Abstract
FANCM, the most highly conserved component of the Fanconi Anaemia (FA) pathway can resolve recombination intermediates and remodel synthetic replication forks. However, it is not known if these activities are relevant to how this conserved protein activates the FA pathway and promotes DNA crosslink repair. Here we use chicken DT40 cells to systematically dissect the function of the helicase and nuclease domains of FANCM. Our studies reveal that these domains contribute distinct roles in the tolerance of crosslinker, UV light and camptothecin-induced DNA damage. Although the complete helicase domain is critical for crosslink repair, a predicted inactivating mutation of the Walker B box domain has no impact on FA pathway associated functions. However, this mutation does result in elevated sister chromatid exchanges (SCE). Furthermore, our genetic dissection indicates that FANCM functions with the Blm helicase to suppress spontaneous SCE events. Overall our results lead us to reappraise the role of helicase domain associated activities of FANCM with respect to the activation of the FA pathway, crosslink repair and in the resolution of recombination intermediates.
INTRODUCTION
A fundamental defect in DNA repair causes Fanconi Anaemia (FA), a genetic illness that leads to abnormal development, bone marrow failure and cancer. As many as 13 genes are known to be mutated in FA, most of these physically and genetically interact to form the FA genome stability pathway (1,2). A key question concerns how such a pathway stimulates DNA repair. The identification of FANCM, a highly conserved gene in the FA pathway, may shed light on this crucial question. Vertebrate FANCM contains highly conserved amino (N) terminal helicase and carboxy (C) terminal nuclease like domains, which suggest that it participates directly in DNA repair (3,4). This domain arrangement is identical to that of the archaeabacterial FANCM orthologue Hef, in which the activities of both domains are coordinated to unwind and cleave a replication fork (5). Recently, FANCM N-terminal helicase and C-terminal nuclease domains have been shown to interact with HCLK2 to promote the activation of DNA damage checkpoints (6). The C-terminal nuclease domain of FANCM interacts with a 24 kDa protein (FAAP24) and this protein complex binds to synthetic replication fork-like DNA structures, preferentially to ssDNA, splayed-arm, and 3′-flap DNA substrates (7). FAAP24 knock down abolishes DNA damage inducible FANCD2 monoubiquitination and leads to sensitivity to DNA crosslinking agents (7).
Biochemical studies of vertebrate and yeast FANCM orthologues, FML (S. pombe) and MPH1 (S. cerevisiae), have revealed that these proteins can bind Holliday junctions and replication fork-structured DNA. Yeast and human FANCM orthologues also promote fork regression, Holliday junction migration and can also unwind RPA-stabilized D-loops (8–11). Furthermore, genetic studies performed in yeast show that the FANCM orthologues promote replication restart and suppress crossovers following sister chromatid recombination after DNA damage (9,12). It is important to emphasize here that only translocase but not helicase activity has been reported for the full length purified FANCM. In addition close sequence analysis of the nuclease like domain does suggest that key catalytic residues seem to be missing leading to the conclusion that this region does not confer nuclease activity. Nevertheless taken together, these findings have led to the proposal that FANCM may move along double-stranded DNA, sensing, remodelling and restarting stalled replication forks.
While some progress has been achieved towards defining the biochemical functions of FANCM in different organisms, evidence about how these activities promote DNA repair following DNA damage in vivo is limited. The yeast orthologues, FML and MPH1, contain only the N terminal helicase-like domain and lack both the middle and C-terminal sections found in vertebrate FANCM. Additionally, no other FA genes have been identified in yeast to date. Therefore, it has not been possible to extrapolate the function of FANCM in the FA DNA repair pathway from the reported in vivo activities of yeast FANCM orthologues.
To circumvent the difficulties encountered in expressing transfected FANCM in either human or chicken FANCM deficient cell lines, Wang and colleagues have used a transient siRNA approach to knock down endogenous FANCM expression in a human cell line. This cell line was simultaneously complemented with siRNA resistant FANCM cDNA. Using this approach they showed that HeLa cells harbouring a point mutation in the ATPase Walker A sequence of FANCM were defective in crosslink repair despite being competent in FANCD2 monoubiquitination (13). This result therefore strongly suggests that the translocase activity of FANCM imparts crosslink repair function.
This work addresses the functions of the distinct domains of chicken FANCM in DNA repair. We systematically dissect how different domains of this large protein function to activate the FA pathway and promote DNA crosslink repair. Furthermore, our genetic analyses uncover two new functions of FANCM that do not overlap with its role in the FA pathway: UV and camptothecin induced DNA damage tolerance. Finally we discover a role for FANCM in the suppression of crossover recombination with the Bloom's (BLM) helicase.
METHODS
Gene disruption of the chicken FANCM locus
The chicken FANCM locus was identified by BLAST search using the human protein sequence against the ENSEMBL draft chicken genome sequence. An ENSEMBL predicted transcript encompassed most of the FANCM gene, although the 5′ end, containing some of the helicase motifs, was missing. The complete 5′ end of the gene was amplified by PCR and all exons encoding the Mph1 like domain were identified. Sequence from the predicted and PCR amplified genomic DNA was used to design the two gene disruption cassettes (A and B). PCR oligos used to amplify 5′ and 3′ arms of construct A were TCGTGACACTTAATC CCTCGGTGCTCTGAGC/GAAGGGC GACAAAACCAACTTCCC and AGGCTGTGCA GCAGGTTGTTTCCAA/TGCAACACACAGCGTGA CACCCAGT, respectively. Those for the knockin construct B were 5′ arm: AGCGGCGCTGGTCT ATATCTTTAGGG/GCAGACAGCG CTGAGGGGAATACA and 3′arm ACCAAAAGCTGAT GGACATCCAAATCC/AGTTGCCATAGGAAATGATGT TGGTGTCAG. PCR products were cloned into pCR2.1-TOPO (Invitrogen) and then the D203A point mutation was introduced using Stratagene QuickChange XL site directed mutagenesis kit and the following primers: GTTAAATGCTTGGTTG TTGCCGCGGCCCACAAAGCTCTGGG/CCCAGAGC TTTGTGGGCTTCGGCAACAACCAAGCATTTAAC. Transfections, selection and Southern analyses of targeted DT40 clones were carried out as described previously (4,14). The lox drug resistance cassette was removed by transiently transfecting cell lines with the Cre recombinase NLS-expression plasmid. Cells were cloned by limiting dilution and then tested individually for loss of drug resistance. To confirm the appropriate disruptions of the GgFANCM locus the following PCR oligos were used; F1, TTTGGGACGGGAATAGAGC; R1, GCCACCTCCTTTCCTCTAT. Generation of ΔBLM FANCM-D203A knockin cell line was carried out by targeting sequentially FANCM-D203A Knockin and FANCM-hel constructs into the ΔBLM strain [kind gift from Professor Enomoto (19)]. 5′ and 3′ arms of the FANCC in situ TAP-tagged construct were generated as described before (4).
