 promotes communication with basal splicing machinery across the exon 5 to enhance complex A assembly.
Figure 3.
Enhanced complex A formation of cTNT exon 5 by CUGBP2 requires both introns 4 and 5. (A) Diagram of E46NB, E45NA and E56NB RNAs. (B) 32P-UTP labeled E46NB, E45NA and E56BD RNAs were incubated for the indicated times under splicing conditions with or without CUGBP2. Spliceosome complexes were resolved on 1.5% native agarose gels and visualized by autoradiography.
CUGBP2 enhances binding of U2 snRNA to the pre-mRNA
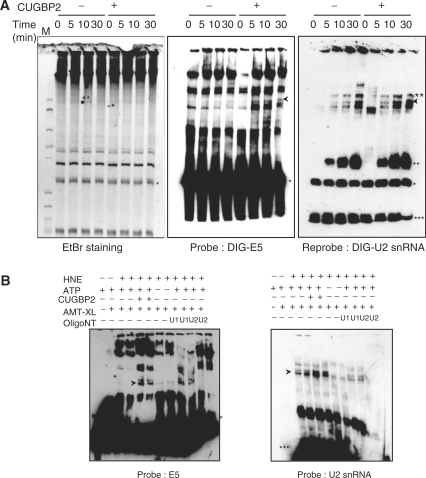
Base-pairing of U2 snRNA to the branch site leads to formation of complex A. We used psoralen UV-cross-linking to determine whether enhanced complex A formation by CUGBP2 is associated with enhanced binding of U2 snRNA to the pre-mRNA. For this assay, the single exon MSE1-4 substrate was used (Figure 2A). The unlabeled MSE1-4 RNA was incubated with HeLa nuclear extract using splicing conditions with and without CUGBP2. AMT-psoralen was added followed by UV irradiation (see ‘Materials and methods’ section). The cross-linked RNA products were separated electrophoretically on urea-acrylamide gels and RNAs were identified by northern blotting using digoxigenin (DIG)-linked probes complementary to either exon 5 or to the U2 snRNA (Supplementary Figure 2). RNAs found to hybridize to both exon 5 and U2 snRNA probes were considered to be cross-linked RNA products of U2 snRNA base-paired with the branch site of cTNT intron 4. These RNAs are indicated with a black arrow throughout Figure 4. Consistent with this interpretation, the indicated RNAs were found to require ATP (Figure 4B, lanes 7 and 8) and to be sensitive to RNase H-mediated digestion using DNA oligos complementary to U2 (Figure 4B, lanes 11 and 12) but not U1 snRNA (Figure 4B, lanes 9 and 10). Importantly, the U2 cross-linked band was consistently enhanced by addition of CUGBP2 to the splicing reaction (Figure 4A and B). From these results, we conclude that CUGBP2 enhances complex A formation by directly increasing U2 snRNA base-pairing with the pre-mRNA.
Figure 4.
CUGBP2 increases U2 snRNA base-pairing with cTNT pre-mRNA. (A) Splicing reactions wih and without CUGBP2 performed for 0, 5, 10 and 30 min at 30°C were used for UV cross-linking with psoralen on ice. RNA was extracted and resolved on a 5% acrylamide/8 M urea gel and stained with ethidium bromide (EtBr, left). Northern analysis was performed to identify the U2 snRNA cross-linked to the cTNT exon 5 MSE1-4 pre-mRNA using digoxigenin (DIG)-labeled oligonucleotide probes recognizing exon 5 (DIG-E5) and U2 snRNA (DIG-U2 snRNA). Hybridized oligos were detected by western blotting using anti-digoxigenin-HRP antibodies. The arrow indicates the cross-linked U2 snRNA with cTNT exon 5 MSE1-4 pre-mRNA. Ethidium bromide staining shows even loading of pre-mRNA and total RNA from nuclear extract. (B) Nuclear extracts were incubated with oligonucleotides complementary to U1 snRNA and U2 snRNA, respectively in the presence of RNase H at 30°C for 30 min. Splicing reactions were done without (lanes 1 and 2) or with (lanes 3–12) nuclear extract in the presence (lanes 1–6 and 9–12) or absence (lanes 7 and 8) of ATP followed by northern analysis. *, cTNT exon 5 MSE 1–4 pre-mRNA; **, U2 snRNA cross-linked with unknown RNA; ***, U2 snRNA.
