Abstract
The mini-chromosome maintenance (MCM) proteins serve as the replicative helicases in archaea and eukaryotes. Interestingly, an MCM homolog was identified, by BLAST analysis, within a phage integrated in the bacterium Bacillus cereus (Bc). BcMCM is only related to the AAA region of MCM-helicases; the typical amino-terminus is missing and is replaced by a segment with weak homology to primases. We show that BcMCM displays 3′→5′ helicase and ssDNA-stimulated ATPase activity, properties that arise from its conserved AAA domain. Isolated BcMCM is a monomer in solution but likely forms the functional oligomer in vivo. We found that the BcMCM amino-terminus can bind ssDNA and harbors a zinc atom, both hallmarks of the typical MCM amino-terminus. No BcMCM-catalyzed primase activity could be detected. We propose that the divergent amino-terminus of BcMCM is a paralog of the corresponding region of MCM-helicases. A divergent amino terminus makes BcMCM a useful model for typical MCM-helicases since it accomplishes the same function using an apparently unrelated structure.
INTRODUCTION
The DNA-replication systems of eukaryotes, archaea and bacteria employ hexameric DNA helicases to unwind DNA ahead of advancing replication forks (1,2). These enzymes unwind duplex DNA by assembling on and translocating along single-strand (ss) DNA at the fork, while displacing the complementary strand. In eukaryotes and archaea, the mini-chromosome maintenance (MCM) ensemble serves as the replicative helicase. This view is supported by several observations including that: an MCM subcomplex exhibits ATP-dependent helicase activity in vitro; the complete MCM complex, along with several accessory proteins, also demonstrates ATP-dependent helicase activity; the MCM assembly travels with the replication fork; and that ATPase-inactivating mutations in MCM proteins prevent DNA unwinding (3–6). Moreover, these MCM genes have proven to be essential to viability in yeast (1).
MCM helicases are DNA-stimulated ATPases that unwind DNA from the 3′ to 5′ direction (1,2). In eukaryotes, the MCM ensemble is a large (Mw ≈ 600 kDa) hetero-oligomer composed of six homologous subunits (MCM2-7). Most archaeal genomes encode a single MCM protein, which assembles into an active homo-oligomer. The eukaryotic MCM2-7 ensemble displays ATPase activity, though helicase activity has only been observed under certain solution conditions in vitro (7–9). On the other hand, the archaeal MCM helicase assembly shows potent ATPase and helicase activity in vitro, allowing it to serve as a simpler and more tractable model to study the hetero-oligomeric eukaryotic MCM complex.
Sequence, biochemical, and structural analyses suggest that MCM helicases share a common core structural domain that extends for ∼500–600 amino acids (1,2). The MCM domain can be sub-divided into two smaller structures: a conserved amino-terminal domain (MCM-N); and a more-highly conserved AAA module, with elements unique to MCM helicases (MCM-AAA). MCM-N binds DNA, is involved in oligomerization, and harbors a zinc-coordination module. Structural and biochemical analysis of the archaeal MCM-N region reveals a homohexameric ring that binds DNA within its central cavity (10,11). β-hairpins, tipped with positively charged residues, project from the walls into the MCM-N cavity and are shown to contact DNA (10). The zinc-coordination module, which is conserved in virtually every MCM helicase subunit [among the eukaryotic MCM subunits (1) and the archaeal orthologs], is essential for activity and appears to serve a structural role (10–13).
The MCM-AAA domain is located at the carboxy terminus and contains AAA-style (ATPases Associated with various cellular Activities) nucleotide-binding elements (e.g. P-loop/Walker A, Walker B, Sensor I, Arginine Finger and Sensor II) (Figure 1) (14). Electron microscopy of full-length MCM proteins shows that the MCM-AAA segment also forms a ring structure, likely by subunits interacting through composite nucleotide-binding sites at subunit interfaces (15). The MCM-AAA contains a second β-hairpin, which is inserted before the AAA Sensor 1 motif (referred to as the ‘pre-sensor 1 β-hairpin’) and is implicated in both contacting DNA and in nucleotide-dependent conformational changes (16). Additional elements, such as the helix-2-insert, help distinguish the MCM-AAA module from other proteins in the AAA ATPase superfamily and likely participate in the unwinding reaction (17). Additionally, archaeal MCMs have a conserved putative winged helix-turn-helix (wHTH) motif at the extreme C-terminus (Figure 1A) (2).
Figure 1.
The Bacillus cereus MCM-helicase is only homologous to the MCM-AAA domain. (A) The MCM-helicase structure can be sub-divided into a conserved amino-terminal domain (MCM-N) and a highly conserved carboxy-terminal domain MCM-AAA. Archaeal orthologues contain a winged helix sub-domain that does not appear in its eukaryotic counterparts. DNA-binding β-hairpins and the zinc coordination site are indicated. Conservation of MCM-helicase proteins is depicted graphically by plotting the percent identity of a large group of homologues against the primary sequence of the MCM-helicase from M. Thermautotrophicus. Positions with a percent identity of 85% or higher are shown as magenta dots. Amino-acid sequences from this group, which includes orthologues from S. cerevisiae (MCM2-7), S. pombe (2–7), H. sapiens (2–9), B. taurus (2–7), D. melanogaster (2–7), D. rerio (2–7), E. cuniculi (2–7), G. gallus (2–6, 8), M, musculus (2–9), P. troglodytes (2–7), R. norvegicus (2–7, 8), X. laevis (9), S. solfataricus, S. acidocaldarius, T. acidophilum, M. acetivorans, P. aerophilum, T. kodakarensis, M. mazei, A. pernix, A. fulgidus, M. Thermautotrophicus and Sulfolobus neozealandicus (pTAU4 plasmid) were aligned with MUSCLE (47) using its default parameters. Percent sequence identity was calculated by averaging similarity scores at each position for all possible pairs of sequences [sum of pairs measure in (48), DJ, unpublished software]. Equivalence of non-identical residues in this calculation was established through a normalized BLOSUM62 amino-acid substitution matrix (49). (B) Sequence analysis of BcMCM reveals regions related to the archaeo-eukaryotic family of primases (AEP, positions 73–182), the large subunit of eukaryotic primase (positions 187–417) and archaeal/eukaryotic MCM subunits (positions 560–900). Percent sequence identity, which represents three separate sequence analyses, are projected on the primary sequence of BcMCM. Regions of the plot that correspond to archaeo-eukaryotic primases and the primase large subunits was calculated from Figure 2 and Figure 8 of (29). The portion of the plot that encompasses MCM-AAA was calculated using all the sequences from part A and the corresponding region of BcMCM. Sequence identity was calculated as described in part A. Positions with a percent identity of 85% or higher are shown as magenta dots.
The complete MCM-helicase domain is, therefore, composed of two sub-structures (MCM-N and MCM-AAA). Oligomerization of MCM proteins generates an ensemble with a long tunnel through which DNA passes, guided, in part, by these two β-hairpins. Two recently described members of the MCM-helicase family, MCM8 and MCM9, retain the features seen in MCM2-7 and archaeal orthologs (18,19).
The structural basis for the unwinding activity by helicases in general, and especially MCM helicases, remains incompletely understood. Structural analysis by electron microscopy (EM) has shown eukaryotic and archaeal MCMs as ring-shaped hexamers, heptamers, or double-hexamers (15,20–22). The observed double-hexamers appear to be two single hexamers that are assembled head-to-head via contacts between MCM-N segments (10). These studies have also offered a glimpse of the richness of the structural changes of the MCM complex as it associates with various cofactors (15,22). Such EM studies, in combination with X-ray structures, enable modeling of the hexameric entity. These analyses now set the stage for a more complete understanding of the structural basis for DNA unwinding (23–27).
