Abstract
To investigate mtDNA recombination induced by multiple double strand breaks (DSBs) we used a mitochondria-targeted form of the ScaI restriction endonuclease to introduce DSBs in heteroplasmic mice and cells in which we were able to utilize haplotype differences to trace the origin of recombined molecules. ScaI cleaves multiple sites in each haplotype of the heteroplasmic mice (five in NZB and three in BALB mtDNA) and prolonged expression causes severe mtDNA depletion. After a short pulse of restriction enzyme expression followed by a long period of recovery, mitochondrial genomes with large deletions were detected by PCR. Curiously, we found that some ScaI sites were more commonly involved in recombined molecules than others. In intra-molecular recombination events, deletion breakpoints were close to or upstream of ScaI cleavage sites, confirming the recombinogenic character of DSBs in mtDNA. A region adjacent to the D-loop was preferentially involved in recombination of all molecules. Sequencing through NZB and BALB haplotype markers in recombined molecules enabled us to show that in addition to intra-molecular mtDNA recombination, rare inter-molecular mtDNA recombination events can also occur. This study underscores the role of DSBs in the generation of mtDNA rearrangements and supports the existence of recombination hotspots.
INTRODUCTION
DNA is constantly exposed to damaging agents from both endogenous and exogenous sources. In order to maintain the integrity of the genome, a sophisticated network of DNA repair pathways remove the majority of harmful lesions (1). Double strand breaks (DSBs) generated by reactive oxygen species (ROS), replication stalling or radiation, represent a highly dangerous form of DNA damage. Bailey et al. (2) have proposed that the POLG mutator mouse displays elevated replication pausing at specific sites in the mtDNA, leading to accumulation of partially replicated linear mtDNA molecules. MtDNA is closely associated with the mitochondrial inner membrane, which is a major site of ROS generation. This is thought to make mtDNA prone to ROS damage as compared to the nuclear genome. Accordingly, mitochondrial genes are highly sensitive to mutations (3). It is now clear that mitochondria are capable of repairing most DNA lesions (1). Repair of DSBs in mtDNA may be conducted by homologous recombination (HR) or non-homologous end-joining (NHEJ), as occurs in the nucleus. The presence of multiple copies of mtDNA make HR one likely system for repairing DSBs in mitochondria (1). A mitochondrial DSBs repair mechanism has been shown to be active in Drosophila melanogaster after the induction of DSBs with Bleomycin (4). Recombination of mtDNA in mammalian cells is less understood despite the fact that molecules with deletions, resulting from intra-molecular recombination, accumulate throughout life (5,6). The events triggering this recombination are not known, but recent evidence suggests that DSBs could have a major role as a trigger (7–9). In the present study, we took advantage of cells harboring different mtDNA haplotypes to investigate the consequences of both intra- and inter-molecular DSBs induced recombination.
MATERIALS AND METHODS
Animal handling
Mice harboring two mtDNA haplotypes (NZB/BALB) were produced as described (10) and mtDNA heteroplasmy was characterized by PCR/RFLP analysis of total DNA obtained from tail biopsy as described (11). Prior to injection the mouse used in this study carried 64% NZB haplotype mtDNA in liver. Mice were maintained in a virus- and antigen-free (VAF) facility in accordance with Institutional and IUCAC committee regulations.
Mito–ScaI–HA construct
The ScaI gene (from an Escherichia coli strain carrying the cloned ScaI gene from Streptomyces caespitosus, a kind gift from New England Biolabs) was cloned downstream of the mitochondrial targeting sequence from human COX8A and a C-terminal HA-tag was added (12). The GeneSwitch system (Invitrogen) was used for the mammalian expression of the mitochondrial targeted ScaI restriction endonuclease because of its absolute low un-induced expression. The expression vector provides a minimal promoter, GAL4–E1b, consisting of the binding sites for the yeast Gal4 DNA-binding protein followed by a TATA sequence from the adenovirus E1b promoter. Without additional factors, the GAL1–E1b promoter is transcriptionally silent. To activate the transcription of GAL4–E1b promoter, the gene Switch regulatory protein is expressed from a minimal TK promoter and is formed by a fusion protein containing the Gal4 DNA-binding domain (Gal4–DBD) to bind the regulatory protein to the GAL4–E1b promoter, the truncated human progesterone receptor-binding domain (hPR–LBD) that undergoes a conformational change when its binds to the progesterone antagonist, mifepristone; and a transcriptional activation factor p65 (p65AD) to activate transcription from the silent GAL4–E1b promoter. Mifepristone is a synthetic 19-norsteroid that binds with high affinity to the human progesterone receptor and the glucocorticoid receptor acting as an atagonist (13). When mifepristone is added into the medium and binds the hPR–LBD region, the Gene Switch fusion protein assumes a conformation that permits it to stimulate transcription from the GAL4–E1b promoter. Mito–ScaI–HA was cloned into this chimeric vector composed of pGene/V5 (Invitrogen) and pSwitch (Invitrogen) kindly donated by Dr R. Voellmy (University of Miami).
rAd–Mito–ScaI–HA construct
Mito-ScaI–HA was cloned into the pAdTrack5 adenoviral vector encoding the insert and eGFP under the control independent CMV promoters. rAd–Mito–ScaI–HA and control rAd–eGFP adenovirus stocks were prepared by the Colorado State University Virus Core Facility (Ft Collins, CO, USA). The adenovirus titers were estimated by OD260: rAd–Mito–ScaI–HA: 5.4 × 1012 particles/ml; rAd–eGFP: 9.2 × 1012 particles/ml (14).