CUGBP2 interacts with the U2 snRNP components, SF3b49 and SF3b145
In a separate set of experiments, we investigated the mechanism of CUGBP2 mediated splicing activation by identifying CUGBP2 interacting proteins using tandem affinity purification (TAP) (31). Tetracycline-inducible C-terminal TAP-tagged CUGBP2 (CUGBP2-TAP) was stably expressed in HeLa cells. CUGBP2-TAP was demonstrated to activate cTNT exon inclusion (data not shown). Nuclear extract was prepared from doxycycline-induced HeLa cell lines and proteins that co-purified with CUGBP2-TAP were isolated using modified two-step purification (Figure 5A and ‘Materials and Methods’ section). RNase A was included in the purifications to exclude co-purification of non-specific proteins via RNA tethering. Following the final purification procedure, putative CUGBP2 interacting proteins were visualized on SDS–PAGE gels (Figure 5B). The bands that were repeatedly observed in four independent experiments were isolated and identified by LC–MS/MS. Identified proteins included RNA-binding proteins, transcription factors and translation regulators (Table 1).
Figure 5.
Identification of CUGBP2 interacting proteins. Nuclear extracts were prepared from HeLa cells stably expressing CUGBP2-TAP and induced by doxycycline treatment for 48 h. A two-step affinity purification was performed as described in A and ‘Materials and Methods’ section. Putative CUGBP2 interacting proteins were separated on 4–20% gradient gel and visualized by Coomassie blue staining (B). Bands (marked with ◂) consistently observed in four experiments were isolated and identified by LC–MS/MS.
Table 1.
List of putative CUGBP2 interacting proteins identified by LC–MS/MS
| Biological function | Gene name | Biological function | Gene name |
|---|---|---|---|
| RNA-binding protein | SF3B2 (19) | Transcription | GTF2I (23) |
| DDX42 (13) | SPT6H (15) | ||
| Matr3 (8) | SET (9) | ||
| TADBP (7) | SNW1 (7) | ||
| PRP31 (6) | MEF2D (6) | ||
| SRP68 (4) | HDAC4 (5) | ||
| SF3B4 (3) | NELFE (4) | ||
| PRP4 (3) | NCoA3 (3) | ||
| RNA/DNA helicase | DDX1 (8) | Signaling | PPP1R13BL (10) |
| DHX9 (5) | ARNT (5) | ||
| Exoribonuclease | XRN2 (10) | CAM2D (3) | |
| DNA repair | ERCC5 (18) | Cytokinesis | ANLN (11) |
| DNA replication | MCM7 (13) | SCY-like protein 2 (8) | |
| MCM3 (8) | Elongation | EF1A (9) | |
| MCM6 (4) | Unknown function | LRPPRC (11) | |
| MCM2 (1) | CAP23 (9) | ||
| PRCC (7) |
Proteins identified by LC–MS/MS were categorized according to their biological functions. Numbers in parentheses indicate the number of peptides identified from given using LS–MS/MS. Gene names refer to the UniProt Knowledgebase.
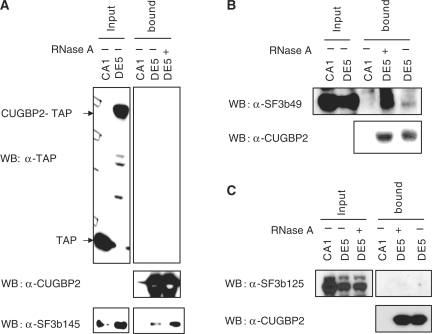
Of particular interest were three components of the 17S U2 snRNP and two U4/U6 snRNP associated splicing factors. Because CUGBP2 enhanced complex A assembly which requires binding of U2 to the branch site, we were particularly interested in validating interactions between CUGBP2 and U2 snRNP components which included two proteins of the 17S U2 snRNP [SF3B2 (SF3b145), SF3B4 (SF3b49)] and the U2 snRNP-associated protein, DDX42 (SF3b125). To validate interactions between CUGBP2 and SF3b49, SF3b125 and SF3b145, we first repeated TAP purification of CUGBP2 and associated proteins in doxycycline-induced cultures of stable HeLa cell lines expressing CUGBP2-TAP (DE5, Figure 6) and TAP alone (CA1, Figure 6) as a control. Purifications were performed with and without RNase A treatment and eluted proteins were assayed by western blots using anti-TAP antibodies to confirm enrichment (Figure 6A). Since the TAP tag is removed in the second purification step, anti-CUGBP2 antibodies were used to determine whether the released CUGBP2 was present in the bound fraction (Figure 6A). The same eluted proteins were also assayed for SF3b145, SF3b125, or SF3b49 to determine whether these proteins co-purified with released CUGBP2. The results demonstrated that SF3b49 and SF3b145 co-purified with CUGBP2 in an RNase A-independent manner (Figure 6A and B) suggesting either direct interaction or co-assembly with components of a complex. In contrast, SF3b125 did not copurify with CUGBP2 (Figure 6C).
