Abstract
HMGA proteins are not translated in normal human somatic cells, but are present in high copy numbers in pluripotent embryonic stem cells and most neoplasias. Correlations between the degree of malignancy, patient prognostic index and HMGA levels have been firmly established. Intriguingly, HMGA2 is also found in rare tumor-inducing cells which are resistant to chemotherapy. Here, we demonstrate that HMGA1a/b and HMGA2 possess intrinsic dRP and AP site cleavage activities, and that lysines and arginines in the AT-hook DNA-binding domains function as nucleophiles. We also show that HMGA2 can be covalently trapped at genomic abasic sites in cancer cells. By employing a variety of cell-based assays, we provide evidence that the associated lyase activities promote cellular resistance against DNA damage that is targeted by base excision repair (BER) pathways, and that this protection directly correlates with the level of HMGA2 expression. In addition, we demonstrate an interaction between human AP endonuclease 1 and HMGA2 in cancer cells, which supports our conclusion that HMGA2 can be incorporated into the cellular BER machinery. Our study thus identifies an unexpected role for HMGA2 in DNA repair in cancer cells which has important clinical implications for disease diagnosis and therapy.
INTRODUCTION
Mammalian high mobility group AT-hook (HMGA) proteins are non-histone chromatin factors encoded by two genes, HMGA1 and HMGA2. Alternative mRNA splicing gives rise to at least four protein isoforms which can be chemically modified (1–3). HMGA proteins are characterized by small sizes, an acidic C-terminal tail and the presence of three individual DNA-binding domains, so-called AT-hooks. Each hook is composed of nine amino acids containing an invariant RGRP repeat flanked by lysine and arginine residues. NMR analyses indicated that this domain is unstructured in solution and adopts a crescent-shaped configuration upon binding to the minor groove of short stretches of AT-rich DNA (4).
The biological roles of HMGA2 were initially studied in the mouse where it is expressed in embryonic stem (ES) cells and during embryogenesis. The protein is classified as a transcriptional regulator that modulates local chromatin structure near promoters critical for cell growth, spermatogenesis and adipogenesis. HMGA2 knock-out mice exhibit a pygmy phenotype with greatly reduced fat tissues, and male mice are infertile (5–8). In contrast, tissue-specific over-expression of full-length or ubiquitous expression of a truncated protein lacking the C-terminal tail resulted in gigantism, lipomatosis and mesenchymal tumors (9,10).
In humans, HMGA2 is not detected in normal adult somatic cells, but it is found in some fetal tissues (11). We have shown recently that HMGA2 remains associated with chromatin throughout the cell cycle in pluripotent human ES (hES) cells, and that expression levels are further elevated during a short time window in embryoid bodies (12). In addition, we demonstrated that HMGA2 is involved in the regulation of key human genes linked to mesenchymal cell lineage differentiation, adipogenesis and hES cell proliferation control (13).
Besides these critical roles in early mouse and human development, clinicians have known for some time that high HMGA protein levels are associated with most malignant human neoplasias, such as breast cancer, sarcomas, pancreas, oral squamous cell carcinomas and lung cancer (14–18). In addition, HMGA2 rearrangements are frequently found in benign tumors of mesenchymal origin (19–21). In fact, the HMGA genes are the only known proto-oncogenes coding for DNA-architectural chromosomal proteins consistently over-expressed in nearly all types of naturally occurring cancers. Their expression level correlates directly with the degree of malignancy and metastatic potential (3,21–23). In line with these observations, recent microarray analyses showed that both HMGA genes belong to a small class of genes whose expression is critical to the cancer phenotype in cells that carry two key oncogenic mutations in p53 and Ras (24).
The miRNAs let-7 and miRNA-98 are directly involved in the regulation of HMGA2 during oncogenic transformation (25,26). Interestingly, the let-7/HMGA2 linkage was recently identified as a candidate signature of cancer stem cells derived from primary human cancer tissues. These cells are resistant to a therapy that introduces chemical modifications of DNA bases, such as oxidation (27–29). There is evidence that HMGA2 is involved in cancer stem cell differentiation and proliferation control (27), but due to the pleiotropic functions of HMGA2 as a DNA-architectural chromatin factor; the molecular mechanism(s) which connect the let-7/HMGA2 linkage with chemotherapy-resistant cancer stem cells remain elusive. It is noteworthy here that the presence of HMGA1 is also functionally linked to chemoresistance of certain types of human carcinomas (30).
During our studies of DNA lyase activities associated with proteins belonging to the Escherichia coli DNABII family of DNA-binding proteins (unpublished result), we discovered that recombinant human HMGA proteins efficiently cleaved DNA containing apurinic/apyrimidinic (AP) sites in vitro. We present here a biochemical analysis of these associated lyase activities and demonstrate with cell-based experiments that HMGA2 is directly involved in base excision repair (BER) and protects cancer cells from DNA-damage that occur, for example, during chemotherapy. We discuss our findings with respect to genome stability in both cancer and hES cells, and point out important immediate clinical implications for cancer diagnosis and therapy.
MATERIALS AND METHODS
DNA substrates and proteins
Oligonucleotides (31-mers) containing uracil were obtained from AitBiotech, and 32P-labeled either at the 5′ end with T4 polynucleotide kinase and [γ32P]ATP (Perkin Elmer) or at the 3′ end with Klenow Fragment (3′-5′ exo−) and [α32P]ddATP. Double-stranded substrates were prepared by annealing the 32P-labeled uracil containing 31-mer with its complementary strand at a 1:1.5 stoichiometry in a buffer containing 20 mM Tris–HCl (pH 7.4), 50 mM NaCl. The AP site was generated by incubating the double-stranded DNA with an excess amount of uracil DNA glycosylase for 1 h at 37°C. The substrate for dRP lyase is the 3′-32P-labeled 31-mer containing an AP site which was pre-incubated with an excess of E. coli endonuclease IV, thus generating a 3′ end-labeled fragment (18-mer) containing a 5′phosphodexoyribosyl moiety. HMGA2 was expressed from pET17b-hHMGA2-His as previously described (12) and HMGA1a and b proteins were expressed from pET21a-HMGA1a/b-His vectors (a kind gift of Dr R. Hock). All HMGA proteins were expressed in E. coli BL21 (DE3) pLys strain harboring previously mentioned plasmids. Bacteria were grown in LB medium to OD600 nm > 0.8 and induced with 0.1 mM IPTG (1st Base) for 4 h. Cells were harvested by centrifugation (10 000 × g) for 20 min. Cell pellets were dissolved in Lysis buffer [300 mM NaCl, 10 mM Imidazole, 50 mM Na Phospate pH 8.0, protease inhibitor cocktail (Roche)] and sonicated. The lysate was centrifuged at 48 500 × g for 1 h. Cleared lysate was loaded on a HisTrap column (GE) and equilibrated with the same buffer. The proteins were eluted in a linear gradient with buffer A (300 mM NaCl, 250 mM Imidazole, 50 mM Na phosphate pH 8.0) between 100–150 mM Imidazole. The protein containing fractions were pooled together and diluted with 50 mM Na phosphate buffer pH 7.3 to have a final concentration of 100 mM NaCl. The protein was further purified by cation exchange chromatography on a HiTrap SP HP column (GE) equilibrated with buffer B (50 mM Na Phosphate pH 7.3, 100 mM NaCl, 5 mM EDTA). The protein was eluted running a linear gradient to 100% buffer C (same as B but 1 M NaCl) at around 300 mM NaCl. Protein containing fractions were confirmed via 15% reducing SDS–PAGE and Maldi-mass spectrometry. The purified proteins were dialysed against 50 mM Tris pH 8.0. Proteins were aliquoted, flash frozen and stored at −80°C. The yield ranged from 2–4 mg/l culture. Gel images were scanned using a Bio-Rad Quantity One GS-800 calibrated densitometer.
In vitro lyase assays
DNA cleavage assays employing a 5 kb supercoiled plasmid (pCMV-Int) were performed with either full-length HMGA2 or a peptide comprising AT-hook 3 (KRPRGRPRK). A 50 μl reaction contained either 9.18 nM abasic or normal DNA substrate (control), was incubated in a buffer made up of 25 mM Tris–HCl, pH 7.5; 2 mM EDTA; 100 mM NaCNBH3 with 4.92 μM HMGA2 at 37°C for 40 min. NaCl (100 mM) was used in place of NaCNBH3 in controls. Reactions were quenched by 2% SDS final concentration and heated at 65°C for 15 min. Samples were purified by Qiagen QIAquick® PCR Purification Kit. For each sample, elution used 25 μl pH 7.4 TE buffer containing 0.5% SDS. Cleavage was analyzed by agarose gel electrophoresis. To detect HMGA2–DNA complexes, DNA was subsequently transferred to a Nylon membrane. The presence of HMGA2 was detected by immunoblotting using anti-HMGA2 polyclonal antibodies (S15, Santa Cruz) at a 1:500 dilution, and bovine anti-goat HRP-conjugated secondary antibodies (Santa Cruz) at a 1:5000 dilution.
