Abstract
The discovery of roles for arginine methylation in intracellular transport and mRNA splicing has focused attention on the methylated arginine–glycine (RG)-rich domains found in many eukaryotic RNA-binding proteins. Sequence similarity among these highly repetitive RG domains, combined with interactions between RG-rich proteins, raises the question of whether these regions are general interaction motifs or whether there is specificity within these domains. Using the essential Saccharomyces cerevisiae mRNA-binding protein Npl3 (ScNpl3) as a model system, we first tested the importance of the RG domain for protein function. While Npl3 lacking the RG domain could not support growth of cells lacking Npl3, surprisingly, expression of the RG domain alone supported partial growth of these cells. To address the specificity of this domain, we created chimeric forms of ScNpl3 with RG-rich domains of S. cerevisiae nucleolar proteins, Gar1 and Nop1 (ScGar1, ScNop1), or of the Candida albicans Npl3 ortholog (CaNpl3). Whereas the CaNpl3 RG chimeric protein retained nearly wild-type function in S. cerevisiae, the ScGar1 and ScNop1 RG domains significantly reduced Npl3 function and self-association, indicating RG domain specificity. Nuclear localization of Npl3 also requires specific RG sequences, yet heterologous RG domains allow similar modulation of Npl3 transport by arginine methylation.
INTRODUCTION
Eukaryotic gene expression requires the coordination of a complex array of messenger RNA-binding proteins, which chaperone and modify mRNAs from the time they are transcribed in the nucleus until they are translated and eventually degraded in the cytoplasm. While individual RNA-binding proteins perform specific tasks within the cell, many RNA-binding proteins share similar sequence modules, including RNA-recognition motifs (RRMs), hnRNP K-homology (KH) domains, zinc fingers and arginine–glycine (RG)-rich regions (1,2). Such RNA-binding domains (RBDs) are not limited to mRNA-binding proteins, but are also shared by proteins involved in ribosome assembly. In addition, multiple RBDs have been implicated in protein-protein as well as protein–RNA interactions (1,3–5).
Most RNA-binding domains are defined by specific amino-acid sequences and structures that interact with the phosphodiester backbone and specific nucleobases of RNA ligands (1,6). In contrast, arginine–glycine-rich regions [which are also referred to as glycine–arginine-rich (GAR) motifs and RG or RGG domains] are characterized by the presence of multiple RG and RGG peptides, but vary widely in overall domain length and spacing between these short motifs (7). No 3D structure has been determined for any RG-rich domain, potentially due to the high glycine content of these regions. The RG-rich domain of Fragile X mental retardation protein FMRP binds to specific G-quartet-containing mRNAs (8), but many RG domains may confer non-specific RNA-binding activity to proteins with additional RBD modules, or mediate protein–protein interactions (2,4). RG-rich proteins are also frequent targets for arginine methylation; this modification can impact protein function through altering molecular interactions (4,9). The ubiquity of RG-rich domains, combined with the lack of a clear consensus sequence even among some likely orthologous proteins, raises the question of whether these regions are general interaction motifs or whether there is any specificity inherent to these domains.
The Saccharomyces cerevisiae mRNA-binding protein Npl3 serves as an excellent model to address this question. This protein has been implicated in multiple aspects of gene expression including transcription elongation and termination, 3′ end formation, splicing, mRNA export and translation (10–15). Whereas mutations in the two RRMs of Npl3 affect mRNA 3′ end processing and nuclear export (10,14), mutations in the extensive RG domain and methylation of this domain influence Npl3 transport (16,17). The effects of the RG domain on transport correlate with protein–protein interactions. Arginine methylation or arginine-to-lysine mutations in the RG domain decrease nuclear export of Npl3, Npl3 self-association and the interaction of Npl3 with Tho2, a protein involved in transcription elongation and mRNA export (16,18–20). A two-hybrid screen for proteins that interact with the RG-rich region of Npl3 revealed five RG-rich proteins (21); this result suggests that RG-rich domains might act as general interaction motifs among RNA-binding proteins. To test this hypothesis, we have replaced the long RG-rich domain of the S. cerevisiae mRNA-binding protein Npl3 with extensive RG domains from three other yeast proteins and have tested these chimeric proteins for function in S. cerevisiae.
Initially we demonstrate that the RG-rich domain of Npl3 is not only necessary for in vivo function, but that this domain alone can partially replace Npl3 function. Whereas replacement of the Npl3 RG-rich domain with that of the C. albicans Npl3 ortholog maintains Npl3 function, the extensive RG-rich domains of two nucleolar proteins, Gar1 and Nop1, only partially support Npl3 function. This result correlates with the ability of the chimeric Npl3 proteins to bind to endogenous Npl3. This RG-domain specificity is not due to differences in domain length, since S. cerevisiae Npl3 with a deletion in its RG-rich domain allows better growth of npl3Δ cells than a chimeric Npl3 protein bearing the slightly longer Nop1 RG domain. Gar1 and Nop1 RG domains increase the cytoplasmic steady-state localization of GFP-Npl3, suggesting the importance of specific sequences within the Npl3 RG-rich domain for transport. Whereas deletion of Hmt1 slows nuclear export of Npl3 (17), it does not affect nucleolar localization of Gar1 and Nop1 (22). We demonstrate that all chimeric GFP-Npl3 proteins are substrates for Hmt1 and that Hmt1 deletion results in increased nuclear localization of chimeric GFP-Npl3 bearing Gar1 and Nop1 RG domains. Therefore, although these heterologous RG domains do not confer full function on Npl3, arginine methylation can modulate transport of Npl3 proteins bearing alternative RG domains.