Figure 6.
CUGBP2 binds to SF3b145 (SF3B2) and SF3b49 (SF3B4) but not SF3b125. Nuclear extracts were prepared from HeLa cells stably expressing CUGBP2-TAP (DE5) or TAP alone (CA1) as a control. Affinity purification using IgG sepharose with or without RNase A treatment was performed. Proteins bound to IgG Sepharose were released by TEV cleavage and precipitated by TCA and labeled as bound fractions. Western blots were performed using anti-SF3b145 (A), SF3b49 (B) and SF3b125 (C). For inputs, 10% of the total was loaded.
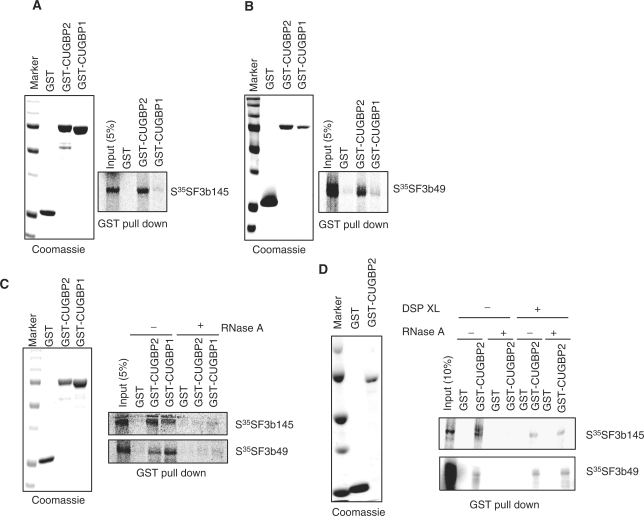
We next tested whether interactions between CUGBP2 and SF3b145 or SF3b49 are direct or indirect using a GST pull down assay. Since CUGBP1 and CUGBP2 have been shown to regulate several of the same splicing events (21,25), we tested binding of CUGBP1 as well as CUGBP2. Bacterially expressed recombinant GST-CUGBP1 and GST-CUGBP2 were incubated with [35S] methionine/cysteine-labeled SF3b145 or SF3b49. Both SF3b145 and SF3b49 were shown to bind to GST-CUGBP1 and GST-CUGBP2 (Figure 7A and B). However, interactions of GST-CUGBP1 and GST-CUGBP2 with SF3b145 or SF3b49 were dramatically reduced by RNase A treatment (Figure 7C). These results were surprising given the result that interactions between exogenous CUGBP2 and endogenous SF3b49 and SF3b149 were RNA independent (Figure 7A and B). We considered that the disparity reflected differences in binding affinities between CUGBP2 and the SF3b components when the SF3b components were assembled as an RNP in HeLa cells compared to individual proteins translated in vitro. We also considered that the presence of RNA, even non-specific bacterial RNA co-expressed with GST fusion proteins, could provide a scaffold enhancing natural interactions between one or both SF3b proteins and CUGBP2. To test this possibility, we repeated the binding assays and included the reversible protein cross-linker, DSP (dithiobis[succinimidylpropionate]), prior to RNase A treatment and the GST pull down, and then cross-linking was reversed prior to SDS–PAGE. Interestingly, interactions of GST-CUGBP1 with SF3b49 and SF3b149 were detected in the presence of RNase A following treatment with the protein cross-linker. The protein cross-linker did not promote non-specific interactions between these proteins and GST (Figure 7D). This result indicates that both SF3b49 and SF3b149 are in contact with CUGBP2 during the binding reaction. The RNase sensitivity of this interaction suggests that non-specific bacterial RNA promotes these protein–protein interactions, perhaps by maintaining a conformation that is critical for interactions.
