Abstract
Lin28 has been shown to block the processing of let-7 microRNAs implicated in the regulation of cell growth and differentiation. Here, we show that Lin28 also specifically associates with ribonucleoprotein particles containing the replication-dependent histone H2a mRNA in mouse embryonic stem cells. We further show that the coding region of H2a mRNA harbors high affinity binding sequences for Lin28 and that these sequences stimulate the expression of reporter genes in a Lin28-dependent manner. We suggest that a key function of Lin28 in the maintenance of pluripotency is to promote the expression of the H2a gene (and perhaps also other replication-dependent histone genes) at the posttranscriptional level in order to coordinate histone production with the unique proliferative properties of embryonic stem cells.
INTRODUCTION
Abundantly expressed in human and mouse embryonic stem (ES) cells, Lin28 is among four factors (including Oct4, Sox2 and Nanog) that together reprogram human fibroblasts to pluripotency (1–3). Despite its apparent critical role in ES cells, the molecular function and mode of action of Lin28 are just beginning to be elucidated. Multiple studies have demonstrated that Lin28 functions to block the production of mature let-7 microRNAs implicated in the regulation of cell growth and differentiation, although the mechanism by which it does so remains controversial (4–8). In addition, let-7 microRNA represses the expression of Lin28, creating a feedback loop [(9) and references therein]. However, other evidence exists that Lin28 may regulate gene expression through multiple mechanisms. Indeed, Polesskaya et al. (2) reported that Lin28 associates with ribonucleoprotein particles (RNPs) containing IGF-2 mRNA and stimulates its translation in muscle cells. Similarly, our recent studies suggest that Lin28 acts to modulate mouse ES cell growth likely in part by effecting the expression of cell cycle genes (including cyclins A and B and cdk4) at the translational level (10). Thus, a major role Lin28 may play in ES cells is to regulate the expression of genes involved in cell cycle progression. This is conceivable given that pluripotency is thought to be mechanistically linked to the unique proliferative properties of ES cells (11,12).
The class of genes that provide the massive amounts of histones required for chromosome packaging during the S-phase of cell cycle are the replication-dependent histone genes. Those encoding the four core histones (H2a, H2b, H3 and H4) and the linker histone H1, are unique in that they do not contain introns and that their mRNAs end in a highly conserved stem-loop structure instead of a poly(A) tail (13–15). As histone synthesis outside the S-phase is highly toxic to the cell, multiple levels of regulation are involved to restrict histone production exclusively to the S-phase. Owing to the structure of the nucleosome, it is critical that the replication-dependent histones are stoichiometrically expressed in cells. However, the sequences and mechanisms that regulate the transcription of the individual histone genes are well known to be diverse, reflecting different regulatory factors and modes of regulation present in different cell types (13). Thus, it is possible that the four histone subtypes (H2a, H2b, H3 and H4) are differentially regulated at the posttranscriptional level depending on cell types. In this report, we suggest that Lin28 plays a role in regulating the production of histones, in particular the H2a subtype, in mouse ES cells.
RESULTS
Lin28 stimulates ES cell proliferation
We have recently reported that Lin28 plays a role in mouse ES cell proliferation (10). Figure 1 recapitulates this observation. Mouse ES cells were transfected with a siRNA specific for Lin28. In parallel, the cells were mock-transfected or transfected with a control siRNA. mRNA and protein levels were measured 24 and 48 h after transfection, respectively. The level of Lin28 mRNA in cells transfected with the Lin28-specific siRNA (Lin28 siRNA) was reduced to ∼10% of those in cells that were mock transfected or transfected with a control siRNA (Control siRNA) (Figure 1a). Importantly, the level of untargeted beta-actin mRNA was not affected, indicating that the siRNA-mediated gene silencing effect was specific. The Lin28 protein level decreased by ∼75% after Lin28 siRNA treatment (Figure 1b, top panel, compare lanes 2 to 1). The cells transfected with Lin28 siRNA exhibited a retarded growth phenotype, as indicated by the lower cell numbers compared to those transfected with control siRNA (Figure 1c). Treatment of ES cells with siRNAs targeted to different regions of Lin28 showed similar effects (Supplementary Figure S1). On the other hand, elevated expression of Lin28 by transfection of a Flag-tagged Lin28 expression vector (Flag-Lin28) resulted in stimulation of cell growth (Figure 1d). The level of Flag-Lin28 expression was estimated to be ∼5-fold higher than that of endogenous Lin28, given a plasmid DNA transfection efficiency of ∼50% (Figure 1b, top panel, compare lane 4 to 3, and data not shown).
Figure 1.