Below, we describe a phage MCM-homolog found within the genome of the bacterium Bacillus cereus (Bc), initially identified by computational methods (28,29), as an alternate model for studying DNA unwinding by MCM helicases. We show that this homolog (BcMCM) possesses ATP-dependent helicase activity in vitro. Unwinding is exclusively in the 3′→5′ direction, similar to eukaryotic and archaeal MCM homologs. BcMCM harbors an ATPase activity that is localized to its MCM-AAA domain and is stimulated by ssDNA. We show that the full-length protein is a monomer in solution, but higher-order oligomers are evident when the BcMCM amino-terminal and MCM-AAA sub-domains are isolated from each other. Both of these isolated sub-domains can bind ssDNA. BcMCM also contains an amino-terminal zinc-coordination site.
MATERIALS AND METHODS
Protein expression constructs
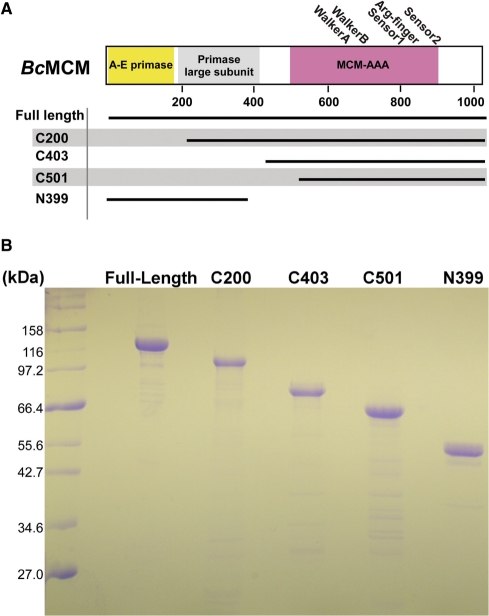
The gene for B. cereus MCM (BcMCM) was amplified from genomic DNA (ATCC 14579) and inserted into the SphI and SacII sites of the plasmid pQE2 (Qiagen). DNA sequencing of the resulting plasmid clone revealed two sequence changes relative to the published sequence (nucleotides C663A and T2775A, resulting in the amino-acid substitutions S221R and D925E, respectively). These two differences may have arisen from sequencing different isolates or from sequencing errors. Fortuitously, the two changes seem relatively conservative. In addition to the full-length gene, we prepared several shorter constructs to isolate the predicted helicase and the primase activities (Figure 1A and Figure 2). Several such constructs were prepared owing to our inability to reliably establish domain boundaries. The C200, C403, C501 and N399 truncations, in addition to the full-length protein, have proven suitable for study (Figure 1B). Each construct contains an amino-terminal hexahistidine tag. We also prepared two full-length mutant proteins: one with a disrupted ATP-binding site (Walker A: K653A); and a second with a disrupted zinc-binding site (C261A) to serve as controls in biochemical studies. PCR primers for the constructs prepared for this study appear in Supplementary Table 1.
Figure 2.
Constructions of BcMCM-helicase used in this study. Three basic types of constructions of BcMCM were prepared. These are (i) full-length; (ii) amino-terminal constructs that include the putative primase domains; and (iii) carboxy-terminal constructs that include the MCM-AAA domain. The isolated MCM and primase domain constructs were devised so that the two activities could be studied separately. Several versions were prepared owing to our inability to reliably establish domain boundaries. (B) Coomassie blue stained SDS–Page analysis of typical preparations of full-length and various shorter constructions of BcMCM.
Protein expression and purification
Full-length and truncated constructs of BcMCM were expressed and purified using an identical sequence of steps. Proteins were expressed in E coli M15 cells in the following media: 90 mM potassium phosphate/pH 7.4; 2.4% yeast extract; 1.2% tryptone; 0.8% glycerol, supplemented with 50 μg/ml kanamycin and 100 μg/ml ampicillin. The culture was grown at 37°C until an OD600 of ∼4.5 was reached. Protein expression was then induced at 22°C for full-length (or at 15°C for the shorter truncations) by addition of 1 mM isopropyl thiogalactopyranoside (IPTG). Sixteen hours later, the cells were harvested by centrifugation (4000 RPM for 30 min), resuspended in lysis buffer (50 mM sodium phosphate/pH 8.0, 1 M NaCl, 10 mM imidazole and 10% sucrose) and frozen at −80°C.
Freshly thawed cells were lysed by sonication and the soluble fraction was applied to a NiNTA column (Qiagen) pre-equilibrated in Buffer A (20 mM sodium phosphate/pH 8.0, 200 mM NaCl, 60 mM imidazole and 10% glycerol). The protein was eluted from the column using a linear gradient from Buffer A to Buffer B (20 mM sodium phosphate/pH 8.0, 200 mM NaCl, 500 mM imidazole and 10% glycerol). The fractions were analyzed using 12% SDS–PAGE, and those containing the protein were collected, pooled and then dialyzed against Buffer C (20 mM Tris–HCl/pH 7.5, 100 mM NaCl, 5 mM β-mercaptoethanol and 10% glycerol). The pool of BcMCM was then applied to an SP-sepharose column pre-equilibrated in Buffer C, and a linear gradient was run on the column from Buffer C to Buffer D (20 mM Tris–HCl/pH 7.5, 1 M NaCl, 5 mM β-mercaptoethanol and 10% glycerol). The eluted fractions containing the protein were then collected and pooled and dialyzed in Buffer C. The protein was next applied to a Q-sepharose column pre-equilibrated in Buffer C and eluted using a linear gradient from Buffer C to Buffer D. BcMCM-containing fractions were pooled and concentrated by ultrafiltration (VivaSpin VivaScience; 10 kDa MWCO). The identity of all purified proteins was verified by mass-spectrometry.
Preparation of the Helicase DNA substrate
A single-strand of DNA was labeled with [γ32P] ATP (Amersham, 10 mCi/ml). The labeling reaction consists of incubating 10 pmol of oligonucleotide (DF50; Supplementary Table 2) with 2.5 μl 10× T4 polynucleotide kinase reaction buffer (New England Biolabs (NEB)), 1 μl T4 polynucleotide kinase (NEB), and 5 μl [γ32P]ATP in a total volume of 25 μl at 37°C for 75 min. The reaction was stopped by adding 1 μl of 0.5 M EDTA–NaOH/pH 8, and then purified by centrifuging the oligonucleotide through G50 desalting beads twice at 0.7RCF for 1 min each time.
A second, unlabeled oligonucleotide (DF61; Supplementary Table 2) was then hybridized to the first in a 3:1 ratio: 30 pmol of DF61 was added to the labeled oligo in 60 mM HEPES–NaOH/pH 7.5 and 50 mM NaCl before being brought to a boil for 3 min and then allowed to cool to room temperature. The resulting helicase substrate was then gel purified via a mini 8% polyacrylamide, 1× TBE native gel that was run at 80 V for 55 min. The radioactive band was then excised from the gel and extracted in 0.5 M ammonium acetate, 10 mM magnesium acetate and 1 mM EDTA; shaking at 400 RPM at room temperature overnight. After elution from the gel slice, the DNA was ethanol precipitated and finally resuspended in 20 mM Tris–HCl/pH 8.5. The final concentration of the substrate was determined by comparing the specific activity of this purified substrate to an earlier sample taken after the labeling reaction. In assays using substrates with biotin–streptavidin beads, 10 fmol of biotinylated DNA substrate was incubated with 1 pmol of streptavidin for 10 min at 30°C (30).