Flag-tag constructs were amplified using chicken FANCM cDNA using forward primers CCGAGATCTCACCATGG ATTACAAGGATGACGACGATAAGGACTATAAGGACGAT GATGACAAGAGCGGCGG CCGGCAGCGCACCCTGCCC for the +HEL construct and CCGGGATCCCACCA TGGATTACAAGGATGACGACGATA AGGACTATAAGGACGATGATGACAAG GGGGATT GCAGCTATGAACTGGAGCTT for either +LMS or +NUC constructs and reverse primers CCGAGATCTTCCTTCAGCAGGAAACACACGTGAG for the +HEL construct, CCGGGATCCTCCTTCA GCAGGAAACACACGTGAG for the +LMS construct and CCGGGATCCAGCGGCCGCTCAGCTCCTGCTG for the +NUC construct. These PCR products were cloned into pCR-TOPO and verified by sequencing. These fragments were cloned from pCR-TOPO into the expression plasmid pEXPRESS and checked for orientation. Expression cassettes were cloned into drug resistance cassette-containing pLOX plasmids. Proteins expression was confirmed by western blot.
Mutagen sensitivity assays
For cisplatin survival assays cell lines were plated into 96-well plates at a density of 3000 cells per plate. A range of doses of cisplatin was added to wells, and the plates were returned to the incubator for five complete cell-doubling times. After this the cells were pulsed with MTS (Promega) and incubated for another 1 h. Cell viability was measured by luminometry according to manufacturer's instructions (Promega), and each dose point was assayed in triplicate. The colony survival values plotted are relative to an untreated control.
For UV sensitivity, cells were irradiated with the indicated doses, diluted in medium and grown for 2 weeks in methylcellulose containing plates. For camptothecin sensitivity, cells were grown for 2 weeks in CPT-containing methylcellulose plates. Plots represent the average of three independent experiments. Error bars represent standard deviation.
FANCC-TAP precipitation
Treated or untreated cells (109) were lysed in lysis buffer (50 mM Tris–HCl, pH 8.0, 0.5 mM EDTA, 0.1% Triton X-100, 200 mM NaCl and protease inhibitor cocktail (Roche Molecular Biochemicals)) with a Dounce homogenizer. Lysates were cleared by centrifugation using a Beckman rotor Ti45, at 4°C and 35 000 r.p.m. for 60 min. Extracts (9 mg total protein) were incubated for 2 h at 4°C with 50 µl IgG-Sepharose beads (Amersham). Precipitated material was extensively washed with lysis buffer containing 500 mM NaCl. Precipitated material was resolved by 4–12% Bis/Tris–SDS–PAGE (Invitrogen) and detected by immunoblot with antibodies to Flag, GgFANCG, or TAP. Western blot analyses for FANCD2 were done as described previously (14). Detection of chicken FANCM was carried out using antisera directed to the large middle section of the chicken FANCM gene. The details of this reagent are published in Mosedale et al. (4).
Cellular subfractionation
Two hundred million DT40 cells were treated with 1 mM cisplatin for 16 h, harvested, washed once with PBS and lysed in hypotonic buffer (10 mM Tris–HCl pH 7.4, 10 mM KCl, 1.5 mM MgCl2, 10 mM 2-mercaptoethanol) by pushing cells through a 19G needle. Nuclei were collected by centrifugation (2700 g, 10 s) washed twice with hypotonic buffer and nuclear proteins were extracted with a high-salt buffer (15 mM Tris–HCl, pH 7.4, 1 mM EDTA, 500 mM NaCl, 1 mM MgCl2, 10% glycerol, 10 mM 2-mercaptoethanol and protease inhibitor cocktail). To enrich for chromatin-associated proteins, salt-extracted pellets were treated with 1500 units micrococcal nuclease (Amersham) for 20 min at room temperature in nuclease reaction buffer (20 mM Tris–HCl, pH 7.4, 100 mM KCl, 2 mM MgCl2, 1 mM CaCl2, 0.3 M sucrose, 0.1% Triton X-100 and protease inhibitor cocktail). Comparable amounts (relative to each fraction) were resolved on 4–16% Tris–glycine gels (Invitrogen) by SDS–PAGE and analysed by immunoblotting using anti-FLAG (sigma), anti PAP or anti-histone H3 (Abcam) antibodies.
Gel-filtration analyses
High salt nuclear extracts were prepared with a few modifications as described previously (4). Briefly, proteins were extracted from nuclei (isolated from 4 × 109 DT40 cells as indicated) with nuclear extraction buffer [15 mM Tris–HCl, pH 8.0, 0.2 mM EDTA, 420 mM NaCl, 1.5 mM MgCl2, 25% glycerol, 0.5 mM DTT and protease inhibitor cocktail (Roche Molecular Biochemicals)]. Cleared nuclear extracts (3.5 mg) were directly applied to a Superose 6 HR16/50 column (Amersham) equilibriated with column buffer (15 mM Tris–HCl pH, 8.0, 0.2 mM EDTA, 420 mM NaCl, 1.5 mM MgCl2, 10% glycerol, 0.5 mM DTT). Fractions (4 ml) were collected and aliquots were resolved by 4–12% Bis/Tris–SDS–PAGE (Invitrogen). FANCC-TAP was detected by immunoblot analyses with an antibody to TAP (Sigma). FANCM and FANCG were detected by immunoblot analyses with antibodies raised against GgFANCM and GgFANCG, respectively. The Superose 6 column was calibrated with a high molecular weight calibration kit (Amersham).
SCE analysis in DT40 cells
We carried out SCE assays as previously described (14) using 10 µM BrdU for two cell cycles and adding 0.1 µg (per ml) of colcemid 2.5 h before harvesting cells. The slides were coded (therefore blinded to the observer) and approximately 50 metaphases for each cell line were scored.