Lin28 promotes cell proliferation. (a) ES cells were transfected with Lin28 siRNA, mock transfected, or transfected with control siRNA. Total cellular RNAs were extracted 24 h later, and the indicated mRNA levels determined by reverse transcription and quantitative real-time PCR (RT-qPCR). (b) ES cells were transfected with control siRNA (lane 1), Lin28 siRNA (lane 2), empty vector (lane 3) or Flag-Lin28 (lane 4). Cellular proteins were extracted 48 h (lanes 1 and 2) or 24 h (lanes 3 and 4) later, and protein levels determined by Western blot. Upper panel, Western blot using a Lin28-specific antibody. The band marked by an asterisk in lane 4 indicates Flag-Lin28, while bands below in lanes 1–4 indicate endogenous Lin28. Bottom panel, Western blot of the same membrane using an antibody specific for mouse beta-actin. (c and d) ES cells were transfected with control siRNA, Lin28 siRNA, empty vector, or Flag-Lin28, and cell numbers counted 48 h (c) or 24 h (d) following transfection. (e) Caspase-3/7 activities of ES cells were measured 24 and 48 h following the various transfections. Each bar represents mean ± SD (n = 3–4).
The altered cell growth rates observed when Lin28 expression was reduced or increased were unlikely due to changes in cell death rates, as caspase activity assays (measurements of cell apoptosis) showed no significant differences between the differently treated cells (Figure 1e). In addition, trypan-blue exclusion assays also revealed <10% non-viable cells in all cases (data not shown). However, we show that these were likely resulted from cell cycle structure changes indicated by the flow cytometric analysis of propidium iodide-stained cells (Supplementary Figure S2). Taken together, the above results are consistent with the notion that Lin28 in its natural context may function to promote progression from S to G2/M phase, which is in line with our previous observation using BrdU incorporation flow cytometric analysis (10).
Lin28 binds histone H2a mRNA in vivo
To explore the possibility that Lin28 may regulate specific genes involved in cell cycle progression, we searched for mRNA targets of Lin28. Using immunoprecipitation (IP) and reverse transcription and quantitative real-time PCR (RT-qPCR), we identified cyclins A and B, and cdk4 mRNAs as the putative targets of Lin28 (10). Here, we report the replication-dependent histone H2a mRNA as a new target of Lin28 regulation. An interaction between Lin28 and histone mRNAs was originally missed by us, because these mRNAs are not polyadenylated and thus could not be detected by our IP and RT-qPCR assays where oligo(dT) was used to generate cDNAs from precipitated RNAs in reverse transcription reactions. Ironically, we were using histone H2a mRNA sequences as negative controls for Lin28 binding. To our surprise, these sequences consistently showed the highest affinity for Lin28 in our in vitro RNA–protein interaction assays. These observations prompted us to explore the possible link between Lin28 and histone mRNAs.
Thus, we performed IP using a monoclonal anti-Flag antibody to isolate RNPs from ES cells transfected with Flag-Lin28. RNA samples extracted from IP complexes were used to generate cDNAs, followed by qPCR to identify associated mRNAs. In the RT reactions, primers specific for the four core histone (H2a, H2b, H3 and H4) mRNAs as well as oligo(dT) were used. Figure 2 presents representative results of multiple independent experiments. The amounts of mRNAs present in the anti-Flag IP complexes relative to those in pre-immune IgG IP complexes (which were arbitrarily set as 1) are shown in Figure 2a. Tubulin mRNA was used as a control for non-specific RNA binding. H2a mRNA exhibited the most dramatic enrichment (∼5-fold) among the four histone mRNAs in the anti-Flag versus pre-immune complexes. The rest (H2b, H3 and H4), however, showed only marginal enrichment (∼1.5–2-fold). Confirming our previous findings (10), mRNAs for cyclins A and B and cdk4 were also significantly enriched. Notably, the enrichment of Oct4 was also reproducibly observed, albeit to a lesser extent (∼2-fold). The possibility that Oct4 mRNA may be a target for Lin28 regulation is currently under investigation. Importantly, these results were mimicked by IP using a polyclonal anti-Lin28 antibody (6) (Figure 2b). Furthermore, the relative fold enrichment did not reflect levels of the mRNAs in the cell extract (Figure 2c), suggesting that the relative enrichments observed were not due to high amounts of the particular mRNAs present in the extracts. The apparently higher levels of H2b and H4 mRNAs compared to those of H2a and H3 in the cell extract were likely resulted from higher primer efficiencies in the RT and PCR reactions. The primers were designed such that at least 12 mRNAs in each subtype of histone genes could be detected in our RT and PCR reactions (see ‘Materials and Methods’ section). Taken together, the preferential enrichment of H2a mRNA in Lin28-containing RNPs indicates that it may be an in vivo target for Lin28 regulation.
Figure 2.