The nature of the 5′ and 3′ termini generated by HMGA2 on DNA containing an AP site was determined by comparing the electrophoretic mobility of products generated by HMGA2 with those created by E. coli enzymes Endo III, Endo IV, or Fpg. The AP lyase activity was assayed in a 10 μl reaction containing 1× reaction buffer (10 mM Tris–HCl, pH 7.5; 1 mM EDTA; 50 mM KCl) and 1.5 μM of 5′-32P-labeled 31-mer-AP substrate plus 15 μM of HMGA proteins or hook 3. For control reactions, eight units of Endonuclease III and FPG (NEB) was used. The Endocnuclease IV (Fermentas) reaction had two units in a 10 μl reaction. Additionally the HMGA and hook 3 reactions were treated afterwards for 15 min with Endo IV. Reactions were stopped after 30 min by the addition of 10 μl loading buffer (90% formamide, 1 mM EDTA, 0.1% xylene cyanol and 1% SDS) and heated at 70°C for 10 min. Total 5 μl of the reaction mixture were loaded onto a 12.5% denaturing polyacrylamide gel. For quantification, a 4% acrylamide-stacking gel was prepared and the substrate concentration was 0.5 μM. Radioactive bands were quantified with a Quantity one FX scanner (Bio-Rad).
In order to identify the nucleophile(s), various AT-hook 3 mutant peptides, as indicated in Figure 3D, were purchased from Research Biolabs or produced in-house. Each sample contained 1× reaction buffer, 0.5 μM peptide and 0.33 μM of the 31-mer, 5′-Cy5dye-labeled uracil-containing substrate (AIT Biotech), pretreated with UDG to generate the AP site as described above. Then, 10 μl reactions were incubated at 37°C for 45 min and stopped by adding 10 μl of gel-loading buffer (90% formamide, 1 mM EDTA, 10% glycerol and 1% SDS). An amount of 5 μl each reaction were heated to 70°C for 10 min and loaded on a 15% denaturing PAGE. Gels were scanned using a Typhoon Variable Mode Imager (GE) and data analyzed using Image Quant TL software (GE). The trapping experiments were performed identically to the AP cleavage assays, except that 50 mM of NaCNBH3 (final conc.) was included before the reaction was initiated by the addition of substrate.
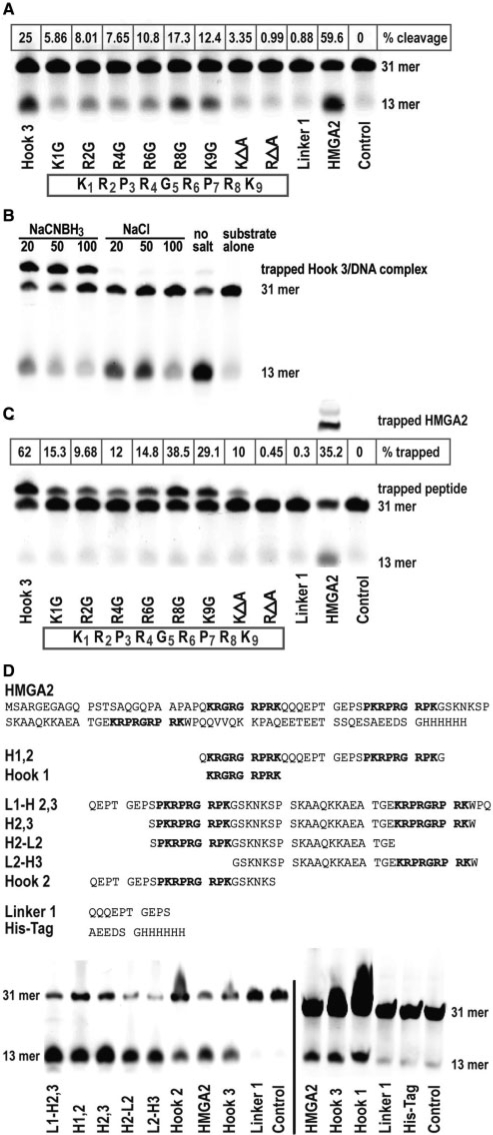
Figure 3.
Identification of nucleophiles in AT-hook 3. (A) Comparison of AP site cleavage activities of various hook 3 peptides with a linker peptide and full-length HMGA2. The hook 3 aa sequence with residues numbered is displayed in the box. The control reaction lacked any protein. The positions of the substrate (31-mer) and the cleavage product (13-mer) are indicated on the right side. The percentage cleavage was calculated based on the combined signal intensities from substrate and product (100%), after subtraction of the background value obtained with the control. (B and C) Trapping of peptides in a covalent intermediate complex with the 31-mer AP site substrate. Trapping conditions were first optimized in (B) with unmodified hook 3 and various concentrations of the trapping salt NaCNBH3, using NaCl as control. The percentage of trapped complexes was calculated in (C) based on the combined signal intensities from the 31-mer substrate and the shifted complex (100%), after subtraction of the background value obtained with the control. (D) Comparison of AP site cleavage activities of various peptides containing hook sequences. The aa sequences of full-length HMGA2 and of the peptides are shown on top of the cleavage assays. Note that we included two controls; a linker (Linker 1) and his-tag peptide.
The dRP lyase activity of HMGA proteins and hook 3 was assayed as described previously (31) with the following modifications: The 5′-dRP moiety was stabilized by the addition of O-benzyl hydroxylamine (BA) (Sigma) prior to electrophoresis. The formation of 5′-BA-dRP also allows for a better separation on a more conventional 16% denaturing polyacrylamide gel. The dRP lyase substrate was generated by preincubation of 3′-labeled 31-mer containing an AP site with two units of E. coli Endo IV for 15 min in 1× reaction buffer and 0.5 μM DNA. The dRP lyase activity was assayed by addition of 5 μM protein and incubated for 15 min at 37°C. The reaction was stopped by the addition of 1 µl 1M BA, followed by further incubation for 10 min. Modification of 5′-dRP by BA was completed within 1 min. Total 10 µl of loading buffer (90% formamide, 1 mM EDTA, 0.1% xylene cyanol, 10% glycerol and 1% SDS) was added and heated at 70°C for 10 min. An amount of 3 µl was loaded onto a 16% denaturing polyacrylamide gel. Radioactive bands were quantified with a Quantity one FX scanner (Bio-Rad).
Generation of HMGA2 cell lines
The hHMGA2 c-DNA was cloned from mRNA isolated from hES2 cells (12) and a histidine affinity tag affixed at the C-terminus by RT–PCR using primers (HMGA2Kpn1F) 5′-CGG GGT ACC ATG AGC GCA CGC GGT GAG GGC-3′ and (HMGA2HisEcoR1R) 5′-GGA ATT CTT ACT AGT GGT GGT GGT GGT GGT GAG CGC TGT CCT CTT CGG CAG ACT C-3′. The expression vector pEF1-hHMGA2-His-Neo was constructed by inserting the hHMGA2 coding sequence into pEF1-mycA-Neo (Invitrogen) using KpnI and EcoRI (New England Biolabs). DNA constructs were confirmed by sequencing. pEF1-hHMGA2-His-Neo was transfected into HeLa (ATCC No. CCL-2) cells using lipofectamine (Invitrogen) and into A549 (ATCC No. CCL-185) cells via electroporation. Neomycin resistant cells were selected at 500 µg/ml G418 and individual colonies were picked and tested for HMGA2 expression by western analysis.
Immunoblotting
Harvested cells were lysed in a NP40 buffer containing 1% SDS. Lysates were sonicated on ice (5 W; 10 × 3 s). Protein concentrations were determined with the BCA protein Assay Kit (Pierce Biotechnology). Lysates equivalent to 15 µg of total protein (or 1 × 105 cells) were analyzed through 15% SDS–PAGE and transferred to PVDF membrane with 20 µm pore size (Bio-Rad). The blot was blocked with 5% nonfat milk and incubated with goat polyclonal antibody against HMGA2 (S15, Santa Cruz) at a 1:500 dilution. Bound antibodies were detected by secondary anti-goat HRP-conjugated antibodies (Santa Cruz) and revealed with Lumi-light (Roche Diagnostic). β-actin was employed as an internal standard using monoclonal antibodies raised against human β-actin (Sigma).
Trapping of HMGA2 on genomic DNA
For in vitro trapping of HMGA2 on genomic DNA isolated from cells challenged with low pH, A549 cells were first treated on culture dishes with pH 2 in PBS, or physiological PBS as control, for 10 min at 37°C, followed by a wash with PBS. Cells were harvested in the same buffer and genomic DNA was purified with DNeasy Blood & Tissue Kit (Qiagen). An amount of 3 μg HMGA2 was incubated with 1 μg of DNA in a buffer containing 25 mM Tris–HCl, pH 7.5, 2 mM EDTA and 100 mM NaCNBH3 at 37°C for 40 min. DNA was purified with QIAquick® PCR Purification Kit (Qiagen) and spotted onto PVDF membrane for HMGA2 detection by immunoblotting. For in vivo trapping, adherent transgenic A549-HMGA2 (1.6) cells were first treated with PBS at pH 2 for 30 min at 37°C, followed by a wash with physiologic PBS containing 100 mM NaCNBH3 (Sigma), or 100 mM NaCl as a control. Cells were harvested in the same buffers and genomic DNA purified with DNeasy Blood & Tissue Kit (Qiagen). Purification involved SDS–based lysis with a short protease treatment for 10 min at 70°C. NaCNBH3 or NaCl was present during elution, and the same amount of genomic DNA was spotted onto a PVDF membrane for HMGA2 detection.