RESULTS
Hypomorphic alleles of FANCM in DT40 show different phenotypes
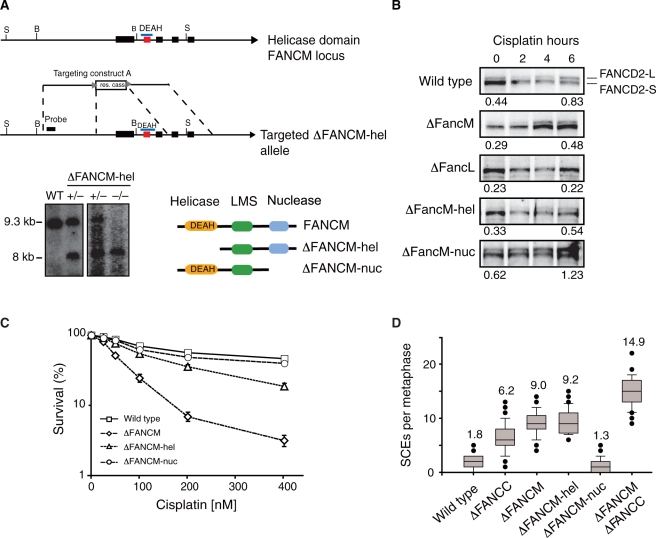
As a first step to distinguish between the functions of the distinct domains of FANCM, we disrupted the exons encoding either the helicase (FANCM-Δhel) (Figure 1A) or the nuclease domain (FANCM-Δnuc) (Mosedale 2004). DT40 strains carrying homozygous deletion of either of these domains were then compared to the full-length deleted FANCM (ΔFANCM) strain in terms of FANCD2 monoubiquitination and sensitivity to DNA crosslinking agents (Figure 1B and C). When these cell lines were exposed to high doses of cisplatin, we noted that FANCD2 ubiquitination in the FANCM null cell line was defective compared to wild-type, but importantly was clearly detectable. However, deletion of FANCL a member of FA core complex, completely abolished FANCD2 monoubiquitination. In the FANCM-Δhel cell line, FANCD2 monoubiquitination levels were also diminished, but were unperturbed in the FANCM-Δnuc cell line. We also measured the sensitivity to cisplatin of the FANCM deficient cell lines. The FANCM null cell line was more sensitive to cisplatin than FANCM-Δhel, whilst FANCM-Δnuc is not sensitive to this agent (Figure 1C). This sensitivity partially correlates with the ability of these cell lines to monoubiquitinate FANCD2. A feature of all chicken DT40 FA pathway knockouts is the presence of raised spontaneous sister chromatid exchange events (SCEs) (14–17). FANCM deficient DT40 cells display increased numbers of spontaneous sister chromatid exchanges (SCEs) (4). We therefore compared the levels of SCEs in each FANCM mutant strain. The ΔFANCM (mean = 9.0 SCEs per metaphase) and the FANCM-Δhel (mean = 9.2 SCEs per metaphase) strains show elevated SCEs, whilst FANCM-Δnuc (mean = 1.3 SCEs per metaphase) shows wild-type SCEs levels (Figure 1D). Double mutants of genes involved in the FA repair pathway such as FANCC/FANCG, FANCC/FANCA and FANCC/FANCJ do not lead to an increase in SCEs compared to respective single mutants, indicating that SCEs arise by virtue of a common process (14,16). As FANCM is a component of the FA core complex, we compared SCEs levels in the double mutant cell line ΔFANCM ΔFANCC and the respective single mutant strains. Strikingly, the double ΔFANCM ΔFANCC strain (mean = 14.9 SCEs per metaphase) has more spontaneous SCEs than either single knockout strain, indicative of an additive impact on this phenotype. Cumulatively, these results suggest that the helicase but not the nuclease domain of FANCM is required for crosslink repair and SCE suppression.
Figure 1.
Disruption and characterization of FANCM helicase and nuclease domain deficient DT40 strains. (A) Map of the FANCM locus containing the exons encoding the helicase domain. The gene disruption strategy removes three exons coding for the conserved helicase domain. Genomic Southern blot confirms disruption of the helicase domain. Cartoon depicting predicted products generated by hypomorphic alleles in FANCM deficient cell lines (LMS—Large Middle Section). (B) Time course experiment of FANCD2 monoubiquitination in FANCM deficient cell lines. Cells were exposed to 5 µM cisplatin for up to 6 h. The ratios for D2-L (monoubiquitinated FANCD2) over D2-S (unmodified FANCD2) are shown for 0-h and 6-h time point. FANCD2 monoubiquitination activation in FANCM-Δhel and ΔFANCM cell lines is deficient, but normal in FANCM-Δnuc deficient cell line. FANCD2 monoubiquitination in ΔFANCL cell line is completely abolished even after 6 h of treatment. (C) Graph showing cisplatin sensitivity determined by cell viability (MTS assay) of FANCM cell lines. (D) Spontaneous SCE frequencies determined for FANCC and FANCM knockout cell lines. The elevated SCEs in the ΔFANCM strain are additive compared to ΔFANCC in the ΔFANCMΔFANCC mutant.