Adenovirus injections and sample preparation
Six- to eight-week-old adult mice were used for all experiments. Liver biopsies were obtained after anesthesia prior to virus injection on Day 0 and at various time points following injection. Mice were euthanized by perfusion with chilled PBS under anesthesia, after which terminal liver samples were taken. Mice were injected in the right external jugular vein with 5 µl of rAd–Mito–ScaI–HA or rAd–eGFP (2.5 × 1010 particles) diluted in 30–50 μl of PBS with 30-gauge dental needle attached to a 100 ul gastight Hamilton syringe via 30-gauge tygon tubing as described (14).
Cell lines, transfections and generation of inducible clones
The NZB/BALB heteroplasmic mouse hepatocyte cell line, HP202B, was grown as described (15). Plasmid transfections were performed with FuGENE 6 (Roche Applied Science, Indianapolis) and clones isolated using ring-cloning. Stable mito–ScaI–HA clones were selected with Hygromycin B (400 mg/ml, Invitrogen) and Mito–ScaI–HA expression was assayed in multiple clones following induction with 10 nM mifepristone for 24 h. Stable clones derived from HP202B tended to have low levels of NZB mtDNA. To generate more useful inducible clones with higher levels of NZB mtDNA, highly inducible HP202B clones were chemically enucleated with actinomycin D (4 µg/ml for 24 h) and fused with hepatocytes carrying 100% of NZB mtDNA treated with Rhodamine-6G (2.5 µg/ml for 2 or 3 days) using polyethylene glycol (PEG), as described (16,17). After subcloning, individual clones were characterized for mtDNA heteroplasmy by RFLP and induced to check expression of Mito–ScaI–HA. Clone 22 was selected for experiments, hereafter HP202B.22; it carries 51% NZB mtDNA and was shown to be highly inducible for Mito–ScaI–HA expression by immunofluorescence after induction with mifepristone (10 nM, 30 min).
Quantification of heteroplasmy by RFLP
Total DNA from samples of interest was phenol–chloroform-extracted and PCR-amplified using primers corresponding to mouse mtDNA positions 5310–5333 and 5665–5690. Final PCR products were subjected to last cycle hot PCR (18) followed by a digestion with ApaLI. This enzyme cleaves BALB mtDNA at position 5461 but does not have cleavage sites in the NZB mtDNA in this amplicon. After digestion, RFLP fragments were resolved in 8% polyacrylamide Tris-Taurine EDTA gels and radioactive signals, corresponding to the two mtDNA haplotypes, were quantified using a Cyclone storage phosphorimager system (Perkin Elmer).
Immunofluorescent studies
HP202B.22 cells grown on glass coverslips were induced with 5 nM of mifepristone for 30 min and then cultured for up to 72 h prior to staining. On the day of staining, mitochondria were visualized by staining with Mitotracker Red (200 nM, Invitrogen) prior to fixation as recommended by the manufacturer. Immunofluorescent staining was carried as described (14). Rat anti-HA primary antibody was purchased from Roche (clone 3F10) and Alexa Fluor 488-conjugated anti-Rat immunoglobulin secondary antibody was purchased from Invitrogen. Images were recorded using a Zeiss LSM 510 confocal microscope.
Isolation of mitochondria and determination of respiratory chain enzyme activities
Crude mitochondrial preparations were obtained using differential centrifugation as described (19) and stored at –80°C until needed. COX activity and citrate synthase activity were determined spectrophotometrically in crude mitochondrial preparations as described (20). Protein concentrations were estimated by the method of Bradford using bovine serum albumin (BSA) as a standard (21).
Southern blots
Five micrograms of total DNA was digested with SacI, which linearizes murine mtDNA by cleaving at nt 9047 and produces an 8 kb fragment of the murine rDNA repeat unit containing the 18S rRNA gene. Digested samples were resolved on 0.8% agarose gels and transferred onto zeta-probe membranes (BioRad). mtDNA was detected by hybridization with random primed probes generated from a 10 kb PCR product corresponding to nt 10–10 145 of the mouse mtDNA. To quantify the nuclear DNA, the same blots were hybridized with random-primed probes generated against a 1-kb PCR product corresponding to nt 502–1515 of the murine rDNA repeat unit encompassing the 18S rRNA gene (accession no. BK000964). Radioactive signal was quantified using a Cyclone storage phosphorimager system (Perkin Elmer).
Real-time quantitative PCR
Total RNA was extracted from cells (RNeasy Plus Mini kit, QIAGEN) followed by treatment with RQ1 DNAse (Promega). cDNA was synthesized using the SuperScript First Strand Kit (Invitrogen) and quantitative real-time PCR reactions were performed on the cDNAs using TaqMan Gene Expression assays (Applied Biosystems) for Lig3 (ligase III, Mn00521933.m1), Tfam (mitochondrial transcription factor A, 00447485.m1) and Polg (polymerase gamma 00553315.m1). The results were normalized against beta actin (NM_007393.3).