Lin28 associates with a specific subset of mRNAs in mouse ES cells. (a) ES cells were transfected with Flag-Lin28 and RNPs isolated using anti-Flag. (b) RNPs were isolated from untransfected ES cells using anti-Lin28 antibody. (c) Relative mRNA levels after normalization against gapdh mRNA levels in the cell extract.
Lin28 binds H2a mRNA in vitro
To ask whether H2a mRNA contains high affinity binding sites for Lin28, we performed in vitro UV-crosslinking (XL) experiments. XL allows the detection of direct contact between RNA and protein based on the natural photo-reactivity of nucleic acids and amino acids upon UV irradiation. Our pilot experiments revealed that, like many other RNA-binding proteins [i.e. the fragile X mental retardation protein FMRP (16)], Lin28 can be crosslinked non-specifically to almost any RNA tested even under very stringent conditions (Figure 3c and data not shown). This was not surprising since the specific affinity of Lin28 for let-7 microRNA precursors has been reported to be relatively low, in the micromolar range (5). Nonetheless, we have been able to show a specific and direct interaction between Lin28 and the coding region of H2a mRNA using a crosslinking and competition strategy similar to that described by Schaeffer et al. (16). These sorts of assays are quite sensitive and largely alleviate the contribution of non-specific binding.
Figure 3.
XL and competition assays. Flag-Lin28 was transfected into HEK293 cells and cell extract prepared. (a) XL were carried out using radioactively labeled H2a coding region RNA in the absence (lane 2) or presence of increasing amounts of unlabeled H4 (lanes 3–5), H3 (lanes 6–8), H2b (lanes 9–11), B1U1 (lanes 12–14) or H2a (lanes 15–17) RNA, followed by IP. Total crosslinked product (5%) and immunoprecipitates were resolved by SDS–PAGE, followed by autoradiography. The bands marked with asterisk are Flag-Lin28. (b) Amounts of crosslinked Flag-Lin28 plotted against unlabeled competitor RNA in molar excess. Each point represents three independent experiments. Numbers are mean ± SD. (c) XL of radioactively labeled B1U1 RNA in the presence of increasing amounts of unlabeled B1U1 (lanes 3–5) or H2a (lanes 6–8) RNA.
To characterize the interaction between H2a mRNA and Lin28, a fixed amount of radioactively labeled H2a RNA (coding region, 393-nt long) was incubated with an extract of HEK293 cells transfected with Flag-Lin28, in the presence of increasing amounts of unlabeled RNA fragments of similar sizes (see ‘Materials and Methods’ section). The unlabeled competitor RNAs used were from the coding regions of histones H4, H3, H2b and H2a, and fragment B1U1. B1U1 was derived from a part of the 3′-untranslated region (UTR) of cyclin B mRNA that does not contain high affinity binding sequences for Lin28 [(10) and see Figure 3], and thus served as a control for non-specific Lin28 binding. The reactions were UV-irradiated, followed by RNase A digestion. Crosslinked (thus radioactively labeled) Lin28 was captured by IP using an anti-Flag antibody and visualized by autoradiography. As shown in Figure 3a, Lin28 binding to the radioactively labeled H2a RNA was competed far more efficiently by the unlabeled H2a RNA than by any of the others (compare lanes 15–17 with lanes 3–14). Under the conditions used, the affinity of H2a RNA for Lin28 is ∼8-fold higher than that of H4 RNA, and ∼5-fold higher than those of B1U1, H2b and H3 RNAs (Figure 3b). The apparent better competition at the 7–molar excess concentration using unlabeled B1U1 versus unlabeled H2a (Figure 3a, compare lanes 12–13 with lanes 15–16) simply reflects intrinsic experimental variation, which is reflected in the error bar shown in Figure 3b at the 7-molar excess concentration. When labeled B1U1 was used, more efficient competition of H2a compared to B1U1 RNA itself was also observed (Figure 3c). Taken together, these results strongly suggest that the H2a coding region contains high affinity binding sites for Lin28.