Complementation of the FANCM knockout with FANCM domain-specific cDNA
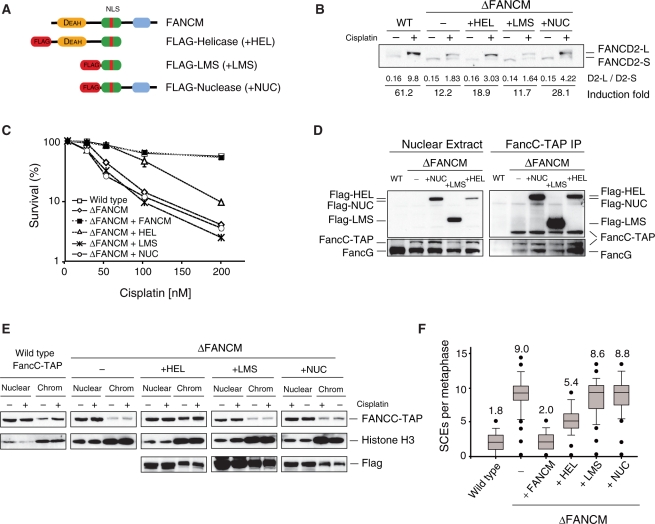
Immunoblot analyses failed to detect residual FANCM protein in any of these strains indicating that the respective disruptions did result in reduced protein expression. However, since the phenotypes of FANCM-Δhel and FANCM-Δnuc cells clearly differed we believe that this most likely reflects the creation of hypomorphic alleles of FANCM (data not shown). To confirm this, we generated FANCM Flag-tagged domain specific over-expression constructs (Figure 2A). These constructs consist of the helicase domain with the large middle section (LMS) of FANCM (+HEL construct), the LMS alone (+LMS construct), or the LMS with the nuclease domain (+NUC construct). LMS was included in all constructs since this region contains the putative bipartite nuclear localization signal. To monitor the FA core complex dynamics, we expressed these cDNAs in a ΔFANCM strain that carries a FANCC-TAP tagged allele (ΔFANCM FANCC-TAP). Drug resistant clones were then tested for expression and complementation of the various ΔFANCM associated defects, such as DNA damage induced FANCD2 monoubiquitination (Figure 2B), cisplatin sensitivity (Figure 2C), ability to interact with the FA core complex (Figure 2D), chromatin targeting of the core complex (Figure 2E) and finally SCEs (Figure 2F). The construct containing the helicase domain (+HEL) partially rescues the FANCD2 monoubiquitination defects (Figure 2B, compare 18.9 monoubiquitination induction fold in +HEL cell line to 12.2 shown by ΔFANCM cell line) and cisplatin sensitivity seen in ΔFANCM cell line (Figure 2C), it is able to form a stable FA core complex and facilitates accumulation of this complex on chromatin (Figure 2D and E). Moreover, the FANCM helicase domain also partially suppresses elevated SCEs in the FANCM null mutant (Figure 2F). The construct containing the nuclease domain of FANCM partially rescues the FANCD2 monoubiquitination defect observed in ΔFANCM (Figure 2B, +NUC cell line shows 28.1 monoubiquitination induction fold compared to 12.2 shown by ΔFANCM cell line) indicating that this function is redundant to the helicase domain. It can also assemble into the FA core complex (Figure 2D), however it does not rescue cisplatin sensitivity or the elevated levels of SCE detected in ΔFANCM (Figure 2C and F). Finally, +LMS does not complement any of the defects observed in FANCM null cell line but importantly, it can interact with the FA core complex (Figure 2D). These results indicate that the helicase and nuclease domains play redundant roles in FANCD2 ubiquitination process, however, only the helicase domain partially rescues ΔFANCM cells from sensitivity to cisplatin and to suppress elevated levels of SCEs observed in the FANCM null cell line. As all the constructs can interact with the FA core complex, we also conclude that LMS region is required to anchor FANCM into this complex.
Figure 2.
Complementation of ΔFANCM deficient cell line with FANCM domain specific cDNA. (A) The cartoon depicts the three cDNA constructs used for over-expressing three different FANCM domains in the ΔFANCM cell line. (B) Suppression by the helicase and nuclease domains of FANCD2 monoubiquitination defects observed in ΔFANCM cell line. (−) and (+) indicates untreated and treated cells exposed to 5 µM cisplatin for 17 h. Shown below is the ratio of FANCD2-L over FANCD2-S. Induction fold was calculated dividing FANCD2 ratio after by the ratio before treatment. (C) Graph showing cisplatin sensitivity as determined by cell viability (MTS assay) for all the strains shown. (D) Pull down of FANCC-TAP from nuclear extracts. Anti-FLAG immunoblot for detecting expression of FANCM domains. FANCC-TAP was detected by anti-PAP immunoblot. Left—Nuclear extract, right—FANCC-TAP immunoprecipitation (25). (E) Cellular fractionation of ΔFANCM cell lines expressing the three different FANCM domains. Above—Accumulation of FANCC-TAP into the chromatin fractions in the absence (−) or presence (+) of DNA damage. TAP blot to detect FANCC-TAP, middle—Histone H3 blot, and below—FLAG blot to detect FLAG tagged FANCM domains. (F) Spontaneous SCEs frequencies in the ΔFANCM cell line expressing the different FANCM domains.
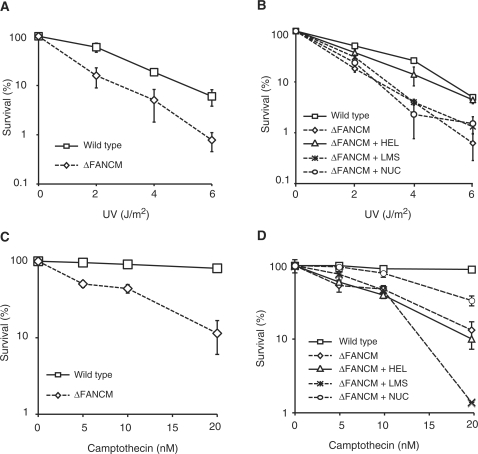
In the course of our studies of the ΔFANCM strain we noticed that this mutant was also sensitive to UV light and camptothecin (CPT). This is not a feature shared with any of the other FA gene knockouts (data not shown). We decided to investigate this further by identifying the section of FANCM that may be important for either UV light or CPT tolerance. As can be seen in Figure 3A, the ΔFANCM strain is sensitive to UV light (D10 value – WT = 5.1 J/m2 ΔFANCM = 2.8 J/m2). However, this sensitivity can be complemented with the construct containing the helicase domain (D10 value- + HEL = 4.7 J/m2) (Figure 3B). A similar analysis was also carried out for CPT sensitivity. Again we noted that ΔFANCM is sensitive to this agent (Figure 3C). CPT sensitivity, in contrast to UV sensitivity, is partially complemented by the construct containing the nuclease domain but not by the helicase domain (Figure 3D). Cumulatively, the results indicate that the region encompassing the helicase domain of FANCM promotes repair of UV light damage, whilst that encompassing the nuclease domain appears to be critical.
Figure 3.
Sensitivity of ΔFANCM cell line to UV and camptothecin. (A) UV sensitivity curve of ΔFANCM cell line. (B) Complementation of UV light sensitivity shown by the ΔFANCM cell line harbouring +HEL but not +NUC construct. (C) Camptothecin sensitivity curve of ΔFANCM cell line. (D) CPT sensitivity of ΔFANCM cell line is partially complemented by +NUC but not +HEL construct.