Figure 7.
CUGBP2 binds directly to SF3b145 and SF3b49. (A) GST-CUGBP2 and GST-CUGBP1 (Coomassie staining gels left side or each panel) coupled on glutathione-sepharose were incubated with [35S] Methionine/Cysteine-labeled SF3b145 (A) and SF3b49 (B). Bound proteins were separated on 10% SDS–PAGE gel and visualized with Cyclone Phosphorimager. (C) RNase A (50 μg/ml) reduced interactions between CELF and SF3b proteins in vitro. (D) Bound proteins in GST pull down were treated with the reversible protein cross-linker (DSP) prior to RNase A treatment. M: protein marker.
We conclude, based on CUGBP2-TAP co-purification analysis in vivo and GST pull down analysis in vitro, that both SF3b49 and SF3b145 bind directly to CUGBP2. Furthermore, we propose that CUGBP2 promotes inclusion of cTNT exon 5 by binding downstream of the exon and acting across the exon to promote binding of U2 snRNP to the branch site via direct interactions with SF3b49 and SF3b145.
DISCUSSION
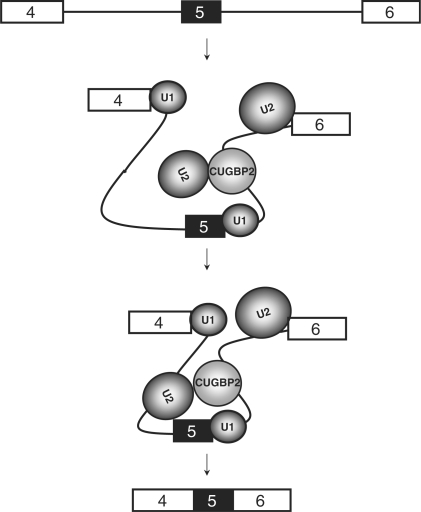
In this study, we investigated the mechanism by which CUGBP2 activates chicken cTNT exon 5 inclusion. Our results strongly support a model (Figure 8) in which CUGBP2 recruits or stabilizes binding of the U2 snRNP to the branch site via direct interactions with two proteins within the activated 17S U2 complex. Furthermore, our results suggest that CUGBP2 enhances cTNT exon 5 recognition by stimulating interactions across the exon, consistent with enhanced exon definition. First, we demonstrated that CUGBP2 increases complex A formation of cTNT pre-mRNAs containing exon 5. Of particular interest, CUGBP2 stimulated assembly of an A-like complex on the MSE1-4 substrate containing exon 5 and segments of the flanking introns including the CUGBP2-binding sites within the downstream intron. Second, stimulation of complex A assembly by CUGBP2 requires the upstream intron containing the branch site since CUGBP2 does not stimulate complex A assembly of the two-exon substrate with just the downstream intron containing the CUGBP2-binding site. Third, CUGBP2 was shown to enhance binding of U2 snRNA on the single exon RNA, MSE1-4. The requirement for the upstream intron combined with the demonstration that CUGBP2 enhances binding of the U2 snRNA strongly suggests that binding of CUGBP2 to its binding sites located 24 and 74 nt downstream from exon 5 promotes binding of the U2 snRNP by interactions across the exon. Fourth, results from tandem affinity purification of CUGBP2-associated proteins in HeLa nuclear extracts identified two proteins, SF3b145 and SF3b49, which were shown to directly interact with CUGBP2 using GST pull-down assays. Taken together, our results strongly suggest that CUGBP2 promotes or stabilizes binding of U2 snRNP to the branch site to stimulate complex A assembly.
Figure 8.
Model of CUGBP2 mediated splicing activation of cTNT exon 5. CUGBP2 binding to intron 5 recruits U2 snRNP across exon 5 without affecting 5′ splice site recognition of U1 snRNP recognition of intron 5. Increased exon definition by CUGBP2 leads to enhanced inclusion of exon 5.