Cloning and sequencing of recombined mtDNA
Total DNA was isolated from HP202B.22 and control hepatocytes before induction and at 2, 6 or 9 days post induction with mifepristone 5 nM for 30 min. Total DNA was isolated from liver before injection and at 10 or 14 weeks post injection with rAd–mito–ScaI–HA or rAd–eGFP. Fragments for analysis were amplified using Takara LA Taq Long PCR system (Takara Bio Inc) with several primers combinations: (i) between mtDNA nt 5399 and either 15 065, 15 690 or 1078. (ii) Between nt 8300 and either 15 065 or 1078. (iii) Between nt 8629 and either 15 065 or 1078. No primers bound polymorphic nucleotides between each mtDNA haplotype. Short extension times were used to increase amplification of recombinant molecules and reduce competitive amplification of full size fragments (short-extension PCR). PCR products were resolved on 0.8% agarose TAE gels and bands that were present only in Mito–ScaI–HA expressing cells or tissues were excised and DNA extracted using Qiaex II gel purification kit (Qiagen). Isolated DNA was cloned using the Topo TA Cloning kit (Invitrogen) and inserts from positive clones were sequenced using vector-specific primers. BALB and NZB mtDNA (accession numbers AJ512208 and L07095, respectively) differ at 90 loci: 85 single nucleotide variants, two di-nucleotide variants and three single-nucleotide insertions corresponding to 99.4% identity (10,22). These differences were used to distinguish between the two haplotypes of mDNA in sequenced fragments.
Statistical analysis
All results are expressed as means ± SEM. Unpaired Student's t-test was performed for significance.
RESULTS
Extensive DSBs cause rapid mtDNA depletion in heteroplasmic hepatocytes
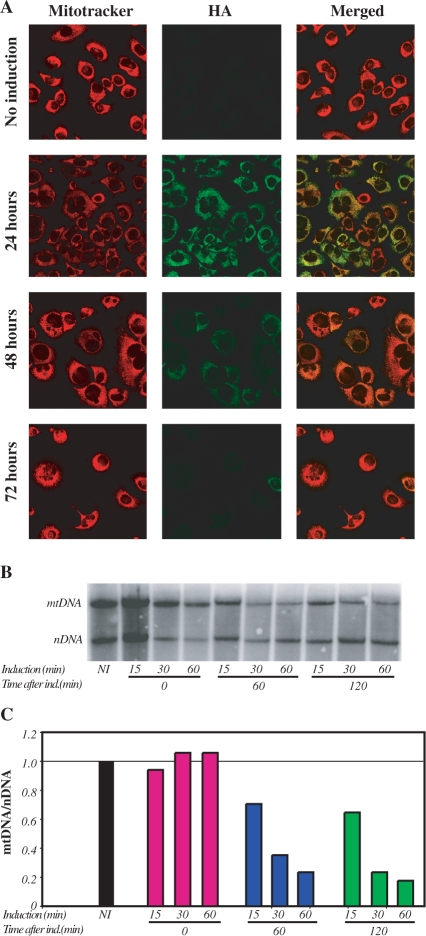
To evaluate mitochondrial localization of the Mito–ScaI–HA, HP202B.22 cells (hepatocyte-derived cells) were transiently trasfected with plasmid containing a mifepristone-inducible Mito–ScaI–HA as described in Methods. After induction with 5 nM mifepristone for 30 min, the levels of Mito–ScaI–HA expression and mitochondrial localization were assessed by immunofluorescent staining for HA and co-localization with Mitotracker (Figure 1A). Mito–ScaI–HA expression was undetectable at 48 h post induction and lower levels of expression were obtained when HP202B.22 cells were induced with lower concentrations of mifepristone (data not shown). Re-induction of HP202B.22 cells with 5 nM mifepristone for 30 min at 2 or 4 days following the first induction had only a small effect on further expression of Mito–ScaI–HA as determined by immunocytochemistry staining (data not shown). It is not clear why re-induction is not as robust as the initial one.
Figure 1.
Inducible expression of Mito–ScaI–HA leads to a fast mtDNA depletion. (A) The mitochondrial localization of ScaI was confirmed by confocal microscopy of HP202B.22 cells. Colocalization of the anti-HA signal with mitotracker red was evident after induction of Mito–ScaI–HA. Cells were analyzed 24, 48 and 72 h after induction. No Mito–ScaI–HA expression was detected after 72 h induction. (B) mtDNA levels were estimated by Southern blot using two probes: one homologous to most of the mtDNA and a second one homologous to the nuclear-coded 18S rDNA. (C) The relative levels of mtDNA were expressed as a ratio to the nuclear encoded 18S rDNA. Cells were induced for 15, 30 and 60 min and collected at 0, 60 and 120 min after induction.
mtDNA/nDNA ratios were evaluated by Southern blot, (Figure 1B). The combination of the induction time and time after the removal of the drug showed that a short 15 min induction followed by 60 min incubation after the removal of the mifepristone is necessary for a 30% reduction in mtDNA content. Sixty minutes of continuing induction with 5 nM mifeprisone were not enough to cause a mtDNA depletion (Figure 1B and C). Interestingly, there appeared to be a subpopulation of mtDNA that was resistant to degradation and depletion did not result in the total loss of mtDNA and production of ρ0 (mtDNA-less) genotype.
MtDNA depletion precedes the decrease in mitochondrial enzymatic activity
Respiratory chain activity is tightly connected to mtDNA copy number; however, the half-life of respiratory complexes has not been well studied. We tested the correlation between the COX activity and the expression of Mito–ScaI–HA. Using constant induction, we observed that decreased mtDNA content precedes the decline of the COX activity in mitochondria by more than 5 h. Even after 24 h induction, the residual COX activity was significantly higher than the residual mtDNA levels (Figure 2A and B).
Figure 2.