A point mutation in the helicase Walker B motif separates SCE suppression from crosslink repair functions of FANCM
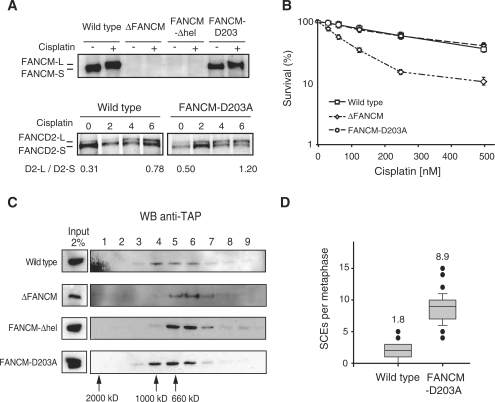
We then focused our studies on the raised spontaneous SCEs in the ΔFANCM and ΔFANCM-hel strains. Genetic epistasis results indicate that these raised spontaneous SCEs are not simply due to a defective FA pathway (Figure 1D). To determine whether the putative helicase activity of FANCM suppresses SCEs, an inactivating point mutation (DEAD to AEAD) was introduced into the Walker B motif of this domain. The isolated strain, FANCM-D203A (helicase domain disruption in one allele and knockin into the second allele), contains the relevant mutation in the genomic sequence (Supplementary Figure 1) and importantly expresses full length FANCM protein (Figure 4A). To our surprise, we noted that the point mutant strain was proficient in monoubiquitinating FANCD2 (Figure 4A) and not sensitive to cisplatin (Figure 4B) indicating that the Walker B motif is dispensable for crosslink repair. Next we knocked in a TAP tag into the last exon of the FANCC allele. This results in the expression of endogenously TAP tagged FANCC which enables us to determine the stability of the FA core complex in our point mutant cell line (Figure 4C). In WT cells, the FA core complex migrated in a peak corresponding to 1 MDa. In ΔFANCM and FANCM-Δhel cell lines, this complex eluted at a lower molecular weight when compared to the wild type. However, the gel filtration profile for the FANCM-D203A cell line was identical to the wild type profile, indicating that the FA core complex is intact (Figure 4C). Strikingly, this mutant shows high levels of SCEs (Figure 4D) demonstrating that the suppression of SCEs can be attributed to the enzymatic activity of the helicase domain of FANCM. Therefore, the Walker B box mutation D203A dissociates the role of FANCM in crosslink repair and FANCD2 monoubiquitination from that in suppressing SCEs. Since FANCM and both yeast FANCM orthologues also suppress crossover recombination (9), this indicates that the SCEs suppression function of FANCM is conserved in evolution.
Figure 4.
A Walker B point mutation in the helicase domain separates the role for FANCM in SCEs suppression from that in crosslink repair. (A) FANCM immunoblot of nuclear extracts without (−) or with (+) DNA damage. A full length FANCM protein is expressed in the knock in strain that is phosphorylated following DNA damage. Below—the FANCD2 monoubiquitination process after DNA damage is not defective in FANCM-D203A cell line. (B) Cisplatin sensitivity curve as determined by cell viability (MTS assay). The strain expressing the knockin point mutation FANCM-D203A is not sensitive to cisplatin. (C) Gel filtration analysis of endogenously modified FANCM cell lines. Fractions were blotted for FANCC-TAP to detect the FA core complex. ΔFANCM and FANCM-Δhel knockout mutants but not FANCM-D203A show a smaller FA core complex size compared to wild-type. (D) High levels of spontaneous SCEs in FANCM-D203A mutant strain compared to wild-type.
FANCM functions with the Bloom helicase to suppress SCEs
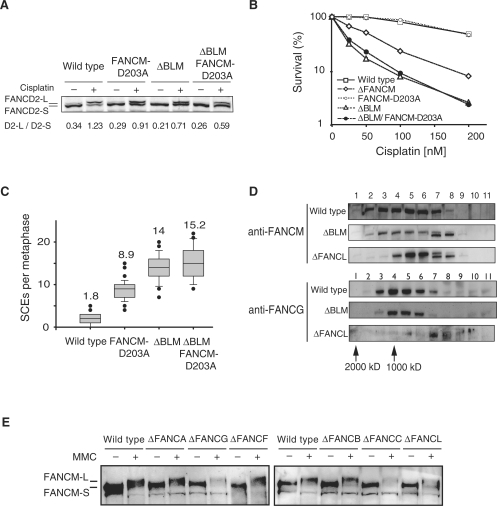
The Blm helicase is the main guardian against SCEs in eukaryotes (18). The yeast FANCM orthologues function in a distinct SCE suppression pathway to their respective Blm orthologues (9). In order to test whether vertebrate FANCM functions in the same or a distinct SCE suppression pathway to Blm, we made the helicase point mutation D203A in a DT40 BLM knockout cell line (ΔBlm) (19) to obtain the cell line ΔBlm FANCM-D203A (Supplementary Figure 2). The double mutant strain displayed the same viability as the single ΔBlm strain (Supplementary Figure 2). This strain was proficient at FANCD2 monoubiquitination although with slightly reduced efficiency when compared to either single mutant strain (Figure 5A). We also determined the crosslink sensitivity and particularly noted that the ΔBlm FANCM-D203A strain retained the cisplatin sensitivity of the ΔBlm strain (Figure 5B). Most importantly, the number of SCEs in the ΔBlm FANCM-D203A strain (mean = 15.2 SCEs per metaphase) was equivalent to the ΔBlm strain (mean = 14 SCEs per metaphase) (Figure 5C).
Figure 5.
FANCM functions with Blm to suppress a subset of SCE events. (A) FANCD2 monoubiquitination in the ΔBlm FANCM-D203A mutant cell line. Shown below are the ratios of FANCD2-L over FANCD2-S and fold induction before (−) and after 6 h treatment (+) with 1 µM cisplatin. (B) Cisplatin sensitivity curve as determined by cell viability (MTS assay). (C) Spontaneous SCE levels in the ΔBlm FANCM-D203A strain resemble those in the ΔBlm strain. (D) Gel filtration analisys of FANCM and FANCG in ΔBlm and ΔFANCL strains. Above—Anti FANCM immunoblot of nuclear extract fractions obtained from gel filtration. FANCM fractionates as a large molecular mass peak that does not change in ΔBlm but shifts in ΔFANCL strains. Bottom—The same fractions were also blotted with an anti-FANCG antibody to monitor the FA core complex. (E) FANCM modification after DNA damage in FA deficient cell lines. FANCM becomes modified in the absence of a functional FA core complex.