H2a coding region contains sequences that stimulate gene expression in a Lin28-dependent manner
As a predominantly cytoplasmic RNA-binding protein, Lin28 may affect its target mRNAs at the translational and/or stability level. This question can be addressed by overexpression or downregulation of Lin28, followed by assessing the effects on levels of proteins and RNAs encoded by the target genes (10). However, this type of approach is not feasible for histone genes due to the intimate coupling between histone expression and the cell cycle. Histone mRNA and protein levels rise in early S-phase and decline dramatically at the end of S-phase (13,14,17). Therefore, differences in histone mRNA and protein levels cannot be discerned using asynchronous cell populations. On the other hand, due to the fast ES cell cycle (mouse ES cells divide every ∼8–10 h) and the requirement for at least 24 h to synchronize ES cells (11,18), experiments using synchronized ES cells are not feasible either. To circumvent these problems, we employed a luciferase reporter system (10). Thus, sequences derived from H2a, H2b, H3 or H4 coding regions were inserted into the 3′UTR of a firefly luciferase reporter construct (19), and the abilities of the sequences to stimulate the reporter gene expression measured. The parental (FFL) and a construct containing the B1U1 sequence were included as negative controls. As shown in Figure 4a, while the H2a sequence exhibited a strong stimulatory effect on firefly luciferase activity in a Lin28-dependent manner, those from H2b, H3 and H4 had a modest stimulatory effect, and B1U1 showed no effect. RT-qPCR analysis confirmed that the Lin28-dependent luciferase activity changes observed were not due to changes in luciferase mRNA levels when Lin28 was introduced into cells at 10 and 20 ng levels (Figure 4b). This is consistent with a role of Lin28 in regulating the translation of the reporter mRNA. However, when the Lin28 expression vector was added at higher amounts, leading to higher expression levels of Lin28 protein (data not shown), the level of H2a-containing mRNA was reproducibly slightly increased over that of the other mRNAs (40 ng, Figure 4b). A simple interpretation for these observations is that the H2a sequence has the ability to enhance both translation and mRNA stability in a Lin28-dependent manner. Experiments are currently underway to understand the molecular basis of the different effects exerted by Lin28 in this system.
Figure 4.
Luciferase activity assays. The indicated reporter constructs were transfected into NIH/3T3 cells that do not express endogenous Lin28, together with increasing amounts of Flag-Lin28. In addition, a Renilla reporter was included in all transfections for normalization purposes. Luciferase activities and mRNA levels were measured 24 h after transfection. Firefly luciferase activities [after normalization against Renilla luciferase (a and c)] and firefly mRNA levels (b) from cells without Flag-Lin28 transfection were arbitrarily set as 1. Numbers are mean ± SD (n = 3). F3-1X and F3-2X, firefly luciferase reporter containing one and two tandem copies of F3, respectively, inserted at its 3′UTR.
We next took a step further to identify by XL and competition assays a 135-nt region (designated as F3, nts from 259 to 393, relative to the translational start site of H2a) in the 3′-end region of the H2a open reading frame that contains binding elements for Lin28 (Figure 4c and data not shown). While a single copy insertion had no effect, the insertion of two tandem copies of the elements recapitulated the translational stimulatory phenomenon observed with the full-length H2a insertion (Figure 4c). That one copy of the element did not produce an intermediate stimulatory effect indicates that H2a may contain more than one Lin28 high affinity binding element and that binding to multiple elements may produce synergistic and not just additive effects. This phenomenon is commonly seen in RNA-based studies (20–23). Taken together, these results suggest that binding of Lin28 to specific sites within the H2a coding region may enhance translation and/or mRNA stability.
DISCUSSION
We have shown here that Lin28 positively influences the mouse ES cell cycle, likely by facilitating the progression from S to G2/M phase (Figure 1 and Supplementary Figure S2). In addition to the key cell cycle regulatory genes cyclins A and B and cdk4 (10), we have identified histone H2a as a new putative target of Lin28. This is based on its preferential association with Lin28 in the context of RNPs (Figure 2) and its ability to directly bind Lin28 in our in vitro assays (Figure 3). Furthermore, we show that sequences derived from the coding region of H2a are able to stimulate reporter gene expression at the posttranscriptional level in a Lin28-dependent fashion (Figure 4). We thus postulate that Lin28 may play a role in regulating the translation and/or stability of H2a mRNA, in addition to a subset of key cell-cycle regulator genes, as part of ES cell-specific mechanisms. As can be seen from the data presented in Figure 4, Lin28 appears to be able to influence not only translation, but also mRNA stability. Interestingly, other studies on Lin28 have implicated this protein in a variety of steps of gene regulation. For example, there have been multiple reports that Lin28 can block the production of mature let-7 microRNAs (4–8). However, the mechanism by which it does so remains highly controversial. Two studies (5,6) found that Lin28 specifically binds to the terminal loop regions of let-7 precursors in vitro and blocks microRNA processing at the Microprocessor step in the nucleus. Rybak et al. (4), on the other hand, concluded that inhibition occurs in the cytoplasm at the Dicer step and that both the loop and stem regions of let-7 precursors bind Lin28. They also found that Lin28 was able to bind mature let-7 that does not contain the terminal loop sequences. Furthermore, Heo et al. (7) reported that Lin28 induces terminal uridylation of pre-let-7 in the cytoplasm, leading to inhibition of Dicer processing and pre-let-7 degradation. Taken together, these studies suggest that binding of Lin28 to its target RNAs may lead to multiple outcomes, perhaps influenced by protein–protein interactions, by other cis-acting elements lying near to Lin28-binding sites, or by the intracellular location.