Changes in cytochrome c oxidase activity, mtDNA/nDNA ratios and mtDNA-associated transcripts after induction of DSBs. (A) COX enzyme activity normalized against citrate synthase at 5 h, 24 h and 7 days post induction; non-induced (NI) control. (B) mtDNA/nDNA ratios determined by Southern blot, time points and controls as in panel A. (C) The mtDNA/nDNA ratios (dark gray bars) were correlated to the transcript levels of POLG, LIG3 and TFAM (filled circles in each sub-panel). All transcript levels were normalized to β-actin. The DNA and RNA samples in (C) were obtained from hepatocytes after the induction of Mito–ScaI–HA expression for 30 min and culturing for different times. The data are expressed as changes from non-induced samples (NI).
Activation of DNA repair mechanisms after the induction of DSBs in mtDNA
To better understand the cellular response to DSBs in mtDNA, we analyzed the transcript levels of genes involved in mtDNA repair mechanisms. RNA samples were obtained from HP202B.22 cells after mifepristone induction (30 min, 5 nM). Two days post induction we observed decreased mtDNA content (black bars on Figure 2C). We examined whether factors associated with mtDNA replication, and potentially involved in repair, were altered by the mtDNA damage. The transcript levels of Polg and Lig3 (solid circles on Figure 2), evaluated by Real Time PCR, were unchanged compared to the levels observed in non-induced (NI) samples; however, at 9 days post induction, the levels of both these transcripts were significantly decreased. The levels of Tfam mRNA did not change during this period (Figure 2C). The levels of TFAM are known to correlate with mtDNA levels (23,24). The levels of POLG and LIG3 proteins were not evaluated due to the lack of adequate antibodies.
Expression of Mito–ScaI–HA generated mtDNA with large deletions
To investigate the effects of DSBs on mtDNA recombination, short-extension PCR products amplified from liver DNA of rAd–Mito–ScaI–HA and rAd–eGFP–injected mice or Mito–ScaI–HA induced and non-induced HP202B.22 cells were resolved on agarose gels. A number of amplification products corresponding to large deletions was observed in the induced samples (Figure 3). With one pair of primers (Figure 3C), smaller amplification products were also observed in the non-induced and 2-day-induced samples, suggesting that they correspond to amplifications from lower abundance, pre-existing rearrangements. The larger bands, which were not present in respective control samples, were gel purified, cloned and sequenced. The location of the recombination breakpoints is shown in Figure 4.
Figure 3.
mtDNA deleted molecules are generated in liver and cells after expression of Mito–ScaI–HA. (A) Agarose gel of short-extension PCR products amplified from liver of non-injected mice (NI) or at 2, 6, 10 or 14 weeks post injection of either rAd–Mito–ScaI–HA or rAd–eGFP using primers 5399F and 1078B. (B) Agarose gel of short-extension PCR products amplified from liver of non-injected mice or at 10 or 14 weeks post injection of rAd–mito–ScaI–HA using alternate primer sets indicated at the bottom of the image. (C) Agarose gel of short-extension PCR products amplified from non-induced and induced HP202B.22 cells at 2, 6 or 9 days post induction using primers 5399F and 1078B. Regions of the gels in A, B or C that were cloned and sequenced are indicated with dashed brackets.
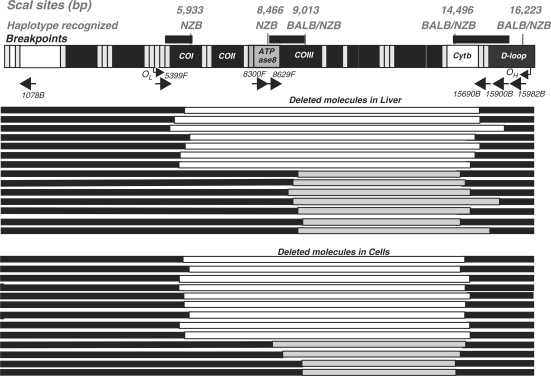
Figure 4.
Schematic representation of the identified mtDNA deletions after multiple DSBs. Two groups of recombined molecules were identified after expression of Mito–ScaI–HA. Recombined molecules (deletions) identified with primers between 5399 and 1078 are shown in white and those identified with primers between 8300 and 15 065 are shown in gray. The preserved regions of the mtDNA are depicted as solid bars. Deletions found in liver are shown in the upper panel and those identified in cells in the lower panel. The mtDNA genetic map on top depicts the location of ScaI sites in both NZB and BALB mtDNA haplotypes, the origins of light strand (OL) and heavy strand replication (OH) and primer binding sites (arrows with primer names).
The sequence from PCR products generated using forward primer 5399 and reverse primers between 15 065 and 1078 revealed breakpoints corresponding to the partially-deleted molecules depicted as white bars in Figure 4. The recombination associated with the generation of these deletions occurred upstream of the NZB-specific ScaI site at nt 5933 and downstream of the shared ScaI site at nt 14 496 regardless of the primers used. When primers amplifying between 8300 and 1078 were used, recombination was observed upstream of the shared ScaI site at nt 9014 and downstream of the shared ScaI site at nt 14 496 (partially deleted molecules shown as gray bars in Figure 4). Interestingly, some ScaI sites appeared to be preferentially involved in recombination events, as the ScaI sites at nt 8466 (NZB specific) and 16 223 (shared) were consistently preserved in the amplification products (not shown).