Since FANCM functions within the FA core complex and also with Blm to carry out distinct functions, we set out to determine if the distribution of FANCM protein complex(es) is/are altered in the absence of Blm or the FA core complex. Nuclear extracts from wild-type, ΔBlm and ΔFANCL DT40 strains were fractionated by size exclusion chromatography and blotted for FANCM or the FA core complex protein FANCG. The high molecular mass FANCM complex is unperturbed in ΔBlm cells but shifts to a slightly smaller mass when the FA complex integrity is disrupted, as seen in ΔFANCL cells (Figure 5D). In addition, FANCM is stable and it can be phosphorylated in most chicken FA knockout cell lines (Figure 5E). Together, these studies point to the existence of a distinct stable FANCM complex that does not contain BLM or the FA core complex. Cumulatively, these experiments indicate that FANCM contributes only in a subset of Blm suppressible SCE events.
DISCUSSION
In conclusion our systematic genetic dissection of chicken FANCM reveals how this protein promotes activation of the FA pathway and crosslink repair. In addition we discovered that FANCM is not only required for tolerance of UV and camptothecin induced DNA damage, but also for the suppression of crossover recombination.
Monoubiquitination of FANCD2 is crucial for the activation of the FA pathway. This process requires an intact FA core complex, Ube2t and FANCI, occurring only during the S and G2 phases of the cell cycle (20). The core enzymatic basis of this reaction is probably modulated by the E2 ligase (Ube2t) interacting with the E3 (FANCL embedded in the core complex) and the substrate (FANCD2 complexed to FANCI) (21–23). Studies using siRNA knockdown of FANCM or its nuclease domain binding partner FAAP24 in human cell lines show that FANCD2 monoubiquitination is severely impaired. In contrast, although chicken FANCM knockouts have reduced FANCD2 monoubiquitination, the defect in this modification is less marked than that observed in human cell lines. Moreover, when the ΔFANCM strain is exposed to large doses of cisplatin we can still see DNA damage induced FANCD2 monoubiquitination. Despite this discrepancy, a question that remains is how FANCM stimulates DNA damage inducible FANCD2 monoubiquitination. This cannot simply be due to assembly of the residual FA core complex existing in the ΔFANCM strain. One possibility is that FANCM mobilizes the FA core complex to chromatin by recognizing damaged DNA replication forks (4,24). Indeed both the helicase and nuclease domains can bind fork structured DNA (4,7,8) and we show here that both domains can independently stimulate FANCD2 monoubiquitination. However if stalled replication forks degenerate into double strand breaks in FANCM null cells, then this may enable a residual FANCM deficient core complex to stimulate FANCD2 monoubiquitination.
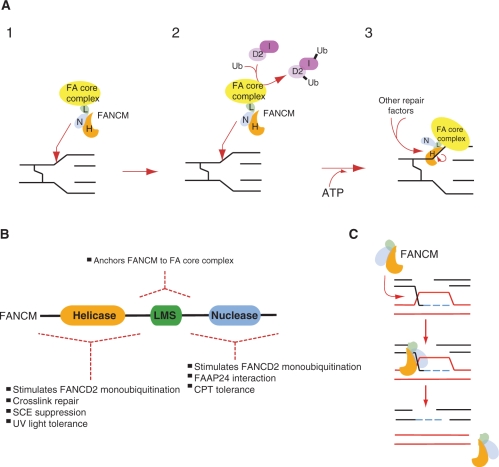
A key function of the FA pathway is to promote DNA crosslink repair. Genetic studies indicate that the FA proteins work upstream of a combined HR and TLS process to remove crosslinks (14). A recent report using Xenopus extract provides biochemical proof of such a combined process (25). We currently do not know the molecular basis of the putative upstream role of the FA pathway in such a combined process. However, FANCM clearly plays a key role in this process and it may now be possible using the genetic and biochemical evidence to integrate this into a working model. Our study clearly shows that the N-terminal helicase domain is crucial for effective repair, although a predicted inactivating mutation in the helicase domain has no impact on crosslink repair. The biochemical activities of FANCM, such as branch migration, translocase activity, D-loop dissolution and fork regression, all require ATPase activity and are very likely to be significantly impacted by mutation of the DEAD box. This suggests that these activities may not confer crosslink repair activity to FANCM. On the other hand, the siRNA complementation based approach does suggest that ATP binding is required for crosslink repair (13). It is therefore plausible that the ATP binding in the context of an intact helicase domain is crucial for crosslink repair, although this may not simply be used to drive the helicase or translocase functions. We propose that the helicase domain of FANCM recognizes and binds to stalled replication forks. DNA dependent ATP binding activity may then induce a conformational change that recruits other repair proteins to the site (Figure 6A). Such a model would predict that apart from FAAP24 other proteins might interact with FANCM.
An unexpected finding reported here is the marked sensitivity to both UV light and camptothecin, interestingly this is a feature it shares with hamster cell line mutant of FANCG (26). FANCM seems to utilize different domains to protect against the genotoxic effects of either of these agents, revealing a new level of complexity in the repair functions of this protein (Figure 6B). The helicase domain, but not its translocase activity (data not shown), is required to protect against UV light. However, it is not possible from our genetic studies to deduce how the FANCM helicase domain facilitates repair of UV light induced DNA damage, creating double mutants where FANCM disruption is combined with either NER or TLS genes may shed important light into this question. The marked sensitivity to camptothecin in the FANCM knockout requires the region emcompassing the nuclease domain. Camptothecin exerts its toxicity by covalently trapping type I topoisomerases-DNA complexes. Such complexes impede replication and are converted into lethal double strand breaks. The nuclease domain appears to have no obvious activity apart from binding to FAAP24 and HCLK2, and to fork structured DNA. Therefore, it could be that this FANCM domain can target repair enzymes to replication forks stalled at locked type I topoisomerases-DNA complexes. It is interesting to note that ΔBlm cells are also sensitive to CPT and it has been suggested that the Blm-Topo III complex helps resolve damage caused by this agent (27). Since FANCM co-purifies with the Blm-Topo III complex (28) it is conceivable that the C terminus of FANCM recruits the Blm-Topo III complex to resolve such stalled forks.
Figure 6.