MATERIALS AND METHODS
Plasmid construction
Oligonucleotides were synthesized at Integrated DNA Technologies, Inc. (Table 1). Plasmids used in this study are summarized in Table 2. To construct N-terminal domain deletions in Protein A (PrA)-Npl3, C-terminal domains were amplified from pPS811 (14) with the following oligo pairs: RRM1-RRM2-RG (codons R126-R414) with AM159/AM166, RRM2-RG (codons P196-R414) with AM162/AM166 and RG (codons P276-R414) with AM165/AM166. PCR fragments were digested with NdeI and BamHI and inserted into pNOPPATA (23) to create pAM369, pAM367 and pAM368. The first three domains of Npl3 (codons M1-N275) were amplified from pPS811 with oligos AM156 and AM164. This fragment and the PrA-Npl3 plasmid pPS2389 (18) were digested with NdeI and NsiI and ligated to create pAM372, in which the first three domains of Npl3 are fused to the conserved C-terminus after the RG domain (R398-R414).
Table 1.
Oligonucleotides used in this study
| Oligo | Sequence (5′–3′) |
|---|---|
| AM61 | GGCTGCAGGAATTCGATATCCC |
| AM137 | CAGAGGTTCTGTCATTACTGTTGAAAGAGATGACA ATCCTCCACCAagaagtggtgccccaggtggccgtgg |
| AM138 | GGCTTACCTGGTTGGTGATCTTTCACGTG GAGCATCTCTGGTtcttctacctcctctgaaaccaccacg |
| AM139 | CAGAGGTTCTGTCATTACTGTTGAAAGAG ATGACAATCCTCCACCAagaccaggtagcagaggtggttcccg |
| AM140 | GGCTTACCTGGTTGGTGATCTTTCACGTG GAGCATCTCTGGtaacgaccttggcaccaccacgggcacc |
| AM156 | cccccatATGTCTGAAGCTCAAGAAACTCACG |
| AM159 | cccccatatgAGATTGTTTGTTAGACCTTTCCC |
| AM162 | cccccatatgCCTGCCAAGAGATACCGTATCACC |
| AM164 | cccatgcatcATTGTCATCTCTTTCAACAGTAATGAC |
| AM165 | cccccatatgCCTCCACCAATCAGAAGATC |
| AM166 | cccggatccTTACCTGGTTGGTGATCTTTCACG |
| AM219 | cccccgagctcTACGGTGGCTATTCCAGAGG |
| AM220 | ACCTCgagctcCACCTCTATTTGATCTTCTG |
| AM221 | CCAGAgagctcTATGGTGGTCCAAGAAATG |
| AM223 | GCCACgagctcCACCTCTGGAATAGCCAC |
| AM225 | CGCCCGGAATTAGCTTGGCTGC |
Lowercase indicates nucleotide differences from endogenous genes.
Table 2.
Plasmids used in this study
| Plasmid | Features | Source |
|---|---|---|
| pAM362 | CEN LEU pNop-PrA-ScNPL3-CaNplRGG AmpR | (24) |
| pAM363 | CEN LEU pNop-PrA-ScNPL3-ScGarRGG AmpR | This study |
| pAM364 | CEN LEU pNop-PrA-ScNPL3-ScNopRGG AmpR | This study |
| pAM367 | CEN LEU pNop-PrA-RRM2-RGG AmpR | This study |
| pAM368 | CEN LEU pNop-PrA-RGG AmpR | This study |
| pAM369 | CEN LEU pNop-PrA-RRM1-RRM2-RGG AmpR | This study |
| pAM372 | CEN LEU pNop-PrA-APQE-RRM1-RRM2 AmpR | This study |
| pAM382 | 2 µ URA3 pGal GFP-ScNPL3-CaNplRGG AmpR | (24) |
| pAM383 | 2 µ URA3 pGal GFP-ScNPL3-ScGarRGG AmpR | This study |
| pAM384 | 2 µ URA3 pGal GFP-ScNPL3-ScNopRGG AmpR | This study |
| pAM447 | CEN LEU pNop-PrA-ScNPL3-ΔRGG2-8 AmpR | This study |
| pAM449 | CEN LEU pNop-PrA-ScNPL3-ΔRGG8-14 AmpR | This study |
| pAM463 | CEN LEU pNop-PrA-ScNPL3-ApaI AmpR | (24) |
| pNOPPATA | CEN LEU pNop-PrA AmpR vector | (23) |
| pPS811 | 2 µ URA3 pGal GFP-NPL3 AmpR | (14) |
| pPS1872 | CEN LEU2 ScHMT1 AmpR | (25) |
| pPS2389 | CEN LEU pNop-PrA-ScNPL3 AmpR | (18) |
| pRS315 | CEN LEU2 AmpR vector | (28) |
Construction of plasmids expressing chimeric Npl3 proteins was performed as described for pAM362, which expresses a Protein A (PrA)-tagged S. cerevisiae Npl3 protein bearing the C. albicans Npl3 RG domain (24). Briefly, the coding region for the longest RG-rich domain of S. cerevisiae Gar1 or Nop1 was amplified from wild-type (FY23) genomic DNA using oligos AM137/AM138 (Gar1) or AM139/AM140 (Nop1). This fragment was co-transformed into wild-type cells with ApaI-linearized pAM463 (24) and plasmids were rescued from Leu+ cells. Plasmids were sequenced at the University of Maine, Orono DNA sequencing facility to verify proper fusion. Heterologous RG domains were removed from plasmids pAM362 (CaNpl3), pAM363 (ScGar1) and pAM364 (ScNop1) with HindIII and inserted into pPS811 to create chimeric GFP-Npl3 plasmids pAM382, pAM383 and pAM384.
To create an N-terminal deletion within the RG domain of PrA-Npl3, nucleotides 860–970, encoding F287–G323 near the N-terminus of the ScNpl3 RG domain, were removed from the PrA-Npl3 plasmid pPS2389 as follows: a 5′ fragment was amplified by PCR from pPS2389 with oligos AM61 and AM220 and cleaved with NdeI and SacI; a 3′ fragment was amplified with oligos AM219 and AM225 and cleaved with Sac I and SalI. These fragments were inserted into NdeI/SalI-cut pPS2389, which was linearized with Sac I, treated with T4 DNA polymerase and religated to create an in-frame deletion within the RG domain (pAM447 = Δ287–323). Fragments for the removal of C-terminal RG-rich sequences were generated by amplification with AM61/AM223 (5′) and AM221/AM225 (3′) and treated similarly. The resulting plasmid, pAM449 (Δ317–365), contained an in-frame deletion including nucleotides 948–1094.