Spliceosome assembly is a stepwise process and each step of assembly can be stimulated or blocked by splicing activators or repressors. In most cases, splicing regulators are involved in the early stages of spliceosome formation and often modulate U1 snRNP association with the 5′ splice site. For example, when TIA1 binds a U-rich sequence downstream of an alternative exon, it increases 5′ splice site recognition by recruiting U1 snRNP through an interaction with U1-C, a U1 snRNP-associated protein (18). Similarly, RBM25 enhances U1 snRNP binding to the 5′ splice site (14) while exonic binding of Nova inhibits U1 snRNP recruitment to the 5′ splice site (13). Splicing regulators affect early steps of spliceosome assembly by other mechanisms. For example, PTB represses c-src exon N1 inclusion by blocking U2AF binding to 3′ splice site of the downstream exon resulting in decreased complex E formation (15). In the case of Fas exon 5, PTB inhibits the association of U2AF and U2 snRNP to the upstream 3′ splice site (32). HuR also inhibits U2AF recognition of the 3′ splice site without decreasing 5′ splice site recognition (33). RS domain containing proteins engage in direct interaction with either the branch site or 5′ splice site and promote U snRNA base-pairing with pre-mRNA (34,35).
In AMT-psoralen cross-linking studies, CUGBP2 does not influence the U1 snRNP recruitment to the 5′ splice site (data not shown). Instead, our results indicate that CUGBP2 increases U2 snRNA binding to the pre-mRNA to enhance assembly of complex A by interacting with U2 snRNP proteins, SF3b49 and SF3b145, lead to enhancement of U2 snRNP recruitment to the branch site.
In addition to being a splicing activator, CUGBP2 and its paralogue, CUGBP1 are also splicing repressors of other substrates such as the insulin receptor exon 11 (G:\Inprocess\Oup\XML-Headers\nar\14-05-09\xml). This is in common with many splicing regulators that can activate some splicing events and repress others. Their roles as an activator or repressor are determined by different features, depending on the regulator and can include post-translational modifications and the position of the binding site relative to the alternative exon. For instance, phosphorylation of SRp38 induces a switch from a general splicing repressor to a sequence-specific activator (38). Binding of Nova to YCAY clusters in exons is associated with exon skipping while binding to downstream intronic YCAY clusters is associated with increased exon inclusion (13). Results suggesting similar position-dependent effects have been found for Fox1/Fox2 and CUGBP2/CUGBP1 in analyses using splicing sensitive microarrays (39–41). It is interesting that different positioning of a splicing regulator has such a dramatic effect on whether exon inclusion is enhanced or repressed. It is also interesting that different regulators appear to promote skipping or inclusion by making contact with different spliceosome components yet binding downstream of the exon is commonly associated with exon inclusion while binding upstream of the exon is associated with repression. It will be interesting to compare the associations that CUGBP2 makes with basal splicing machinery to repress exon inclusion.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
American Heart Association postdoctoral fellowship (Texas Affilliate) [0625119Y to Y-H.G]; National Institutes of Health [R01HL45565 to T.C]. Funding for open access charge: R01HL45565.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Jin Han for establishing TAP and CUGBP2 TAP cell lines, Nicolas Charlet-Berguerand for analysis of U1 binding in response to CUGBP2, Dr James P Orengo and Ioannis Grammatikakis for helpful discussions on the manuscript and Dr Robin Reed, Dr Reinhard Luhrmann and Cindy Will for antibodies.