Helicase assay
Purified proteins were assayed for in vitro helicase activity by incubating them with the helicase DNA substrate. 10 fmol of the DNA substrate were incubated with enzyme in 20 mM Tris–HCl/pH 8.5, 10 mM MgCl2, 2 mM DTT and 0.1 mg/ml BSA for 60 min at 37°C in a 15 μl reaction. The reaction was terminated by addition of 5 μl of 4× stop buffer (50% glycerol, 0.1 M EDTA, 0.1% Bromophenol Blue, 0.1% Xylene Cyanol). The results were then visualized on 8% polyacrylamide, 0.5× TBE native mini-gels that were run for 50 min at 90 V, and then exposed to a phosphor screen. The phosphor screen was read with a PhosphorImager (Molecular Dynamics), and visualized using the ImageQuant® software.
ATPase assay
The rate of ATP hydrolysis was measured using a steady-state, NADH-coupled spectrophotometric assay. The purified proteins were incubated in a reaction buffer consisting of 50 mM HEPES–KOH/pH 7.5, 150 mM potassium acetate and 8 mM magnesium acetate. Each assay was repeated eight times in a column on a UV-transparent, 96-well plate (Costar 3635). Rates were measured using a Molecular Devices, SpectraMax M5 plate reader and quantified using SoftMax Pro v5. Experiments that included ssDNA employed a 61-mer sequence (Supplementary Table 2). The ATP turnover rates were calculated from the equation: ATPase rate = −(dA340/dt) × (1/Kpath) × (1/mol of ATPase); where ‘ATPase rate’ is measured in terms of ATP turned over per minute; ‘dA340/dt’ is expressed in OD/min; and ‘Kpath’ is the molar absorption coefficient for NADH at a given optical pathlength (31).
Tryptophan-quenching assay
The ability to bind single-stranded, synthetic DNA oligonucleotides (Supplementary Table 2) was tested by measuring the quenching of BcMCM's intrinsic tryptophan fluorescence. Binding constants were determined by keeping the protein concentration fixed (1 μM) and titrating DNA from 0 to 50 μM. The reactions were incubated in 20 mM Tris–HCl/pH 7.5, 125 mM NaCl and 10 mM MgCl2. The reactions were conducted in 96-well, UV-transparent trays (CoStar 3635), repeated four times per tray, and the fluorescent quenching was read using the SoftMax Pro v5 software. Dissociation constants (Kd) were calculated using the equation: dF/dFmax = [DNA]/([DNA] + Kd); where ‘dF/dFmax’ represents the change in the protein's fluorescence relative to its maximal change in fluorescence. Calculations were performed using the programs Microsoft Excel and Kaleidagraph.
Primase assay
Full length BcMCM was assayed for in vitro primase activity by providing a ssDNA substrate and observing whether the enzyme is competent to make an RNA primer that enables subsequent DNA synthesis. The enzymes were incubated with 7.5 nM of single-stranded ϕX174 plasmid (NEB); 100 μM each of ATP, CTP, GTP and UTP; 50 μM each of dCTP, dGTP and dTTP; 1.32 μM [α32P] dATP (Amersham, 20 mCi/ml); and 1 μl Klenow in a buffer of 50 mM Tris–HCl/pH 7.5, 5 mM DTT, 10% glycerol and 0.1 mg/ml BSA. Reactions were carried out at 30°C for 10 min and terminated by spotting 1 μl of the 25 μl reaction onto DE81 filter paper (Whatman). The filters were then washed four times in a buffer of 0.3 M ammonium formate and 10 mM sodium pyrophosphate and left to dry overnight before counting their scintillation (32).
Multi-angle laser light scattering
Purified proteins were applied to a Shodex KW-804 column equilibrated in 25 mM Tris–HCl/pH 7.5 and 350 mM ammonium sulfate. Light scattering and refractive index signals were measured using a Wyatt Optilab and Dawn EOS system. Scattering curves were processed using the provided Astra software package. The protein concentration for each experiment was 5 mg/ml.
Zinc analysis by inductively coupled plasma mass spectrometry
Purified protein samples were prepared for zinc analysis by dialysis against 20 mM Tris–HCl/pH 7.5, 400 mM sodium chloride, 5% glycerol and 0.5 mM EDTA. Samples were flash frozen and submitted to the National Institute of Standards and Technology (NIST), Analytical Chemistry Division. Some trace solids, which were apparent upon thawing, were re-solubilized by treatment with high-purity nitric acid. The zinc content was measured using inductively coupled plasma–mass spectrometry (ICP–MS) (12). The zinc content measurements were calibrated against UvrA, whose crystal structure reveals three-bound zinc atoms per mole of protein (33).
RESULTS
BcMCM is homologous to the MCM-AAA subdomain
Computational analysis of the B. cereus genome revealed a prophage harboring a gene with a segment (spanning BcMCM residues 500–900) that is homologous to the MCM-AAA region of the complete MCM-helicase domain (28) (Figures 1 and 2A). The MCM-AAA portion of eukaryotic and archaeal MCM proteins is highly conserved and contains the ATP binding and hydrolysis functions along with a number of structural elements unique to MCM-helicases. By contrast, residues 1–499 of BcMCM align poorly with the MCM-N segment of the MCM-helicase fold, as confirmed in an extensive analysis of MCM helicase sequences (Figure 1B). Others have noted that the amino-terminus of BcMCM shows weak homology to the archaeo-eukaryotic family of primases at residues 1–183 and to the large subunit of the eukaryotic primase at residues 183–417 (29) (Figure 2A). Iyer et al. (29) predicts that there is a zinc-coordination site within this latter segment (BcMCM residues C261, C342, C364 and C369), serving as a key aspect of the region's homology to the primase large subunit. It is interesting to note that this site is located relative to the MCM-AAA region in a similar manner to the zinc site seen in archaeal and eukaryotic MCM helicases.
BcMCM displays ATP-dependent DNA unwinding activity
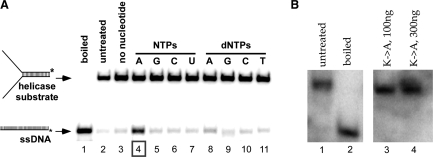
BcMCM was identified within a prophage in the genome of B. cereus using computational methods (28). To characterize its biochemical activity, we have used helicase assays to show that BcMCM harbors ATP-dependent DNA unwinding activity. In Figure 3A, lane 4, we show that BcMCM is an ATP-dependent DNA helicase, since it can convert the duplex substrate into ssDNA product. On the other hand, no unwinding was observed upon mutation of the ATP-binding site (Walker A: K635A), confirming that activity is due to BcMCM (Figure 3B). ATP dependence is established in lanes 3 and 5–11, where no activity is seen with any other nucleotide (compare lane 4 to lanes 5–11). This experiment also rules out contamination by a bacterial nuclease as an explanation for the appearance of ssDNA in lane 4, since the substrate remains intact in all other lanes. Taken together, our data implies: (i) that BcMCM is a helicase (when comparing the wild-type activity to that of the Walker A mutant); and (ii) that the helicase product band is not a result of nucleolytic activity given the absence of such a band when ATP is withheld from the reaction and when the Walker A mutant is employed.
Figure 3.
BcMCM displays ATP dependent helicase activity. (A) Reaction mixture contained 2.7 pmol wild-type BcMCM (as monomer) except for lanes 1 and 2. Lane 1, boiled substrate; lane 2, substrate only; lane 3, without NTPs or dNTPs; lane 4, with 0.67 mM ATP; lane 5, with 0.67 mM GTP; lane 6, with 0.67 mM CTP; lane 7, with 0.67 mM UTP; lane 8, with 0.67 mM dATP; lane 9, with 0.67 mM dGTP; lane 10, with 0.67 mM dCTP; lane 11, with 0.67 mM dTTP. Asterisk represents 32P-label. (B) Reaction mixture contained 0.9 pmol and 2.7 pmol Walker A (K635A) BcMCM in lanes 3 and 4, respectively, in the presence of 0.67 mM ATP. Lane 1, boiled substrate; lane 2, substrate only.