Npl3 function, self-association and methylation studies
Saccharomyces cerevisiae strains were manipulated as previously described (16,24,25). Growth assays were performed by transforming npl3Δ (PSY814) (26) or npl3Δcbp80Δ (YAM505) (16) cells with LEU2 plasmids that express PrA-tagged proteins then plating mid-log-phase cells on media lacking leucine and containing 5-fluoro-orotic acid, to select for loss of a URA3 NPL3 maintenance plasmid. This assay therefore tested the function of the PrA-tagged Npl3 proteins as the sole copy of the essential Npl3 protein.
Protein A (PrA) pulldown assays were performed as described (16). Briefly, cells expressing Npl3-myc (YAM533) and PrA fusion proteins were grown at 30°C to mid-log-phase and lysed using a FastPrep cell disruptor (Bio101) in lysis buffer (150 mM KCl, 5 mM MgCl2, 20 mM Tris–HCl, pH 8.0) containing protease inhibitors and 0.5% Triton X-100. PrA fusion proteins and interacting proteins were precipitated with IgG-Sepharose (Pharmacia) and analyzed by immunoblotting with polyclonal anti-myc antiserum (sc-789, Santa Cruz Biotechnology; 1:2000 dilution) (16). GFP-Npl3 proteins were expressed from plasmids pPS811, pAM382, pAM383, and pAM384 in hmt1Δ cells (PSY865) that either expressed Hmt1 from pPS1872 or contained the vector plasmid pRS315, which lacks HMT1. GFP-Npl3 methylation was tested by inducing GFP-Npl3 expression in mid-log-phase cells grown at 30°C with 2% galactose for 1.5 h prior to lysis in RIPA buffer (150 mM NaCl, 1% NP-40, 0.5% DOC, 0.1% SDS, 50 mM Tris–HCl, pH 8.0) supplemented with protease inhibitors (25). Protein expression and arginine methylation were detected by resolving these whole cell lysates by SDS-10% PAGE (4 µg total protein for anti-GFP immunoblots and 40 µg total protein for anti-dimethylarginine immunoblots), followed by immunoblotting with monoclonal anti-GFP (Roche; 1:1000) and anti-dimethylarginine (Ab412, Abcam; 1:500) antibodies.
Fluorescence microscopy
Cells lacking Hmt1 (PSY865) (27) bearing either a vector (pRS315) (28) or an HMT1 expression plasmid (pPS1872) (25) and different GFP-expression plasmids were grown at 30°C to mid-log phase in synthetic dropout medium lacking uridine. After induction of GFP-Npl3 fusion proteins as above, cells were washed with phosphate-buffered saline (PBS), and visualized by fluorescence microscopy as described (24). Exposure times were equivalent for all experiments.
RESULTS
The RG-rich domain is essential for Npl3 function and self-association
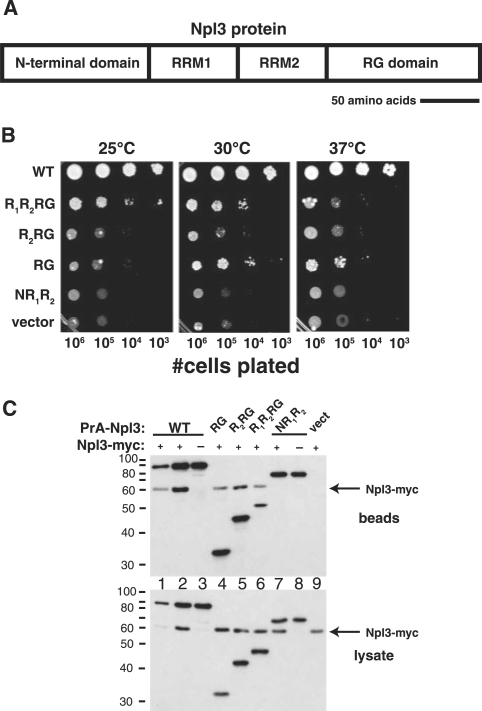
The mRNA-binding protein Npl3 comprises four regions: an N-terminal region, two RNA-recognition motifs (RRMs), and a C-terminal RG-rich domain (Figure 1A). We created domain deletions within Npl3 proteins fused to Protein A to determine the importance of the RG domain for Npl3 function (Figure 1B). Intriguingly, the PrA fusions that contain either both RRMs and the RG domain or the RG domain alone partially support growth of an npl3Δ strain (Figure 1B). Whereas domain deletions have been used to define determinants for cellular localization (29,30), this is the first evidence that the RG domain on its own can provide some Npl3 function in vivo.
Figure 1.
Role of the RG-rich domain in Npl3 self-association and function. (A) Schematic diagram of Npl3 protein primary structure including two RRMs and the arginine–glycine (RG)-rich C-terminal domain (total length = 414 amino acids, scale bar = 50 amino acids). (B) The RG domain alone can support partial growth of cells lacking Npl3. PSY814 (npl3Δ) bearing the plasmids described in (C) below was grown to mid-log-phase, plated on medium containing 5-FOA and lacking leucine, and grown for 4 days at 25°C, 3 days at 30°C or 3 days at 37°C. (C) The RG-rich domain is required for Npl3 self-association. YAM533 (hmt1ΔNPL3-myc) was transformed with plasmids expressing Protein A (PrA) fused to: full-length Npl3p (WT; lanes 1 and 2), the RG domain of Npl3 (RG; lane 4), the second RRM and the RG domain (R2RG; lane 5), both RRMs and the RG domain (R1R2RG; lane 6), or Npl3 lacking the RG domain (NR1R2; lane 7). Mid-log-phase cells were lysed, the PrA fusion protein isolated with IgG-sepharose, and co-precipitated Npl3-myc was detected (beads) by immunoblotting with a polyclonal anti-myc antibody (which also recognizes the PrA fusion protein). Controls included cells lacking Npl3-myc (lanes 3 and 8) and cells expressing only PrA (vect; lane 9). Expression of PrA fusion proteins and Npl3-myc was verified by anti-myc immunoblot analysis of the whole cell lysates used for purification (lysates).