Model for the role of FANCM in the DNA damage response. (A) Model for the role of FANCM in the FA pathway and crosslink repair in the DNA damage response. (1) The helicase or nuclease domain can target the FA core complex to a replication block at a DNA crosslink. (2) The core complex can stimulate FANCD2 monoubiquitination, perhaps by being in close proximity to its substrates FANCD2 and FANCI. (3) An ATP driven conformational change may enable the FANCM complex to recruit additional DNA repair proteins that carry out crosslink repair. (B) A cartoon of the FANCM protein divided into the three domains systematically dissected in this article. Each of the domains confers distinct functions to FANCM in the DNA damage response. (C) FANCM helicase suppresses crossovers by dissolving D-loops, by acting early to suppress D loop formation FANCM may prevent the formation of Holliday junction, which are then resolved by crossover.
FANCM loss in DT40 results in an increase in spontaneous SCE events. These events could reflect the presence of spontaneous DNA damage that is then resolved by HR. In such a scenario it is possible that replication fork breakage occurs more frequently in the absence of FANCM resulting in the accumulation of double strand breaks. Unrestrained homologous recombination may then repair such lesions and since some of such events are resolved by crossover this may result in raised SCE. Alternatively and more likely the SCE events could be due to a bias towards resolving recombination intermediates by crossover. How might FANCM act to suppress crossover recombination? Our studies clearly show that this requires the Walker DEAD box motif, and this is a distinct activity from the other FANCM based DNA damage tolerance responses reported here. In addition, our genetic studies show that FANCM functions in only a subset of Blm dependent non-crossover events. It is also noteworthy that both yeast FANCM orthologues—MPH1 and FML1 also suppress crossover recombination. However, in contrast to FANCM, both yeast FANCM orthologues function in an independent pathway to their respective Blm orthologues. Recombinant FANCM, Mph1 and Fml1 can all dissolve D loops (Figure 6C) (9,10,12). These recombination intermediates precede crossover-resolving steps. By resolving such early recombination intermediates, FANCM may indirectly reduce crossover events. What might then be the function of Blm in such situations? One possibility is that FANCM synergises the function of Blm in dissolving D loops. The fact that Blm seems to be more important than FANCM in suppressing crossovers may reflect its key role in decatenating double Holliday junctions (18). It will therefore be important to test whether Blm augments the anti-recombinogenic biochemical activities of FANCM. Multiple DNA helicases in both budding yeast (Sgs1, Srs2, Mph1) (29,30) and vertebrates (Blm, Wrn, Fbh1, FANCM, RecQL1, RecQL5) (31–33) defend their genomes from crossovers. The fact that mitotic eukaryotic cells display very low levels of crossovers is a testament to the evolutionary conserved function of these pathways. Their inactivation leaves genomes vulnerable to loss of heterozygosity, chromosome translocations and thereby to inevitable consequences on health.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Personal long-term FEBS fellowship to I.V.R.; personal postdoc fellowship from the AICR to W.N. and A.A. Funding for open access charge: MRC.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Paul Pace, Georgina Mosedale and Alicja Trebinska for valuable advice and creation of key reagents, Professors Enomoto, S.Takeda for DT40 knockout strains. We also thank to J.P. de Winter for communicating us unpublished results, and all the members in K.J. Patel lab for critical reading of the manuscript. W.N. is currently a recipient of an AICR investigator award.
REFERENCES
- 1.Patel KJ, Joenje H. Fanconi anemia and DNA replication repair. DNA Rep. 2007;6:885–890. doi: 10.1016/j.dnarep.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 2.Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat. Rev. 2007;8:735–748. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- 3.Meetei AR, Medhurst AL, Ling C, Xue Y, Singh TR, Bier P, Steltenpool J, Stone S, Dokal I, Mathew CG, et al. A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat. Genet. 2005;37:958–963. doi: 10.1038/ng1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mosedale G, Niedzwiedz W, Alpi A, Perrina F, Pereira-Leal JB, Johnson M, Langevin F, Pace P, Patel KJ. The vertebrate Hef ortholog is a component of the Fanconi anemia tumor-suppressor pathway. Nat. Struct. Mol. Biol. 2005;12:763–771. doi: 10.1038/nsmb981. [DOI] [PubMed] [Google Scholar]
- 5.Komori K, Hidaka M, Horiuchi T, Fujikane R, Shinagawa H, Ishino Y. Cooperation of the N-terminal Helicase and C-terminal endonuclease activities of Archaeal Hef protein in processing stalled replication forks. J. Biol. Chem. 2004;279:53175–53185. doi: 10.1074/jbc.M409243200. [DOI] [PubMed] [Google Scholar]
- 6.Collis SJ, Ciccia A, Deans AJ, Horejsi Z, Martin JS, Maslen SL, Skehel JM, Elledge SJ, West SC, Boulton SJ. FANCM and FAAP24 function in ATR-mediated checkpoint signaling independently of the fanconi anemia core complex. Mol. Cell. 2008;32:313–324. doi: 10.1016/j.molcel.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 7.Ciccia A, Ling C, Coulthard R, Yan Z, Xue Y, Meetei AR, Laghmani el H, Joenje H, McDonald N, de Winter JP, et al. Identification of FAAP24, a Fanconi anemia core complex protein that interacts with FANCM. Mol. Cell. 2007;25:331–343. doi: 10.1016/j.molcel.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 8.Gari K, Decaillet C, Stasiak AZ, Stasiak A, Constantinou A. The Fanconi anemia protein FANCM can promote branch migration of Holliday junctions and replication forks. Mol. Cell. 2008;29:141–148. doi: 10.1016/j.molcel.2007.11.032. [DOI] [PubMed] [Google Scholar]