BcMCM unwinds DNA in the 3′→5′ direction
Many helicases unwind DNA in a specific direction (either 3′→5′ or 5′→3′). This polarity depends upon which DNA strand the helicase travels along as it displaces the complementary strand. At the replication fork, a helicase that runs in the 3′→5′ will be traveling along the leading strand, whereas a 5′→3′ helicase will travel along the lagging strand. Experimentally, helicase polarity can be determined by observing activity on a series of substrates with a single ssDNA tail (either 3′ or 5′). In these experiments, the addition of a biotin-streptavidin group serves two roles: it blocks assembly and translocation on one strand as it also increases unwinding efficiency (30).
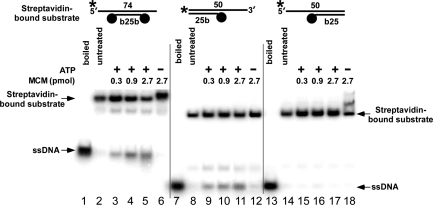
Three different substrates were used to determine the unwinding polarity of BcMCM: (i) a molecule with an available 3′ tail; (ii) one with an available 5′ tail; and (iii) one with both 5′ and 3′ tails. Biotin-streptavidin groups (represented by black dots in Figure 4) restrict the helicase to the available strand and was succesfully used in the study of the directionality of other helicases (30). The pattern of activity of these substrates suggests that BcMCM unwinds DNA from 3′→5′ as BcMCM shows robust unwinding activity on substrates with only an accessible 3′ tail (Figure 4, lanes 9–11). No activity is seen on substrates with only an accessible 5′ tail (lanes 15–17). Naturally, a substrate with both 3′ and 5′ tails are unwound (lanes 1–6). This substrate establishes (in comparison to Figure 3) that altering the substrate by adding biotin-streptavidin does not, in and of itself, inhibit helicase activity. In lanes 6 and 18, we note a supershift of the substrate when it is incubated with enzyme in the absence of ATP. Other hexameric helicases, specifically Escherichia coli DnaB, have also been shown to bind DNA substrates more tightly when inactive (34–36). The lighter bands on this gel (e.g. lanes 3–5 below the substrate band) represent DNA molecules that have not been modified by streptavidin. This experiment establishes that the polarity of BcMCM is 3′→5′, which is identical to that of other MCM helicases (1,2).
Figure 4.
BcMCM unwinds DNA with 3′→5′ polarity. Helicase assays were performed using the substrates shown in the top of figure. In each substrate the longer oligonucleotide is 32P-labeled (asterisk) and streptavidin is marked with a closed circle. Lanes 1, 7 and 13: boiled substrate; lanes 2, 8 and 14: substrate only; lanes 3, 9 and 15: 0.3 pmol of BcMCM as monomer; lanes 4, 10 and 16: 0.9 pmol of BcMCM; lanes 5, 11 and 17: 2.7 pmol of BcMCM; lanes 6, 12 and 18: 2.7 pmol of BcMCM but without addition of ATP.
BcMCM is an ATPase that is stimulated by DNA
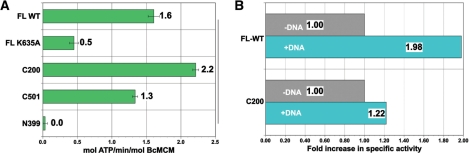
DNA helicases (specifically, the bacterial replicative helicase DnaB) were initially identified as ssDNA-stimulated ATPases (37). We therefore sought to investigate BcMCM's ATPase activity to more fully characterize its helicase activity. ATPase activity was measured using an NADH-coupled assay that allowed for steady-state kinetics measurements. Using this assay, we observed that BcMCM exhibits a basal specific activity of 1.6 mol ATP/min/mol of enzyme in the absence of DNA (Figure 5). We note that ATP is hydrolyzed in AAA+ ATPases at subunit interfaces (14), suggesting that BcMCM is oligomeric. Mutation of the ATP-binding site (Walker A: K653A) reduced activity to ∼30% of wild-type (Figure 5A). This drop in ATPase activity corresponds to an abrogation of helicase activity observed in the same mutant (Figure 3B). A parallel analysis with our BcMCM truncations (C200 and C501), which both harbor the MCM-AAA subdomain, revealed that the ATPase activity is indeed localized to the carboxy-terminal region. Despite observing ATPase activity in the C200 and C501 truncations, we were not able to reliably demonstrate helicase activity for these constructs (data not shown). No activity was seen with the isolated amino-terminal, putative primase domain (N399). Thus, BcMCM is an active ATPase, and this activity is restricted to the MCM-AAA module.
Figure 5.
BcMCM is an ATPase, which is stimulated by ssDNA. (A) Shown is a summary of ATP hydrolytic activity by various BcMCM constructs in the absence of ssDNA. (B) Inclusion of ssDNA (61-mer) led to a 2-fold stimulation of activity by both the full-length, wild-type, enzyme and by a shorter construct (C200). Activity has been normalized to that of the isolated enzyme (values taken from part A). The BcMCM concentration in these experiments is 1 µM.
We next examined whether ssDNA could stimulate this ATPase activity. Figure 5B shows that titration of a 61-nt long oligonucleotide molecule stimulated the ATPase of full-length BcMCM by 1.5-fold when present at a DNA:protein ratio of 1:5. Additional stimulation (2.8-fold) could be obtained by using a 4:1 excess of DNA over protein (data not shown). The C200 truncation, which lacks the putative primase domain, was also modestly stimulated by ssDNA.
Both amino- and carboxy-termini of BcMCM can bind ssDNA
Having measured the ability of BcMCM to be stimulated by ssDNA, we next sought to measure the enzyme's binding affinity for ssDNA. This measurement employed tryptophan quenching and exploited our observation that addition of DNA quenches the intrinsic fluorescence of BcMCM. Experiments that titrated ssDNA into protein solutions were analyzed according to the following equation: (ΔF/ΔFmax) = [DNA]/(Kd + [DNA]), where ΔF is the amount of fluorescence quenching at a given DNA concentration and ΔFmax is the maximum possible quenching. The dissociation constant (Kd) was determined by fitting measurements to the equation above using the Kaleidagraph program. Our fits yielded R2 values of 0.94 or higher (Figure 6 inset).
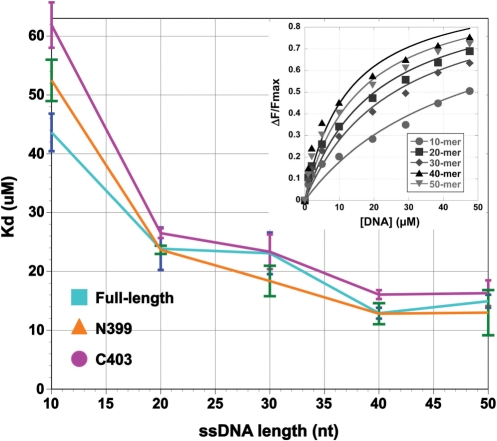
Figure 6.