MtDNA molecules undergo predominantly intra-molecular recombination, but inter-molecular recombination is also observed after DSBs
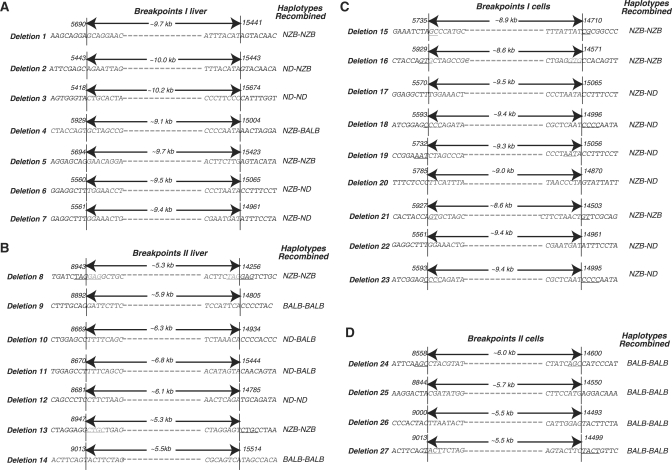
Because of the signature polymorphisms present in the BALB and NZB mtDNA haplotypes we were able to determine the origin of either end of each recombination event. Figure 5A and C show the recombination breakpoints for molecules that recombined upstream of the ScaI site at 5399 and downstream of the ScaI site at nt 14 496. Such recombination events generated deletions in the range of 8.6–10.2 kb and could be identified as principally NZB–NZB haplotype events, most likely reflecting intra-molecular recombination (Figure 5, deletions 1–3, 5–7 and 15–23). A single NZB–BALB haplotype event was identified that must have arisen as the result of an inter-molecular recombination event (Figure 5, deletion 4).
Figure 5.
Intramolecular and intermolecular recombination after DSBs. Twenty-seven different patterns of deleted molecules were identified in liver (panels A and B) and cells (panels C and D) after expression of Mito–ScaI–HA. Arrows indicate the extent of the deletion, nucleotide numbering is shown above each sequence. mtDNA haplotypes involved in each recombination event are indicated on the right, NZB, BALB or ND, not determined. Deletions 1–7 (A) and 8–14 (B), respectively, correspond to the white and gray bars in the upper panel if Figure 4; deletions 15–23 (C) and 24–27 (D), respectively, correspond to the white and gray bars the lower panel of Figure 4.
Recombination events that occurred upstream of the ScaI site at nt 9013 and downstream of the ScaI site at nt 14 496 generated deletions of 5.3–6.8 kb. In this, set of amplicons BALB–BALB and NZB–NZB intra-molecular recombination events were identified in liver (Figure 5, deletions 8–14,) while only BALB–BALB recombination was found in the hepatocytes (Figure 5, deletions 24–27). No inter-molecular recombination events were observed in this set of amplicons. Small direct repeats were identified in some recombination breakpoints from each group (underlined sequences in Figure 5).
DISCUSSION
In the present study we analyzed the effects of multiple DSBs on mtDNA levels, expression of genes linked to mtDNA metabolism and mtDNA recombination. DSBs were induced in mtDNA via the expression of a mitochondria-targeted form of ScaI in heteroplasmic cells and mice. We have previously demonstrated that mtDNA molecules cleaved by mitochondria-targeted restriction enzymes are rapidly degraded by endogenous nucleases prior to repopulation via replication of residual mtDNA (11). Both haplotypes of mtDNA present in our models had multiple ScaI restriction sites and in agreement with other studies mito–ScaI–HA expression led to a rapid decline in mtDNA levels followed by a recovery as expression levels decreased. A functional consequence of the drop in mtDNA levels was confirmed by a reduction of COX activity but this was delayed, reflecting the half life of the enzyme complex (25,26). The expression of three genes involved in mtDNA metabolism were investigated in the same model system, Polg, Lig3 and Tfam. POLG is the catalytic subunit of DNA polymerase-γ, the only DNA-directed DNA polymerase present in mammalian mitochondria. Polymerase-γ is capable of filling small gaps in dsDNA and due to the proofreading function of POLG also plays a role in mitochondrial base excision repair (BER) (3). DNA ligase III, encoded by Lig3, is thought to function along side polymerase-γ in mitochondrial BER because the final step of BER pathway requires a DNA ligase activity and a mitochondrial form of LIG3 has been identified and shown to interact with POLG. A number of studies have found a positive correlation between mtDNA copy number and the levels of transcripts involved in mtDNA repair (24,27–29). Our initial expectation was that genes linked to mtDNA metabolism would be upregulated after the induction of DSBs; however, we found that levels of Polg and Lig3 transcripts were unaltered as mtDNA levels dropped and later reduced as mtDNA levels returned to normal. The pattern of change in transcripts levels that we observed in independent experiments, were not found in skeletal muscle mRNA where mitochondrial biogenesis was promoted after continuous neural activation (30) and our observed decline of expression is contrary to the stable expression of mtDNA metabolic genes seen in an ethidium bromide model of mtDNA depletion (31). This suggests that the decrease in expression of Polg and Lig3 was related to the type of mtDNA damage i.e. DSBs and not mtDNA depletion per second. The levels of Tfam transcripts did not change during the depletion-recovery cycle induced by mito–ScaI–HA in agreement with other studies (23,24). It remains unclear why Polg and Lig3 RNA levels were decreased while Tfam transcript levels were unchanged. It may be that Polg and Lig3 expression reflects the lower mtDNA levels apparent during the recovery phase and that mtDNA packaging is altered during this process leading to no change in Tfam levels.