Cell survival assays
To challenge cells with low pH, cell lines were treated with PBS (pH 2) at 37°C for 6 min. HeLa and the respective transgenic cell lines were challenged at pH 3. Cells were allowed to recover under normal culture conditions for 24 h before harvest. Cells were treated with 100 mM hydroxyurea (Sigma) under culture conditions for 48 h before harvest. To challenge cells with paclitaxel (Sigma), A549 cell lines were treated with 50 nM paclitaxel for 48 h before harvest, while HeLa cell lines were treated with 20 nM paclitaxel for the same period. A549 cell lines were treated with 20 μM cisplatin (Sigma) for 48 h before harvest, and HeLa cell lines were exposed to 20 μM cisplatin for 24 h. To challenge cells with MMS (Sigma), A549 cell lines were treated with MMS for 1 h followed by a recovery period for 48 h before harvest. In some experiments, we added 5 mM BA (O-benzyl hydroxylamine) to the cell culture media throughout the recovery period. HeLa cell lines were treated with 4 mM MMS. After recovery, cells were trypsinized and harvested, including those from the culture media and wash buffer. Cells were collected at room temperature by centrifugation at 600 × g for 3 min, washed once with PBS and stained with Annexin-V fluorescence and Propidium Iodide (PI) using Annexin-V-FLUOS Staining Kit (Roche) according to the manufacturer's protocol. Cells were analyzed by FACS Calibur (BD). Cells with negative staining for both Annexin-V and PI were counted as life cells; those with positive staining for both markers were qualified as necrotic cells; and those with positive staining for Annexin-V, but negative staining for PI, counted as apoptotic cells.
In MMS challenge experiments with transient HMGA2 transfected cells, 5 × 104 A549 or HeLa cells were seeded in each well of a 24-well plate before 1 μg of pEF1-HMGA2-His vector was transfected. Forty-eight hours after transfection, cells were treated with various amount of MMS (0.5 and 1 mM for A549 cells; 0.2 and 0.5 mM for HeLa cells) for 1 h. Cells were then recovered for 24 h before being trypsinized. Total 2000 cells from each sample were counted and plated in a 10 cm culture dish and left untouched until single cell colonies were formed and become large enough to count. CMV-EGFP cells were used as controls in these experiments, and transfection efficiency was determined before MMS treatment by flow cytometry. Each experiment was performed in triplicate.
Protein co-affinity precipitation
A total of 2 × 107 of each A549, A549-HMGA2 (1.3), HeLa and HeLa-HMGA2 (P2) cells were collected and lysed with 0.5 ml native lysis buffer containing 50 mM Tris–Cl, pH 8.0, 150 mM NaCl, 0.1% NP-40, 2 mM DTT, 5% glycerol and protease inhibitors (Roche). Sonication was applied to each lysate at output power of 2 W for 15 × 5 s in iced water bath. Each sample was topped up to 1 ml with lysis buffer and centrifuged at 16 100 × g for 10 min at room temperature, and the supernatant was incubated overnight rolling at 4°C with 50 μl Ni+ sepharose beads (GE Healthcare) which was pre-incubated with lysis buffer containing 1% BSA. An amount of 50 μl supernatant was kept as cell lysate control. The beads were collected at 100 × g for 2 min at room temperature and washed with lysis buffer for 4 × 10 min with rolling. Elution was done with 50 μl of buffer containing 20 mM NaH2PO4, 20 mM Na2HPO4, 500 mM NaCl and 500 mM Imidazole, pH 8.0. The eluted material, together with the cell lysate controls were then analyzed on SDS–PAGE and later subjected to western blotting analysis for HMGA2 and APE1. Detection of HMGA2 was done as described above, and detection of APE1 was performed using rabbit polyclonal antibody against APE1 (Santa Cruz) followed by HRP-conjugated goat anti rabbit secondary antibody.
Comet assays
Cells (106 cells/ml) were collected in DMEM without serum. An amount of 100 µl cell suspension were mixed with 0.1 mM MMS (30 µl of 10 mM MMS in DMEM) and 2.87 ml serumfree medium for 2 h at 37°C. Cells were collected, washed in PBS, and resuspended in 10 µl PBS. The standard alkaline assay (pH > 13) for detecting DNA damage in single cells was performed according to the method developed by Singh (32), with minor modifications. Briefly, 105 cells/10 µl cell suspension in PBS was mixed with 50 µl of 0.7% low melting point agarose (LMP; Fulka), layered on a microscopic slide previously coated with 1% normal melting point agarose (NMA; Bio-Rad). After the agarose solidified, slides were immersed in pre-chilled lysis solution (2.5 M NaCl, 100 mM EDTA, 10 mM Tris, pH 10 and 1.5% Triton X-100) for 1 h at 4°C. Slides were then washed with distilled water and incubated in a horizontal gel electrophoresis tank with fresh alkaline electrophoresis buffer (300 mM NaOH, 1 mM EDTA, pH 13.5) for 40 min at 4°C. Electrophoresis was conducted at 4°C for 30 min at 25 V (0.8 V/cm) and 300 mA. Once completed, the gels were neutralized by three washes with 0.4 M Tris–HCl, pH 7.5 for 5 min each. After neutralization, the agarose gels were dehydrated by immersing the slides in absolute ethanol for 5 min, and stained overnight with DAPI (500 ng/ml) in PBS. Images were obtained at 400-fold magnification using a fluorescence microscope (Olympus). Fluorescence was visualized using a chroma filter at excitation 385 nm and emission 450 nm. Slides were analyzed by Comet Score Version 1.5 Software (http://www.autocomet.com/products_cometscore.php). DNA damage was quantified by Olive tail moment as introduced by Olive (33), which is defined as multiplying the total intensity of the comet tail by the tail length, measured from the centre of the comet head.
RESULTS
HMGA2 has an intrinsic AP lyase activity
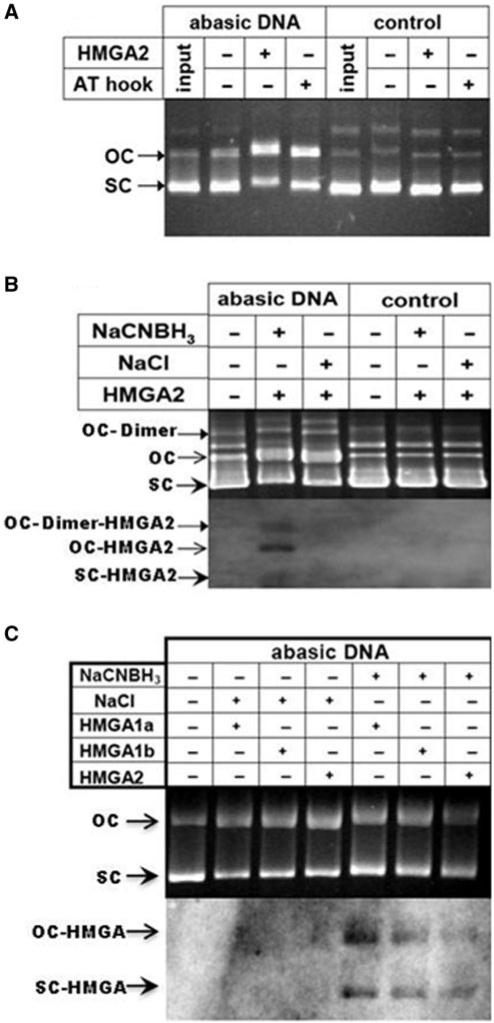
Our initial screen for AP lyase activities employed supercoiled plasmids which contained an undefined number of AP sites that resulted from depurination during exposure to acidic pH. Incubation with HMGA2 converted the majority of supercoiled AP plasmids into the open circular form, while control plasmids remained supercoiled (Figure 1A). If the observed cleavage of AP site-containing plasmids was due to an associated AP lyase activity, Schiff′base intermediates should have been generated by nucleophilic attack of a primary amine group to the aldehyde form of the AP site. Therefore, we tested whether covalent DNA–HMGA2 complexes could be trapped by the reducing agent NaCNBH3 (31). HMGA2 was incubated either with AP site plasmids or control substrates under trapping or non-trapping conditions, and reactions were stopped with SDS and heated to 65°C. DNA was resolved by agarose gel electrophoresis and transferred to a nylon membrane. Western blotting revealed that HMGA2 could be trapped during incubation only on AP plasmids, and that the protein could be detected on both the supercoiled and relaxed DNA forms, as expected for substrates with multiple AP sites (Figure 1B).
Figure 1.
HMGA2 acts as an AP lyase on abasic supercoiled DNA. (A) Cleavage assay with full-length HMGA2 or a peptide comprising the third AT-hook with abasic supercoiled (sc) DNA. AP site cleavage converts the topology of sc DNA into the open circular (oc) form. Note that depurinated input DNA is mostly sc and does not change during incubation without HMGA2. (B) Detection of in vitro trapped HMGA2 on AP site plasmid DNA using western blotting. The positions of HMGA2–DNA complexes detected on the western blot membrane are marked by arrows. (C) Cleavage and in vitro trapping assay with HMGA1a, HMGA1b and HMGA2 on abasic sc DNA.
Since the three independent AT-hooks of HMGA2 are mainly composed of arginine and lysine residues which penetrate deeply into the minor DNA grove, and are nearly identical in amino-acid sequence (4), we hypothesized that the primary amine(s) responsible for the cleavage activity likely resided within DNA-binding domains. We employed a short peptide comprising the third AT-hook of HMGA2, which exhibits the lowest DNA-binding affinity, and incubated it with AP site or control plasmids. As observed with the full-length protein, the peptide cleaved only AP site-containing plasmids (Figure 1A). These results indicate that a nucleophile(s) that attacks the C′1 of deoxyribose at AP sites could reside within the AT-hooks.