The finding that either arginine methylation or mutations in the RG region of Npl3 disrupt Npl3 self-association (16,18) suggests that the RG domain plays a role in Npl3–Npl3 interactions. To test this possibility, the PrA-Npl3 fusion proteins were co-expressed in cells expressing full-length Npl3-myc and lacking the major arginine methyltransferase Hmt1. The PrA-Npl3 was precipitated with IgG sepharose beads and co-purifying Npl3-myc was detected by immunoblot analysis (beads; Figure 1C). Whereas Npl3-myc co-purified with all fusion proteins containing the RG-rich C-terminal domain (Figure 1C upper panel, lanes 4–6), it did not co-purify with a protein containing only the first three domains of Npl3 (lane 7). Thus the RG domain is required for Npl3 self-association.
Heterologous RG domains and Npl3 function
Multiple eukaryotic RNA-binding proteins contain methylated RG domains of varying lengths (7). The RG-rich C-terminus of Npl3 can bind to at least five RG-domain containing proteins in a two-hybrid assay (21). This assay also suggested that the interaction between the Npl3 RG-rich domain and the other RG-rich proteins was the same or greater in cells lacking the arginine methyltransferase Hmt1 than in wild-type cells (21). This result indicates that arginine methylation may decrease these putative RG–RG interactions, similar to its disruption of Npl3 self-association (18). These results raise the question of whether RG-rich domains can serve a general purpose in directing binding to other RG-rich proteins.
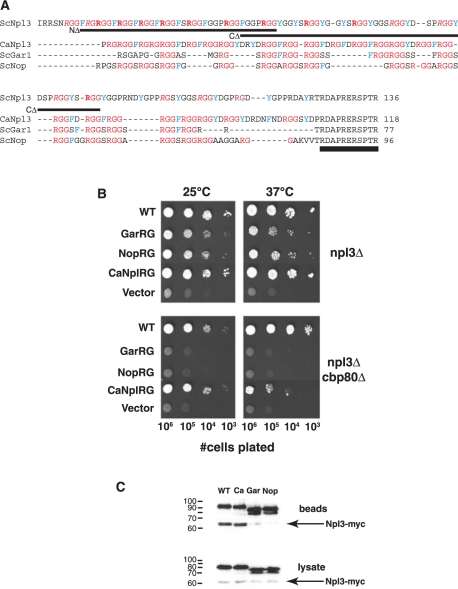
To test this possibility, the first three domains of S. cerevisiae Npl3 were fused to three extensive RG-rich regions from other yeast proteins: two nucleolar S. cerevisiae proteins (ScGar1 and ScNop1) and the Candida albicans Npl3 ortholog [CaNpl3 (24)]. Given that phosphorylation of a serine within a conserved heptapeptide at the C-terminus influences Npl3 nuclear transport and autoregulation (31–34) and also affects transcription elongation (12), this region was included in all chimeric proteins (Figure 2A). These chimeric proteins partially support growth of an npl3Δ strain (Figure 2B). The major arginine methyltransferase Hmt1, which methylates Npl3, is not essential in otherwise wild-type cells, yet this gene becomes essential in a strain lacking the 80 kDa mRNA cap-binding protein, Cbp80 (17), potentially due to the increased importance of Npl3 methylation (16). In this sensitized strain background (npl3Δcbp80Δ), the chimeric proteins bearing the Gar1 and Nop1 RG domains are not functional (Figure 2B). In contrast, the RG domain of C. albicans Npl3, which shares more sequence similarity with that of S. cerevisiae Npl3, promotes greater Npl3 function than the RG domains of ScGar1 and ScNop1 (Figure 2B). These results support the idea that specific sequences within the RG domain are important for Npl3 function. When the chimeric proteins were tested for binding to wild-type Npl3-myc, the chimeric protein containing the CaNpl3 RG domain co-precipitated ScNpl3-myc whereas those Npl3 proteins with Gar1 and Nop1 RG domains showed severely reduced binding to Npl3-myc (Figure 2C). Therefore an Npl3 RG domain is required both for overall function of Npl3 and for Npl3 self-association.
Figure 2.
Specificity of the RG-rich domain in Npl3 function. (A) Comparison of RG domains of four yeast RNA-binding proteins. C-terminal sequences of chimeric Npl3 proteins containing RG domains from the C. albicans Npl3 ortholog and S. cerevisiae nucleolar proteins Gar1 and Nop 1 were aligned with ClustalW. Lines indicate the regions removed from the ScNpl3 RG domain in the N-terminal (NΔ) and C-terminal (CΔ) deletion mutants used in Figure 3. Note the presence of the C-terminal 11 amino acids of S. cerevisiae Npl3 on each chimeric protein (double underline). In ScNpl3, arginines that were previously found to be exclusively dimethylated are noted in bold and arginines with variable levels of methylation are italicized (16). (B) Heterologous RG domains can partially support growth of cells lacking Npl3. PSY814 (npl3Δ) and YAM505 (npl3Δcbp80Δ) were transformed with plasmids expressing chimeric Npl3 proteins containing the RG regions described in (A). Cells were tested for chimera function as described for Figure 1, with growth for 3 days (npl3Δ, 37°C) or 4 days (npl3Δ, 25°C; npl3Δcbp80Δ, 25°C, 37°C). (C) RG-domain specificity in Npl3 self-association. Plasmids in (B) were transformed into YAM533 and the ability of Npl3-myc to co-precipitate with PrA-tagged chimeric proteins was tested as in Figure 1.