MtDNA recombination after DSBs in homoplasmic cells and tissues has been previously reported by our group using mitochondria-targeted PstI restriction enzyme that cleaves the murine mtDNA at two sites (7,9). In the PstI model, DSBs generated recombined molecules preferentially between a free end and a specific region in the D-loop. In the case of ScaI, which has multiple cleavage sites in mouse mtDNA, recombination followed a similar pattern involving regions upstream or close to ScaI sites distal from the D-loop and specific regions adjacent to the D-loop. This propensity for recombination close to the D-loop may be due to the fact that the D-loop has a loose triple-strand conformation that may allow easy access for single-strand annealing and recombination (32) or because replication fork arrest, which may be more common in this region, is known to stimulate recombination events (33). Krishnan et al. (8) have proposed a model whereby mtDNA deletions are generated during repair of damaged mtDNA. They also suggested that a region around nt 16 070 bp in the human mtDNA D-loop is a hot spot for intra-molecular recombination deletion formation. Likewise, in the mouse, we have found that specific areas of the D-loop are preferentially involved in recombination regardless of the locations of restriction endonuclese cleavage (7,9). Both the 8466 ScaI site, only present in the NZB haplotype, and the shared 16 223 ScaI site present in the D-loop were preserved in all the identified recombinant molecules, suggesting that either some mtDNA regions are less susceptible to the activity of ScaI or that these specific free ends are less able to recombine than other free ends. The D-loop area contains the heavy strand and light strand mtDNA promoters (HSP, 16 255; LSP, 16 180, respectively). Binding of TFAM has been found upstream of these two sites in the D-loop (34–37). The ScaI site located at the D-loop (position 16 223) is close to this regulatory region and might be protected from digestion due to the binding of factors to mtDNA (37). Other protein-binding sites may be associated with the protection of the 8466 ScaI site, but further experimentation is required to differentiate between protection and preferential recombination.
We have found breakpoints involving small direct repeats (2–6 bp) in the recombinant molecules, as well as breakpoints not associated with obvious homologies. These results are in agreement with those obtained previously in our lab after DSBs (7,9) and other labs on naturally occurring mtDNA deletions (38–42). After induction of DSBs with ScaI, recombined molecules underwent mostly recombination between homologous mtDNA haplotypes, suggesting intramolecular recombination, although we cannot exclude that some of these events occurred between two different mtDNA molecules. Accordingly, we identified only one deletion with a clear signature of intermolecular recombination. MtDNA recombination, including intermolecular recombination, has been reported in mammalian cells both in vitro and in vivo (43,44). Mammalian mitochondria can rejoin blunt-ended and cohesive plasmid DNA molecules at low levels (45). D’Aurelio and colleagues also found evidence of low levels of intermolecular recombination between human cybrids harboring two haplotypes of mtDNA (44). Our results lend further support to the existence of rare intermolecular recombination events on mammalian system and demonstrate that DSBs can stimulate such events.
In summary, we have shown that the induction of multiple DSBs in mtDNA promotes recombination resulting in large deletions. However, there are both preferential free ends and regions relatively distant from free ends (i.e. a specific region of the D-loop) involved in the recombination, suggesting that mtDNA topology is a major contributor to the type of rearrangement. We also found, that even though infrequent, intermolecular recombination is one of the potential consequences of DSBs. These findings provide support for the concept that DSBs can be the originator of the different types of naturally occurring mtDNA rearrangements observed during ageing, disease and evolution.
FUNDING
National Institutes of Health (EY10804, NS041777, CA85700); and the Muscular Dystrophy Association. SB was supported by a supplement to the National Institutes of Health grant EY10804. Funding for open access charge: NIH EY10804.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We are grateful to Sofia Garcia for technical assistance and Dr Alexander Marcillo for surgical assistance. We are indebted to Brendan Battersby and Eric A Shoubridge for the NZB/BALB heteroplasmic mice and to New England Biolabs for the ScaI construct.
REFERENCES
- 1.Larsen NB, Rasmussen M, Rasmussen LJ. Nuclear and mitochondrial DNA repair: similar pathways? Mitochondrion. 2005;5:89–108. doi: 10.1016/j.mito.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 2.Bailey LJ, Cluett TJ, Reyes A, Prolla TA, Poulton J, Leeuwenburgh C, Holt IJ. Mice expressing an error-prone DNA polymerase in mitochondria display elevated replication pausing and chromosomal breakage at fragile sites of mitochondrial DNA. Nucleic Acids Res. 2009;37:2327–2335. doi: 10.1093/nar/gkp091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berneburg M, Kamenisch Y, Krutmann J, Rocken M. ‘To repair or not to repair—no longer a question': repair of mitochondrial DNA shielding against age and cancer. Exp. Dermatol. 2006;15:1005–1015. doi: 10.1111/j.1600-0625.2006.00508.x. [DOI] [PubMed] [Google Scholar]