- 9.Sun W, Nandi S, Osman F, Ahn JS, Jakovleska J, Lorenz A, Whitby MC. The FANCM ortholog Fml1 promotes recombination at stalled replication forks and limits crossing over during DNA double-strand break repair. Mol. Cell. 2008;32:118–128. doi: 10.1016/j.molcel.2008.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gari K, Decaillet C, Delannoy M, Wu L, Constantinou A. Remodeling of DNA replication structures by the branch point translocase FANCM. Proc.Natl Acad. Sci. USA. 2008;105:16107–16112. doi: 10.1073/pnas.0804777105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prakash R, Krejci L, Van Komen S, Anke Schurer K, Kramer W, Sung P. Saccharomyces cerevisiae MPH1 gene, required for homologous recombination-mediated mutation avoidance, encodes a 3' to 5' DNA helicase. J. Biol. Chem. 2005;280:7854–7860. doi: 10.1074/jbc.M413898200. [DOI] [PubMed] [Google Scholar]
- 12.Prakash R, Satory D, Dray E, Papusha A, Scheller J, Kramer W, Krejci L, Klein H, Haber JE, Sung P, et al. Yeast Mph1 helicase dissociates Rad51-made D-loops: implications for crossover control in mitotic recombination. Genes Dev. 2009;23:67–79. doi: 10.1101/gad.1737809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xue Y, Li Y, Guo R, Ling C, Wang W. FANCM of the Fanconi anemia core complex is required for both monoubiquitination and DNA repair. Hum. Mol. Genet. 2008;17:1641–1652. doi: 10.1093/hmg/ddn054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niedzwiedz W, Mosedale G, Johnson M, Ong CY, Pace P, Patel KJ. The Fanconi anaemia gene FANCC promotes homologous recombination and error-prone DNA repair. Mol. Cell. 2004;15:607–620. doi: 10.1016/j.molcel.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 15.Yamamoto K, Hirano S, Ishiai M, Morishima K, Kitao H, Namikoshi K, Kimura M, Matsushita N, Arakawa H, Buerstedde JM, et al. Fanconi anemia protein FANCD2 promotes immunoglobulin gene conversion and DNA repair through a mechanism related to homologous recombination. Mol. Cell Biol. 2005;25:34–43. doi: 10.1128/MCB.25.1.34-43.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bridge WL, Vandenberg CJ, Franklin RJ, Hiom K. The BRIP1 helicase functions independently of BRCA1 in the Fanconi anemia pathway for DNA crosslink repair. Nat. Genet. 2005;37:953–957. doi: 10.1038/ng1627. [DOI] [PubMed] [Google Scholar]
- 17.Ling C, Ishiai M, Ali AM, Medhurst AL, Neveling K, Kalb R, Yan Z, Xue Y, Oostra AB, Auerbach AD, et al. FAAP100 is essential for activation of the Fanconi anemia-associated DNA damage response pathway. EMBO J. 2007;26:2104–2114. doi: 10.1038/sj.emboj.7601666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu L, Hickson ID. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 19.Wang W, Seki M, Narita Y, Sonoda E, Takeda S, Yamada K, Masuko T, Katada T, Enomoto T. Possible association of BLM in decreasing DNA double strand breaks during DNA replication. EMBO J. 2000;19:3428–3435. doi: 10.1093/emboj/19.13.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang W. A major switch for the Fanconi anemia DNA damage-response pathway. Nat. Struct. Mol. Biol. 2008;15:1128–1130. doi: 10.1038/nsmb1108-1128. [DOI] [PubMed] [Google Scholar]
- 21.Alpi A, Langevin F, Mosedale G, Machida YJ, Dutta A, Patel KJ. UBE2T, the Fanconi anemia core complex, and FANCD2 are recruited independently to chromatin: a basis for the regulation of FANCD2 monoubiquitination. Mol. Cell Biol. 2007;27:8421–8430. doi: 10.1128/MCB.00504-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Machida YJ, Machida Y, Chen Y, Gurtan AM, Kupfer GM, D'Andrea AD, Dutta A. UBE2T is the E2 in the Fanconi anemia pathway and undergoes negative autoregulation. Mol. Cell. 2006;23:589–596. doi: 10.1016/j.molcel.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 23.Ishiai M, Kitao H, Smogorzewska A, Tomida J, Kinomura A, Uchida E, Saberi A, Kinoshita E, Kinoshita-Kikuta E, Koike T, et al. FANCI phosphorylation functions as a molecular switch to turn on the Fanconi anemia pathway. Nat. Struct. Mol. Biol. 2008;15:1138–1146. doi: 10.1038/nsmb.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim JM, Kee Y, Gurtan A, D'Andrea AD. Cell cycle-dependent chromatin loading of the Fanconi anemia core complex by FANCM/FAAP24. Blood. 2008;111:5215–5222. doi: 10.1182/blood-2007-09-113092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raschle M, Knipsheer P, Enoiu M, Angelov T, Sun J, Griffith JD, Ellenberger TE, Scharer OD, Walter JC. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell. 2008;134:969–980. doi: 10.1016/j.cell.2008.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tebbs RS, Hinz JM, Yamada NA, Wilson JB, Salazar EP, Thomas CB, Jones IM, Jones NJ, Thompson LH. New insights into the Fanconi anemia pathway from an isogenic FancG hamster CHO mutant. DNA Rep. 2005;4:11–22. doi: 10.1016/j.dnarep.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 27.Rao VA, Fan AM, Meng L, Doe CF, North PS, Hickson ID, Pommier Y. Phosphorylation of BLM, dissociation from topoisomerase IIIalpha, and colocalization with gamma-H2AX after topoisomerase I-induced replication damage. Mol. Cell Biol. 2005;25:8925–8937. doi: 10.1128/MCB.25.20.8925-8937.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meetei AR, Sechi S, Wallisch M, Yang D, Young MK, Joenje H, Hoatlin ME, Wang W. A multiprotein nuclear complex connects Fanconi anemia and Bloom syndrome. Mol. Cell Biol. 2003;23:3417–3426. doi: 10.1128/MCB.23.10.3417-3426.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robert T, Dervins D, Fabre F, Gangloff S. Mrc1 and Srs2 are major actors in the regulation of spontaneous crossover. EMBO J. 2006;25:2837–2846. doi: 10.1038/sj.emboj.7601158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ira G, Malkova A, Liberi G, Foiani M, Haber JE. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115:401–411. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang W, Seki M, Narita Y, Nakagawa T, Yoshimura A, Otsuki M, Kawabe Y, Tada S, Yagi H, Ishii Y, et al. Functional relation among RecQ family helicases RecQL1, RecQL5, and BLM in cell growth and sister chromatid exchange formation. Mol. Cell Biol. 2003;23:3527–3535. doi: 10.1128/MCB.23.10.3527-3535.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Imamura O, Fujita K, Shimamoto A, Tanabe H, Takeda S, Furuichi Y, Matsumoto T. Bloom helicase is involved in DNA surveillance in early S phase in vertebrate cells. Oncogene. 2001;20:1143–1151. doi: 10.1038/sj.onc.1204195. [DOI] [PubMed] [Google Scholar]
- 33.Hu Y, Lu X, Barnes E, Yan M, Lou H, Luo G. Recql5 and Blm RecQ DNA helicases have nonredundant roles in suppressing crossovers. Mol. Cell Biol. 2005;25:3431–3442. doi: 10.1128/MCB.25.9.3431-3442.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.