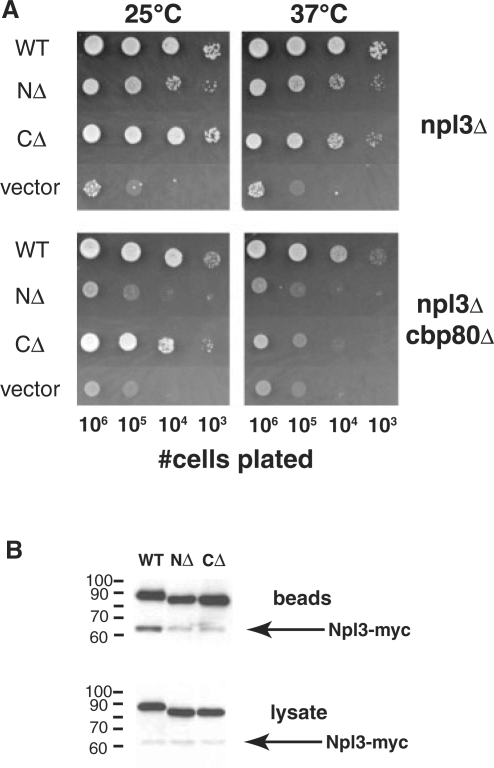
The inability of Gar1 and Nop1 RG-rich domains to support full function of Npl3 could be due to differences in either sequence or length. To address the importance of length and sequence context, N-terminal or C-terminal sequences within the RG domain were deleted from PrA-Npl3 (Figure 2A) and the deletion mutants were tested for function (Figure 3A). Deletion of 49 amino acids in the C-terminal half of the ScNpl3 RG domain, which results in a shorter protein than the Nop1 chimeric protein, had little effect on growth of npl3Δ cells (CΔ, Figure 3A). While this C-terminal deletion supported partial growth of npl3Δcbp80Δ cells at 25°C, deletion of 37 amino acids at the N-terminus of the domain (NΔ) decreased growth of this strain to levels observed in the absence of Npl3 (vector, Figure 3A). These growth patterns of the N-terminal RG deletion therefore mirror the growth of cells expressing the Gar1 and Nop1 chimeric proteins. In contrast, both deletion mutant proteins show reduced interaction with wild-type Npl3-myc (Figure 3B). In combination, the growth and interaction results indicate that the N-terminal sequences in the RG domain are more important than the length of the domain for overall Npl3 function, whereas both the N and C-terminal portions of the RG domain play a role in stabilizing binding to a full-length Npl3 RG domain in the absence of methylation.
Figure 3.
Differential effects of deletions within the RG-rich domain of Npl3. (A) N-terminal deletions within the RG-rich domain of Npl3 decrease growth of a strain lacking Npl3 to a greater extent than C-terminal deletions. PSY814 (npl3Δ) and YAM505 (npl3Δcbp80Δ) were transformed with plasmids expressing S. cerevisiae Npl3 proteins containing the full-length RG domain (WT), a domain lacking amino acids 287–323 (NΔ) or an RG domain lacking amino acids 317–365 (CΔ); the vector lacking Npl3 was used as a control. Cells were tested for Npl3 function as described for Figure 1, with growth at 25°C for 4 days and 37°C for 3 days. (B) N- and C-terminal deletions within the RG domain of Npl3 decrease binding to full-length Npl3. Plasmids in (A) were transformed into YAM533 and the ability of Npl3-myc to co-precipitate with PrA-tagged Npl3 proteins with deletions within the RG domain was tested as in Figure 1.
One defined role for the RG domain of Npl3 is in directing Npl3 localization (30). Methylation within this domain facilitates nuclear export of Npl3 (16,17,35). In contrast, methylation does not affect nucleolar localization of either Gar1 or Nop1 (22). Therefore we tested whether the heterologous RG domains affect localization of GFP-Npl3 in the presence or absence of Hmt1 (Figure 4A). Whereas the chimeric protein containing the C. albicans Npl3 RG domain was predominantly nuclear, the proteins with Gar1 and Nop1 RG domains localized throughout cells expressing Hmt1 (Figure 4A; +Hmt1). This result indicates the importance of Npl3-specific RG domain sequences for steady-state nuclear localization. Notably, these chimeric proteins were predominantly nuclear in cells lacking the arginine methyltransferase (Figure 4A; −Hmt1), suggesting that methylation facilitates export of these proteins as well as of wild-type Npl3. Thus, although these heterologous RG domains do not support full Npl3 function, they do allow Npl3 transport to be modulated by methylation.
Figure 4.
Localization and methylation of chimeric Npl3 proteins. (A) Chimeric Npl3 proteins are predominantly nuclear in the absence of HMT1. PSY865 (hmt1Δ) cells bearing either pPS1872 (+Hmt1) or pRS315 (−Hmt1) were transformed with plasmids expressing GFP-tagged chimeric Npl3 proteins. Localization of GFP-Npl3 was visualized after induction of expression for 1.5 h in mid-log-phase cells grown at 30°C. (B) Chimeric Npl3 proteins are methylated. Aliquots of the cultures shown in (A) were lysed in RIPA buffer, then proteins from these whole cell lysates were resolved by SDS-10% PAGE and analyzed by immunoblotting with monoclonal antibodies. Gels used for anti-GFP immunoblots contained 4 µg total protein in each lane; gels used for anti-dimethylarginine immunoblots contained 40 µg total protein in each lane.
To test whether the effect of Hmt1 on chimera localization could be due to methylation of the chimeric proteins, whole cell lysates from the cells in Figure 4A were probed with anti-GFP and anti-dimethylarginine antibodies (Figure 4B). Both anti-GFP and anti-dimethylarginine antibodies recognized all RG-rich proteins in the presence of Hmt1, but the anti-dimethylarginine antibody did not bind to any of the GFP-fusion proteins in the absence of Hmt1 (Figure 4B). This result indicates that Hmt1 methylates GFP-ScNpl and all three chimeric GFP-fusion proteins. Therefore, the decreased function of Gar1 and Nop1 chimeric proteins is not due to an inability to function as substrates for Hmt1. In addition, methylation of Gar1 and Nop1 RG domains within chimeric proteins may facilitate their export from the nucleus.