- 4.Morel F, Renoux M, Lachaume P, Alziari S. Bleomycin-induced double-strand breaks in mitochondrial DNA of Drosophila cells are repaired. Mutat. Res. 2008;637:111–117. doi: 10.1016/j.mrfmmm.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 5.Wiesner RJ, Zsurka G, Kunz WS. Mitochondrial DNA damage and the aging process: facts and imaginations. Free Radic. Res. 2006;40:1284–1294. doi: 10.1080/10715760600913168. [DOI] [PubMed] [Google Scholar]
- 6.Gorbunova V, Seluanov A, Mao Z, Hine C. Changes in DNA repair during aging. Nucleic Acids Res. 2007;35:7466–7474. doi: 10.1093/nar/gkm756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Srivastava S, Moraes CT. Double-strand breaks of mouse muscle mtDNA promote large deletions similar to multiple mtDNA deletions in humans. Hum. Mol. Genet. 2005;14:893–902. doi: 10.1093/hmg/ddi082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krishnan KJ, Reeve AK, Samuels DC, Chinnery PF, Blackwood JK, Taylor RW, Wanrooij S, Spelbrink JN, Lightowlers RN, Turnbull DM. What causes mitochondrial DNA deletions in human cells? Nat. Genet. 2008;40:275–279. doi: 10.1038/ng.f.94. [DOI] [PubMed] [Google Scholar]
- 9.Fukui H, Moraes CT. Mechanisms of formation and accumulation of mitochondrial DNA deletions in aging neurons. Hum. Mol. Genet. 2008;18:1028–1036. doi: 10.1093/hmg/ddn437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jenuth JP, Peterson AC, Shoubridge EA. Tissue-specific selection for different mtDNA genotypes in heteroplasmic mice. Nat. Genet. 1997;16:93–95. doi: 10.1038/ng0597-93. [DOI] [PubMed] [Google Scholar]
- 11.Bayona-Bafaluy MP, Blits B, Battersby BJ, Shoubridge EA, Moraes CT. Rapid directional shift of mitochondrial DNA heteroplasmy in animal tissues by a mitochondrially targeted restriction endonuclease. Proc. Natl Acad. Sci. USA. 2005;102:14392–14397. doi: 10.1073/pnas.0502896102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Giorgi F, Ahmed Z, Bastianutto C, Brini M, Jouaville LS, Marsault R, Murgia M, Pinton P, Pozzan T, Rizzuto R. Targeting GFP to organelles. Methods Cell Biol. 1999;58:75–85. doi: 10.1016/s0091-679x(08)61949-4. [DOI] [PubMed] [Google Scholar]
- 13.Baulieu EE. Contragestion and other clinical applications of RU 486, an antiprogesterone at the receptor. Science. 1989;245:1351–1357. doi: 10.1126/science.2781282. [DOI] [PubMed] [Google Scholar]
- 14.Bacman SR, Williams SL, Hernandez D, Moraes CT. Modulating mtDNA heteroplasmy by mitochondria-targeted restriction endonucleases in a ‘differential multiple cleavage-site' model. Gene Ther. 2007;14:1309–1318. doi: 10.1038/sj.gt.3302981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Battersby BJ, Shoubridge EA. Selection of a mtDNA sequence variant in hepatocytes of heteroplasmic mice is not due to differences in respiratory chain function or efficiency of replication. Hum. Mol. Genet. 2001;10:2469–2479. doi: 10.1093/hmg/10.22.2469. [DOI] [PubMed] [Google Scholar]
- 16.Bayona-Bafaluy MP, Manfredi G, Moraes CT. A chemical enucleation method for the transfer of mitochondrial DNA to rho(o) cells. Nucleic Acids Res. 2003;31:e98. doi: 10.1093/nar/gng100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bacman SR, Moraes CT. Transmitochondrial technology in animal cells. Methods Cell Biol. 2007;80:503–524. doi: 10.1016/S0091-679X(06)80025-7. [DOI] [PubMed] [Google Scholar]
- 18.Moraes CT, Ricci E, Bonilla E, DiMauro S, Schon EA. The mitochondrial tRNA(Leu(UUR)) mutation in mitochondrial encephalomyopathy, lactic acidosis, and strokelike episodes (MELAS): genetic, biochemical, and morphological correlations in skeletal muscle. Am. J. Hum. Genet. 1992;50:934–949. [PMC free article] [PubMed] [Google Scholar]
- 19.Diaz F, Thomas CK, Garcia S, Hernandez D, Moraes CT. Mice lacking COX10 in skeletal muscle recapitulate the phenotype of progressive mitochondrial myopathies associated with cytochrome c oxidase deficiency. Hum. Mol. Genet. 2005;14:2737–2748. doi: 10.1093/hmg/ddi307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barrientos A. In vivo and in organello assessment of OXPHOS activities. Methods. 2002;26:307–316. doi: 10.1016/S1046-2023(02)00036-1. [DOI] [PubMed] [Google Scholar]
- 21.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 22.Loveland B, Wang CR, Yonekawa H, Hermel E, Lindahl KF. Maternally transmitted histocompatibility antigen of mice: a hydrophobic peptide of a mitochondrially encoded protein. Cell. 1990;60:971–980. doi: 10.1016/0092-8674(90)90345-f. [DOI] [PubMed] [Google Scholar]
- 23.Larsson NG, Oldfors A, Holme E, Clayton DA. Low levels of mitochondrial transcription factor A in mitochondrial DNA depletion. Biochem. Biophys. Res. Commun. 1994;200:1374–1381. doi: 10.1006/bbrc.1994.1603. [DOI] [PubMed] [Google Scholar]
- 24.Poulton J, Morten K, Freeman-Emmerson C, Potter C, Sewry C, Dubowitz V, Kidd H, Stephenson J, Whitehouse W, Hansen FJ, et al. Deficiency of the human mitochondrial transcription factor h-mtTFA in infantile mitochondrial myopathy is associated with mtDNA depletion. Hum. Mol. Genet. 1994;3:1763–1769. doi: 10.1093/hmg/3.10.1763. [DOI] [PubMed] [Google Scholar]