The amino and carboxy terminal regions of BcMCM bind to ssDNA. ssDNA-binding affinities were determined by tryptophan quenching, as described under Materials and Methods section. Three proteins were used: the full-length protein; the N399 truncation that includes the enzyme's amino-terminus; and the C403 truncation, that includes the MCM-AAA subdomain. When added together, the N399 and C403 truncations reconstitute the full-length protein. For each protein, dissociation constant was determined for synthetic ssDNA oligonucleotides 10, 20, 30, 40 and 50 nt long. To determine the dissociation constant for each oligonucleotide, the DNA concentration was titrated from 0–50 µM in reactions containing 1 µM protein. Inset: representative data used to generate this plot. ΔF/ΔFmax data are shown for the full length BcMCM protein.
We titrated ssDNA of various lengths (10, 20, 30, 40 or 50 nt) to allow calculation of Kd as a function of length (Figure 6). In general, full-length BcMCM binds longer oligos more tightly. We measure a higher affinity to a 20-nt than to a 10-nt species, but the trend plateaus for chains longer than 30-nt. In our experiment, the highest affinity is recorded with a 40-nt molecule (Kd = 12.9 μM, SD = 0.9 µM). Both the N399 and C403 constructs also showed an interaction with ssDNA with similar length dependent properties. In general, the N399 construct (Kd = 13.0 μM, SD = 3.8, for 50-nt oligos) bound slightly tighter than C403 (Kd = 16.3 μM, SD = 2.2, for 50-nt oligos). We note that the full-length protein and the smaller constructs displayed similar dissociation constants for ssDNA. The use of tryptophan quenching to measure dissociation constants, however, makes it difficult to compare the data between the full-length protein and the shorter constructs since the number and exposure of tryptophan residues is different. Nevertheless, the observation that both the amino- and carboxy-terminal domains bind ssDNA suggests the presence of either two binding sites or one extended composite-binding surface in the full-length protein.
BcMCM is a monomer in solution
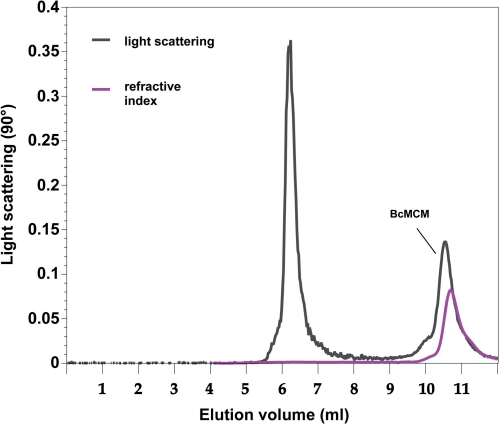
MCM helicases are AAA+ ATPases and consequently their activity is expected to require oligomerization (14). Structural and biochemical studies of MCM helicases have yielded several different oligomerization states: the eukaryotic assembly is a heterohexamer (1,8), while the archaeal MCMs have been observed as hexamers, heptamers and/or double-hexamers (15,20,21). These oligomers are pre-formed and do not require substrates or co-factors to form. In order to understand the oligomeric state of BcMCM, we measured its native molecular weight using size exclusion chromatography paired with static, multi-angle light scattering (38). These measurements reveal that the molecular weight of full-length BcMCM is 127–138 kDa (Figure 7 and Table 1). Since the molecular weight calculated from the sequence is 117 kDa, we conclude that full-length BcMCM is a monomer in solution (the spread in our data could reflect experimental errors or the presence of small amounts of oligomeric species). The oligomerization state of BcMCM was unchanged when ATP and/or the helicase substrate were included (data not shown). The first peak in Figure 7 represents a very small amount (negligible refractive index) of a large unknown aggregate or particulate.
Figure 7.
BcMCM is a monomer in solution as revealed by multi-angle laser light scattering. Full length BcMCM (5 mg/ml) in 50 mM Tris–HCl, pH = 7.5, 200 mM NaCl, 5% glycerol, 0.05 mM EDTA was applied to a Shodex KW-804 column equilibrated in 25 mM Tris–HCl, 350 mM KCl, 5 mM Mg•(OAc)2. Light scattering and refractive index signals were measured using a Wyatt Optilab and Dawn EOS system. Scattering curves were processed using the provided Astra software package.
Table 1.
Molecular masses of full-length and shorter constructs of BcMCM, measured by light scattering
| Construct (kDa) | Measured molecular mass | Interpretation |
|---|---|---|
| Full-length (117) | 127–138 | Monomer |
| 258 | Dimer | |
| C200 (95.3) | 191 | Dimer |
| 286 | Trimer | |
| 388 | Tetramer | |
| C501 (60) | 72–77 | Monomer |
| N399 (45.8) | 44 | Monomer |
| 93 | Dimer | |
| 198 | Tetramer |
One explanation for observing a monomer of BcMCM instead of the expected hexamer is that our experimental conditions do not suitably mimic those found in vivo. To further pursue this question, we sought to determine whether domains of BcMCM might form specific oligomers in isolation. Light scattering measurements with the C200 and C501 constructs (both contain the MCM-AAA domain) and the N399 construct (containing the putative primase domain) revealed that these constructs exist in mixtures of oligomeric states (Table 1). We speculate that the higher-order assemblies formed by discrete domains of BcMCM may recapitulate the contacts in the active oligomeric species. These data imply that the full-length enzyme may operate as an oligomer when unwinding DNA but that oligomerization may be prevented in the context of the full-length protein, in vitro.
It is possible that the relatively high ionic strength of our measurements (350 mM ammonium sulfate) disrupts the oligomer. We used this condition with all our measurements to accommodate the poor behavior of the shorter constructs. To rule out disruption of the oligomer by salt, we performed light scattering measurements with full-length protein in 200 mM sodium chloride and observed only monomers. We note that the Sulfolobus solfataricus (Sso) archaeal MCM-helicases is oligomeric in 100 mM sodium chloride (39) and requires 1M sodium chloride to dissociate into monomers (27). The most probable hindrance to observing a higher-oligomeric form of BcMCM, however, is its divergent amino-terminus, which in its conserved form provides a significant amount of the binding energy holding MCM hexamers together (10,11). The altered BcMCM amino-terminus may still provide enough binding energy to enable oligomerization in vivo, as evident by our in vitro biochemical assays, but may not allow for the observation of oligomerization in solution.
BcMCM binds one zinc atom
An important component of the MCM-N domain is a zinc coordination site, which is conserved in virtually all archaeal and eukaryotic MCM helicases, and which disrupts MCM activity when mutated (10–13). Accordingly, inspection of the amino-acid sequence of BcMCM revealed a number of cysteine residues upstream of the MCM-AAA segment that could form a zinc-coordination with a similar position to other MCM proteins. To help understand the relationship between the amino-terminal domain of BcMCM and MCM-N, we sought to detect the presence of a coordinated zinc ion in BcMCM using analytical methods. The zinc content of wild-type BcMCM was measured using inductively-coupled plasma-mass spectrometry (ICP–MS, National Institute of Standards and Technology). While we sought to investigate whether this region was a component of the MCM-N domain, we note that this region has been suggested by Iyer et al. to be related to the large subunit of eukaryotic primases rather than the MCM family (29). In addition to testing the wild-type protein, we also mutated a prospective amino-terminal cysteine residue (C261A) in hopes of disrupting the putative zinc coordination site.