ES cells proliferate rapidly and indefinitely while maintaining their capacity of differentiating into any cell type found in the body. It has been proposed that pluripotency may be intrinsically coupled to the unique ES cell-cycle structure which is characterized by very rapid proliferation with a truncated G1-phase (11,12,24). As such, the cells devote more than half of the entire cycle length to their S-phase and apparently lack a G1/S checkpoint. Given that histone expression is mechanistically coupled to the S-phase, ES cells may have evolved unique mechanisms controlling histone production in order to coordinate with their unique proliferative properties. Our finding that the replication-dependent histone H2a gene is a likely target for Lin28-mediated regulation highlights this possibility.
As the transcriptional regulation of the individual histone genes are well known to be diverse, (reviewed in 13), it is not surprising that the four histone subtypes (H2a, H2b, H3 and H4) appear to be differentially regulated at the posttranscriptional level with respect to Lin28 in an ES cell-specific manner. Although H2a mRNA is likely the major target for Lin28 regulation relative to H2b, H3 and H4, based on our assays, the possibility exists that the other three may also be regulated by Lin28, albeit to a lesser extent.
The regulation of expression of replication-dependent histone genes in non-ES cells has been extensively studied. SLBP/HBP is among the many well-studied factors involved in histone mRNA metabolism. It binds the 3′-end stem-loop of histone mRNAs with high specificity and regulates the 3′-processing, nucleocytoplasmic transport, stability and translation of these mRNAs (13,14,17). In addition, specific elements that reside in the coding regions of histone genes and that participate in the various steps of histone mRNA metabolism including transcription (25), nucleocytoplasmic transport (26) and histone 3′-end processing (20) have been described. Our finding that the H2a coding region may contain translational/stability stimulatory sequence elements suggests for the first time that there may exist translational/stability ‘modulators’ within the coding regions of histone mRNAs that might be recognized by cell-type-specific factors. It is intriguing to note that the expression of cyclin A and histones are intimately coupled to each other, and that both are coupled to DNA synthesis and cell-cycle progression (27). Our finding that both cyclin A and H2a mRNA are targets of Lin28 regulation underscores the importance of coordinated regulation of gene expression by Lin28.
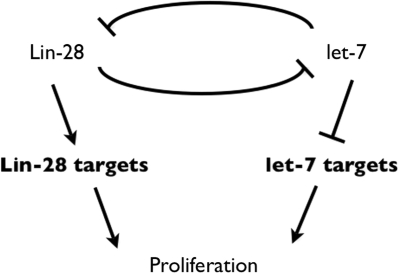
Finally, based on our findings [this report and (10)] as well as those of others (2), growing evidence suggests that Lin28 has important biological functions distinct from the inhibition of let-7 processing. While the reported effects of Lin28 on let-7 microRNA expression are negative, those on mRNA targets are positive. However, the ultimate effect of Lin28 on all known targets is to promote cell proliferation (Figure 5).
Figure 5.
A feedback model for the regulation of cell proliferation by Lin28 and let-7. Lin28 enhances the expression of genes that control proliferation, while let-7 inhibits them. We cannot exclude that at least some Lin28 targets are also regulated by let-7.
MATERIALS AND METHODS
Antibodies, plasmids and siRNAs
The polyclonal anti-Lin28 antibody (Abcam, ab46020), monoclonal anti-beta-actin (Sigma, A2228), anti-Flag (Sigma, F3165), and mouse pre-immune IgG (Chemicon, PP54) were purchased. The rabbit pre-immune sera were previously described (28). Flag-Lin28 was created as previously described (10). Lin28 siRNA (Dharmacon, L-051530-01), Lin28 siRNAb (Integrated DNA Technologies, Inc., NM_145833 duplex 2), and control siRNA (Dharmacon, D-001810-01-05) were purchased.
Cell culture and transfection
Mouse ES cell line ES-C57BL/6 cells (ATCC, SCRC-1002) were cultured on mitomycin-inactivated MEF (ATCC, SCRC-1040) feeder layer as previously described (10). HEK293 and NIH/3T3 cells were cultured using standard protocols provided by ATCC. Cell transfections were carried out as described (29).
Protein extraction and Western blot analyses
These were done as described (10).
Caspase-3/7 assays
Caspase activities were measured using the Apo-ONE Homogeneous Caspase-3/7 Assay kit (Promega) according to the manufacturer's protocols.