- 25.Saikumar P, Kurup CK. Effect of administration of 2-methyl-4-dimethylaminoazobenzene on the half-lives of rat liver mitochondria and cytochrome oxidase. Biochim. Biophys. Acta. 1985;840:127–133. doi: 10.1016/0304-4165(85)90169-2. [DOI] [PubMed] [Google Scholar]
- 26.Aschenbrenner B, Druyan R, Albin R, Rabinowitz M. Haem a, cytochrome c and total protein turnover in mitochondria from rat heart and liver. Biochem. J. 1970;119:157–160. doi: 10.1042/bj1190157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davis AF, Ropp PA, Clayton DA, Copeland WC. Mitochondrial DNA polymerase gamma is expressed and translated in the absence of mitochondrial DNA maintenance and replication. Nucleic Acids Res. 1996;24:2753–2759. doi: 10.1093/nar/24.14.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lakshmipathy U, Campbell C. Antisense-mediated decrease in DNA ligase III expression results in reduced mitochondrial DNA integrity. Nucleic Acids Res. 2001;29:668–676. doi: 10.1093/nar/29.3.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seidel-Rogol BL, Shadel GS. Modulation of mitochondrial transcription in response to mtDNA depletion and repletion in HeLa cells. Nucleic Acids Res. 2002;30:1929–1934. doi: 10.1093/nar/30.9.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schultz RA, Swoap SJ, McDaniel LD, Zhang B, Koon EC, Garry DJ, Li K, Williams RS. Differential expression of mitochondrial DNA replication factors in mammalian tissues. J. Biol. Chem. 1998;273:3447–3451. doi: 10.1074/jbc.273.6.3447. [DOI] [PubMed] [Google Scholar]
- 31.Moraes CT, Kenyon L, Hao H. Mechanisms of human mitochondrial DNA maintenance: the determining role of primary sequence and length over function. Mol. Biol. Cell. 1999;10:3345–3356. doi: 10.1091/mbc.10.10.3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee DY, Clayton DA. Initiation of mitochondrial DNA replication by transcription and R-loop processing. J. Biol. Chem. 1998;273:30614–30621. doi: 10.1074/jbc.273.46.30614. [DOI] [PubMed] [Google Scholar]
- 33.Michel B, Flores MJ, Viguera E, Grompone G, Seigneur M, Bidnenko V. Rescue of arrested replication forks by homologous recombination. Proc. Natl Acad. Sci. USA. 2001;98:8181–8188. doi: 10.1073/pnas.111008798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fisher RP, Clayton DA. A transcription factor required for promoter recognition by human mitochondrial RNA polymerase. Accurate initiation at the heavy- and light-strand promoters dissected and reconstituted in vitro. J. Biol. Chem. 1985;260:11330–11338. [PubMed] [Google Scholar]
- 35.Fisher RP, Topper JN, Clayton DA. Promoter selection in human mitochondria involves binding of a transcription factor to orientation-independent upstream regulatory elements. Cell. 1987;50:247–258. doi: 10.1016/0092-8674(87)90220-0. [DOI] [PubMed] [Google Scholar]
- 36.Fisher RP, Clayton DA. Purification and characterization of human mitochondrial transcription factor 1. Mol. Cell Biol. 1988;8:3496–3509. doi: 10.1128/mcb.8.8.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghivizzani SC, Madsen CS, Nelen MR, Ammini CV, Hauswirth WW. In organello footprint analysis of human mitochondrial DNA: human mitochondrial transcription factor A interactions at the origin of replication. Mol. Cell Biol. 1994;14:7717–7730. doi: 10.1128/mcb.14.12.7717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Samuels DC, Schon EA, Chinnery PF. Two direct repeats cause most human mtDNA deletions. Trends Genet. 2004;20:393–398. doi: 10.1016/j.tig.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 39.Mita S, Rizzuto R, Moraes CT, Shanske S, Arnaudo E, Fabrizi GM, Koga Y, DiMauro S, Schon EA. Recombination via flanking direct repeats is a major cause of large-scale deletions of human mitochondrial DNA. Nucleic Acids Res. 1990;18:561–567. doi: 10.1093/nar/18.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamashita S, Nishino I, Nonaka I, Goto Y. Genotype and phenotype analyses in 136 patients with single large-scale mitochondrial DNA deletions. J. Hum. Genet. 2008;53:598–606. doi: 10.1007/s10038-008-0289-8. [DOI] [PubMed] [Google Scholar]
- 41.Bua E, Johnson J, Herbst A, Delong B, McKenzie D, Salamat S, Aiken JM. Mitochondrial DNA-deletion mutations accumulate intracellularly to detrimental levels in aged human skeletal muscle fibers. Am. J. Hum. Genet. 2006;79:469–480. doi: 10.1086/507132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reeve AK, Krishnan KJ, Elson JL, Morris CM, Bender A, Lightowlers RN, Turnbull DM. Nature of mitochondrial DNA deletions in substantia nigra neurons. Am. J. Hum. Genet. 2008;82:228–235. doi: 10.1016/j.ajhg.2007.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thyagarajan B, Padua RA, Campbell C. Mammalian mitochondria possess homologous DNA recombination activity. J. Biol. Chem. 1996;271:27536–27543. doi: 10.1074/jbc.271.44.27536. [DOI] [PubMed] [Google Scholar]
- 44.D'Aurelio M, Gajewski CD, Lin MT, Mauck WM, Shao LZ, Lenaz G, Moraes CT, Manfredi G. Heterologous mitochondrial DNA recombination in human cells. Hum. Mol. Genet. 2004;13:3171–3179. doi: 10.1093/hmg/ddh326. [DOI] [PubMed] [Google Scholar]
- 45.Lakshmipathy U, Campbell C. Double strand break rejoining by mammalian mitochondrial extracts. Nucleic Acids Res. 1999;27:1198–1204. doi: 10.1093/nar/27.4.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]