The first two AT-hooks are identical in HMGA1 and HMGA2 proteins, which led us to test whether the two main HMGA1 isoforms generated by alternative splicing, i.e. HMGA1a and HMGA1b, also exhibit AP lyase activities that can be trapped by NaCNBH3 (Figure 1C). Furthermore, data obtained with mass spectrometry on trapped HMGA2–plasmid complexes strengthen our conclusion that the primary amine(s) responsible for the lyase activity resides in the AT-hook (Supplementary Figure 1).
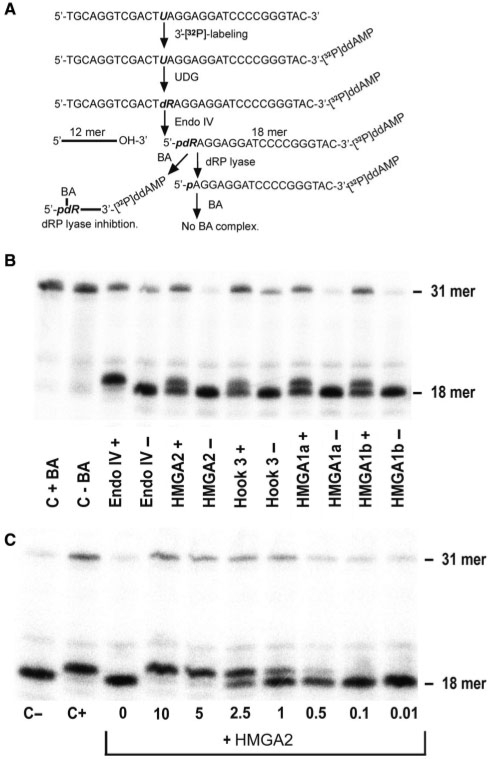
Our finding that HMGA1 and HMGA2 can be covalently trapped on AP substrates by NaCNBH3 indicated that the AP lyase-associated cleavage reaction employed β elimination (34). Hence, in order to determine the chemical nature of DNA ends generated by HMGA proteins and the efficiency of AP site cleavage, we employed a 32P-labeled double-stranded 31-mer oligonucleotide as substrate, which contained a single AP site (Figure 2A). The results show that incubation of purified HMGAs or a peptide comprising HMGA2 AT-hook 3 with the substrate generated cleavage products, which exhibit the same electrophoretic mobility as those produced by endonuclease III, an AP lyase derived from E. coli (Figure 2B and C, lanes 2–6). In addition, products generated by HMGA2 differed from those obtained with E. coli enzymes AP endonuclease IV, which produces 3′OH ends, and from those obtained with formamidopyrimidine N-glycosylase (Fpg), which generates 3′ phosphates during AP site cleavage (Figure 2C, lanes 7 and 12, respectively). Importantly, when we subsequently incubated 32P-labeled products generated by HMGA proteins with AP endonuclease IV, their mobility was increased. This indicates that the 4-hydroxy-2-pentenal moiety generated by HMGA's AP site cleavage activity had been removed from the 3′ end (Figure 2C, compare lanes 3–6 with lane 8–11). Incubation with increasing protein/DNA ratios and quantification of cleavage established that all 3 HMGA proteins have similar activities, which was higher as compared to the isolated AT-hook 3 from HMGA2 (Figure 2D).
Figure 2.
Characterization of the AP lyase activity. (A) Creation of AP substrate containing a single abasic site using uracil DNA glycosylase (UDG). (B) 15% denaturing SDS–PAGE. Four micrograms per lane of each protein was loaded onto the gel. (C) Determination of the chemical nature of DNA ends generated by HMGA2. Cleavage products obtained with the following E. coli proteins: formamidopyrimidine N-glycosylase (Fpg), endonucleases III (endo III) and IV (endo IV), as indicated, can be separated from substrates through denaturing polyacrylamide gel electrophoresis. The 31-mer incubated under the same conditions, but in the absence of protein is shown as control in lane 1. (D) Quantification of AP lyase activities. After electrophoresis and autoradiography, bands corresponding to substrate and products were quantified and plotted against the molar protein to DNA ratio.
Nucleophiles responsible for AP site cleavage reside within AT-hooks
Our data indicate that HMGA proteins possess AP site cleavage activity which involves β elimination. This activity resides within the DNA-binding domain, the AT-hook 3. In an attempt to identify the nucleophile(s) within the hook, we employed a series of peptides according to the peptide sequence of the AT-hook 3 in which each of the lysine or arginine residues was individually replaced by glycine and tested them for cleavage activity using the same 31-mer substrate which had been 5′-Cy5dye-labeled. The result of a representative experiment showed that compared with the unmodified hook 3, substitution of the lysine at position one (K1) substantially reduced cleavage from 25% to about 5.8% (Figure 3A). With the exception of the arginine at position eight (R8), substitutions of arginines R2, R4 and R6 also led to substantially diminished cleavage activities (Figure 3A). The importance of arginines for AP site cleavage was further demonstrated with two additional peptides in which either all arginines or lysines were replaced by alanines. While the latter still catalyzed residual cleavage (3.3%), the activity of these peptides lacking arginines was reduced to that observed with a control peptide representing the linker connecting AT-hook 1 and 2 in HMGA2 (Figure 3A).
These results suggest that the lysine at the N-terminus of the hook might be the crucial nucleophile responsible for the main cleavage activity. However, they also hint at an important role for arginines, which are crucial for DNA binding (4). In order to investigate this further, we employed the same set of peptides under trapping conditions and demonstrated first that the majority of substrate molecules was trapped in a covalent linkage with the hook 3 peptides at 50 mM NaCNBH3 (Figure 3B). Substitutions of residues K1, R2, R4 or R6 substantially reduced but did not completely eliminate complex formation (Figure 3C). Likewise, the peptide lacking lysines still generated a substantial amount of trapped complexes. Compared with the linker peptide used as control, only the peptide without arginines failed to generate detectable complexes, which, taken together, is in agreement with the AP site cleavage data shown in Figure 3A. We conclude that the AT-hook contains more than one nucleophile capable of attacking AP sites, and arginines, in addition to the N-terminal lysine, must be involved.
We have shown so far that hook 3 has AP lyase activity. We next tested whether hooks 1 and 2, which deviate slightly from hook 3, also exhibit this cleavage activity. The results obtained with a second set of peptides clearly demonstrated that this is indeed the case (Figure 3D). In addition, using a peptide representing the histidine tag at the C-terminus of our recombinant HMGA2, we also show that histidines have no AP site cleavage activity under these conditions (Figure 3D, right panel).
HMGA2 is also a dRP lyase
Having established that HMGA2 is an AP lyase, we next examined whether this protein also possesses the related 5′-deoxyribosyl phosphate (dRP) lyase activity. To test this possibility, we employed the same 31-mer oligonucleotide containing a single abasic site. However, the substrate was 3′-labeled and pre-incubation with endonuclease IV produced a 5′-dRP moiety on the labeled strand (Figure 4A). In order to stabilize the chemically labile deoxyribosyl phosphate group and increase separation of cleaved from non-cleaved dRP lyase products during electrophoresis; the dRP moiety was reacted with BA. The presence of BA-adducts leads to retardation during electrophoresis when compared with products that lost the dRP moiety due to a dRP lyase activity (Figure 4B, lanes 3–4). Using this assay, we established that HMGA proteins and the AT-hook 3 peptide efficiently removed the dRP moiety. Thus, HMGA proteins act as a dRP lyase (Figure 4B, lanes 5–12). By contrast the linker peptide connecting HMGA2 hooks 1 and 2 was unable to cleave dRP substrates (data not shown).
Figure 4.
Characterization of the dRP lyase activity. (A) Strategy for the creation of the dRP lyase substrate and expected cleavage products. (B) dRP lyase activity assay. The electrophoretic mobility of the 18-mer substrate containing the dRP moiety is retarted due to BA-adduct formation. The observed shift and visible double band verified that the HMGA proteins and hook 3 cleaved the substrate. (C) Quantitative in vitro analysis of BA as an inhibitor of the HMGA2 lyase activity. BA was first added to the reaction for 5 min, followed by HMGA incubation for 15 min.
Once a genomic AP site is generated, the next step in BER involves AP endonucleases and/or AP/dRP lyases which cleave the DNA backbone at the base lesion. However, the tautomeric open-ring form of deoxyribose produced by DNA glycosylases can react very fast with BA (Figure 4B). It has been shown that BA-adducts inhibit DNA cleavage by mammalian AP endonucleases and AP lyases (35), and we confirmed that 5 mM BA completely blocked the dRP lyase activity of HMGA2 in vitro (Figure 4C).
HMGA2 can be covalently trapped at genomic abasic sites in vivo
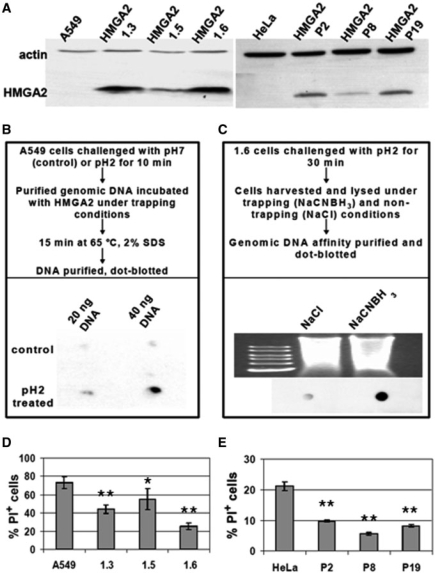
After demonstrating that HMGA2 is an AP and a dRP lyase, we investigated whether these BER functions play a bona fide role in human cervical and lung cancer cells, which exhibit a much reduced expression level of p53 and contain an oncogenic mutation in RAS, respectively. We employed HeLa and A549 cells, and first generated stable transgenic cell lines which constitutively expressed HMGA2. Western blot analysis showed that HMGA2 fell below the detection limit in extracts prepared from parental cells, but was present in transgenic cells. Since these were generated by random genome insertion of expression vectors, we chose three transgenic lines of each parental line for further analysis to eliminate the possibility that a particular phenotype is due to insertional mutagenesis (Figure 5A).