Flow cytometry analyses
ES cells were harvested 24 h (plasmid DNA transfection) or 48 h (siRNA transfection) post-transfection and fixed in 70% ethanol at 4°C for 30 min. After being washed once with PBS and resuspended in PBS (5 × 105 cells/100 µl of PBS), cells were treated with RNase A at a final concentration of 100 µg/ml for 10 min at room temperature. The cell suspension was then diluted at a 1:5 ratio with cold PBS, and propidium iodide added at a final concentration of 16 µg/ml. After passing through a 75 μm mesh (to remove cell clumps), cell suspension was kept on ice for 30 min, followed by flow cytometric analyses. The measurement of cell cycle parameters was performed in a CALIBUR FACScan using Cellquest software and data processed using the Modfit software from Verity Software House (verity@vsh.com).
Immunoprecipitation, RNA extraction and RT-qPCR
These were carried out based on protocols as previously described (10). The RT primers specific for the four mouse histone genes are: H2a, 5′-CTTGTTGAGCTCCTCGTCGT; H2b, 5′-GGTCGAGCGCTTGTTGTAAT; H3, 5′-GACGGGCCAGCTGGATGTCCT; and H4, 5′-TAACCGCCGAATCCGTAGA. The PCR primers for the individual genes are: Gapdh forward: 5′-TTAGCACCCCTGGCCAAGG; Gapdh reverse: 5′-CTTACTCCTTGGAGGCCATG; Beta-actin forward: 5′-GTGGGCCGCTCTAGGCACCAA; Beta-actin reverse: 5′-CTCTTTGATGTCACGCACGATTTC; Tubulin forward: 5′-CGTGTTCGGCCAGAGTGGTGC; Tubulin reverse: 5′-GGGTGAGGGCATGACGCTGAA; Cdk4 forward: 5′-TGTGGAGCGTTGGCTGTATC; Cdk4 reverse: 5′-TGGTCGGCTTCAGAGTTCC; Oct4 forward: 5′-TGGAGAAGGTGGAACCAACTCCC; Oct4 reverse: 5′-ACACGGTTCTCAATGCTAGTTCGC; Lin28 forward: 5′-GTCTTTGTGCACCAGAGCAA; Lin28 reverse: 5′-CTTTGGATCTTCGCTTCTGC; Cyclin B (B1) forward: 5′-TCCCTCGGTGGGATTCAAGTGC; Cyclin B (B1) reverse: 5′-CAGGAGTGGCGCCTTGGTATGG; Cdk1 forward: 5′-TTGGAGAAGGTACTTACGGTGTGGTG; Cdk1 reverse: 5′-CCAGGAGGGATGGAGTCCAGGT; Cyclin A (A2) forward: 5′-GCTCAAGACTCGACGGGTTGC; Cyclin A (A2) reverse: 5′-GCTGCATTAAAAGCCAGGGCATC; H2a forward: 5′-GGCGGTGCTGGAGTACCTA; H2a reverse: 5′-GATGATGCGCGTCTTCTTG; H2b forward: 5′-GAGAGCTACTCGGTGTACGTG; H2b reverse: 5′-CGCTCGAAGATGTCGTTCAC; H3 forward: 5′-AAACAGATCTGCGCTTCCAG; H3 reverse: 5′-TTGTTACACGTTTGGCATGG; H4 forward: 5′-AACATCCAGGGCATCACGAA; H4 reverse: 5′-TCTTGCGCTTGGCGTGCT; Firefly luciferase forward: 5′-GCTGGGCGTTAATCAGAGAG; Firefly luciferase reverse: 5′-GTGTTCGTCTTCGTCCCAGT; Renilla forward: 5′-GCAAATCAGGCAAATCTGGT; Renilla reverse: 5′-GGCCGACAAAAATGATCTTC.
Firefly reporter constructs and Luciferase activity assays
The various firefly reporter constructs were created by inserting open reading frames (ORFs) of mouse Hist2h2aa1 (NM_013549), Hist1h2bc (NM_023422), Hist1h3g (NM_145073) and Hist1h4h (NM_153173), respectively, into the 3′ UTR of the parental firefly reporter (19) opened at Not I and XhoI. The ORFs were obtained by RT-PCR of total RNA from mouse ES cells. F3-1X was created by inserting a PCR fragment containing a 135-nt region of H2a (nts 259–393, relative to the translational start site of H2a) into the 3′ UTR of the firefly reporter opened at Not I and Xho I sites. To make F3-2X, the 135-nt region of H2a (generated by PCR with Not I sites at both ends) were inserted into F3-1X opened at Not I. The resulting clones were confirmed by sequencing. The B1U1 luciferase construct was described previously (10). The constructs were transfected into NIH/3T3 cells, together with increasing amounts of Flag-Lin28 DNA. In addition, a Renilla reporter was included in all transfections for normalization purposes. Transfection was carried out in a 48-well plate scale. The amount of total plasmid DNA per well was 400 ng that included 100 ng of firefly reporter DNA, 2 ng of Renilla DNA, and 0, 10, 20 or 40 ng of Flag-Lin28. Luciferase activities were determined using a TD 20/20n (Turner BioSystems) and the Dual Luciferase Assay System (Promega) according to the manufacturer's instructions. Luciferase mRNA levels were determined by RT-qPCR and levels plotted after normalization against beta-tubulin and Renilla mRNAs.