DISCUSSION
The goal of this work was to determine whether RG-rich regions act as general interaction motifs or whether specific sequences within the RG domain of RNA-binding proteins influence their function. Our results clearly indicate that RG domains contribute specifically to protein function. Saccharomyces Npl3 can support growth of an npl3Δ strain with its own RG domain or with the RG domain of the Candida Npl3 ortholog, whereas the RG domains of nucleolar proteins Nop1 and Gar1 only support partial Npl3 function. This assay reflects a complex combination of in vivo roles of Npl3, as this protein has been implicated in almost all aspects of gene expression from transcription elongation and termination, to 3′ end processing, splicing, mRNA export and translation (10–15). To explore how RG domain specificity affects Npl3 function, we therefore tested the effect of heterologous RG domains on two specific characteristics of Npl3 that are linked to its RG domain, self-association and intracellular localization.
The presence of the CaNpl3 RG domain conferred PrA-Npl3 binding to wild-type ScNpl3-myc, but this interaction was significantly decreased by the substitution of RG domains from nucleolar proteins. Similarly, the CaNpl3 RG domain directed nuclear localization of GFP-ScNpl3 whereas the presence of nucleolar RG domains in GFP-ScNpl3 led to mislocalization within the cytoplasm. In contrast, the nucleolar RG domains did confer one key role of the Npl3 RG domain: the modulation of Npl3 localization through methylation. Nuclear export of GFP-ScNpl3 is decreased in the absence of the major arginine methyltransferase, Hmt1 (17), whereas nucleolar localization of Gar1 and Nop1 is not influenced by methylation (22). Although chimeras with RG domains from Gar1 and Nop1 are distributed throughout the cell at steady state, in the absence of methylation they become increasingly nuclear. This effect may be direct, through methylation of the heterologous RG domain, or indirect, through methylation of another protein (35). These results indicate that, while RG domains do not act as interchangeable general interaction motifs, some functions of RG domains can be transferred between proteins.
Partial function of an RG domain in isolation
Before testing ScNpl3 proteins with heterologous RG domains in our assays, we wished to confirm that the RG domain was required for Npl3 self-association, as suggested by earlier mutational analysis (16). We have demonstrated not only that the RG domain is required for Npl3 self-association and growth of npl3Δ cells, but that expression of the RG domain alone can support partial growth of npl3Δ cells (Figure 1B). Given the effects of point mutations within the RRMs on multiple steps in gene expression (10,12–14), it is unlikely that the ability of PrA-RG to support growth is due to facilitating any of these processes through direct binding to mRNA. Alternatively, the C-terminal RG-rich domain alone may help direct essential protein–protein interactions: RG domains play roles in many protein–protein interactions (4) and physical interactions have been detected between Npl3 and over 70 other proteins in S. cerevisiae, which likely reflect both direct interactions and association as members of macromolecular complexes (Saccharomyces Genome Database, http://www.yeastgenome.org).
Significance of sequence versus length among RG domains
Many RG-rich proteins contain similar subsets of other amino acids within their RG domains including phenylalanine, tyrosine, proline, serine and alanine and aspartate (7). In spite of these similarities, both the arrangement of these amino acids within RG domains and the length of the domains vary among proteins. In testing whether the Npl3 RG domain contains specific sequences that are crucial for function, we therefore considered the length and sequences of heterologous RG domains used to make chimeric Npl3 proteins. Only three S. cerevisiae proteins contain single RG domains longer than 50 amino acids (aa): Npl3 (109 aa), Gar1 (59 aa) and Nop1 (76 aa). Npl3 and these nucleolar proteins share similar spacing between arginines in these domains (Npl3 = 5.2 ± 2.6 aa, Gar1 = 3.4 ± 1.3 aa, Nop1 = 3.6 ± 0.3 aa) and all three of these RG domains contain eight or more serine residues, many within SR dipeptides (Figure 2A). The RG domain of the C. albicans Npl3 ortholog, while sharing similar RG spacing (3.0 ± 1.8 aa), only contains one serine residue, but has nine aspartate–arginine dipeptides. In contrast, the RG domains of both Npl3 proteins are significantly longer than those of the nucleolar proteins and contain many more aromatic (F/Y) residues.
The greater ability of the CaNpl3 RG domain than those of Gar1 and Nop1 to confer Npl3 function in S. cerevisiae suggests a key role for aromatic residues in Npl3 function in vivo as well as in Npl3 self-association (Figure 2). In previous work we showed that accumulation of arginine-to-lysine substitutions within the RG domain decreased Npl3 function. Similarly, it is likely that multiple aromatic residues can support Npl3 function (16), since simultaneous substitution of alanine for aromatic residues at four of nineteen positions (293, 301, 310, 340) did not affect growth of strains that require Npl3 or binding to Npl3-myc (unpublished data). In addition, our results indicate that, if serines within the RG domain play a role in Npl3 function, non-phosphorylatable acidic residues such as the asparates in CaNpl3 can also support Npl3 function.
Since the Gar1 and Nop1 RG domain chimeric Npl3 proteins also differed from ScNpl3 and CaNpl3 in length, we tested whether this difference could explain our results. A C-terminal deletion within the ScNpl3 RG domain, decreasing its length to shorter than the Nop1 chimeric protein, had minimal effects on overall Npl3 function (Figure 3A), indicating the greater importance of sequence than overall length. The reduced growth of npl3Δ cells expressing ScNpl3 with a deletion within the N-terminus of the RG domain, which results in a longer protein than the C-terminal deletion, points to the importance of residues in this part of the RG domain. Similarly, we have shown previously that N-terminal arginine-to-lysine (RK) substitutions decrease Npl3 function more than substitutions in the C-terminus of the RG domain (16). In addition, Lukasiewicz and colleagues demonstrated that an N-terminal deletion, but not a C-terminal deletion, within the RG domain altered the kinetics of Npl3 phosphorylation by Sky1 (36), a post–translational modification that has been implicated in transcription, autoregulation and Npl3 transport (12,31–34). In contrast to the different effects on overall Npl3 function, both deletion mutants show reduced binding to full-length Npl3 (Figure 3B). This result suggests that full self-association is not required for the essential functions of Npl3.