Figure 5.
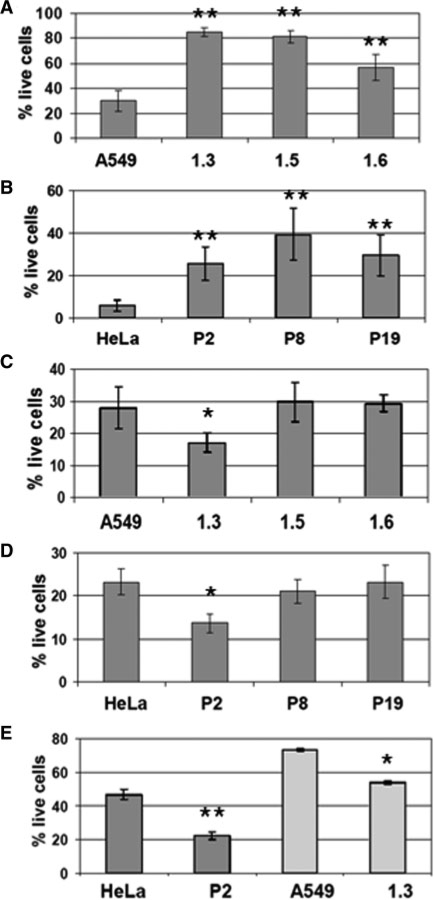
Transgenic HMGA2-overexpressing cancer cell lines and cell viablity after depurination at low pH. (A) Western blot analysis. HMGA2 transgenic cells (A549 derived 1.3, 1.5 and 1.6; and HeLa derived P2, P8 and P19) show various expression levels, using β-actin as internal control. (B) Dot-blot showing in vitro trapping of HMGA2 on purified A549 genomic DNA after depurination in vivo. The experimental set-up is described in the upper panel. (C) Dot-blot showing in vivo trapping of HMGA2 on 1.6 cell genomic DNA due to depurination. The experimental set-up is described in the upper panel. A control for the amount of DNA loaded is shown above the blot. (D) FACS analysis of A549 necrotic cells (PI+) determined after 6 min depurination at pH 2 and 24 h recovery. Data analyzed with Student's t-test. *P < 0.005. **P < 0.001. (E) FACS analysis of HeLa necrotic cells (PI+) determined after 6 min depurination at pH 3 and 24 h recovery. Bars represent mean values with standard deviations obtained from biological triplicates. Data analyzed with Student's t-test. **P < 0.001.
To investigate whether HMGA2 can be covalently trapped at genomic AP sites using NaCNBH3, parental A549 cells were first challenged with low pH or physiological pH as control. DNA purified from these cells was then incubated with HMGA2 under trapping conditions, affinity purified and dot-blotted. The results show that HMGA2 could only be detected in complex with genomic DNA isolated from cells that were challenged with low pH (Figure 5B). This demonstrates that AP sites introduced in vivo can be cleaved by HMGA2 in vitro. In order to directly show that HMGA2 was trapped at AP sites in vivo, we employed A549 (1.6) cells and exposed them to low pH. Treated cells were morphologically indistinguishable from untreated controls, and were harvested and lysed at neutral pH under either trapping or non-trapping conditions. Purified DNA was dot-blotted and the results clearly showed that a significant amount of HMGA2–DNA complexes was detectable only with genomic fragments harvested under trapping conditions (Figure 5C). We conclude that cells exposed to low pH challenge must contain a significant number of genomic AP sites that can be cleaved by HMGA2 in vitro and in vivo.
We next analyzed cytotoxic effects that might result from depurination in parental and transgenic cells, and challenged them for 6 min with low pH. After recovery, FACS analysis of necrotic cells revealed that all transgenic cell lines were substantially more resistant to low pH challenge than parental cells (Figure 5D and E).
HMGA2 displays compound selectivity to protect cancer cells from genotoxicants
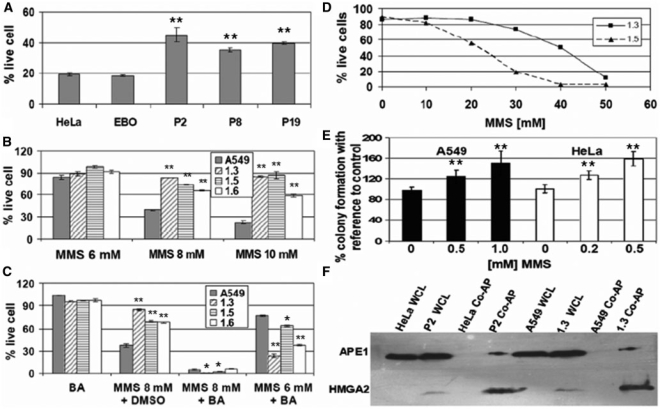
Hydroxyurea (Hu) is a frequently used chemotherapeutic agent for the treatment of proliferative disorders and solid tumors, and is able to induce base oxidation and depurination (36). We exposed parental and transgenic cells to Hu for a period of 48 h, and determined the fraction of live cells using FACS analysis. Expression of HMGA2 resulted in significant protection against cell death, leading to a 2–8-fold increase in cell survival (Figure 6A and B). In order to demonstrate that transgenic cells were not impaired per se in entering apoptosis, we next exposed cells to the chemotherapeutic agent paclitaxel, which is an inhibitor of mitosis and targets tubulin and the Bcl-2 oncoprotein, instead of DNA (37). When compared with parental cell lines, none of the HMGA2 transgenic lines displayed a protective phenotype (Figure 6C and D).
Figure 6.
Cell viability of parental and HMGA2 expressing cancer cell lines after challenge with various compounds. Percentage of live cells was determined by FACS after staining with Annexin-V fluorescence and Propidium Iodide. Data analyzed with Student's t-test. *P < 0.005. **P < 0.001. (A and B) Cells exposed to 100 mM Hydroxyurea for 48 h. (C) A549 cells and derivatives exposed treated with 50 nM paclitaxel for 48 h. (D) HeLa cells and derivatives treated with 20 nM paclitaxel for 48 h. (E) Cisplatin treatment. Bars charts represent mean values with standard deviations indicated and obtained from biological assays in triplicate.
Expression of HMGA2 was recently shown to increase the cytotoxic effects of DNA double strand breaks induced by certain topoisomerase type II inhibitors and the chemotherapeutic agent cisplatin (38). Cisplatin DNA cross-links are mainly repaired by nucleotide excision repair (NER), and not through BER. We therefore tested whether our HMGA2 transgenic lines exhibited a similar phenotype when treated with cisplatin, and the results showed that this is indeed the case (Figure 6E). Hence, the protective effects exerted by HMGA2 seem to be limited to particular repair pathways.
HMGA2-mediated cellular protection from MMS is blocked by lyase inhibition
Both AP and dRP lyase activities play central roles in the initial steps of BER. The DNA methylating agent methyl methanesulphonate (MMS) has been used frequently to study this particular repair pathway in mammalian cells. MMS produces genomic AP sites through the action of DNA glycosylases, which remove the chemically modified bases. In a first set of experiments, we exposed parental HeLa and the respective transgenic cells to 4 mM MMS for 1 h, followed by 48 h recovery before FACS. We included here a second HeLa cell line as a control (EBO) which, like the HMGA2 transgenic lines, expressed a neomycin marker but lacked the HMGA2 transgene. The results show that the presence of HMGA2 led to a significant increase in the number of live cells in all transgenic cell lines (Figure 7A). Likewise, treatment of parental and transgenic A549 cells with various MMS concentrations revealed that HMGA2 always conferred strong protection against MMS-induced cytotoxicity (Figure 7B). This result was confirmed by colony forming assays after MMS treatment (Supplementary Table 1).
Figure 7.
HMGA2 expression confers resistance against MMS which can be reversed by co-treatment with the lyase inhibitor BA, and physical interaction of HMGA2 with APE 1. (A) Percentage of live cells determined by FACS after challenging HeLa cells and derivatives with 4 mM MMS for 1 h, followed by 48 h recovery. Data analyzed with Student's t-test. **P < 0.001. (B and C) Percentage of live cells determined by FACS after challenging A549 cells with MMS at the indicated concentrations for 1 h, followed by 48 h recovery. Data were normalized to untreated parental and transgenic cells (100%). BA was added to an additional set of wells during recovery, and DMSO, the solvent for BA, was added to a control set. Note that BA alone does not affect cell viability. Data analyzed with Student's t-test. *P < 0.005. **P < 0.001. (D) Percentage of live cells determined by FACS after challenging 1.3 and 1.5 cell lines with increasing concentration of MMS for 1 h, followed by 48 h recovery. (E) Percentage of surviving colonies from HMGA2-transfected cells upon treatment with MMS, with reference to cells transfected with control vector. Data analyzed with Student's t-test. **P < 0.001. (F) Western blotting analysis of co-affinity precipitation of cellular APE1 via HMGA2-His in transgenic cells (lanes 3, 4, 7 and 8). Whole cell lysates (wcl) were used as controls (lanes 1, 2, 5 and 6).