In vitro transcription and UV-crosslinking (XL) assays
5′ T7 promoter-containing transcription templates were created by PCR using the various elements-containing firefly reporter constructs described above as templates. The resulting PCR fragments were gel purified and used to generate H2a, H2b, H3, H4 and B1U1 RNAs using MEGAscript T7 (Ambion, AM1334) according to the manufacturer's instructions. The resulting RNA fragments were gel purified and used in the XL and competition assays. The sizes of the RNA fragments in nucleotides are: H2a, 393; H2b, 381; H3, 411; H4, 282; and B1U1, 297. To prepare cell extract for XL, Flag-Lin28 was transfected into HEK293 cells and cell extracts prepared 24 h later by incubating cells in 10 cell volumes of lysis buffer [0.5% Triton X-100, 10 mM NaCl, 10 mM Tris–HCl, pH 7.5, 10 mM EDTA, 0.5 mM PMSF, 1 mM DTT, 1× protease inhibitor cocktail (Calbiochem)] on ice for 20 min, followed by centrifugation to remove insoluble material. In a 40 ul of XL reaction, there were 2 ul of extract, 20 nM of in vitro transcribed and 32P-UTP labeled H2a or B1U1 RNA, 6.5 ul of XL buffer (1 mM MgCl2, 280 mM KCl, 1 mg/ml of yeast total RNA, 35 mg/ml of heparin, 20 mM HEPES, pH 7.9, 3% glycerol and 1 mM DTT), and 140, 420 and 1260 nM of the indicated unlabeled competitor RNA. The mixture was incubated at 30°C for 5 min and then exposed to UV light (254 nm) on ice for 5 min. After RNase A (working concentration 1 mg/ml) treatment at 37°C for 30 min, cross-linked product was immunoprecipitated in 300 ul of IP buffer (10 mM Tris–HCl, pH 7.5, 150 mM NaCl, 10 mM EDTA, 0.5% Triton X-100, 0.5 mM PMSF, 1 mM DTT, 1X protease inhibitor cocktail) containing 10 ul of protein A Sepharose pre-bound to 10 ug of anti-Flag antibody. Immunoprecipitation was carried out at 4°C overnight. After washing five times with 1 ml of cold IP buffer, bound proteins were eluted using 3× SDS-sample buffer and resolved on 12% SDS–PAGE, followed by autoradiography.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online
FUNDING
Technology Initiatives for Connecticut Innovations. Funding for open access charge: Technology Initiatives for Connecticut Innovations grant 06SCA02.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Kexiong Zhang for flow cytometric analysis.
REFERENCES
- 1.Richard M, Tan SP, Tan JH, Chan WK, Bongso A. The transcriptome profile of human embryonic stem cells as defined by SAGE. Stem Cells. 2004;22:51–64. doi: 10.1634/stemcells.22-1-51. [DOI] [PubMed] [Google Scholar]
- 2.Polesskaya A, Cuvellier S, Naguibneva I, Duquet A, Moss EG, Harel-Bellan A. Lin-28 binds IGF-2 mRNA and participates in skeletal myogenesis by increasing translation efficiency. Genes Dev. 2007;21:1125–1138. doi: 10.1101/gad.415007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 4.Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, Wulczyn FG. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat. Cell Biol. 2008;10:987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 5.Piskounova E, Viswanathan SR, Janas M, Lapierre RJ, Daley GQ, Sliz P, Gregory RI. Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. J. Biol. Chem. 2008;283:21310–21314. doi: 10.1074/jbc.C800108200. [DOI] [PubMed] [Google Scholar]
- 6.Newman MA, Thomson J, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heo I, Joo C, Cho J, Ha M, Han J, Kim NV. Lin28 mediates the terminal uridylation of let-7 precursor microRNA. Mol. Cell. 2008;32:276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 8.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin-28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Büssing I, Slack FJ, Großhans H. let-7 microRNAs in development, stem cells and cancer. Trends Mol. Med. 2008;14:400–409. doi: 10.1016/j.molmed.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 10.Xu B, Zhang K, Huang Y. Lin28 modulates cell growth and associates with a subset of cell cycle regulator mRNAs in mouse embryonic stem cells. RNA. 2009;15:357–361. doi: 10.1261/rna.1368009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujii-Yamamoto H, Kim JM, Arai K, Masai H. Cell cycle and developmental regulations of replication factors in mouse embryonic stem cells. J. Biol. Chem. 2005;280:12976–12987. doi: 10.1074/jbc.M412224200. [DOI] [PubMed] [Google Scholar]