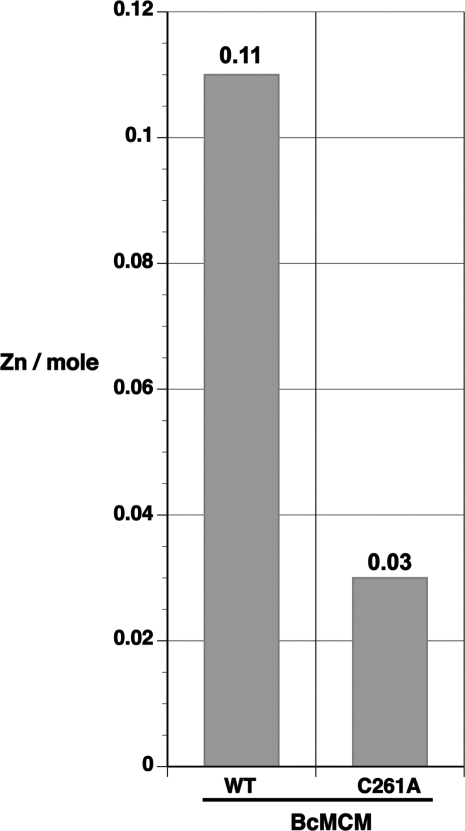
This analysis revealed that BcMCM contains 0.11 zinc atoms per mole, while one mole of the C261A mutant contains only 0.03 atoms (Figure 8). These raw values were scaled using control measurements carried out with Geobacillus stearothermophilus UvrA (0.32 zinc atoms/mole), whose structure indicates coordination of three zinc ions per monomer (33), and hen egg white lysozyme (0.01 zinc atoms/mole), which is free of zinc. We conclude from our study that BcMCM coordinates one zinc ion (0.11 × 3/0.32 = 1.03), whereas the C261A mutant bound only 0.28 zinc ions per mol of protein (= 0.03 × 3/0.32), indicating that cysteine 261 contributes to the coordination site. Scaling the data in this way is justified owing to the fact that low concentrations of zinc in the protein samples are a challenge to measure by ICP–MS due to contamination and instrument background.
Figure 8.
Full-length BcMCM binds a zinc atom. Purified protein samples were prepared for zinc analysis by dialysis against 20 mM Tris–HCl/pH 7.5, 400 mM sodium chloride, 5% glycerol and 0.5 mM EDTA. Samples were subsequently analyzed for zinc using inductively coupled plasma-mass spectrometry (ICP–MS). Shown here are results for full-length, wild-type BcMCM and full-length, C261A BcMCM mutant.
BcMCM has no detectible primase activity
An intriguing prediction made by the informatic analysis of the BcMCM amino-acid sequence is the presence of segments homologous to the archaeo-eukaryotic family of primases (residues 1–183) and to the large subunit of eukaryotic primase (residues 183–417) (Figure 2A). The assay that we used to evaluate this prediction calls for incubation of BcMCM with ssDNA (ϕX174, NEB) and ribonucleotides. If a primer is synthesized, then the Klenow fragment is able to synthesize a DNA complement to the ϕX174plasmid using radiolabeled dNTPs. Using this assay and buffer conditions described for the E. coli DnaG primase, we did not observe any primase activity while using EcDnaG itself as the positive control (data not shown) (32).
DISCUSSION
The main goal of this study is to develop the phage BcMCM-helicase as a tractable model for the more elaborate heterohexameric helicases found in eukaryotes. The study of MCM helicases is important given their critical roles in both initiating eukaryotic DNA replication and in fork progression after replication begins. Here, we describe the initial biochemical analysis of an unusual MCM helicase found in a phage genome integrated within that of the bacterium B. cereus. Phage proteins such as the DNA, RNA polymerases and helicases have long proven to be excellent models to study core functions of more complicated eukaryotic entities and we expect that BcMCM may play a similar role (40,41).
The BcMCM-helicase is interesting for two reasons. First, MCM-helicases are typical to eukaryotes and archaea, but have not been previously described in bacteria (although a homolog has now also been found in Exiguibacterium species AT1b, NCBI accession #ZP 02990802). Second, only the carboxy-terminal AAA domain of BcMCM (the MCM-AAA domain) is related to eukaryotic and archaeal MCM proteins, whereas the amino-terminal portion of BcMCM is divergent in sequence. The typical amino-terminus of the MCM family (MCM-N) appears to be missing from BcMCM. This is especially puzzling given the importance of MCM-N to the unwinding reaction (13,42,43). Rather than the conserved MCM-N domain, BcMCM contains a stretch of 500 amino acids that exhibits weak similarity to the archaeo-eukaryotic primase family and the large subunit of eukaryotic primase subunit (Figures 1 and 2) (28,29).
In this study, we have shown that BcMCM unwinds DNA in an ATP-dependent manner with 3′→5′ directionality. These properties resemble those of conventional MCM-helicases despite the lack of a standard MCM amino-terminus. From our experiments, we conclude that BcMCM helicase activity arises from the cooperation between its conserved MCM-AAA domain and a paralog of the MCM-N domain. We speculate that the divergent amino-terminus may allow BcMCM to serve as an alternate model for MCM function, since study of different structures (i.e. the MCM-N versus the BcMCM amino-terminus) that perform the same function often proves illuminating.
Previous studies have established that the MCM-N domain regulates the DNA unwinding activity and influences the ATPase activity (13,42,43). Our results suggest that an intact amino-terminus of BcMCM is required for unwinding activity. By contrast, the MCM-N domain of the archaeal MCMs can stimulate unwinding activity, but is not in and of itself necessary. Our work with BcMCM truncations indicates that the amino-terminus also diminishes the intrinsic ATPase activity of the MCM-AAA (compare the activity of the C200 construct to that of the full-length protein in Figure 5). These results are consistent with those with the Aeropyrum pernix and the Archaeoglobus fulgidus MCM proteins, in which amino-terminal truncations also demonstrated increased ATPase activity (13,42).
Our studies suggest that BcMCM binds ssDNA with both the MCM-AAA domain and the amino-terminus (Figure 6). Within error, our measurements suggest that both isolated segments display a length-dependent affinity to ssDNA. The affinity displayed by the amino-terminus and MCM-AAA domains is virtually identical to that of full-length. The tryptophan-binding assay makes it complicated to directly compare these results due to the differing number of tryptophan residues and their degrees of exposure. Nevertheless, our data argues for DNA-binding sites within the amino-terminal and MCM-AAA segments and these may be organized into one composite-binding site in the full-length chain. It should also be noted that the MCM-N and MCM-AAA domains of the SsoMCM protein bind ssDNA with similar affinities (16).
The presence of a zinc-coordination module in the MCM-N region is another hallmark of MCM helicase subunits. Several studies have implicated this structural feature in contacts to DNA (12,13), though the molecular details of these contacts are not well understood. We have measured the zinc content of full-length, wild-type BcMCM and obtained evidence for the presence of a zinc atom as well as identifying cysteine 261 as one of the coordinating residues (Figure 8). The finding that BcMCM harbors a zinc ion in its divergent amino-terminus lends support to the idea that this element substitutes for the missing MCM-N region.
Instead of the expected MCM-N domain found in MCM helicases, the amino-terminus of BcMCM displays weak similarities to eukaryotic/archaeal primases (29). Efforts to measure primase activity using a standard assay did not reveal primer synthesis. While we may have failed to detect primase activity due to sub-optimal buffer conditions or the indirect nature of the primase assay, an alternate view suggests that the sequence in question is no longer competent to synthesize RNA primers. While sequence analysis shows the presence of two aspartate residues (D74 and D76), which would be expected to lie in the primase active site, sequence divergence throughout the rest of the region does not allow us to discern whether other residues required by a primase are truly present (Figure 1).
Taken together, our experiments suggest that the crucial role played by the MCM-N domain of conventional MCM-helicases in ATP hydrolysis, DNA binding and unwinding is being carried out by an alternate structure formed at the amino-terminus of BcMCM. This conclusion relies on three observations: the ability of the amino-terminus to modulate both the helicase and ATPase activity of the MCM-AAA domain; its role in binding DNA; and its ability to coordinate zinc.