In order to demonstrate that protection from MMS-induced DNA damage observed with transgenic cells involves HMGA2 lyase activities, we inhibited these early steps in BER by sequential exposure to MMS and BA. The results show that BA alone had no effect on the survival of parental or transgenic A549 cells (Figure 7C). However, pre-treatment with 6 mM or 8 mM MMS followed by exposure to BA not only negated the protection seen with transgenic cells but sensitized these cells to MMS treatment (Figure 7B and C). This result was confirmed by colony forming assays (Supplementary Table 2).
We noted that the degree of protection against MMS did not directly correlate with HMGA2 levels. We reasoned that this might be due to the fact that even in transgenic A549 (1.5) cells, which express HMGA2 at a comparatively low level, the intracellular amount of HMGA2 protein might be sufficient to efficiently cope with the number of AP sites generated by 10 mM MMS. If so, differences in protection against MMS between low- and high-level HMGA2 cells might become apparent only at higher MMS doses. We therefore exposed A549 (1.5) and A549 (1.3) cells to increasing concentrations of MMS and measured cell survival as before. The results clearly reveal that the protective effect directly correlates with HMGA2 expression at MMS concentrations above 10 mM (Figure 7D).
To confirm that the protective effect against MMS is not due to the long-term expression of HMGA2 in our transgenic cell models, colony-forming assays were performed with A549 and HeLa cells exposed to MMS challenge 48 h after transient transfection with HMGA2 expression vectors. HMGA2 transfected cells gave rise to at least 20% more colonies as compared to CMV-EGFP-transfected control cells and this difference increases with higher concentration of MMS (Figure 7E). In addition, these differences originated from the 60% to 70% of cells successfully transfected, as judged by FACS using CMV-EGFP-transfected cells before MMS treatment (data not shown). Together, these results showed that even a transient presence of HMGA2 protects cells against MMS-induced DNA damage.
HMGA2 interacts with the BER protein APE1
We next tested the possibility whether HMGA2 interacts with the cellular BER machinery in vivo. In this context, it is worth noting that HMGA2 physically interacts with DNA repair proteins AP endonuclease 1 (APE1) and Ku70 in vitro (39). The former is a major BER component in human cells. We used Ni+ sepharose co-affinity precipitation and were able to detect APE1 signals enriched in 1.3 and P2 cell lysates under native conditions, whereas no signal was detected in lysates from parental cells lacking tagged HMGA2 (Figure 7F).
HMGA2 protects cells against MMS-induced DNA strand breaks
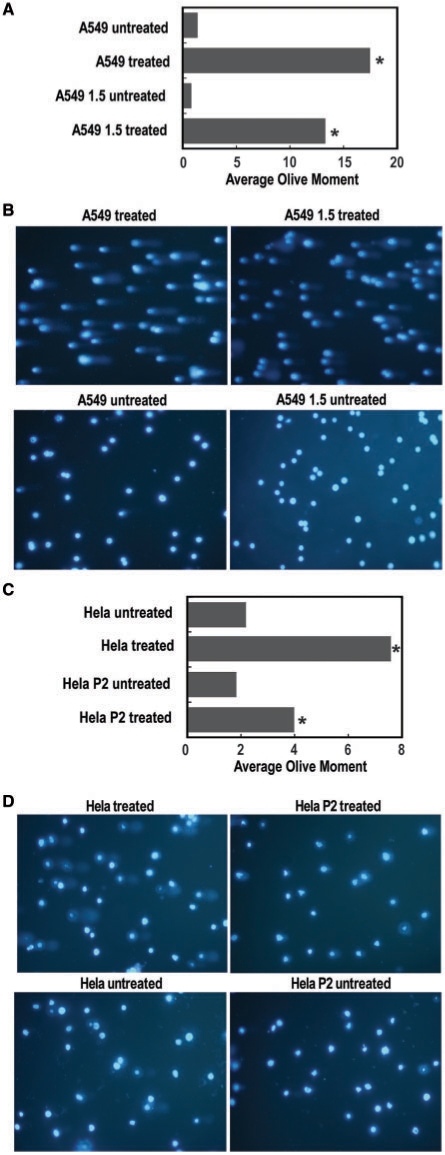
The protective effect of HMGA2 against MMS-induced cytotoxicity and its reversal by the inhibition of AP site processing through BA aducts indicated that the protein is directly involved in BER. Thus, HMGA2 should reduce the number of genomic DNA strand breaks induced by MMS. To demonstrate this directly, we performed Comet assays comparing parental A549 with transgenic 1.5 cells, and parental HeLa with P2 cells after treatment with 0.1 mM MMS in suspension culture for 2 h without a subsequent recovery period. The results clearly show that HMGA2 significantly reduced the extent of DNA damage in both transgenic cell lines as measured by tail moments (Figure 8).
Figure 8.
Quantification of DNA damage via Comet assays. (A and C) The Olive tail moment from every cell present in a given sample image was determined until 100 cells were scored from each sample. The average tail moment values were then calculated for each sample and comparisons between the two treated and untreated samples analyzed with Student's t-test, which revealed significant differences only between treated samples (P < 0.05). (B and D) Representative images obtained with treated and untreated A549 and HeLa samples, respectively, as indicated.
DISCUSSION
We show in this study that HMGA2 and the homologous HMGA1 proteins have intrinsic dRP/AP lyase activities. Furthermore, we identified the N-terminal lysine within the DNA-binding domains as a crucial nucleophile that attacks AP sites during β elimination. Neighboring arginines can fulfill this function as well. Substitutions within hooks which could compromise or eliminate lyase activities would almost certainly also affect DNA binding and, in turn, the functions of HMGA2 as a DNA-architectural chromatin factor. Based on the fact that the AT-hooks are almost identical in sequence, substitutions would have to be introduced into all three hooks simultaneously. In addition to DNA binding, some of these substitutions may also interfere with nuclear import and HMGA2–pRB protein interactions (20,40). Hence, a mutational screen did not seem a productive approach to distinguish the lyase activities from the other critical biological functions of HMGA2.
We presented here several lines of evidence that the lyase activities do indeed have important biological functions and play a direct role in conferring resistance against certain DNA-damaging agents. First, HMGA2 was efficiently trapped in a covalent complex with genomic AP sites in vivo. This indicated that the HMGA2 protein recognizes and cleaves AP sites within the background of chromatin. Subsequent to MMS challenge, treatment with BA sensitized transgenic cells to MMS, thus negating the protection that was observed when cells were exposed to MMS only. BA reacts with the deoxyribose moiety at AP sites and inhibits mammalian AP endonuclease and AP/dRP lyase activities. We showed that HMGA2 interacts with APE1 and conclude that HMGA2 confers resistance to MMS and other AP site-inducing genotoxicants through a role in BER, which at least in part, involves HMGA2 lyase activities. This protective effect is directly correlated with the expression level of HMGA2 at elevated MMS concentrations and protection was also observed when HMGA2 was only transiently expressed. Further evidence for an involvement of HMGA2 in BER repair stems from the result of the Comet assays. Expression of HMGA2 significantly reduced the number of genomic DNA strand breaks after MMS treatment.
The steady-state number of AP sites in the genome of mammalian cells is at least 10 000. Most of these sites appear to be processed quickly by APE1, which generates the 5′-dRP moiety (41). Its removal by dRP lyase seems to be the rate-limiting step in single nucleotide BER. Consequently, the level of dRP lyase activity will directly affect the cellular repair capacity and sensitivity against DNA damage that leads to AP sites (42). The level of DNA polymerase β, the main dRP lyase activity in human cells, varies substantially in human cancer cells (43). This scenario is in agreement with our finding that an elevated level of HMGA2 protects cancer cells from DNA-damage-induced cytotoxicity. Mechanistically, BER involves protein-protein and protein–DNA interactions, which are most likely coordinated to some extent. It is clear that the level of other BER factors, such as OGG1, APE1, or the scaffold protein XRCC1, may also affect the repair capacity in a situation where there is an abundance of dRP lyase molecules (44,45). In agreement with a recent report on HMGA2 and APE1 interaction in vitro (39), we demonstrate here that both proteins also interact with each other inside cells.
Exposure of cells to the chemotherapeutic agent Hu has been shown to induce base oxidation and depurination (36). However, the main cytotoxic effect of Hu is attributed to the inhibition of the enzyme ribonucleotide reductase, which, in turn, inhibits DNA synthesis during replication and repair (46). Our results showed that HMGA2 protects cells from Hu-induced cytotoxicity and it is conceivable that this involves HMGA2's novel activity in AP site processing during BER. However, we were surprised by the magnitude of protection. This might indicate that HMGA2 could participate in other aspects of genome stability; for example, rescue of stalled replication forks, or the chemical processing of single strand breaks which will also result from Hu treatment.