- 12.White J, Dalton S. Cell cycle control of embryonic stem cells. Stem Cell Rev. 2005;1:131–138. doi: 10.1385/SCR:1:2:131. [DOI] [PubMed] [Google Scholar]
- 13.Jaeger S, Barends S, Giege R, Eriani GE, Martin F. Expression of metazoan replication-dependent histone genes. Biochimie. 2005;87:827–834. doi: 10.1016/j.biochi.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 14.Dominski Z, Marzluff WF. Formation of the 3′ end of histone mRNA: getting closer to the end. Gene. 2007;396:373–390. doi: 10.1016/j.gene.2007.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilmartin GM. Eukaryotic mRNA 3′ processing: a common means to different ends. Genes & Dev. 2005;19:2517–2521. doi: 10.1101/gad.1378105. [DOI] [PubMed] [Google Scholar]
- 16.Schaeffer C, Bardoni B, Mandel J-L, Ehresmann B, Ehresmann C, Moine H. The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif. EMBO J. 2001;20:4803–4813. doi: 10.1093/emboj/20.17.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marzluff WF, Wagner EJ, Duronio RJ. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat. Genet. 2008;9:843–854. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stead E, White J, Faast R, Coon S, Goldstone S, Rathjen J, Dhingra U, Rathjen P, Walker D, Dalton S. Pluripotent cell division cycles are driven by ectopic Cdk2, cyclin A/E and E2F activities. Oncogene. 2002;21:8320–8333. doi: 10.1038/sj.onc.1206015. [DOI] [PubMed] [Google Scholar]
- 19.Vasudevan S, Steitz JA. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell. 2007;128:1105–1118. doi: 10.1016/j.cell.2007.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friend K, Lovejoy AF, Steitz JA. U2 snRNP binds intronless histone pre-mRNAs to facilitate U7-snRNP-dependent 3′ end formation. Mol. Cell. 2007;28:240–252. doi: 10.1016/j.molcel.2007.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu X, Mertz JE. HnRNP L binds a cis-acting RNA sequence element that enables intron-dependent gene expression. Genes Dev. 1995;9:1766–1780. doi: 10.1101/gad.9.14.1766. [DOI] [PubMed] [Google Scholar]
- 22.Huang Y, Wimler KM, Carmichael GG. Intronless mRNA transport elements may affect multiple steps of pre-mRNA processing. EMBO J. 1999;18:1642–1652. doi: 10.1093/emboj/18.6.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang Y, Steitz JA. Splicing factors SRp20 and 9G8 promote the nucleocytoplasmic export of mRNA. Mol. Cell. 2001;7:899–905. doi: 10.1016/s1097-2765(01)00233-7. [DOI] [PubMed] [Google Scholar]
- 24.Becker KA, Ghule PN, Therrien JA, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Self-renewal of human embryonic stem cells is supported by a shortened G1 cell cycle phase. J. Cell Physiol. 2006;209:883–893. doi: 10.1002/jcp.20776. [DOI] [PubMed] [Google Scholar]
- 25.Eliassen KA, Baldwin A, Sikorski EM, Hurt MM. Role for a YY1-binding element in replication-dependent mouse histone gene expression. Mol. Cell Biol. 1998;18:7106–7118. doi: 10.1128/mcb.18.12.7106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang Y, Steitz JA. Splicing factors SRp20 and 9G8 promote the nucleocytoplasmic export of mRNA. Mol. Cell. 2001;7:899–905. doi: 10.1016/s1097-2765(01)00233-7. [DOI] [PubMed] [Google Scholar]
- 27.Zhao X, Mckillop-Smith S, Muller B. The human histone gene expression regulator HBP/SLBP is required for histone and DNA synthesis, cell cycle progression and cell proliferation in mitotic cells. J. Cell Sci. 2004;117:6043–6051. doi: 10.1242/jcs.01523. [DOI] [PubMed] [Google Scholar]
- 28.Lai D, Sakkas D, Huang Y. The fragile X mental retardation protein interacts with a distinct mRNA nuclear export factor NXF2. RNA. 2006;12:1–4. doi: 10.1261/rna.94306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang M, Guller S, Huang Y. Method to enhance transfection efficiency of cell lines and placental fibroblasts. Placenta. 2007;28:779–782. doi: 10.1016/j.placenta.2007.01.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.