Therefore, although deletions within the RG domain may decrease Npl3 function by altering overall protein structure, these studies, combined with the RK-mutagenesis results (16), suggest that N-terminal and C-terminal sequences within the long Npl3 RG domain differentially affect overall protein function, while both regions help mediate Npl3 self-association. Interestingly, seven of the ten arginines that we previously identified as being exclusively dimethylated in Npl3 are within the N-terminal region that was deleted, which suggests that methylation of this region may positively influence Npl3 function.
RG-rich proteins and eukaryotic RNA metabolism
RG-rich RNA-binding proteins participate in complex processes that require the coordination of many proteins and RNAs. Just as Npl3 interacts with a plethora of proteins as it aids in mRNA transcription, processing and transport in yeast, RG-rich heterogeneous nuclear ribonucleoprotein particle proteins hnRNPA1 and hnRNPK play roles in mammalian mRNA processing and transport. Ribosome assembly also relies on a complex series of events involving multiple proteins, including nucleolin and fibrillarin in mammals and Nop1 and Gar1 in S. cerevisiae. Given the flexibility of extensive RG domains, as suggested by the abundance of glycine residues, the structure of these domains is likely influenced by their dynamic interactions with other proteins and RNAs. RG domains may promote remodeling of complexes as their interactions with different binding partners change over time and subtle differences in the sequences within long RG-rich domains may help modulate these transitions.
Thus, in contrast to the repetitive nature and similar amino-acid contents of RG-rich domains, our work highlights the importance of specific sequences within the RG-rich domain for function of the major mRNA-binding protein, Npl3. In particular, N-terminal sequences and the presence of multiple aromatic residues within this domain are likely to influence protein function. Interestingly, while many arginines within the RG domain are fully dimethylated in vivo, several arginines are variably methylated, including two arginines at the N-terminus of the domain (16). In combination, these results suggest that differential methylation of specific sequences within RG domains may modulate function of RNA-binding proteins.
FUNDING
National Science Foundation [grant MCB-0235590 to AEM]; Howard Hughes Medical Institute summer research fellowship (to AKC and CA); National Institutes of Health [Grant Number P20 RR-016463 from the INBRE Program of the National Center for Research Resources]. Funding for open access charge: Bowdoin College.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank members of the McBride lab for thoughtful discussions throughout this work and Anita Corbett, Bruce Kohorn and Michael Yu for critical reading of the manuscript.
REFERENCES
- 1.Lunde BM, Moore C, Varani G. RNA-binding proteins: modular design for efficient function. Nat. Rev. Mol. Cell Biol. 2007;8:479–490. doi: 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Godin KS, Varani G. How arginine-rich domains coordinate mRNA maturation events. RNA Biol. 2007;4:69–75. doi: 10.4161/rna.4.2.4869. [DOI] [PubMed] [Google Scholar]
- 3.Sakashita E, Sakamoto H. Protein-RNA and protein-protein interactions of the Drosophila sex-lethal mediated by its RNA-binding domains. J. Biochem. 1996;120:1028–1033. doi: 10.1093/oxfordjournals.jbchem.a021495. [DOI] [PubMed] [Google Scholar]
- 4.Bedford MT, Frankel A, Yaffe MB, Clarke S, Leder P, Richard S. Arginine methylation inhibits the binding of proline-rich ligands to Src homology 3, but not WW, domains. J. Biol. Chem. 2000;275:16030–16036. doi: 10.1074/jbc.M909368199. [DOI] [PubMed] [Google Scholar]
- 5.Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;272:2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- 6.Auweter SD, Oberstrass FC, Allain FH. Sequence-specific binding of single-stranded RNA: is there a code for recognition? Nucleic Acids Res. 2006;34:4943–4959. doi: 10.1093/nar/gkl620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gary JD, Clarke S. RNA and protein interactions modulated by protein arginine methylation. Prog. Nucleic Acid Res. Mol. Biol. 1998;61:65–131. doi: 10.1016/s0079-6603(08)60825-9. [DOI] [PubMed] [Google Scholar]
- 8.Stetler A, Winograd C, Sayegh J, Cheever A, Patton E, Zhang X, Clarke S, Ceman S. Identification and characterization of the methyl arginines in the fragile X mental retardation protein Fmrp. Hum. Mol. Genet. 2006;15:87–96. doi: 10.1093/hmg/ddi429. [DOI] [PubMed] [Google Scholar]
- 9.Boisvert FM, Chenard CA, Richard S. Protein interfaces in signaling regulated by arginine methylation. Sci. STKE. 2005;2005:re2. doi: 10.1126/stke.2712005re2. [DOI] [PubMed] [Google Scholar]
- 10.Bucheli ME, Buratowski S. Npl3 is an antagonist of mRNA 3′ end formation by RNA polymerase II. EMBO J. 2005;24:2150–2160. doi: 10.1038/sj.emboj.7600687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bucheli ME, He X, Kaplan CD, Moore CL, Buratowski S. Polyadenylation site choice in yeast is affected by competition between Npl3 and polyadenylation factor CFI. RNA. 2007;13:1756–1764. doi: 10.1261/rna.607207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dermody JL, Dreyfuss JM, Villen J, Ogundipe B, Gygi SP, Park PJ, Ponticelli AS, Moore CL, Buratowski S, Bucheli ME. Unphosphorylated SR-like protein Npl3 stimulates RNA polymerase II elongation. PLoS ONE. 2008;3:e3273. doi: 10.1371/journal.pone.0003273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kress TL, Krogan NJ, Guthrie C. A single SR-like protein, Npl3, promotes pre-mRNA splicing in budding yeast. Mol. Cell. 2008;32:727–734. doi: 10.1016/j.molcel.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee MS, Henry M, Silver PA. A protein that shuttles between the nucleus and the cytoplasm is an important mediator of RNA export. Genes Dev. 1996;10:1233–1246. doi: 10.1101/gad.10.10.1233. [DOI] [PubMed] [Google Scholar]