Our data showed that HMGA2 confers cellular protection to three different genotoxicants, i.e. Hu, MMS and low pH. This is not a consequence of the role of HMGA2 as a chromatin factor and transcriptional regulator which could affect the ability of transgenic cells to enter apoptosis. Exposure to the chemotherapeutic agent paclitaxel revealed no difference in cytotoxicity compared with parental cells. In addition, expression of HMGA2 was recently shown to increase the cytotoxic effects of DNA double strand breaks induced by certain topoisomerase inhibitors and the chemotherapeutic agent cisplatin (38). The latter effect was attributed to the inhibition of the nucleotide excision repair (NER) gene ERCC1, which is negatively regulated by HMGA2 (47). It was furthermore hypothesized that HMGA2 masks certain DNA lesions from recognition by the NER machinery (48). We confirmed in this study that HMGA2 sensitized cancer cells to cisplatin, thus providing further proof that the ability of our transgenic cell models to enter apoptosis was not compromised. Taken together, a major conclusion from our data is that even transient expression of HMGA2 has a direct supporting role in BER, which involves dRP/AP lyase activities. This leads to resistance of cancer cells against certain genotoxicants. In contrast, the presence of HMGA2 inhibits NER and, thereby, sensitizes cells to a different class of compounds.
It is well established that unrepaired AP sites are mutagenic (49). Since HMGA2 is found at high levels in many human neoplastic and pluripotent hES cells, the interesting question emerges whether HMGA2, through its associated lyase activities, contributes to lower mutation rates. From an evolutionary point of view, this is obviously most important for metabolically active ES cells which replicate and transcribe their genomes at a high rate and eventually produce germ cells. In fact, recent data showed that genes involved in different types of DNA repair are significantly upregulated in hES cells when compared with differentiated progeny cells (50). We found that the expression level of HMGA2 in hES cells is comparable with that of our transgenic A549 (1.5) and HeLa (P8) cells (Li and Dröge, unpublished results). In this context it is perhaps interesting to note that HMGB1, HMGN1 and HMGN2 proteins are also expressed in hES cells (12). Prasad et al. (31) recently showed that HMGB1 possesses dRP lyase activity, and we detected very robust AP and dRP lyase activities for HMGN1 and HMGN2 (Summer and Dröge, unpublished results). It emerges, therefore, a possible scenario in which these abundant non-histone chromatin-modifying factors play a hitherto unrecognized, direct role in the maintenance of genome stability.
Our finding that HMGA2 protects cancer cells from certain DNA-damaging agents used in cancer treatment has important implications for disease diagnosis, choice of treatment regimens, and the future development of anti-cancer drugs (51). First, we think it will now become even more important than previously suggested (21) to type malignant tumors, and, whenever possible, the replenishing cancer stem cell compartment with respect to the HMGA2 status. This knowledge will impact on the choice of the most suitable chemotherapeutic agents. Since we have shown that HMGA1 isoforms also exhibit dRP/AP lyase activities in vitro, we anticipate that the protective effect observed for HMGA2-expressing cancer cells will also be detectable for tumors that show an elevated level of HMGA1 proteins. In that respect, it is noteworthy that HMGA1 was recently shown to belong to a class of proteins that are specifically phosphorylated upon ATM/ATR activation through DNA damage (52). In the future it will be important to investigate how posttranslational modifications affect the newly discovered lyase activities of HMGA proteins.
We have provided first evidence obtained with MMS and BA treatment that a combination therapy, which targets HMGA2's role in BER and includes established genotoxicants could prove particularly effective against cancer cells otherwise resistant to chemotherapy. Secondly, anti-cancer therapy targeting HMGA2 will likely not affect normal adult cells which lack detectable HMGA2 levels, but exclusively target HMGA2 expressed in malignant cells. This novel class of anti-cancer compounds could act by interfering with DNA-binding (21) and/or protein–protein interactions in BER involving HMGA2.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Singapore Academic research council (ARC) [grant number 90/07] and NTU graduate scholarships to PD. NSERC (Natural Sciences and Engineering Research Council of Canada) and MHRC (Manitoba Health Research Council) (to T.K.). Funding for open access charge: Academic research council 90/07.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Randong Yuan for help with MS analyses, Yoga Dirga Cahya for help with colony forming assays, and R. Hock for HMGA1 expression vectors. Very special thanks go to Gabriela and Curt Davey for critical reading of the manuscript and the provision of several peptides.
REFERENCES
- 1.Bustin M, Reeves R. High-mobility-group chromosomal proteins: architectural components that facilitate chromatin function. Prog. Nucleic Acid Res. Mol. Biol. 1996;54:35–100. doi: 10.1016/s0079-6603(08)60360-8. [DOI] [PubMed] [Google Scholar]
- 2.Reeves R. Molecular biology of HMGA proteins: hubs of nuclear function. Gene. 2001;277:63–81. doi: 10.1016/s0378-1119(01)00689-8. [DOI] [PubMed] [Google Scholar]
- 3.Cleynen I, van de Ven WJ. The HMGA proteins: a myriad of functions (Review) Int. J. Oncol. 2008;32:289–305. [PubMed] [Google Scholar]
- 4.Huth JR, Bewley CA, Nissen MS, Evans JN, Reeves R, Gronenborn AM, Clore GM. The solution structure of an HMG-I(Y)-DNA complex defines a new architectural minor groove binding motif. Nat. Struct. Biol. 1997;4:657–665. doi: 10.1038/nsb0897-657. [DOI] [PubMed] [Google Scholar]
- 5.Zhou X, Benson KF, Ashar HR, Chada K. Mutation responsible for the mouse pygmy phenotype in the developmentally regulated factor HMGI-C. Nature. 1995;376:771–774. doi: 10.1038/376771a0. [DOI] [PubMed] [Google Scholar]
- 6.Hirning-Folz U, Wilda M, Rippe V, Bullerdiek J, Hameister H. The expression pattern of the Hmgic gene during development. Genes Chromosomes Cancer. 1998;23:350–357. [PubMed] [Google Scholar]
- 7.Chieffi P, Battista S, Barchi M, Di Agostino S, Pierantoni GM, Fedele M, Chiariotti L, Tramontano D, Fusco A. HMGA1 and HMGA2 protein expression in mouse spermatogenesis. Oncogene. 2002;21:3644–3650. doi: 10.1038/sj.onc.1205501. [DOI] [PubMed] [Google Scholar]
- 8.Caron L, Bost F, Prot M, Hofman P, Binetruy B. A new role for the oncogenic high-mobility group A2 transcription factor in myogenesis of embryonic stem cells. Oncogene. 2005;24:6281–6291. doi: 10.1038/sj.onc.1208781. [DOI] [PubMed] [Google Scholar]
- 9.Battista S, Fidanza V, Fedele M, Klein-Szanto AJ, Outwater E, Brunner H, Santoro M, Croce CM, Fusco A. The expression of a truncated HMGI-C gene induces gigantism associated with lipomatosis. Cancer Res. 1999;59:4793–4797. [PubMed] [Google Scholar]
- 10.Zaidi MR, Okada Y, Chada KK. Misexpression of full-length HMGA2 induces benign mesenchymal tumors in mice. Cancer Res. 2006;66:7453–7459. doi: 10.1158/0008-5472.CAN-06-0931. [DOI] [PubMed] [Google Scholar]
- 11.Gattas GJ, Quade BJ, Nowak RA, Morton CC. HMGIC expression in human adult and fetal tissues and in uterine leiomyomata. Genes Chromosomes Cancer. 1999;25:316–322. [PubMed] [Google Scholar]
- 12.Li O, Vasudevan D, Davey CA, Droge P. High-level expression of DNA architectural factor HMGA2 and its association with nucleosomes in human embryonic stem cells. Genesis. 2006;44:523–529. doi: 10.1002/dvg.20242. [DOI] [PubMed] [Google Scholar]
- 13.Li O, Li J, Droge P. DNA architectural factor and proto-oncogene HMGA2 regulates key developmental genes in pluripotent human embryonic stem cells. FEBS Lett. 2007;581:3533–3537. doi: 10.1016/j.febslet.2007.06.072. [DOI] [PubMed] [Google Scholar]
- 14.Rogalla P, Drechsler K, Kazmierczak B, Rippe V, Bonk U, Bullerdiek J. Expression of HMGI-C, a member of the high mobility group protein family, in a subset of breast cancers: relationship to histologic grade. Mol. Carcinog. 1997;19:153–156. doi: 10.1002/(sici)1098-2744(199707)19:3<153::aid-mc2>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 15.Berner JM, Meza-Zepeda LA, Kools PF, Forus A, Schoenmakers EF, Van de Ven WJ, Fodstad O, Myklebost O. HMGIC, the gene for an architectural transcription factor, is amplified and rearranged in a subset of human sarcomas. Oncogene. 1997;14:2935–2941. doi: 10.1038/sj.onc.1201135. [DOI] [PubMed] [Google Scholar]