REFERENCES
- 1.Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- 2.Valadkhan S. snRNAs as the catalysts of pre-mRNA splicing. Curr. Opin. Chem. Biol. 2005;9:603–608. doi: 10.1016/j.cbpa.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 3.Will CL, Lührmann R. Spliceosome structure and function. In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA World. 3rd. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press; 2006. pp. 369–400. [Google Scholar]
- 4.Berget SM. Exon recognition in vertebrate splicing. J. Biol. Chem. 1995;270:2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- 5.Robberson BL, Cote GJ, Berget Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol. Cell Biol. 1990;10:84–94. doi: 10.1128/mcb.10.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 7.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blencowe BJ. Alternative splicing: new insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 9.Yeo G, Holste D, Kreiman G, Burge CB. Variation in alternative splicing across human tissues. Genome Biol. 2004;5:R74. doi: 10.1186/gb-2004-5-10-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sugnet CW, Srinivasan K, Clark TA, O’Brien G, Cline MS, Wang H, Williams A, Kulp D, Blume JE, Haussler D, et al. Unusual intron conservation near tissue-regulated exons found by splicing microarrays. PLoS Comput. Biol. 2006;2:e4. doi: 10.1371/journal.pcbi.0020004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonnal S, Martinez C, Forch P, Bachi A, Wilm M, Valcarcel J. RBM5/Luca-15/H37 regulates Fas alternative splice site pairing after exon definition. Mol. Cell. 2008;32:81–95. doi: 10.1016/j.molcel.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 12.Sharma S, Kohlstaedt LA, Damianov A, Rio DC, Black DL. Polypyrimidine tract binding protein controls the transition from exon definition to an intron defined spliceosome. Nat. Struct. Mol. Biol. 2008;15:183–191. doi: 10.1038/nsmb.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ule J, Stefani G, Mele A, Ruggiu M, Wang X, Taneri B, Gaasterl T, Blencowe BJ, Darnell RB. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006;444:580–586. doi: 10.1038/nature05304. [DOI] [PubMed] [Google Scholar]
- 14.Zhou A, Ou AC, Cho A, Benz E.J., Jr, Huang SC. Novel splicing factor RBM25 modulates Bcl-x pre-mRNA 5′ splice site selection. Mol. Cell Biol. 2008;28:5924–5936. doi: 10.1128/MCB.00560-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharma S, Falick AM, Black DL. Polypyrimidine tract binding protein blocks the 5′ splice site-dependent assembly of U2AF and the prespliceosomal E complex. Mol. Cell. 2005;19:485–496. doi: 10.1016/j.molcel.2005.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.House AE, Lynch KW. Regulation of alternative splicing: more than just the ABCs. J. Biol. Chem. 2008;283:1217–1221. doi: 10.1074/jbc.R700031200. [DOI] [PubMed] [Google Scholar]
- 17.Zhou HL, Lou H. Repression of prespliceosome complex formation at two distinct steps by Fox-1/Fox-2 proteins. Mol. Cell Biol. 2008;28:5507–5516. doi: 10.1128/MCB.00530-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forch P, Puig O, Martinez C, Seraphin B, Valcarcel J. The splicing regulator TIA-1 interacts with U1-C to promote U1 snRNP recruitment to 5′ splice sites. EMBO J. 2002;21:6882–6892. doi: 10.1093/emboj/cdf668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Charlet BN, Logan P, Singh G, Cooper TA. Dynamic antagonism between ETR-3 and PTB regulates cell type-specific alternative splicing. Mol. Cell. 2002;9:649–658. doi: 10.1016/s1097-2765(02)00479-3. [DOI] [PubMed] [Google Scholar]
- 20.Leroy O, Dhaenens CM, Schraen-Maschke S, Belarbi K, Delacourte A, Andreadis A, Sablonniere B, Buee L, Sergeant N, et al. ETR-3 represses Tau exons 2/3 inclusion, a splicing event abnormally enhanced in myotonic dystrophy type I. J. Neurosci. Res. 2006;84:852–859. doi: 10.1002/jnr.20980. [DOI] [PubMed] [Google Scholar]
- 21.Faustino NA, Cooper TA. Identification of putative new splicing targets for ETR-3 using sequences identified by systematic evolution of ligands by exponential enrichment. Mol. Cell Biol. 2005;25:879–887. doi: 10.1128/MCB.25.3.879-887.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang W, Liu H, Han K, Grabowski PJ. Region-specific alternative splicing in the nervous system: implications for regulation by the RNA-binding protein NAPOR. RNA. 2002;8:671–685. doi: 10.1017/s1355838202027036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anderson PA, Malouf NN, Oakeley AE, Pagani ED, Allen PD. Troponin T isoform expression in humans. A comparison among normal and failing adult heart, fetal heart, and adult and fetal skeletal muscle. Circ. Res. 1991;69:1226–1233. doi: 10.1161/01.res.69.5.1226. [DOI] [PubMed] [Google Scholar]