- 15.Windgassen M, Sturm D, Cajigas IJ, Gonzalez CI, Seedorf M, Bastians H, Krebber H. Yeast shuttling SR proteins Npl3p, Gbp2p, and Hrb1p are part of the translating mRNPs, and Npl3p can function as a translational repressor. Mol. Cell Biol. 2004;24:10479–10491. doi: 10.1128/MCB.24.23.10479-10491.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McBride AE, Cook JT, Stemmler EA, Rutledge KL, McGrath KA, Rubens JA. Arginine methylation of yeast mRNA-binding protein Npl3 directly affects its function, nuclear export, and intranuclear protein interactions. J. Biol. Chem. 2005;280:30888–30898. doi: 10.1074/jbc.M505831200. [DOI] [PubMed] [Google Scholar]
- 17.Shen EC, Henry MF, Weiss VH, Valentini SR, Silver PA, Lee MS. Arginine methylation facilitates the nuclear export of hnRNP proteins. Genes Dev. 1998;12:679–691. doi: 10.1101/gad.12.5.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu MC, Bachand F, McBride AE, Komili S, Casolari JM, Silver PA. Arginine methyltransferase affects interactions and recruitment of mRNA processing and export factors. Genes Dev. 2004;18:2024–2035. doi: 10.1101/gad.1223204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strasser K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, Rondon AG, Aguilera A, Struhl K, Reed R, et al. TREX is a conserved complex coupling transcription with messenger RNA export. Nature. 2002;417:304–308. doi: 10.1038/nature746. [DOI] [PubMed] [Google Scholar]
- 20.Piruat JI, Aguilera A. A novel yeast gene, THO2, is involved in RNA pol II transcription and provides new evidence for transcriptional elongation-associated recombination. EMBO J. 1998;17:4859–4872. doi: 10.1093/emboj/17.16.4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inoue K, Mizuno T, Wada K, Hagiwara M. Novel RING finger proteins, Air1p and Air2p interact with Hmt1p and inhibit the arginine methylation of Npl3p. J. Biol. Chem. 2000;275:32793–32799. doi: 10.1074/jbc.M004560200. [DOI] [PubMed] [Google Scholar]
- 22.Xu C, Henry PA, Setya A, Henry MF. In vivo analysis of nucleolar proteins modified by the yeast arginine methyltransferase Hmt1/Rmt1p. RNA. 2003;9:746–759. doi: 10.1261/rna.5020803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hellmuth K, Lau DM, Bischoff FR, Kunzler M, Hurt E, Simos G. Yeast Los1p has properties of an exportin-like nucleocytoplasmic transport factor for tRNA. Mol. Cell Biol. 1998;18:6374–6386. doi: 10.1128/mcb.18.11.6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McBride AE, Zurita-Lopez C, Regis A, Blum E, Conboy A, Elf S, Clarke S. Protein arginine methylation in Candida albicans: role in nuclear transport. Eukaryot. Cell. 2007;6:1119–1129. doi: 10.1128/EC.00074-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McBride AE, Weiss VH, Kim HK, Hogle JM, Silver PA. Analysis of the yeast arginine methyltransferase Hmt1p/Rmt1p and its in vivo function. Cofactor binding and substrate interactions. J. Biol. Chem. 2000;275:3128–3136. doi: 10.1074/jbc.275.5.3128. [DOI] [PubMed] [Google Scholar]
- 26.Henry M, Borland CZ, Bossie M, Silver PA. Potential RNA binding proteins in Saccharomyces cerevisiae identified as suppressors of temperature-sensitive mutations in NPL3. Genetics. 1996;142:103–115. doi: 10.1093/genetics/142.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henry MF, Silver PA. A novel methyltransferase (Hmt1p) modifies poly(A)+-RNA-binding proteins. Mol. Cell Biol. 1996;16:3668–3678. doi: 10.1128/mcb.16.7.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flach J, Bossie M, Vogel J, Corbett A, Jinks T, Willins DA, Silver PA. A yeast RNA-binding protein shuttles between the nucleus and the cytoplasm. Mol. Cell Biol. 1994;14:8399–8407. doi: 10.1128/mcb.14.12.8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Senger B, Simos G, Bischoff FR, Podtelejnikov A, Mann M, Hurt E. Mtr10p functions as a nuclear import receptor for the mRNA-binding protein Npl3p. EMBO J. 1998;17:2196–2207. doi: 10.1093/emboj/17.8.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yun CY, Fu XD. Conserved SR protein kinase functions in nuclear import and its action is counteracted by arginine methylation in Saccharomyces cerevisiae. J. Cell Biol. 2000;150:707–718. doi: 10.1083/jcb.150.4.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gilbert W, Siebel CW, Guthrie C. Phosphorylation by Sky1p promotes Npl3p shuttling and mRNA dissociation. RNA. 2001;7:302–313. doi: 10.1017/s1355838201002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gilbert W, Guthrie C. The Glc7p nuclear phosphatase promotes mRNA export by facilitating association of Mex67p with mRNA. Mol. Cell. 2004;13:201–212. doi: 10.1016/s1097-2765(04)00030-9. [DOI] [PubMed] [Google Scholar]
- 34.Lund MK, Kress TL, Guthrie C. Autoregulation of Npl3, a yeast SR protein, requires a novel downstream region and serine phosphorylation. Mol. Cell Biol. 2008;28:3873–3881. doi: 10.1128/MCB.02153-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu C, Henry MF. Nuclear export of hnRNP Hrp1p and nuclear export of hnRNP Npl3p are linked and influenced by the methylation state of Npl3p. Mol. Cell Biol. 2004;24:10742–10756. doi: 10.1128/MCB.24.24.10742-10756.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lukasiewicz R, Nolen B, Adams JA, Ghosh G. The RGG domain of Npl3p recruits Sky1p through docking interactions. J. Mol. Biol. 2007;367:249–261. doi: 10.1016/j.jmb.2006.12.031. [DOI] [PubMed] [Google Scholar]