Efforts to establish the oligomeric state of the active entity revealed that BcMCM is present in solution predominantly as a monomer. These results are unchanged when ATP and magnesium are included in the analysis. The finding that BcMCM is a monomer in solution is unexpected and distinguishes it from other MCM helicases, which are active as oligomers (usually hexamers), and do not require co-factors or substrates to form. We suspect that our inability to detect the active hexameric form is due to experimental conditions that do not suitably mimic those found in vivo.
We fully expect that the active BcMCM helicase is an oligomer, most likely a hexamer, which assembles under the right conditions. We rest this conclusion on three observations. First, BcMCM exhibits the sequence motifs that are hallmarks of the AAA+ family of ATPases. The active AAA+ ATPase is always an oligomer since nucleotides are bound and hydrolyzed at subunit interfaces (14). Indeed, isolated subunits of the yeast heterohexameric MCM2-7 do not hydrolyze nucleotide until provided with the appropriate neighboring subunit (7). Moreover, the ssDNA-ATPase stimulation is also indicative of oligomerization. Recent crystallographic studies of closed ring, hexameric helicases (similar in form to the MCM helicases) indicate that DNA stimulates ATPase activity by properly orienting neighboring ATPase domains within the hexamer to optimize nucleotide hydrolysis (25,44). Second, all characterized MCM-helicases unwind DNA with identical polarity to BcMCM (3′→5′) and do so as oligomers (usually as hexamers). Lastly, oligomerization is strongly suggested by light scattering analysis of the shorter constructions, which could be recapitulating the subunit interactions present in the active helicase (Table 1).
The idea that an active ring helicase can be assembled from monomers has precedent. One example is the SV40 large T antigen helicase, which can only form the active oligomer by assembling on DNA from monomers (45). The bacterial helicase DnaC serves as another example (46).
The question of the biological function, if any, of the BcMCM helicase in the physiology of B. cereus—with its clear evolutionary implications—is beyond the scope of this study. We note that the B. cereus genome harbors a complete set of the typical bacterial-replication initiation and replisome proteins (DnaA, DnaB, DnaI, etc.) and thus we do not discern any obvious reason why the presence of additional genes from an inserted phage would provide a selective advantage.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [grant number GM084162 to D.J.]; National Science Foundation (grant number MCB-0815646 to Z.K.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank past and present members of the Jeruzalmi laboratory for useful discussions, Stanley Lo and Nozomi Sakakibara for assistance with some of the experiments. We thank Mike O’Donnell, Matt Michael, Brian Chait and Danaya Pakotiprapha for providing reagents and advice. Some of the mutants in this study were prepared by the Harvard College Class, LS100R. We are grateful to Nicole Francis for help with experiments and with comments on the manuscript.
REFERENCES
- 1.Forsburg SL. Eukaryotic MCM proteins: beyond replication initiation. Microbiol. Mol. Biol. Rev. 2004;68:109–131. doi: 10.1128/MMBR.68.1.109-131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sakakibara N, Kelman LM, Kelman Z. Unwinding the structure and function of the archaeal MCM helicase. Mol. Microbiol. 2009;72:286–296. doi: 10.1111/j.1365-2958.2009.06663.x. [DOI] [PubMed] [Google Scholar]
- 3.Ying CY, Gautier J. The ATPase activity of MCM2-7 is dispensable for pre-RC assembly but is required for DNA unwinding. EMBO J. 2005;24:4334–4344. doi: 10.1038/sj.emboj.7600892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc. Natl Acad. Sci. USA. 2006;103:10236–10241. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pacek M, Tutter AV, Kubota Y, Takisawa H, Walter JC. Localization of MCM2-7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol. Cell. 2006;21:581–587. doi: 10.1016/j.molcel.2006.01.030. [DOI] [PubMed] [Google Scholar]
- 6.Ishimi Y. A DNA helicase activity is associated with an MCM4, -6, and -7 protein complex. J. Biol. Chem. 1997;272:24508–24513. doi: 10.1074/jbc.272.39.24508. [DOI] [PubMed] [Google Scholar]
- 7.Davey MJ, Indiani C, O'Donnell M. Reconstitution of the mcm2-7p heterohexamer, subunit arrangement, and ATP site architecture. J. Biol. Chem. 2003;278:4491–4499. doi: 10.1074/jbc.M210511200. [DOI] [PubMed] [Google Scholar]
- 8.Kanter DM, Bruck I, Kaplan DL. Mcm subunits can assemble into two different active unwinding complexes. J. Biol. Chem. 2008;283:31172–31182. doi: 10.1074/jbc.M804686200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bochman ML, Schwacha A. The Mcm2-7 complex has in vitro helicase activity. Mol. Cell. 2008;31:287–293. doi: 10.1016/j.molcel.2008.05.020. [DOI] [PubMed] [Google Scholar]
- 10.Fletcher RJ, Bishop BE, Leon RP, Sclafani RA, Ogata CM, Chen XS. The structure and function of MCM from archaeal M. Thermoautotrophicum. Nat. Struct. Biol. 2003;10:160–167. doi: 10.1038/nsb893. [DOI] [PubMed] [Google Scholar]
- 11.Lium W, Pucci B, Rossi M, Pisani FM, Ladenstein R. Structural analysis of the Sulfolobus solfataricus MCM protein N-terminal domain. Nucleic Acids Res. 2008;36:3235–3243. doi: 10.1093/nar/gkn183. [PMCID: 2425480] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poplawski A, Grabowski B, Long SE, Kelman Z. The zinc finger domain of the archaeal minichromosome maintenance protein is required for helicase activity. J. Biol. Chem. 2001;276:49371–49377. doi: 10.1074/jbc.M108519200. [DOI] [PubMed] [Google Scholar]
- 13.Atanassova N, Grainge I. Biochemical characterization of the minichromosome maintenance (MCM) protein of the crenarchaeote Aeropyrum pernix and its interactions with the origin recognition complex (ORC) proteins. Biochemistry. 2008;47:13362–13370. doi: 10.1021/bi801479s. [DOI] [PubMed] [Google Scholar]
- 14.Duderstadt KE, Berger JM. AAA+ ATPases in the initiation of DNA replication. Crit. Rev. Biochem. Mol. Biol. 2008;43:163–187. doi: 10.1080/10409230802058296. [DOI] [PubMed] [Google Scholar]
- 15.Costa A, Pape T, van Heel M, Brick P, Patwardhan A, Onesti S. Structural basis of the Methanothermobacter thermautotrophicus MCM helicase activity. Nucleic Acids Res. 2006;34:5829–5838. doi: 10.1093/nar/gkl708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McGeoch AT, Trakselis MA, Laskey RA, Bell SD. Organization of the archaeal MCM complex on DNA and implications for the helicase mechanism. Nat. Struct. Mol. Biol. 2005 doi: 10.1038/nsmb974. [DOI] [PubMed] [Google Scholar]