- 24.Cooper TA, Ordahl CP. A single cardiac troponin T gene generates embryonic and adult isoforms via developmentally regulated alternate splicing. J. Biol. Chem. 1985;260:11140–11148. [PubMed] [Google Scholar]
- 25.Ladd AN, Charlet N, Cooper TA. The CELF family of RNA binding proteins is implicated in cell-specific and developmentally regulated alternative splicing. Mol. Cell Biol. 2001;21:1285–1296. doi: 10.1128/MCB.21.4.1285-1296.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dignam JD, Martin PL, Shastry BS, Roeder RG. Eukaryotic gene transcription with purified components. Methods Enzymol. 1983;101:582–598. doi: 10.1016/0076-6879(83)01039-3. [DOI] [PubMed] [Google Scholar]
- 27.Seiwert SD, Steitz JA. Uncoupling two functions of the U1 small nuclear ribonucleoprotein particle during in vitro splicing. Mol. Cell Biol. 1993;13:3135–3145. doi: 10.1128/mcb.13.6.3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gould KL, Ren L, Feoktistova AS, Jennings JL, Link AJ. Tandem affinity purification and identification of protein complex components. Methods. 2004;33:239–244. doi: 10.1016/j.ymeth.2003.11.019. [DOI] [PubMed] [Google Scholar]
- 29.Strathdee CA, McLeod MR, Hall JR. Efficient control of tetracycline-responsive gene expression from an autoregulated bi-directional expression vector. Gene. 1999;229:21–29. doi: 10.1016/s0378-1119(99)00045-1. [DOI] [PubMed] [Google Scholar]
- 30.Reed R. Mechanisms of fidelity in pre-mRNA splicing. Curr. Opin. Cell Biol. 2000;12:340–345. doi: 10.1016/s0955-0674(00)00097-1. [DOI] [PubMed] [Google Scholar]
- 31.Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Seraphin B. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 32.Izquierdo JM, Majos N, Bonnal S, Martinez C, Castelo R, Guigo R, Bilbao D, Valcarcel J. Regulation of Fas alternative splicing by antagonistic effects of TIA-1 and PTB on exon definition. Mol. Cell. 2005;19:475–484. doi: 10.1016/j.molcel.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 33.Izquierdo JM. Hu antigen R (HuR) functions as an alternative pre-mRNA splicing regulator of Fas apoptosis-promoting receptor on exon definition. J. Biol. Chem. 2008;283:19077–19084. doi: 10.1074/jbc.M800017200. [DOI] [PubMed] [Google Scholar]
- 34.Shen H, Green MR. A pathway of sequential arginine-serine-rich domain-splicing signal interactions during mammalian spliceosome assembly. Mol. Cell. 2004;16:363–373. doi: 10.1016/j.molcel.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 35.Shen H, Green MR. RS domains contact splicing signals and promote splicing by a common mechanism in yeast through humans. Genes Dev. 2006;20:1755–1765. doi: 10.1101/gad.1422106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Han J, Cooper TA. Identification of CELF splicing activation and repression domains in vivo. Nucleic Acids Res. 2005;33:2769–2780. doi: 10.1093/nar/gki561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ho TH, Charlet BN, Poulos MG, Singh G, Swanson MS, Cooper TA. Muscleblind proteins regulate alternative splicing. EMBO J. 2004;23:3103–3112. doi: 10.1038/sj.emboj.7600300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feng Y, Chen M, Manley JL. Phosphorylation switches the general splicing repressor SRp38 to a sequence-specific activator. Nat. Struct. Mol. Biol. 2008;15:1040–1048. doi: 10.1038/nsmb.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kalsotra A, Xiao X, Ward A, Castle JC, Johnson JM, Burge CB, Cooper TA. A post natal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc. Natl Acad. Sci. USA. 2008;105:20333–20338. doi: 10.1073/pnas.0809045105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang C, Zhang Z, Castle J, Sun S, Johnson J, Krainer AR, Zhang MQ. Defining the regulatory network of the tissue-specific splicing factors Fox-1 and Fox-2. Genes Dev. 2008;22:2550–2563. doi: 10.1101/gad.1703108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Castle JC, Zhang C, Shah JK, Kulkarni AV, Kalsotra A, Cooper TA, Johnson JM. Expression of 24,426 human alternative splicing events and predicted cis regulation in 48 tissues and cell lines. Nat. Genet. 2008;40:1416–1425. doi: 10.1038/ng.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.