- 16.Abe N, Watanabe T, Suzuki Y, Matsumoto N, Masaki T, Mori T, Sugiyama M, Chiappetta G, Fusco A, Atomi Y. An increased high-mobility group A2 expression level is associated with malignant phenotype in pancreatic exocrine tissue. Br. J. Cancer. 2003;89:2104–2109. doi: 10.1038/sj.bjc.6601391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miyazawa J, Mitoro A, Kawashiri S, Chada KK, Imai K. Expression of mesenchyme-specific gene HMGA2 in squamous cell carcinomas of the oral cavity. Cancer Res. 2004;64:2024–2029. doi: 10.1158/0008-5472.can-03-1855. [DOI] [PubMed] [Google Scholar]
- 18.Meyer B, Loeschke S, Schultze A, Weigel T, Sandkamp M, Goldmann T, Vollmer E, Bullerdiek J. HMGA2 overexpression in non-small cell lung cancer. Mol. Carcinog. 2007;46:503–511. doi: 10.1002/mc.20235. [DOI] [PubMed] [Google Scholar]
- 19.Subramanian D, Griffith JD. Interactions between p53, hMSH2-hMSH6 and HMG I(Y) on Holliday junctions and bulged bases. Nucleic Acids Res. 2002;30:2427–2434. doi: 10.1093/nar/30.11.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fedele M, Visone R, De Martino I, Troncone G, Palmieri D, Battista S, Ciarmiello A, Pallante P, Arra C, Melillo RM, et al. HMGA2 induces pituitary tumorigenesis by enhancing E2F1 activity. Cancer Cell. 2006;9:459–471. doi: 10.1016/j.ccr.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 21.Fusco A, Fedele M. Roles of HMGA proteins in cancer. Nat. Rev. Cancer. 2007;7:899–910. doi: 10.1038/nrc2271. [DOI] [PubMed] [Google Scholar]
- 22.Tallini G, Dal Cin P. HMGI(Y) and HMGI-C dysregulation: a common occurrence in human tumors. Adv. Anat. Pathol. 1999;6:237–246. [PubMed] [Google Scholar]
- 23.Di Cello F, Hillion J, Hristov A, Wood LJ, Mukherjee M, Schuldenfrei A, Kowalski J, Bhattacharya R, Ashfaq R, Resar LM. HMGA2 participates in transformation in human lung cancer. Mol. Cancer Res. 2008;6:743–750. doi: 10.1158/1541-7786.MCR-07-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McMurray HR, Sampson ER, Compitello G, Kinsey C, Newman L, Smith B, Chen SR, Klebanov L, Salzman P, Yakovlev A, et al. Synergistic response to oncogenic mutations defines gene class critical to cancer phenotype. Nature. 2008;453:1112–1116. doi: 10.1038/nature06973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hebert C, Norris K, Scheper MA, Nikitakis N, Sauk JJ. High mobility group A2 is a target for miRNA-98 in head and neck squamous cell carcinoma. Mol. Cancer. 2007;6:5. doi: 10.1186/1476-4598-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, Huang Y, Hu X, Su F, Lieberman J, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 28.Park SM, Shell S, Radjabi AR, Schickel R, Feig C, Boyerinas B, Dinulescu DM, Lengyel E, Peter ME. Let-7 prevents early cancer progression by suppressing expression of the embryonic gene HMGA2. Cell Cycle. 2007;6:2585–2590. doi: 10.4161/cc.6.21.4845. [DOI] [PubMed] [Google Scholar]
- 29.Droge P, Davey CA. Do cells let-7 determine stemness? Cell Stem Cell. 2008;2:8–9. doi: 10.1016/j.stem.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 30.Liau SS, Whang E. HMGA1 is a molecular determinant of chemoresistance to gemcitabine in pancreatic adenocarcinoma. Clin. Cancer Res. 2008;14:1470–1477. doi: 10.1158/1078-0432.CCR-07-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prasad R, Liu Y, Deterding LJ, Poltoratsky VP, Kedar PS, Horton JK, Kanno S, Asagoshi K, Hou EW, Khodyreva SN, et al. HMGB1 is a cofactor in mammalian base excision repair. Mol. Cell. 2007;27:829–841. doi: 10.1016/j.molcel.2007.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh NP, McCoy MT, Tice RR, Schneider EL. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp. Cell Res. 1988;175:184–191. doi: 10.1016/0014-4827(88)90265-0. [DOI] [PubMed] [Google Scholar]
- 33.Olive PL, Banath JP, Durand RE. Detection of etoposide resistance by measuring DNA damage in individual Chinese hamster cells. J. Natl Cancer Inst. 1990;82:779–783. doi: 10.1093/jnci/82.9.779. [DOI] [PubMed] [Google Scholar]
- 34.Verdine GL, Norman DP. Covalent trapping of protein-DNA complexes. Annu. Rev. Biochem. 2003;72:337–366. doi: 10.1146/annurev.biochem.72.121801.161447. [DOI] [PubMed] [Google Scholar]
- 35.Liu L, Gerson SL. Therapeutic impact of methoxyamine: blocking repair of abasic sites in the base excision repair pathway. Curr. Opin. Investig. Drugs. 2004;5:623–627. [PubMed] [Google Scholar]
- 36.Sakano K, Oikawa S, Hasegawa K, Kawanishi S. Hydroxyurea induces site-specific DNA damage via formation of hydrogen peroxide and nitric oxide. Jpn J. Cancer Res. 2001;92:1166–1174. doi: 10.1111/j.1349-7006.2001.tb02136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang TH, Popp DM, Wang HS, Saitoh M, Mural JG, Henley DC, Ichijo H, Wimalasena J. Microtubule dysfunction induced by paclitaxel initiates apoptosis through both c-Jun N-terminal kinase (JNK)-dependent and -independent pathways in ovarian cancer cells. J. Biol. Chem. 1999;274:8208–8216. doi: 10.1074/jbc.274.12.8208. [DOI] [PubMed] [Google Scholar]
- 38.Boo LM, Lin HH, Chung V, Zhou B, Louie SG, O'Reilly MA, Yen Y, Ann DK. High mobility group A2 potentiates genotoxic stress in part through the modulation of basal and DNA damage-dependent phosphatidylinositol 3-kinase-related protein kinase activation. Cancer Res. 2005;65:6622–6630. doi: 10.1158/0008-5472.CAN-05-0086. [DOI] [PubMed] [Google Scholar]
- 39.Sgarra R, Furlan C, Zammitti S, Lo Sardo A, Maurizio E, Di Bernardo J, Giancotti V, Manfioletti G. Interaction proteomics of the HMGA chromatin architectural factors. Proteomics. 2008;8:4721–4732. doi: 10.1002/pmic.200800193. [DOI] [PubMed] [Google Scholar]
- 40.Cattaruzzi G, Altamura S, Tessari MA, Rustighi A, Giancotti V, Pucillo C, Manfioletti G. The second AT-hook of the architectural transcription factor HMGA2 is determinant for nuclear localization and function. Nucleic Acids Res. 2007;35:1751–1760. doi: 10.1093/nar/gkl1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakamura J, Swenberg JA. Endogenous apurinic/apyrimidinic sites in genomic DNA of mammalian tissues. Cancer Res. 1999;59:2522–2526. [PubMed] [Google Scholar]
- 42.Sobol RW, Prasad R, Evenski A, Baker A, Yang XP, Horton JK, Wilson SH. The lyase activity of the DNA repair protein beta-polymerase protects from DNA-damage-induced cytotoxicity. Nature. 2000;405:807–810. doi: 10.1038/35015598. [DOI] [PubMed] [Google Scholar]
- 43.Srivastava DK, Husain I, Arteaga CL, Wilson SH. DNA polymerase beta expression differences in selected human tumors and cell lines. Carcinogenesis. 1999;20:1049–1054. doi: 10.1093/carcin/20.6.1049. [DOI] [PubMed] [Google Scholar]
- 44.Almeida KH, Sobol RW. A unified view of base excision repair: lesion-dependent protein complexes regulated by post-translational modification. DNA Rep. 2007;6:695–711. doi: 10.1016/j.dnarep.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parsons JL, Tait PS, Finch D, Dianova, Allinson SL, Dianov GL. CHIP-mediated degradation and DNA damage-dependent stabilization regulate base excision repair proteins. Mol. Cell. 2008;29:477–487. doi: 10.1016/j.molcel.2007.12.027. [DOI] [PubMed] [Google Scholar]
- 46.Weinberger M, Trabold PA, Lu M, Sharma K, Huberman JA, Burhans WC. Induction by adozelesin and hydroxyurea of origin recognition complex-dependent DNA damage and DNA replication checkpoints in Saccharomyces cerevisiae. J. Biol. Chem. 1999;274:35975–35984. doi: 10.1074/jbc.274.50.35975. [DOI] [PubMed] [Google Scholar]
- 47.Borrmann L, Schwanbeck R, Heyduk T, Seebeck B, Rogalla P, Bullerdiek J, Wisniewski JR. High mobility group A2 protein and its derivatives bind a specific region of the promoter of DNA repair gene ERCC1 and modulate its activity. Nucleic Acids Res. 2003;31:6841–6851. doi: 10.1093/nar/gkg884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reeves R, Adair JE. Role of high mobility group (HMG) chromatin proteins in DNA repair. DNA Rep. 2005;4:926–938. doi: 10.1016/j.dnarep.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 49.Loeb LA, Preston BD. Mutagenesis by apurinic/apyrimidinic sites. Annu. Rev. Genet. 1986;20:201–230. doi: 10.1146/annurev.ge.20.120186.001221. [DOI] [PubMed] [Google Scholar]
- 50.Saretzki G, Walter T, Atkinson S, Passos JF, Bareth B, Keith WN, Stewart R, Hoare S, Stojkovic M, Armstrong L, et al. Downregulation of multiple stress defense mechanisms during differentiation of human embryonic stem cells. Stem Cells. 2008;26:455–464. doi: 10.1634/stemcells.2007-0628. [DOI] [PubMed] [Google Scholar]
- 51.Ding J, Miao ZH, Meng LH, Geng MY. Emerging cancer therapeutic opportunities target DNA-repair systems. Trends Pharmacol. Sci. 2006;27:338–344. doi: 10.1016/j.tips.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 52.Matsuoka S, Ballif BA, Smogorzewska A, McDonald E.R., 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.