- 17.Iyer LM, Leipe DD, Koonin EV, Aravind L. Evolutionary history and higher order classification of AAA+ ATPases. J. Struct. Biol. 2004;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 18.Lutzmann M, Maiorano D, Mechali M. Identification of full genes and proteins of MCM9, a novel, vertebrate-specific member of the MCM2-8 protein family. Gene. 2005;362:51–56. doi: 10.1016/j.gene.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 19.Maiorano D, Cuvier O, Danis E, Mechali M. MCM8 is an MCM2-7-related protein that functions as a DNA helicase during replication elongation and not initiation. Cell. 2005;120:315–328. doi: 10.1016/j.cell.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 20.Yu X, VanLoock MS, Poplawski A, Kelman Z, Xiang T, Tye BK, Egelman EH. The Methanobacterium thermoautotrophicum MCM protein can form heptameric rings. EMBO Rep. 2002;3:792–797. doi: 10.1093/embo-reports/kvf160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pape T, Meka H, Chen S, Vicentini G, Van Heel M, Onesti S. Hexameric ring structure of the full-length archaeal MCM protein complex. EMBO Rep. 2003;4:1079–1083. doi: 10.1038/sj.embor.7400010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Costa A, Pape T, van Heel M, Brick P, Patwardhan A, Onesti S. Structural studies of the archaeal MCM complex in different functional states. J. Struct. Biol. 2006;156:210–219. doi: 10.1016/j.jsb.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 23.Wang G, Klein MG, Tokonzaba E, Zhang Y, Holden LG, Chen XS. The structure of a DnaB-family replicative helicase and its interactions with primase. Nat. Struct. Mol. Biol. 2008;15:94–100. doi: 10.1038/nsmb1356. [DOI] [PubMed] [Google Scholar]
- 24.Bailey S, Eliason WK, Steitz TA. Structure of hexameric DnaB helicase and its complex with a domain of DnaG primase. Science. 2007;318:459–463. doi: 10.1126/science.1147353. [DOI] [PubMed] [Google Scholar]
- 25.Enemark EJ, Joshua-Tor L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature. 2006;442:270–275. doi: 10.1038/nature04943. [DOI] [PubMed] [Google Scholar]
- 26.Bae B, Chen Y, Costa A, Onesti S, Brunzelle J, Lin Y, Cann I, Nair S. Insights into the architecture of the replicative helicase from the structure of an archaeal MCM homolog. Structure. 2009;17:211–22. doi: 10.1016/j.str.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 27.Brewster AS, Wang G, Yu X, Greenleaf WB, Carazo JM, Tjajadia M, Klein M, Chen X. Crystal structure of a near-full-length archaeal MCM: functional insights for an AAA+ hexameric helicase. Proc. Natl Acad. Sci. USA. 2008;105:20191–20196. doi: 10.1073/pnas.0808037105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McGeoch AT, Bell SD. Eukaryotic/archaeal primase and MCM proteins encoded in a bacteriophage genome. Cell. 2005;120:167–168. doi: 10.1016/j.cell.2004.11.031. [DOI] [PubMed] [Google Scholar]
- 29.Iyer LM, Koonin EV, Leipe DD, Aravind L. Origin and evolution of the archaeo-eukaryotic primase superfamily and related palm-domain proteins: structural insights and new members. Nucleic Acids Res. 2005;33:3875–3896. doi: 10.1093/nar/gki702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shin JH, Kelman Z. DNA unwinding assay using streptavidin-bound oligonucleotides. BMC Mol. Biol. 2006;7:43. doi: 10.1186/1471-2199-7-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kiianitsa K, Solinger JA, Heyer WD. NADH-coupled microplate photometric assay for kinetic studies of ATP-hydrolyzing enzymes with low and high specific activities. Anal. Biochem. 2003;321:266–271. doi: 10.1016/s0003-2697(03)00461-5. [DOI] [PubMed] [Google Scholar]
- 32.Rowen L, Kornberg A. Primase, the dnaG protein of Escherichia coli. An enzyme which starts DNA chains. J. Biol. Chem. 1978;253:758–764. [PubMed] [Google Scholar]
- 33.Pakotiprapha D, Inuzuka Y, Bowman BR, Moolenaar GF, Goosen N, Jeruzalmi D, Verdine GL. Crystal structure of Bacillus stearothermophilus UvrA provides insight into ATP-modulated dimerization, UvrB interaction, and DNA binding. Mol. Cell. 2008;29:122–133. doi: 10.1016/j.molcel.2007.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Galletto R, Jezewska MJ, Bujalowski W. Interactions of the Escherichia coli DnaB helicase hexamer with the replication factor the DnaC protein. Effect of nucleotide cofactors and the ssDNA on protein-protein interactions and the topology of the complex. J. Mol. Biol. 2003;329:441–465. doi: 10.1016/s0022-2836(03)00435-2. [DOI] [PubMed] [Google Scholar]
- 35.Learn BA, Um SJ, Huang L, McMacken R. Cryptic single-stranded-DNA binding activities of the phage lambda P and Escherichia coli DnaC replication initiation proteins facilitate the transfer of E. coli DnaB helicase onto DNA. Proc. Natl Acad. Sci. USA. 1997;94:1154–1159. doi: 10.1073/pnas.94.4.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arai K, Kornberg A. Mechanism of dnaB protein action. III. Allosteric role of ATP in the alteration of DNA structure by dnaB protein in priming replication. J. Biol. Chem. 1981;256:5260–5266. [PubMed] [Google Scholar]
- 37.Arai K, Kornberg A. Mechanism of dnaB protein action. II. ATP hydrolysis by dnaB protein dependent on single- or double-stranded DNA. J. Biol. Chem. 1981;256:5253–5259. [PubMed] [Google Scholar]
- 38.Wyatt P. Light scattering and the absolute characterization of macromolecules. Analytica Chimica Acta. 1993;272:1–40. [Google Scholar]
- 39.Pucci B, De Felice M, Rocco M, Esposito F, De Falco M, Esposito L, Rossi M, Pisani FM. Modular organization of the Sulfolobus solfataricus mini-chromosome maintenance protein. J. Biol. Chem. 2007;282:12574–12582. doi: 10.1074/jbc.M610953200. [DOI] [PubMed] [Google Scholar]
- 40.Jeruzalmi D, O'Donnell M, Kuriyan J. Clamp loaders and sliding clamps. Curr. Opin. Struct. Biol. 2002;12:217–224. doi: 10.1016/s0959-440x(02)00313-5. [DOI] [PubMed] [Google Scholar]
- 41.Steitz TA, Yin YW. Accuracy, lesion bypass, strand displacement and translocation by DNA polymerases. Philos Trans R Soc Lond, B, Biol. Sci. 2004;359:17–23. doi: 10.1098/rstb.2003.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grainge I, Scaife S, Wigley DB. Biochemical analysis of components of the pre-replication complex of Archaeoglobus fulgidus. Nucleic Acids Res. 2003;31:4888–4898. doi: 10.1093/nar/gkg662. PMCID: 169903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barry ER, McGeoch AT, Kelman Z, Bell SD. Archaeal MCM has separable processivity, substrate choice and helicase domains. Nucleic Acids Res. 2007;35:988–998. doi: 10.1093/nar/gkl1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singleton MR, Sawaya MR, Ellenberger T, Wigley DB. Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell. 2000;101:589–600. doi: 10.1016/s0092-8674(00)80871-5. [DOI] [PubMed] [Google Scholar]
- 45.Dean FB, Borowiec JA, Eki T, Hurwitz J. The simian virus 40 T antigen double hexamer assembles around the DNA at the replication origin. J. Biol. Chem. 1992;267:14129–14137. [PubMed] [Google Scholar]
- 46.Davey MJ, O'Donnell M. Replicative helicase loaders: ring breakers and ring makers. Curr. Biol. 2003;13:R594–R596. doi: 10.1016/s0960-9822(03)00523-2. [DOI] [PubMed] [Google Scholar]
- 47.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. PMCID: 390337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pei J, Grishin NV. AL2CO: calculation of positional conservation in a protein sequence alignment. Bioinformatics. 2001;17:700–712. doi: 10.1093/bioinformatics/17.8.700. [DOI] [PubMed] [Google Scholar]
- 49.Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc. Natl Acad. Sci. USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.